Abstract
Non-invasive imaging of gene expression can be used to track implanted cells in vivo but often requires the addition of an exogenous contrast agent that may have limited tissue access1. We show that the urea transporter (UT-B) can be used as a gene reporter, where reporter expression was detected using 1H MRI measurements of UT-B-mediated increases in plasma membrane water exchange. HEK cells transfected with the reporter showed an increased apparent water exchange rate (AXR), which increased in line with UT-B expression. AXR values measured in vivo, in UT-B-expressing HEK cell xenografts, were significantly higher (~ 2-fold, p < 0.0001), compared with non-expressing controls and fluorescence imaging of a red fluorescent protein (mStrawberry), co-expressed with UT-B, showed that UT-B expression correlated in a linear fashion with AXR. Transduction of rat brain cells in situ with a lentiviral vector expressing UT-B resulted in a ~ 2-fold increase in AXR at the site of virus injection.
Most imaging gene reporters are receptors, enzymes, or membrane transporters that generate image contrast by interaction with an exogenous imaging agent1. Imaging reporter expression in vivo could be used in the clinic to monitor delivery of gene therapy vectors or for cell tracking in regenerative medicine and in immunotherapy2, 3. Compared with direct labelling approaches that preload cells with contrast agents, gene reporters have the advantage that contrast arises from protein expression, which reports indirectly on cell viability, and that the signal is not diluted after cell division. Moreover, if the reporter is expressed from a tissue specific promoter the location of the cell and its differentiation state can be detected.
Various gene reporters have been described for use with MRI, where these rely on different underlying contrast mechanisms4 with most using exogenous contrast agents that alter longitudinal (T1) or transverse relaxation times (T2, T2*)5. Reporters that produce endogenous contrast are potentially more useful, because they do not require contrast agent delivery. Endogenous reporters include those that accumulate iron, such as tyrosinase, transferrin, MagA and ferritin, although they may need to be supplemented with exogenous iron6, 7. Another endogenous reporter is the lysine-rich protein (LRP), which contains multiple exchangeable amide protons that can be detected using chemical exchange saturation transfer CEST 8, 9. We recently identified a gene reporter based on the urea transporter (UT-B), whose expression was detected in vivo using 13C magnetic resonance spectroscopy measurements of the apparent diffusion coefficient (ADC) of injected hyperpolarized 13C urea10. UT-Bs also function as aqueous channels, transporting water at rates similar to aquaporins11. Therefore we hypothesized that UT-B expression might also be detectable by 1H MRI measurements of water exchange across the plasma membrane, thus allowing imaging of gene expression.
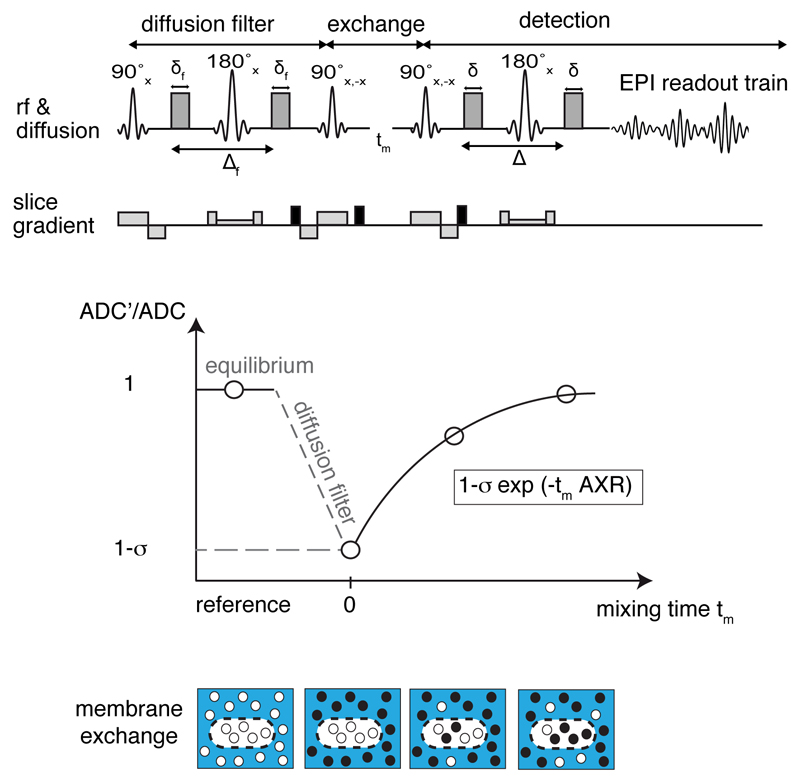
Intracellular water diffusion is restricted by the plasma membrane and hindered by intracellular macromolecules, resulting in a lower apparent diffusion coefficient for intracellular as compared to extracellular water12. Filter exchange spectroscopy (FEXSY) uses a “diffusion filter” to remove magnetization from “fast-diffusing” extracellular water and then measures the return of the magnetization to equilibrium following water exchange between the extra- and intracellular compartments (Fig. 1). An imaging version of FEXSY (Filter-Exchange Imaging (FEXI)) can also be used to measure the apparent exchange rate (AXR) between these compartments13.
