Abstract
Background
Enteropathogenic Yersinia circulate in the pig reservoir and are the third bacterial cause of human gastrointestinal infections in Europe. In West Africa, reports of human yersiniosis are rare. This study was conducted to determine whether pathogenic Yersinia are circulating in pig farms and are responsible for human infections in the Abidjan District.
Methodology/Principal findings
From June 2012 to December 2013, pig feces were collected monthly in 41 swine farms of the Abidjan district. Of the 781 samples collected, 19 Yersinia strains were isolated in 3 farms: 7 non-pathogenic Yersinia intermedia and 12 pathogenic Yersinia enterocolitica bioserotype 4/O:3. Farm animals other than pigs and wild animals were not found infected. Furthermore, 2 Y. enterocolitica 4/O:3 strains were isolated from 426 fecal samples of patients with digestive disorders. All 14 Y. enterocolitica strains shared the same PFGE and MLVA profile, indicating their close genetic relationship. However, while 6 of them displayed the usual phage type VIII, the other 8 had the highly infrequent phage type XI. Whole genome sequencing and SNP analysis of individual colonies revealed that phage type XI strains had unusually high rates of mutations. These strains displayed a hypermutator phenotype that was attributable to a large deletion in the mutS gene involved in DNA mismatch repair.
Conclusions/Significance
This study demonstrates that pathogenic Y. enterocolitica circulate in the pig reservoir in Côte d'Ivoire and cause human infections with a prevalence comparable to that of many developed countries. The paucity of reports of yersiniosis in West Africa is most likely attributable to a lack of active detection rather than to an absence of the microorganism. The identification of hypermutator strains in pigs and humans is of concern as these strains can rapidly acquire selective advantages that may increase their fitness, pathogenicity or resistance to commonly used treatments.
Author Summary
Diarrhea is a major public health problem in developing countries, especially in Africa, but the causative agents are often unknown, impeding the implementation of appropriate therapeutic measures. Although pathogenic Yersinia enterocolitica are a frequent cause of gastroenteritis in developed countries, reports of human yersiniosis are scarce in West Africa. We performed this study to determine whether this pathogen was present in pigs (the natural reservoir of this bacterium) in various swine farms of the Abidjan district, and whether it was causing human gastro-intestinal infections. We show here that this bacterium is indeed circulating among Ivorian pigs and is causing human digestive disorders with a frequency similar to that reported in developed countries. The paucity of reports of this infection in African countries may be explained by the difficulty of isolating Y. enterocolitica from stools and the need for specific and time-consuming procedures. During this study we also made the unexpected observation that some Ivorian strains have acquired the capacity to mutate at a much higher frequency than normal strains. This property may render them better fitted to new environments, more virulent to their host, or capable of resisting some commonly used treatments, which could be of great public health concern.
Introduction
Yersinia enterocolitica is an enteropathogenic bacterium responsible for human gastroenteritis. This species belongs to the genus Yersinia, and to the family Enterobacteriaceae. Clinical presentation of yersiniosis includes diarrhea, abdominal pain, fever, and sometimes vomiting [1]. Although the infection is often mild and self-limiting, more severe clinical presentations such as pseudo-appendicular syndromes mimicking an appendicitis [2] or septicemia in elderly and immuno-compromized patients [3] can occur. Reactive arthritis and erythema nodosum are the most frequent secondary complications [4].
Enteric yersiniosis is a foodborne disease [5,6], which is transmitted through the fecal-oral route. The species Y. enterocolitica is subdivided into 6 biotypes [7]. Biotype 1A is non-pathogenic while the 5 other biotypes (1B, 2–5) cause human and/or animal infections. The biotype the most frequently responsible for human infections worldwide is biotype 4 [8], which is almost systematically associated with serotype O:3 (4/O:3), followed by bioserotype 2/O:9. Pigs are the main reservoir of bioserotype 4/O:3 strains [8]. These animals are asymptomatic carriers of the bacteria in their tonsils and intestinal tract, and they shed the enteropathogen in the environment with their stools. Contamination of pork meat often occurs during pig evisceration at slaughter [9].
Although Y. enterocolitica represents the third cause of bacterial diarrhea in Europe, after campylobacteriosis and salmonellosis [10] reports of human yersiniosis are scarce in West Africa. A systematic survey of children with diarrhea in Ouagadougou (Burkina Faso) allowed the isolation of Yersinia strains from 1.7% of the stools tested [11]. However, no indication of the species or biotype of the strains was provided, making difficult to estimate whether these isolates corresponded to saprophytic or pathogenic species. Y. enterocolitica strains were also isolated from the stools of children with gastroenteritis in Gabon, but their bioserotype was not determined [12]. In Ghana, a few Y. enterocolitica strains were isolated from blood products at transfusion centers [13] or were detected by PCR in the blood of some patients with sepsis [14], indicating that severe cases of yersiniosis occur in this country. The most active West African country for the survey of Yersinia infections is Nigeria. Several studies reported the isolation of pathogenic Y. enterocolitica from the stools of patients presenting with enteric infections [15–19].
An active surveillance of the animal reservoir also revealed the presence of pathogenic bioserotypes of Y. enterocolitica pigs, bovine and sheep in Nigeria [20–23]. In contrast, a survey of the pig reservoir in Senegal [24] and Burkina Faso [25] did not identify any Yersinia strains. The search for these bacteria in water sources in Nigeria also remained negative [26].
These data suggest that Y. enterocolitica is present in the animal reservoir and causes human infections in West Africa. Insufficient public health surveillance and inappropriate isolation procedures may account for the paucity of reports of this infection [27]. The fast-growing demand for milk and meat in urban centers in resource-limited countries is leading to the intensification of livestock production systems, especially in peri-urban areas. However, because efficient zoonosis surveillance and food safety are lacking, the risk for zoonosis transmission is increasing, particularly in rapidly growing urban centers of resource limited countries [28].
In Côte d'Ivoire, there is some indication that Y. enterocolitica circulates and may be a cause of human infections. In 1983, a search for the presence of fecal coliforms in drinking well water and in human stools allowed the isolation of a few Yersinia strains [29]. As their species were not determined, whether they were environmental non-pathogenic strains, or enteropathogens could not be established. More recently, three Y. enterocolitica strains of bioserotype 4/O:3 were isolated from pig carcasses at slaughter [30]. These results thus showed the presence of pathogenic Yersinia in the pig reservoir, suggesting that Y. enterocolitica may be a cause of human gastroenteritis in Côte d'Ivoire.
This study aimed at further investigating the carriage of Y. enterocolitica in the animal reservoir and at estimating the prevalence of enteropathogenic Yersinia in human diarrheal diseases in the Abidjan area of Côte d'Ivoire.
Methods
Ethics statement
The human and animal components of this study were approved by the National Ethical Research Committee of Côte d’Ivoire (Ref 095/MSLS/CNER-kp). Written informed consents were obtained from the patients or their parents, by medical doctors and laboratory teams in charge of surveillance activities, and from farmers and animal owners.
Sample processing
This study was carried out from June 2012 to December 2013. Forty-one swine farms distributed in 4 sub-prefectures of the Abidjan district (Abidjan, Anyama, Bingerville and Songon) were selected based on their high pig production capacity and were visited monthly. Two to 3 stool samples were collected by rectal swab from apparently healthy pigs randomly sampled in each farm and pooled. Fecal samples were also taken from cattle in the infected farms and pooled. Wild animals (rats and gigantic snails) living around the farms were also captured and their intestinal contents were individually analyzed for the presence of Yersinia. Fecal samples were collected from humans with digestive disorders in 8 hospitals from the 4 sub-prefectures. Animal feces and intestinal contents, as well as human stools were collected freshly and stored cold during transportation in an ice box to the laboratory for immediate processing.
Isolation, identification and characterization of Yersinia strains
Yersinia strains were isolated using two stages enrichment procedures as described by the Department of Food and Environmental Hygiene in Finland [31]. This procedure included prior-enrichment at 25°C during 24h of 1 ml of sample in 9 ml of Brain Heart Infusion broth containing 2.5 mg/l novobiocin. This was followed by an enrichment step for 7 and 14 days at 4°C in a modified phosphate buffer saline supplemented with 1% mannitol, 0.15% bile salts, and 0.5% soy peptone. An alkali treatment for 20s of 0.5 ml of the enriched sample in 4.5 ml of 0.25% potassium hydroxide was then performed to reduce the background-contaminating flora as described [32]. A 10 μl volume of the enriched sample was then streaked on Cefsulodin-Irgasan-Novobiocin agar (CIN). The plates were incubated under aerobic conditions at 30°C for 18h–48h. Putative Yersinia colonies were identified with oxidase, Kligler iron and Christensen’s urea. Oxidase-negative, glucose-positive, H2S-negative and urease-positive colonies were finally identified with API 20E strips. Colonies displaying typical patterns were further characterized at the French Yersinia Reference Laboratory (Institut Pasteur, Paris) for species determination, biotyping and serotyping [3]. Phage typing was performed and interpreted as described in [33], except that phage type VIII was characterized by a susceptibility to all phages except phage l (observation from the Yersinia Reference Laboratory).
Antibiotic susceptibility testing
From an 18-24h bacterial culture onto trypticase soy agar, a bacterial suspension in saline was prepared at McFarland 0.5 (equivalent to ≈108 cfu/ml). Antibiotic susceptibilities were determined by the disc diffusion method on Mueller-Hinton agar (Oxoid) according to the procedure described in [34]. The results were interpreted according to the guidelines of the European Committee on Antimicrobial Susceptibility Testing (EUCAST: http://www.eucast.org). The antimicrobial drugs tested and their concentrations on the discs (BioRad) were the following: amoxicillin (25 μg), amoxicillin-clavulanic acid (20 μg/10 μg), cefalotin (30 μg), cefoxitin (30 μg), ceftriaxone (30 μg), ciprofloxacin (5 μg), nalidixic acid (30 μg), trimethoprim (5 μg), sulphonamide (200 μg), tetracycline (30 UI) and ticarcillin (75 μg).
Primers and PCR conditions
Bacterial suspensions from 8-24h bacterial cultures were diluted to 104 cfu/ml in sterile water (Eurobio) and centrifuged for 10 min at 13,300 x g at 4°C. Genomic DNA was extracted using the phenol/chloroform and alkali lysis methods [35]. Primers specific for the chromosomal genes ail (attachment invasion locus) and ystA (Yersinia heat-stable enterotoxin), and for the pYV plasmid-borne gene yadA (Yersinia adhesin) of Y. enterocolitica were used (S1 Table). PCR reactions were carried out in a volume of 50 μl containing 2.5 μl of 5X Green Flexi buffer (Promega), 2.5 μl of 5X Colorless Flexi buffer 3 μl of 25 mM MgCl2; 1 μl of 10 μM nucleotides dATP, dTTP, dGTP and dCTP; 1 μl of a 20 μM solution of each primer (S1 Table); 0.2 μl of 5 U/μl Taq DNA polymerase and 5 μl of DNA. The amplifications were performed in a thermal cycler with the following conditions: denaturation at 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 30s, annealing at 60°C for 30s, and extension at 72°C for 1 min, with a final extension at 72°C for 5 min. PCR products (10 μl) were subjected to electrophoresis in a 2% agarose gel and stained with ethidium bromide.
Genotyping
Pulsed Field Gel Electrophoresis (PFGE) of Y. enterocolitica 4/O:3 isolates was carried out as previously described [36]. The genomic DNA was digested with PmeI and SpeI and subjected to electrophoresis for 24h using pulse times ranging from 1 to 13 s at 14°C for PmeI, and from 1 to 15 s at 17°C for SpeI, with an angle of 120° in a CHEF-DR III apparatus A middle-range PFGE size marker) was used. MLVA (Multiple-locus variable-number tandem-repeat analysis) was based on six loci previously defined [37] and was performed by sequence analysis of the PCR products. The PCR mixtures (40 μl) contained 100 ng of DNA template, 0.2 μM of each primer, 1.25 unit of Taq DNA polymerase (Thermo Scientific), 200 μM of dNTPs, 1.25 mM MgCl2, and a final concentration of 1X Taq buffer. The amplification was carried out in a DNA thermocycler with a pre-denaturation step at 94°C for 10 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and elongation at 72°C for 30s. A final 3 min elongation step at 72°C was done after the last cycle to ensure complete amplicon extension. PCR products were sequenced, and the number of repetitions for each locus was determined by the analysis of each sequence.
Whole genome sequencing and analysis
Two ml of overnight bacterial cultures at 28°C were centrifuged at 5,000 x g for 5 min, and the genomic DNA was extracted from the cell pellet using the Genomic DNA mini kit. The DNA was suspended in 100 μl of elution buffer and quantified with the LUX (Thermo Fisher Scientific). Genomic libraries were prepared with 0.1 ng of DNA as template, using the Nextera XT protocol on the SureCycler 8800 thermal cycler (Agilent). The libraries were purified with the AMPure beads and quality control was performed with the High Sensitivity D1000 kit on Tape station 2200 Inserts were sized (400–900 bp) using the Pippin Prep kit CDF 1510 and enriched with 35 cycles of qPCR with the KAPA kit on the Lightcycler 96 before library quantification and validation. Hybridization of the library to the flow cell and bridge amplification was performed to generate clusters. Paired-end reads of 150 cycles were collected on a NextSeq500 sequencer using the HighOutput kit.
Demultiplexing and generation of FASTQ files from the raw sequence data were performed using the bcl2fastq software. Trimming, clipping, and filtering off exogenous and/or non-confident bases (options:–q 13, -l 30, -p 80) within FASTQ files were performed with AlienTrimmer [38]. Redundant or over-represented reads were reduced using the khmer software package (option:–c 70) [39]. Finally, sequencing errors were corrected using Musket [40] and overlapping paired reads were merged with FLASH [41]. A de novo assembly was performed for each strain with the quality-filtered reads using SPAdes v3.6 (options:-k 21,33,55,77—only-assembler—careful) [42].The alignment of the quality-filtered reads against a reference genome YE1203 (accession number: HF933425) was performed using BWA v0.7.7, and variant calling (SNPs and indels) using SAM tools v1.2 and VarScan v2.3.6 (options:–min-coverage 30 and–min-var-freq 0.8) [43].
Genome sequences are available in European Nucleotide Archive under BioProject PRJEB13626 (sample ID starting with ERS and accession numbers starting with FK) for strains IP134 (ERS1122523, FKKS01000001-FKKS01000141), IP35459 (ERS1122524, FKKM01000001-FKKM01000146), IP35462 (ERS1122525, FKKN01000001-FKKN01000154), IP35463 (ERS1122526, FKKL01000001-FKKL01000153), IP35464 (ERS1122527, FKKW01000001-FKKW01000150), IP35465 (ERS1122528, FKKU01000001-FKKU01000159), IP35466 (ERS1122529, FKKJ01000001-FKKJ01000148), IP35467 (ERS1122530, FKKT01000001-FKKT01000151), IP35470 (ERS1122531, FKKV01000001-FKKV01000149), IP35471 (ERS1122532, FKKP01000001-FKKP01000150), IP35472 (ERS1122533, FKKQ01000001-FKKQ01000151), IP35474 (ERS1122534, FKKO01000001-FKKO01000149), IP35475 (ERS1122535, FKKK01000001-FKKK01000157), IP35477 (ERS1122536, FKLN01000001-FKLN01000190), IP35478 (ERS1122537, FKKR01000001-FKKR01000152).
Hypermutator phenotype
Strains were streaked on TSA agar plates. Ten colonies were picked and were grown in 10 ml Luria Bertani (LB) broth under agitation at 28°C for 24h. OD600 values were recorded and 200 μl of each culture were plated onto LB agar containing 50 μg/ml nalidixic acid or 100 μg/ml rifampicin.All plates were incubated at 28°C for 3 days. Colony enumeration was performed with a Scan500 automatic colony counter. The results were expressed as the number of colonies on antibiotic plates per 109 cfu of the original inoculum.
Results
Search for the presence of Yersinia in pig feces
A total of 781 samples of pooled pig feces collected over the 19 months study period in 41 farms of four sub-prefectures of the Abidjan district were analyzed for the presence of Yersinia. The two steps enrichment procedure used in this study allowed the isolation of 19 strains (Table 1).
Table 1. Samples analyzed for the detection of Yersinia strains in animals and humans.
| Host | District | Yersinia | Total | |||||
|---|---|---|---|---|---|---|---|---|
| Abidjan | Anyama | Bingerville | Songon | Negative | Positive | |||
| Animal | Pig | 209 | 98 | 306 | 168 | 762 | 19 | 781 |
| sheep | 69 | 15 | 35 | 59 | 178 | 0 | 178 | |
| Bovine | 55 | 0 | 22 | 32 | 109 | 0 | 109 | |
| Rodent | 47 | 0 | 62 | 30 | 139 | 0 | 139 | |
| Snail | 17 | 0 | 23 | 19 | 59 | 0 | 59 | |
| Human | Patients with diarrhea | 161 | 14 | 93 | 19 | 285 | 2 | 287 |
| Patients without diarrhea | 87 | 9 | 32 | 11 | 139 | 0 | 139 | |
| Total | 645 | 136 | 573 | 338 | 1671 | 21 | 1692 | |
Seven of them belonged to the non-pathogenic species Yersinia intermedia. All 7 Y. intermedia strains were of biotype 4, but they had different serotypes: 2 were of serotype O:7,8-8-8,19, while the remaining 5 were of serotype O:7,8-8-13-8,19 (S2 Table); indicative of the circulation of different Y. intermedia strains among pigs in this area. These strains were isolated from 3 farms in 2 sub-prefectures (Bingerville and Abidjan, Fig 1) over a ≈1-year period (S2 Table).