Figure 1. Imaging transmembrane water-exchange.
a, The filter-exchange imaging (FEXI) method consists of a diffusion filter module, a storage/exchange module, and signal detection using an EPI-readout. b, At equilibrium, with no diffusion filter, a reference ADC is acquired. When the diffusion filter is applied, signal from fast diffusing extracellular water is suppressed, indicated by the black dots, resulting in a different weighting of the signal contributions from the two compartments and leading to a reduction in the measured ADC’. This decrease in ADC is determined by the filter efficiency σ. With increasing mixing times, tm, between the filter and detection modules, exchange of water between the two compartments results in an increase in the measured ADC. Relaxation back to the equilibrium ADC can be described by the apparent exchange rate constant, AXR.
We used FEXI to detect UT-B transgene expression in vivo (see Supplementary Material S1 and Figs. S1 and S2 for validation of this imaging methodology), and introduce this as a new endogenous MRI gene reporter that produces higher levels of contrast than endogenous reporters described previously.
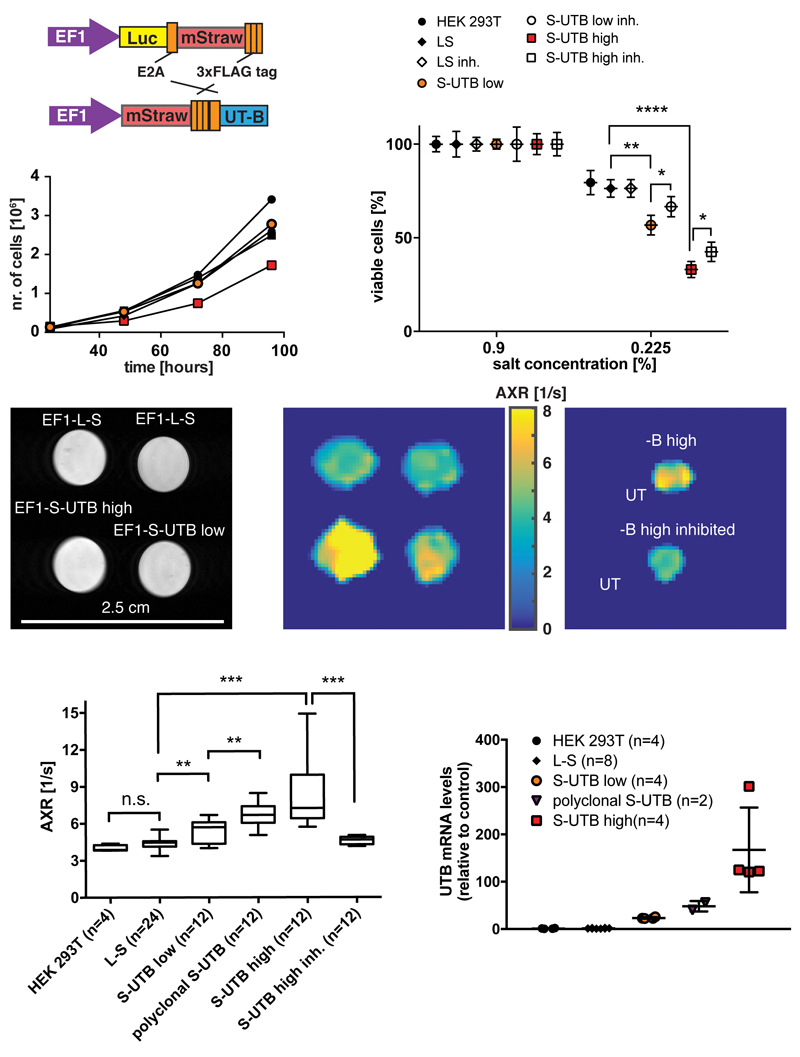
HEK 293T cells were transduced with a vector co-expressing, via an E2A sequence, luciferase and the red fluorescent protein, mStrawberry, or a vector co-expressing mStrawberry and UT-B (Fig. 2 a). The levels of mStrawberry fluorescence provided a surrogate measure of luciferase and UT-B protein expression. We used these cells to derive six different cell lines, five of which expressed UT-B. Lines included a monoclonal control cell line expressing Luciferase and mStrawberry (EF1-L-S), a polyclonal line expressing mStrawberry and UT-B (polyclonal EF1-S-UTB), and monoclonal populations isolated using single-cell-sorting for low and high mStrawberry expression, co-expressing low and high levels of UT-B (EF1-S-UTB low, and EF1-S-UTB high) respectively (Supplementary Fig. S3). We also used the PGK-S-UTB cell line, which we used previously in urea transport studies10. Very high levels of UT-B expression were measured in EF1-S-UTB high cells (Figure 2 h and Supplementary Fig. S3), which inhibited their growth (Fig 2 b). Water transport by UT-B was assessed by measuring decreased cell viability after incubation in hypotonic salt solutions for 5 min (see Methods). In a hypotonic 0.225 % salt solution expression of UT-B resulted in up to 47 % and 24 % decreases in viability of EF1-S-UTB high cells and EF1-S-UTB low cells, respectively, when compared to control cells. This effect was partially reversed by addition of the UT-B inhibitor N,N’-Dimethylthiourea, showing that at these expression levels UT-B mediates relatively rapid water transport across the plasma membrane (Fig 2 c).
Figure 2. Measurements of water transport by UT-B in cells expressing UT-B at different levels.