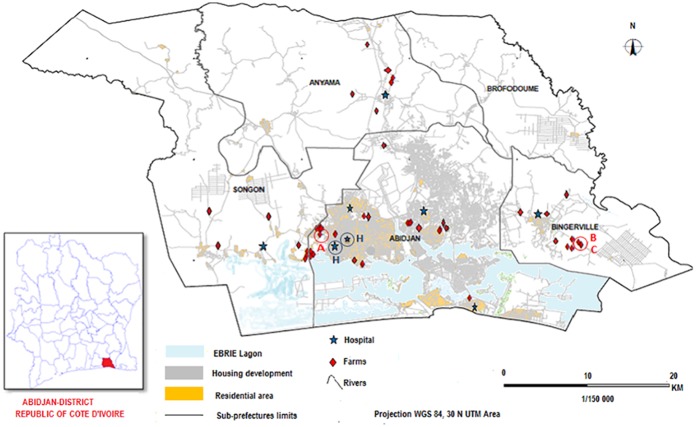
Fig 1. Location of the sampling farms and hospitals in the Abidjan district.
Red circles indicate the location of the farms (A, B and C) from which Yersinia strains were isolated, and blue circles (H) the location of the hospitals where human cases of yersiniosis were identified.
All other 12 strains belonged to the species Y. enterocolitica and were of bioserotype 4/O:3 (Table 2). These pathogenic Y. enterocolitica were recovered from pig feces during a period extending from March to August without isolation during the other 6 months of the year, suggesting a periodicity in the carriage of pathogenic Y. enterocolitica by swine in the Abidjan district. In contrast, some Y. intermedia strains were isolated in February (S2 Table).
Table 2. Characteristics of the Y. enterocolitica strains isolated from human and animal samples.
| Strain # | Host | Bio-type | Serotype | Farm | Sub-prefecture | Phage type | Date of isolation | Virulence genes | ||
|---|---|---|---|---|---|---|---|---|---|---|
| ail | ystA | yadA | ||||||||
| IP35459 | Pig | 4 | O:3 | A | Abidjan | XI | 2012, August 30 | + | + | + |
| IP35462 | Pig | 4 | O:3 | C | Bingerville | VIII | 2013, March 21 | + | + | + |
| IP35463 | Pig | 4 | O:3 | B | Bingerville | VIII | 2013, March 21 | + | + | + |
| IP35464 | Pig | 4 | O:3 | C | Bingerville | VIII | 2013, April 16 | + | + | - |
| IP35465 | Pig | 4 | O:3 | A | Abidjan | XI | 2013, April 18 | + | + | + |
| IP35466 | Pig | 4 | O:3 | A | Abidjan | XI | 2013, May 23 | + | + | - |
| IP35467 | Pig | 4 | O:3 | B | Bingerville | XI | 2013, May 28 | + | + | + |
| IP35470 | Pig | 4 | O:3 | C | Bingerville | VIII | 2013, June 25 | + | + | - |
| IP35471 | Pig | 4 | O:3 | C | Bingerville | XI | 2013, June 25 | + | + | + |
| IP35472 | Pig | 4 | O:3 | A | Abidjan | XI | 2013, July 9 | + | + | + |
| IP35474 | Pig | 4 | O:3 | C | Bingerville | VIII | 2013, July 16 | + | + | - |
| IP35475 | Pig | 4 | O:3 | C | Bingerville | VIII | 2013, August 15 | + | + | - |
| IP35477 | Human | 4 | O:3 | NA | Abidjan | XI | 2013, April 26 | + | + | + |
| IP35478 | Human | 4 | O:3 | NA | Abidjan | XI | 2013, September 05 | + | + | + |
NA: Not applicable
The Y. enterocolitica strains were isolated from the same 3 farms from which the Y. intermedia strains were isolated (Table 2). The isolation rate of Yersinia strains (pathogenic and non-pathogenic) was similar (8 to 9%) in the 3 farms (S3 Table). Farms B and C were close to each other (1.5 km) in the Bingerville sub-prefecture (Fig 1), arguing for a local circulation of Y. enterocolitica in this area. Farm A was located in the Abidjan sub-prefecture and was distant by more than 30 km from the other two, indicating the existence of at least two different geographical foci of yersiniosis in the Abidjan district.
Search for the presence of Yersinia in other animal species living in or around the farms
To determine whether pathogenic Y. enterocolitica strains were circulating among other animal species living in the farms, fecal samples were taken from sheep and bovine and pooled after each visit. None of these samples (178 from cattle and 109 from bovine) were found infected with a Yersinia strain (Table 1). Furthermore, 202 rodents (Rattus, Rattus norvegicus and Thryonomys swinderianus (greater cane rat)) and 95 gigantic snails (Achatina fulica) living around the farms were captured. Their intestinal content was taken during autopsy and analyzed for the presence of Yersinia. No Yersinia strains were recovered from these wild animals. These results confirm the preferential association of Y. enterocolitica 4/O:3 with pigs [44,45]
Analysis of human stools for the presence of Yersinia
Between June 2012 and December 2013, 426 fecal samples were collected from humans in 8 hospitals from the 4 sub-prefectures (Fig 1 and Table 1). They were collected from 287 patients with diarrhea during a childhood diarrhea surveillance program (205 samples), or as part of routine stool cultures in medical microbiology laboratories (82 samples). The 139 other patients presented with digestive disorders (abdominal pain, nausea, vomiting) without diarrhea. Overall the patients aged between 7 months and 55 years with an average of 4 years.
Two Yersinia strains were recovered from the 426 human fecal samples analyzed, indicating a prevalence of 0.47% for all patients with intestinal disorders, and of 0.69% for patients with diarrhea. The two patients were infant females aged 1 year 7 months, and 3 years 2 months who presented with diarrhea and fever. They were seen at the university hospital and at the annex of the hospital (2 km away) in the sub-prefecture of Abidjan (Fig 1).
Both clinical strains were pathogenic Y. enterocolitica 4/O:3 (Table 2).They were isolated in April and September from the 2 patients (Table 2), i.e. at a time of the year where pigs were also found to be carriers of Y. enterocolitica 4/O:3 strains.
Phage typing of Y. enterocolitica 4/O:3 strains
Phage typing is a simple means, routinely used by the French Yersinia Reference Laboratory to subgroup 4/O:3 strains. This subtyping method was applied to the Ivorian Y. enterocolitica strains to determine whether the porcine and human isolates share the same phage type (ΦT). Six strains of pig origin were of ΦT VIII (Table 2), which is by far the most common ΦT among bioserotype 4/O:3 strains worldwide [3,46]. The other 8 strains exhibited ΦT XI (Table 2), which is highly unusual in 4/O:3 strains. These ΦT XI strains were isolated from all 3 farms and from the 2 human patients.
ΦT XI means susceptibility profiles to the 12 lysogenic phages different from the well established ones (VIII, IXa and IXb) for 4/O:3 strains [33]. Actually, a susceptibility or resistance phenotype to each phage was sometimes hard to assign because the growth of ΦT XI strains was heterogeneous within the lysis zone. We often noted a bacterial growth that was less dense in contact to than at distance from the phage drop, or isolated colonies growing within the lysis zone. These suggested mixed bacterial populations composed of colonies susceptible and resistant to each phage.
To test this hypothesis, 3 ΦT XI strains (IP35471, IP35477 and IP35478) were selected and 6 individual colonies from each of them were picked and individually phage typed. Two to 3 susceptibility profiles were observed for each strain (S4 Table), confirming heterogeneity of the bacterial population. Some colonies of strains IP35471 and IP35477 displayed the usual ΦT VIII, arguing for the emergence of variants from this parental phenotype. The 2 other susceptibility profiles were designated ΦT XIa and XIb. ΦT XIa was identified in colonies from the 3 strains, while ΦT XIb colonies were detected in 2 strains (S4 Table). Since a single colony from each biological sample was originally picked and stored, these data show that mutations leading to variable susceptibility or resistance to the set of lysogenic phages occurred in these strains. Altogether these results argue for an unusual propensity of some Ivorian strains to display heterogeneous profiles of resistance to the set of Y. enterocolitica lysogenic phages.
Antibiotic susceptibility and virulence markers of the Y. enterocolitica strains isolated in the Abidjan district
As commonly observed [47,48], the Y. enterocolitica 4/O:3 strains isolated in our study were resistant to amoxicillin, amoxicillin/clavulanic acid, cefalotin, and ticarcillin. However, they were susceptible to cefoxitin, ceftriaxone, ciprofloxacin, nalidixic acid, trimethoprim, sulphonamide and tetracycline, indicating that they had not acquired any unusual antibiotic resistances.
As expected, all 14 Y. enterocolitica 4/O:3 strains carried the ail and ystA chromosomal virulence genes (Table 2), further indicating their pathogenic potential. Only 9 of them (64.3%) harbored the pYV-borne yadA gene, in line with the observation that the pYV plasmid is easily lost upon in vitro subculture of pathogenic Yersinia [9].
Genotypic characterization of the Y. enterocolitica strains isolated
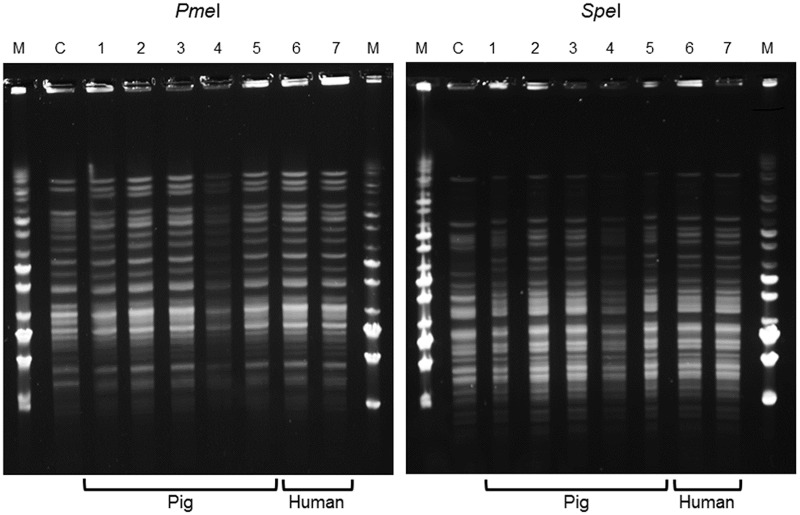
PFGE analysis after digestion with PmeI and SpeI of the 14 Y. enterocolitica 4/O:3 strains isolated from pigs and humans showed a single profile with each enzyme for all strains analyzed (illustrated in Fig 2). This unique profile differed only slightly from that of an epidemiologically unrelated Y. enterocolitica 4/O:3 control strain (IP134, isolated in Sweden).
Fig 2. Pulsed Field Gel Electrophoresis patterns of 7 Y. enterocolitica 4/O:3 strains isolated from animals and humans.
C: Control Y. enterocolitica 4/O:3 strain IP134, M: middle range molecular size marker, 1: IP35470; 2: IP35471; 3: IP35472; 4: IP35474; 5: IP35475; 6: IP35477; 7: IP35478.
As the discriminatory power of PFGE has recently been shown to be low for pathogenic Y. enterocolitica [49], the strains were then subjected to MLVA, which has a higher discriminatory power [36,37]. This MLVA analysis based on six loci also gave a unique pattern (7-8-7-8-8-2) for the 14 Y. enterocolitica strains analyzed. This pattern differed drastically from that of the IP134 control strain (11-2-9-16-6-X), indicating either that the Y. enterocolitica strains isolated in the Abidjan district are genetically closely related, or that the same strain circulated in the 3 pig farms and in humans.
Genetic relationships among the Ivorian Y. enterocolitica strains
To further examine the genetic relatedness of the Y. enterocolitica 4/O:3 strains isolated from pigs and humans in the Abidjan district over the study period, the genomes of the 14 Y. enterocolitica strains collected were sequenced. A pair-wise analysis of Single Nucleotide Polymorphism (SNP) showed that the Ivorian strains displayed between 590 and 675 SNPs with the epidemiologically unrelated Y. enterocolitica 4/O:3 strain IP134 (Table 3). In contrast, the number of SNPs between each pair of Ivorian strains was always <96, indicating that they were genetically much closer to each other than to the epidemiologically unrelated strain. Two isolates (IP35462 and IP35464) had no SNPs between each other. They were both of ΦT VIII and were isolated at one-month interval from the same farm, suggesting that one strain was persisting and isolated twice from farm C. The other strains displayed some degrees of genetic polymorphism (Table 3). These results thus demonstrate that most of the strains that circulate in the pig and human populations in the Abidjan area are distinct from each other.
Table 3. Pair-wise analysis of SNPs among Ivorian Y. enterocolitica strains and an epidemiologically unrelated 4/O:3 isolate.
| Strains | IP 134 | IP 35459 | IP 35462 | IP 35463 | IP 35464 | IP 35465 | IP 35466 | IP 35467 | IP 35470 | IP 35471 | IP 35472 | IP 35474 | IP 35475 | IP 35477 | IP 35478 |
| IP134 | 0 | ||||||||||||||
| IP35459 | 668 | 0 | |||||||||||||
| IP35462 | 595 | 86 | 0 | ||||||||||||
| IP35463 | 598 | 86 | 2 | 0 | |||||||||||
| IP35464 | 596 | 86 | 0 | 2 | 0 | ||||||||||
| IP35465 | 595 | 87 | 5 | 7 | 5 | 0 | |||||||||
| IP35466 | 662 | 42 | 80 | 82 | 80 | 81 | 0 | ||||||||
| IP35467 | 666 | 48 | 86 | 88 | 86 | 85 | 16 | 0 | |||||||
| IP35470 | 591 | 83 | 11 | 13 | 11 | 10 | 77 | 81 | 0 | ||||||
| IP35471 | 674 | 70 | 94 | 96 | 94 | 93 | 64 | 68 | 89 | 0 | |||||
| IP35472 | 675 | 69 | 93 | 95 | 93 | 94 | 63 | 69 | 90 | 45 | 0 | ||||
| IP35474 | 590 | 84 | 12 | 14 | 12 | 13 | 78 | 84 | 3 | 92 | 91 | 0 | |||
| IP35475 | 590 | 83 | 11 | 13 | 11 | 10 | 77 | 81 | 2 | 89 | 90 | 5 | 0 | ||
| IP35477 | 659 | 59 | 81 | 83 | 81 | 80 | 53 | 57 | 76 | 45 | 46 | 79 | 76 | 0 | |
| IP35478 | 673 | 69 | 93 | 95 | 93 | 92 | 63 | 67 | 88 | 55 | 56 | 91 | 88 | 26 | 0 |
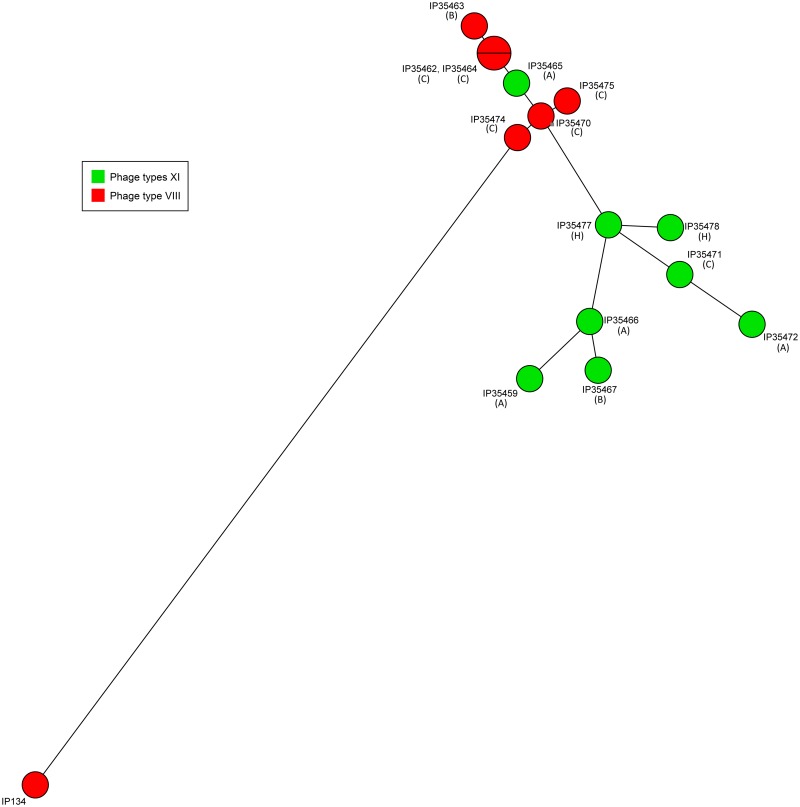
A minimal spanning tree (MST) based on whole genome SNP analysis was constructed to determine the genetic relatedness between the Ivorian Y. enterocolitica isolates. This tree further showed that the Ivorian strains were genetically much closer to each other than to the epidemiologically unrelated Y. enterocolitica 4/O:3/VIII strain IP134 (Fig 3).
Fig 3. Minimal spanning tree of the Y. enterocolitica 4/O:3 strains isolated from pigs and humans in the Abidjan district.
IP134 is a non-Ivorian Y. enterocolitica 4/O:3/VIII strain. Letters into brackets indicate the pig farm (A, B, C) or the human (H) origin of the strains.
The isolates grouped neither according to their geographical location, nor to their date of isolation, arguing for a circulation of strains between the 3 farms, even though farm A was distant from farms B and C. Interestingly, with the exception of strain IP35465, the Ivorian isolates separated into two clusters based on their ΦT (Fig 3). This and the fact that strain IP134 branched with Ivorian strains of ΦT VIII, further strengthen the hypothesis that strains exhibiting the unusual ΦT XI are variants that emerged and gradually diverged from an ancestral ΦT VIII strain.
Genetic bases for the heterogeneity of the phage susceptibility profiles
The lipopolysaccharide O-antigen has been shown to act as a receptor for various Y. enterocolitica phages [50]. Although this has not been demonstrated for the set of phages used for Y. enterocolitica phage typing at the Reference Laboratory, we hypothesized that mutations in the O-antigen gene cluster might have occurred in the ΦT XI branch of the Ivorian isolates and could be responsible for the unusual phage susceptibility patterns observed. However, no polymorphism in the 6.8 kb nucleotide sequence of the O-antigen gene cluster was observed in Ivorian strains, indicating that their unusual susceptibilities to the set of phages are due to other unidentified mutations.
To further understand the genetic bases for the heterogeneity of the phage susceptibility profiles, the genomes of 2 colonies of ΦT XIa (#5 and #6) and of 1 colony of ΦT VIII (#4) from strain IP35471 were sequenced. The aim was to identify genetic modifications that would be common to the 2 ΦT XIa colonies and absent from the ΦT VIII colony, and could thus account for the change in the phage susceptibility profiles. No gene deletions, insertions or mutations (SNPs) having these characteristics were identified among the 3 genomes. However, this analysis revealed an unexpectedly high number of SNPs (9 to 12) between individual colonies of the same strain. To further explore this phenomenon, the number of within-strain SNPs was evaluated on 9 individual colonies from 3 strains of ΦT XI and compared to that of individual colonies from a typical ΦT VIII Y. enterocolitica 4/O:3 strain (IP33927). While the number of SNPs between colonies of strain IP33927 was always ≤2 (average of 1 SNP), the 3 Ivorian strains of ΦT XI exhibited between 9 and 16 within-colonies SNPs (average of 12 SNPs) (S5 Table), indicating a >10-fold higher mutation rate in these strains.
Hypermutator phenotype of ΦT XI Ivorian Y. enterocolitica strains
The higher mutation rate observed in the Ivorian strains of ΦT XI was suggestive of a hypermutator phenotype [51]. To determine whether ΦT XI Y. enterocolitica Ivorian isolates do have a higher rate of mutations than typical ΦT VIII strains, colonies from 3 ΦT XI strains: IP35471#5 (XIa), IP35477#3 (XIa) and IP35478#4 (XIb) were selected along with strain IP35463 that exhibits the usual ΦT VIII. The capacity to grow on nalidixic and rifampicin agar plates was evaluated for 10 colonies from each of these 4 strains. While very few colonies (≤3) of IP35463 ΦT VIII grew on rifampicin and nalidixic acid agar plates, an average of ≈1000 to 2000 spontaneous RifR and NalR mutants were observed in the 3 ΦT XI strains (Table 4). Our results thus demonstrate that some Ivorian Y. enterocolitica strains have a hypermutator phenotype.
Table 4. Average number of spontaneous RifR and NalR mutants in colonies with various phage types.
| Strain | Phage type | RifR mutantsa | NalR mutantsa |
|---|---|---|---|
| IP35463 | VIII | 1 | 3 |
| IP35471#5 | XIa | 1670 | 2071 |
| IP35477#3 | XIa | 1426 | 934 |
| IP35478#4 | XIb | 2002 | 937 |
a cfu number/109 cfu of the initial inoculum, average of 10 colonies.
A hypermutator phenotype has been linked to mutations in several genes involved in the fidelity of DNA replication, and more particularly in mutS [51]. When we looked at this gene in the genome of all 14 Ivorian strains, we observed that the 6 ΦT VIII strains had an intact and identical sequence, while all 8 ΦT XI strains, except IP35465, exhibited a 960 bp deletion at the 3' end of mutS (S1A Fig), corresponding to position 935,820 to 936,779 in the reference genome YE1203. This deletion would lead to the synthesis of a protein truncated of more than one third of its normal size (S1B Fig). Therefore the hypermutator phenotype most likely results from a large deletion of the mutS gene in some Ivorian isolates.
Discussion
Diarrheal diseases are a major public health problem in developing countries, with a high infant mortality rate in Africa [52]. Adapted therapeutic measures and control strategies are essential, but cannot be implemented without a proper identification of the etiological agents. Yersiniosis is the third most frequent bacterial disease causing human enteric infections in Europe [10], but reports of this disease are extremely infrequent in developing countries. In West Africa, only few countries reported the isolation of Yersinia from clinical cases [11–20], most likely because an active search for these bacteria is not performed. This is supported by the observation that in Nigeria, where studies were carried out to specifically look for this pathogen in human and animal samples, Yersinia strains were isolated [15–19]. A major reason for the lack or poor detection of these bacteria is the difficulty to recover them from poly-contaminated samples such as stools, which contain an abundant bacterial flora [53]. Indeed, Yersinia strains differ from other enterobacteria by a slower growth rate (48h instead of 24h) and an optimal growth temperature of 28°C instead of 37°C. Therefore, cultures performed under conditions suitable for most enteropathogens are not effective for the recovery of Yersinia colonies from polycontaminated biological samples. Specific procedures are required to enhance the isolation rate [53–55], but these procedures are time consuming and costly, and therefore are not performed on a routine basis in most laboratories in West Africa.
There were some indications that pathogenic Yersinia are circulating in the swine reservoir in the Abidjan area of Côte d'Ivoire, as a few pathogenic strains of Y. enterocolitica were isolated from pig carcasses at slaughter houses [30]. In this work we wanted to get an estimate of the number and distribution of Yersinia-infected pig farms, and most importantly to determine whether enteric yersiniosis is a cause of human diarrhea in the Abidjan district of Côte d'Ivoire. Using a procedure for the specific isolation of Yersinia strains that included several enrichment steps (growth at 25°C, addition of novobiocin, enrichment at 4°C, and growth on CIN agar), we were able to isolate 19 Yersinia strains from 781 samples of pig stools collected in 41 farms over 19 months.
Seven of these strains belonged to the non-pathogenic species Y. intermedia. This species was also previously recovered form pigs at slaughter in the Abidjan region, but the strains had biotypes or serotypes different from those of this study, and they were isolated from other areas [30]. Y. intermedia strains were also isolated from rectal or tongue swabs of healthy pigs in Nigeria [21,22], suggesting that the environmental conditions in West African countries are favorable for the maintenance of this non-pathogenic species.
The other 12 strains isolated from pig feces were pathogenic Y. enterocolitica. Pigs are regarded as the major reservoir of enteropathogenic Y. enterocolitica in most countries worldwide [56]. Although these animals were also found to be carriers of Yersinia in the Abidjan district, none of the other cattle sampled within these farms and of the rodents captured in the vicinity of the farms were found infected with Y. enterocolitica. Snails were also sampled because giant African snails may be abundant around the farms, they are widely consumed as a source of protein, and it was previously shown that they may carry enteropathogenic Y. enterocolitica for long periods of time [57]. However, none of the 95 snails analyzed were found infected. Therefore our findings support the hypothesis that, as in many other countries, pigs are the main reservoir of enteropathogenic Y. enterocolitica in the Abidjan district.
According to the 2012 annual report of the Department of Animal Production, over 60% of the national pig production is concentrated in the farms of the Abidjan District. At pig slaughterhouses, meat inspection is limited to a search for macroscopic lesions on the carcasses, without any microbiological investigations. The prevalence of infected pigs at slaughterhouses is usually higher than in farms because samplings are performed on tonsils, which are the most reliable tissue to evaluate the carriage of enteropathogenic Yersinia [58], while this cannot be done in live pigs owing to animal welfare. Since excretion of Y. enterocolitica in the feces is transient, the prevalence of infected pig farms (3/41) in the Abidjan district is thus most likely an underestimation of the risk of human exposure to yersiniosis upon consumption of pork meat.
Although some Yersinia strains were previously isolated from human stools in Côte d'Ivoire [29], their species and bioserotype were not determined, so it was not possible to establish whether they were enteropathogenic. Our active search for Yersinia in patients presenting with digestive disorders in the Abidjan district identified two human cases of Y. enterocolitica infections. This is the first demonstration that yersiniosis is a cause of human diarrhea in this country. These patients were two female infants from the same area (Yopougon). However, they were infected at 4 months interval, indicating that their infection was not caused by the consumption of the same contaminated product.
The 2 human strains had the same bioserotype 4/O:3 as the pig strains. Three strains of this bioserotype were also previously recovered from raw pig samples at slaughterhouse in the Abidjan district [30]. This bioserotype is the most frequently isolated from pigs and human cases in most countries worldwide [8], although 2/O:9 was the predominant bioserotype in Y. enterocolitica strains isolated from pigs and human cases in the recent years in Nigeria [18,20]. However, since the selective CIN agar we used for the enrichment procedure is inhibitory for some Y. enterocolitica strains of serotypes O:8 and O:9, and for Y. pseudotuberculosis [59,60], the possibility that other pathotypes of Yersinia circulate in the Abidjan region cannot be excluded.
The frequency of pathogenic Y. enterocolitica isolated from human stools greatly varies depending on the study, the country, the patients recruited, and the isolation procedures. For instance it was reported to be 0.19% (82/41,848 patients) in Finland [61], 4% (24/600 patients) in Palestine [62], 0.74% (36/4,841 patients) in the USA [63], 0.16% (6/3,784 patients) in UK [64], 0.6% (46/7,090 patients) in Crete [65], 2.46% (267/10,838 patients) in Belgium [66], 0.13% (3/2250 patients) in Canada [67], 0.42% (4/956 patients) in China [68], or 0% in Ireland (0/1,189 patients) [69] and the Netherlands (0/857 patients) [70]. In the Abidjan district, the isolation rate was 0.46% (2/427 patients), and therefore equivalent to or higher than those reported in several European countries (Finland, UK, Ireland and the Netherlands), Canada and China. This demonstrates that, although neglected, Y. enterocolitica may be a cause of human diarrhea as frequent in Côte d'Ivoire as in European countries.
As usually observed, the Y. enterocolitica 4/O:3 isolates from the Abidjan district were susceptible to most antibiotics commonly used to treat Gram-negative enteropathogens, and were resistant to penicillin and first and second-generation cephalosporin, due to the presence of the chromosomal blaA and blaB genes [71,72]. If the diagnosis of yersiniosis is not made, these classes of antibiotics may be used to treat patients, thus leading to treatment failure.
Although pigs and human patients were sampled all year round, pathogenic Y. enterocolitica strains were only isolated during a period extending from March to September. This period overlaps both the 2 dry seasons (March to May, and August to September) and the rainy season (May to August), with no major temperature variations along the year in the Abidjan district. The periodicity of the Y. enterocolitica carriage by pigs and consequent human infections may thus have causes independent of the climatic conditions.
The Ivorian Y. enterocolitica 4/O:3 isolates shared an identical PFGE and MLVA profile, and at the whole genome level they were genetically much closer to each other than to a non-Ivorian isolate. The relative clonality of the strains isolated in the Abidjan district is suggestive of a single import followed by the spread and diversification of the introduced strain. However, we noted that the Y. enterocolitica isolates could be phenotypically subdivided into ΦT VIII and ΦT XI, the latter being highly unusual. Indeed, in the collection of the French Yersinia Reference Laboratory, which includes numerous strains of worldwide origins, 7,853 out of the 8,295 Y. enterocolitica 4/O:3 that were phage typed were of ΦT VIII (94.7%), while only 25 of them (0.3%) were of ΦT XI. This unusual ΦT and the existence of ΦT subgroups within colonies of the same strain prompted us to perform a whole genome analysis. This analysis revealed that ΦT XI isolates had actually a hypermutator phenotype most likely caused by a large deletion at the 3' end of the mutS gene. In Escherichia coli, large deletions removing the 3' end of mutS have also been shown to cause an excessive rate of point mutations [73]. mutS is involved in DNA mismatch repair that ensures the fidelity of replication of the bacterial chromosome [74]. The variety of ΦT observed in colonies harboring the mutS deletion thus probably results from random mutations occurring at higher frequencies in various genes, some of them used by phages to enter or kill their bacterial hosts. Of note, only one ΦT XI strain (IP35465) carried an intact mutS gene, and this is the only ΦT XI isolate that grouped with ΦT VIII strains in the minimal spanning tree. It is therefore likely that this strain harbored a mutation in a gene conferring resistance to some phages, but because its genome was not prone to multiple mutations, it was genetically closer to the non-mutator strains of ΦT VIII.
Since the deletion of mutS was identical in all Ivorian isolates, this event probably occurred once in an ancestral strain from which the other mutS strains derived. The ability to rapidly expand mutant cell types is a clear advantage for pathogenic organisms to evade host defenses and drugs, and to adapt to stresses and changing environments [73]. The longer branches between Ivorian strains in the hypermutator cluster, as compared to the non-hypermutator cluster in the minimal spanning tree, are indicative of a faster genetic diversification of the mutator strains. This may thus lead to the expansion of pathogenic Y. enterocolitica strains with new phenotypes that may increase their capacity to multiply in their animal host or to cause severe infections in humans. Mutations leading to resistances to several classes of antibiotics may also arise at higher frequencies. Since the hypermutator phenotype is most likely caused by a large deletion of mutS, the reversion to a non-mutator phenotype is now hardly possible in these strains.
In conclusion, this study demonstrated that pathogenic Y. enterocolitica are circulating in the pig reservoir in Côte d'Ivoire and are causing human infections with a prevalence comparable to that of some European countries. The paucity of reports of this infection in African countries is most likely attributable to a lack of active detection rather than to an absence of the microorganism. The identification of hypermutator strains circulating in the pig reservoir and in humans may be of concern as these strains may acquire at a faster rate selective advantages that may increase their fitness, pathogenesis or resistance to commonly used treatments.
Supporting Information
(PDF)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Acknowledgments
The authors thank Nadine Delarue for her help in bibliography mining and Liliane Martin for phenotypic characterization of the strains. We benefitted from the expertise of the PibNet platform at the Institut Pasteur in Paris for whole genome sequencing of the Y. enterocolitica strains.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was funded by Gouverment of COTE D'IVOIRE through the budget of Institut Pasteur de Cote d'Ivoire (www.pasteur.ci) in the chapiters 600.2 and 620.9. Insitut Pasteur of COTE D'IVOIRE design the study, purchased reagent for Yersinia strains isolation and paid for data collection and sampling. The director gave his decision to publish this study. This study was conducted in collaboration with Pasteur Institute in Paris (France www.pasteur.fr). I received from this Institute a grant (Ref: EC / MAM / No 99/14) for three months stay in Yersinia research unit to perform strains characterization by MLVA PFGE and sequencing. The members of this research unit contributed highly to data analysis and to the drafting of the manuscript.
References
- 1.Rosner BM, Werber D, Hoehle M, Stark K. Clinical aspects and self-reported symptoms of sequelae of Yersinia enterocolitica infections in a population-based study, Germany 2009–2010. BMC Infect Dis. 2013; 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perdikogianni C, Galanakis E, Michalakis M, Giannoussi E, Maraki S, Tselentis Y, et al. Yersinia enterocolitica infection mimicking surgical conditions. Pediatr Surg Int. 2006. [DOI] [PubMed] [Google Scholar]
- 3.Savin C, Carniel E. Les diarrhées d'origine bactérienne: le cas de Yersinia enterocolitica. Rev Francoph Lab. 2008; 400: 49–58. [Google Scholar]
- 4.Bottone EJ. Yersinia enterocolitica: the charisma continues. Clinical Microbiology Reviews. 1997; 10: 257–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fredriksson-Ahomaa M, Korkeala H. Molecular epidemiology of Yersinia enterocolitica 4/O:3. Adv Exp Med Biol. 2003; 529: 295–302. 10.1007/0-306-48416-1_56 [DOI] [PubMed] [Google Scholar]
- 6.Nuorti JP, Niskanen T, Hallanvuo S, Mikkola J, Kela E, Hatakka M, et al. A widespread outbreak of Yersinia pseudotuberculosis O:3 infection from iceberg lettuce. J Infect Dis. 2004; 189: 766–774. 10.1086/381766 [DOI] [PubMed] [Google Scholar]
- 7.Wauters G, Kandolo K, Janssens M. Revised Biogrouping Scheme of Yersinia enterocolitica. Contrib Microbiol Immunol. 1987; 9: 14–21. [PubMed] [Google Scholar]
- 8.Drummond N, Murphy BP, Ringwood T, Prentice MB, Buckley JF, Fanning S. Yersinia enterocolitica: A Brief Review of the Issues Relating to the Zoonotic Pathogen, Public Health Challenges, and the Pork Production Chain. Foodborne Pathog Dis. 2012; 9: 179–189. 10.1089/fpd.2011.0938 [DOI] [PubMed] [Google Scholar]
- 9.Kapperud G. Yersinia enterocolitica in food hygiene. Int J Food Microbiol. 1991; 12: 53–65. [DOI] [PubMed] [Google Scholar]
- 10.Eurosurveillance-Editorial-Team. The 2013 joint ECDC/EFSA report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks published. Eurosurveillance. 2015; 20: 43–43. [DOI] [PubMed]
- 11.Simpore J, Ouermi D, Ilboudo D, Kabre A, Zeba B, Pietra V, et al. Aetiology of acute gastro-enteritis in children at Saint Camille Medical Centre, Ouagadougou, Burkina Faso. Pak J Biol Sci. 2009; 12: 258–263. [DOI] [PubMed] [Google Scholar]
- 12.Gendrel D, Sitbon M, Richard-Lenoble D, Galliot A, Kombila M, Ivanoff B, et al. Etiologies des gastroentérites aigues infantiles au Gabon. Bull Soc Pathol Exot. 1985; 78: 290–295. [PubMed] [Google Scholar]
- 13.Adjei AA, Kuma GK, Tettey Y, Ayeh-Kumi PF, Opintan J, Apeagyei F, et al. Bacterial Contamination of Blood and Blood Components in Three Major Blood Transfusion Centers, Accra, Ghana. Jap J Infect Dis. 2009; 62: 265–269. [PubMed] [Google Scholar]
- 14.Frickmann H, Dekker D, Boahen K, Acquah S, Sarpong N, Adu-Sarkodie Y, et al. Increased detection of invasive enteropathogenic bacteria in pre-incubated blood culture materials by real-time PCR in comparison with automated incubation in Sub-Saharan Africa. Scand J Infect Dis. 2013; 45: 616–622. 10.3109/00365548.2013.777777 [DOI] [PubMed] [Google Scholar]
- 15.Anjorin FI, Sturrock RD, Subbuswamy SG, Lawande RV, Fakunle YM. Yersinia enterocolitica infection from West Africa—a case report. Trans Roy Soc Trop Med Hyg. 1979; 73: 634–635. [DOI] [PubMed] [Google Scholar]
- 16.Agbonlahor DE, Odugbemi TO, Lasi Q. Isolation of Yersinia enterocolitica from stools of acute gastroenteritis cases in Lagos, Nigeria. East Afr Med J. 1981; 58: 520–524. [PubMed] [Google Scholar]
- 17.Agbonlahor DE, Odugbemi TO, Dosunmu-Ogunbi O. Isolation of species of Yersinia from patients with gastroenteritis in Nigeria. J Med Microbiol. 1983; 16: 93–96. 10.1099/00222615-16-1-93 [DOI] [PubMed] [Google Scholar]
- 18.Okwori AEJ, Agada GOA, Olabode AO, Agina SE, Okpe ES, Okopi J. The prevalence of pathogenic Yersinia enterocolitica among diarrhea patients in Jos, Nigeria. Afr J Biotech. 2007; 6: 1031–1034. [Google Scholar]
- 19.Onyemelukwe NF. Yersinia enterocolitica as an aetiological agent of childhood diarrhoea in Enugu, Nigeria. Cent Afr J Med. 1993; 39: 192–195. [PubMed] [Google Scholar]
- 20.Okwori AEJ, Martinez PO, Fredriksson-Ahomaa M, Agina SE, Korkeala H. Pathogenic Yersinia enterocolitica 2/O:9 and Yersinia pseudotuberculosis 1/O:1 strains isolated from human and non-human sources in the Plateau State of Nigeria. Food Microbiol. 2009; 26: 872–875. 10.1016/j.fm.2009.06.001 [DOI] [PubMed] [Google Scholar]
- 21.Lombin LH, Adesiyun AA, Agbonlahor DE, Kwaga JKP. Isolation of Yersinia species from Pigs in Nigeria. Vet Rec. 1985; 117: 364–364. [DOI] [PubMed] [Google Scholar]
- 22.Agbonlahor DE, Adesiyun AA, Kwaga KP, Lombin LH. Colonial, biochemical and serological characteristics of Yersinia species isolated from animals in Nigeria. Rev Elev Méd Vét Pays Trop. 1985; 38: 416–422. [PubMed] [Google Scholar]
- 23.Adesiyun AA, Agbonlahor DE, Lombin LH, Kwaga JK. Occurrence of virulence markers in species of Yersinia isolated from animals in Nigeria. Vet Microbiol. 1986; 12: 289–294. [DOI] [PubMed] [Google Scholar]
- 24.Chambron J, Bourdin M. A propos d'une enquête bactériologique sur les ganglions mésentériques de porcs à Dakar. Recherche négative deYersinia enterocolitica. Intérêt épidémiologique d'une telle recherche. Méd Afr Noire. 1971; 18: 823–826. [Google Scholar]
- 25.Hakalehto E, Nyholm O, Bonkoungou IJ, Kagambega A, Rissanen K, Heitto A, et al. Development of microbiological field methodology for water and food-chain hygiene analysis of Campylobacter spp. and Yersinia spp. in Burkina Faso, West Africa. Pathophysiology. 2014; 21: 219–229. 10.1016/j.pathophys.2014.07.004 [DOI] [PubMed] [Google Scholar]
- 26.Lombin LH, Adesiyun AA, Haruna M, Kwaga JKP, Agbonlahor DE. A survey for Yersinia enterocolitica in water from ponds, streams and wells in Northern Nigeria. Microbiologica. 1986; 9: 95–100. [PubMed] [Google Scholar]
- 27.Savin C, Leclercq A, Laurent E, Carniel E, Vaillant V. National survey on diagnosis of enteropathogenic Yersinia infections in metropolitan France, 2003. Bull Epid Hebd. 2010: 307–311. [Google Scholar]
- 28.Zinsstag J, Schelling E, Roth F, Bonfoh B, de Savigny D, Tanner M. Human Benefits of Animal Interventions for Zoonosis Control. Emerg Infect Dis. 2007; 13: 527–531. 10.3201/eid1304.060381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavoie MC. Identification of strains isolated as total and fecal coliforms and comparison of both groups as indicators of fecal pollution in tropical climates. Can J Microbiol. 1983; 29: 689–693. [DOI] [PubMed] [Google Scholar]
- 30.Atobla K, Kakou-Ngazoa S, Dadié AT, Karou TG, Dosso M, Ahonzo-Niamké LS, et al. Characterization of Yersinia spp. strains isolated from pigs in Abidjan, Côte d’Ivoire, West Africa. Afr J Microbiol Res. 2014; 8: 1909–1915. [Google Scholar]
- 31.Laukkanen R, Hakkinen M, Lunden J, Fredriksson-Ahomaa M, Johansson T, Korkeala H. Evaluation of isolation methods for pathogenic Yersinia enterocolitica from pig intestinal content. J Appl Microbiol. 2010; 108: 956–964. 10.1111/j.1365-2672.2009.04494.x [DOI] [PubMed] [Google Scholar]
- 32.Aulisio CC, Mehlman IJ, Sanders AC. Alkali method for rapid recovery of Yersinia enterocolitica and Yersinia pseudotuberculosis from foods. Appl Environ Microbiol. 1980; 39: 135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicolle P, Mollaret HH, Brault J. Nouveaux résultats sur la lysotypie de Yersinia enterocolitica portant sur plus de 4000 souches d'origines diverses. Rev Epidem Santé Publ. 1976; 24: 479–496. [PubMed] [Google Scholar]
- 34.Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Tech Bull Regist Med Technol. 1966; 36: 49–52. [PubMed] [Google Scholar]
- 35.Ausubel FM, Brent R., Kingston R.E., o D., Seidman J.G., Smith J.A., Struhl K. Current Protocols in Molecular Biology. 1999; 1–2: 2.1.1, 10.11.14. [Google Scholar]
- 36.Martin L, Cabanel N, Lesoille C, Menard T, Carniel E. Investigation of an unusual increase in human yersinioses in Creuse, France. Int J Infect Dis. 2015; 34: 76–78. 10.1016/j.ijid.2015.03.011 [DOI] [PubMed] [Google Scholar]
- 37.Gierczynski R, Golubov A, Neubauer H, Pham JN, Rakin A. Development of multiple-locus variable-number tandem-repeat analysis for Yersinia enterocolitica subsp. palearctica and its application to bioserogroup 4/O3 subtyping. J Clin Microbiol. 2007; 45: 2508–2515. 10.1128/JCM.02252-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Criscuolo A, Brisse S. AlienTrimmer: a tool to quickly and accurately trim off multiple short contaminant sequences from high-throughput sequencing reads. Genomics. 2013; 102: 500–506. 10.1016/j.ygeno.2013.07.011 [DOI] [PubMed] [Google Scholar]
- 39.Crusoe MR, Alameldin HF, Awad S, Boucher E, Caldwell A, Cartwright R, et al. The khmer software package: enabling efficient nucleotide sequence analysis. F1000Res. 2015; 4: 900 10.12688/f1000research.6924.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Schroder J, Schmidt B. Musket: a multistage k-mer spectrum-based error corrector for Illumina sequence data. Bioinformatics. 2013; 29: 308–315. 10.1093/bioinformatics/bts690 [DOI] [PubMed] [Google Scholar]
- 41.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011; 27: 2957–2963. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012; 19: 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012; 22: 568–576. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le Guern AS, Martin L, Savin C, Carniel E. Yersiniosis in France: overview and potential sources of infection. Int J Infect Dis. 2016; 46: 1–7. 10.1016/j.ijid.2016.03.008 [DOI] [PubMed] [Google Scholar]
- 45.Jaakkola K, Somervuo P, Korkeala H. Comparative Genomic Hybridization Analysis of Yersinia enterocolitica and Yersinia pseudotuberculosis Identifies Genetic Traits to Elucidate Their Different Ecologies. Biomed Res Int. 2015; 2015: 760494 10.1155/2015/760494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicolle P, Mollaret HH, Brault J. La lysotypie de Yersinia enterocolitica: Arguments géographiques, zoologiques, antigèniques et biochimiques plaidant en sa faveur. Bull Acad Natl Méd. 1976; 160: 404–408. [Google Scholar]
- 47.Pham JN, Bell SM, Hardy MJ, Martin L, Guiyoule A, Carniel E. Susceptibility to beta-lactam agents of Yersinia entercolitica biotype 4, serotype 0:3 isolated in various parts of the world. J Med Microbiol. 1995; 43: 9–13. 10.1099/00222615-43-1-9 [DOI] [PubMed] [Google Scholar]
- 48.Hammerberg S, Sorger S, Marks MI. Antimicrobial Susceptibilities of Yersinia enterocolitica Biotype 4, Serotype 0:3. Antimicrob Agents Chemother. 1977; 11: 566–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilpin BJ, Robson B, Lin S, Hudson JA, Weaver L, Dufour M, et al. The limitations of pulsed-field gel electrophoresis for analysis of Yersinia enterocolitica isolates. Zoon Publ Health. 2014; 61: 405–410. [DOI] [PubMed] [Google Scholar]
- 50.Skurnik M, Toivanen P. Yersinia enterocolitica lipopolysaccharide: genetics and virulence. Trends Microbiol. 1993; 1: 148–152. [DOI] [PubMed] [Google Scholar]
- 51.Woodford N, Ellington MJ. The emergence of antibiotic resistance by mutation. Clin Microbiol Infect. 2007; 13: 5–18. 10.1111/j.1469-0691.2006.01492.x [DOI] [PubMed] [Google Scholar]
- 52.Boschi-Pinto C, Velebit L, Shibuya K. Estimating child mortality due to diarrhoea in developing countries. World Health Org Bull. 2008; 86: 710–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fredriksson-Ahomaa M, Korkeala H. Low occurrence of pathogenic Yersinia enterocolitica in clinical, food, and environmental samples: a methodological problem. Clin Microbiol Rev. 2003; 16: 220–229. 10.1128/CMR.16.2.220-229.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Denis M, Houard E, Labbe A, Fondrevez M, Salvat G. A Selective Chromogenic Plate, YECA, for the Detection of Pathogenic Yersinia enterocolitica: Specificity, Sensitivity, and Capacity to Detect Pathogenic Y. enterocolitica from Pig Tonsils. J Pathog. 2011; 2011: 296275 10.4061/2011/296275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wauters G, Goossens V, Janssens M, Vandepitte J. New enrichment method for isolation of pathogenic Yersinia enterocolitica serogroup O:3 from pork. Appl Environ Microbiol. 1988; 54: 851–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laukkanen-Ninios R, Fredriksson-Ahomaa M, Korkeala H. Enteropathogenic Yersinia in the Pork Production Chain: Challenges for Control. Compr Rev Food Sci Food Safety. 2014; 13: 1165–1191. [Google Scholar]
- 57.Igumor EO, Ogbimi AI, Agbonlahor DE, Ndip RN. Carriage of Yersinia species by snails (Achachatina marginata): an epidemiological survey. Cent Afr J Med. 1994; 40: 102–104. [PubMed] [Google Scholar]
- 58.Thibodeau V, Frost EH, Chenier S, Quessy S. Presence of Yersinia enterocolitica in tissues of orally-inoculated pigs and the tonsils and feces of pigs at slaughter. Can J Vet Res. 1999; 63: 96–100. [PMC free article] [PubMed] [Google Scholar]
- 59.Fukushima H, Gomyoda M. Inhibition of Yersinia enterocolitica serotype O3 by natural microflora of pork. Appl Environ Microbiol. 1986; 51: 990–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bosi E, Madie P, Wilks CR. Growth of Yersinia pseudotuberculosis on selective media. New Zealand Vet J. 1994; 42: 35. [DOI] [PubMed] [Google Scholar]
- 61.Sihvonen LM, Haukka K, Kuusi M, Virtanen MJ, Siitonen A. Yersinia enterocolitica and Y. enterocolitica-like species in clinical stool specimens of humans: identification and prevalence of bio/serotypes in Finland. Eur J Clin Microbiol Infect Dis. 2009; 28: 757–765. 10.1007/s10096-008-0696-y [DOI] [PubMed] [Google Scholar]
- 62.El Qouqa IA, El Jarou MA, Abu Samaha AS, Al Afifi AS, Al Jarousha AMK. Yersinia enterocolitica infection among children aged less than 12 years: a case-control study. Int J Infect Dis. 2011; 15: E48–E53. 10.1016/j.ijid.2010.09.010 [DOI] [PubMed] [Google Scholar]
- 63.Lee LA, Taylor J, Carter GP, Quinn B, Farmer JJ, Tauxe RV. Yersinia enterocolitica O:3—An Emerging Cause of Pediatric Gastroenteritis in the United States. The Yersinia enterocolitica Collaborative Study Group. J Infect Dis. 1991; 163: 660–663. [DOI] [PubMed] [Google Scholar]
- 64.Greenwood M, Hooper WL. Human carriage of Yersinia spp. J Med Microbiol. 1987; 23: 345–348. 10.1099/00222615-23-4-345 [DOI] [PubMed] [Google Scholar]
- 65.Maraki S, Georgiladakis A, Tselentis Y, Samonis G. A 5-year study of the bacterial pathogens associated with acute diarrhoea on the island of Crete, Greece, and their resistance to antibiotics. Eur J Epidemiol. 2002; 18: 85–90. [DOI] [PubMed] [Google Scholar]
- 66.Walckiers D, Stroobant A, Vandepitte J, Verbist L, Wauters G. Surveillance des maladies infectieuses en Belgique par un réseau de laboratoire vigies. Application à Campylobacter et Yersinia enterocolitica. Expérience de sept années. Méd Mal Infect. 1991; 21: 244–249. [Google Scholar]
- 67.Noble MA, Barteluk RL. Species and serotypes of yersiniae recovered in British Columbia. Can J Publ Hlth. 1988; 79: 130–133. [PubMed] [Google Scholar]
- 68.Zheng XB, Xie C. Isolation, characterization and epidemiology of Yersinia enterocolitica from humans and animals. J Appl Bacteriol. 1996; 81: 681–684. [DOI] [PubMed] [Google Scholar]
- 69.Ringwood T, Murphy BP, Drummond N, Buckley JF, Coveney AP, Redmond HP, et al. Current evidence for human yersiniosis in Ireland. Eur J Clini Microbiol Infect Dis. 2012; 31: 2969–2981. [DOI] [PubMed] [Google Scholar]
- 70.de Wit MA, Koopmans MP, Kortbeek LM, van Leeuwen NJ, Vinje J, van Duynhoven YT. Etiology of gastroenteritis in sentinel general practices in the netherlands. Clin Infect Dis. 2001; 33: 280–288. 10.1086/321875 [DOI] [PubMed] [Google Scholar]
- 71.Seoane A, Garcia Lobo JM. Cloning of chromosomal beta-lactamase genes from Yersinia enterocolitica. J Gen Microbiol. 1991; 137: 141–146. 10.1099/00221287-137-1-141 [DOI] [PubMed] [Google Scholar]
- 72.Bonke R, Wacheck S, Stuber E, Meyer C, Martlbauer E, Fredriksson-Ahomaa M. Antimicrobial susceptibility and distribution of beta-lactamase A (blaA) and beta-lactamase B (blaB) genes in enteropathogenic Yersinia species. Microb Drug Resist. 2011; 17: 575–581. 10.1089/mdr.2011.0098 [DOI] [PubMed] [Google Scholar]
- 73.Horst JP, Wu TH, Marinus MG. Escherichia coli mutator genes. Trends Microbiol. 1999; 7: 29–36. [DOI] [PubMed] [Google Scholar]
- 74.Harfe BD, Jinks-Robertson S. DNA mismatch repair and genetic instability. Annu Rev Genet. 2000; 34: 359–399. 10.1146/annurev.genet.34.1.359 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.