a, Cells were transduced with viral vectors expressing mStrawberry and either UT-B (EF1-S-UTB) or Luciferase (EF1-S-L). Cells were selected that expressed either high (EF1-S-UTB high) or low (EF1-S-UTB low) levels of UT-B. b, Effect of UTB expression on cell growth. Viable cell numbers were determined using an automated ViCell cell counter every 24 h after seeding 1 x 106 cells. The results are the mean of three independent experiments. c, Assay of UT-B function. Viable cells were counted using a ViCell analyser after 5 min of incubation in NaCl concentrations of 0.9 % (isosmotic) and 0.225 % (hypoosmotic). Increased UT-B expression accelerated cell swelling and bursting in the hypoosmotic solution. The loss of viability in cells expressing high levels of UT-B was inhibited by incubating the cells with 170 mM N,N’-Dimethylthiourea, which did not affect cell viability at 0.9 % salt concentration. Each cell line was measured in triplicate. d, Reference proton image of tubes containing dense pellets of the indicated cell lines. e, AXR maps of the cell pellets shown in d). f, AXR map of pellets of EF1-S-UTB high cells. For one of the pellets the cells had been incubated with 170 mM N-N’-Dimethylthiourea. g, Independent experiments show increased AXR with increasing UT-B expression (n = 4-24). h, Real-time PCR measurements of UT-B mRNA expression levels. Data shown in g and h are mean ± SD. Data shown in g are shown in a box-and-whisker representation with the box extending from 25th to 75th percentiles and the whiskers ranging from the minimum to the maximum data point. The line in within the box is the data median. Significance was tested using a Mann-Whitney-Wilcoxon test; significance levels: n.s. p > 0.05, * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001.
FEXI images of pellets of HEK 293T, EF1-L-S, polyclonal, EF1-S-UTB, EF1-S-UTB low and EF1-S-UTB high cell lines, revealed an increase in AXR in parallel with increases in UT-B expression (Fig. 2 d - g). When corrected for background levels of water transport (in HEK 293T and EF1-L-S cells) the increase in AXR correlated with the increase in UT-B expression, as determined from measurements of UT-B mRNA levels using real-time PCR (Fig. 2 h, Supplementary Fig. S4, Spearman r = 1, p = 0.0083; linear regression with R2 = 0.88 and slope significantly different from zero, p = 0.018). Cell viability was > 95% before and after imaging. Inhibition of UT-B transporter activity in the EF1-S-UTB high cell line, with 170 mM N-N’-Dimethylthiourea, decreased AXR (p < 0.001, Mann-Whitney-Wilcoxon test) to a value similar to that observed in control cells (Fig. 2 f, g). To assess whether exchangeable protons on intracellular proteins might contribute to the measured AXR, we performed measurements on solutions of 35% w/w and 4 % w/w bovine serum albumin (BSA). At both high and low BSA concentrations, there was no measurable AXR (Supplementary Fig. S5).
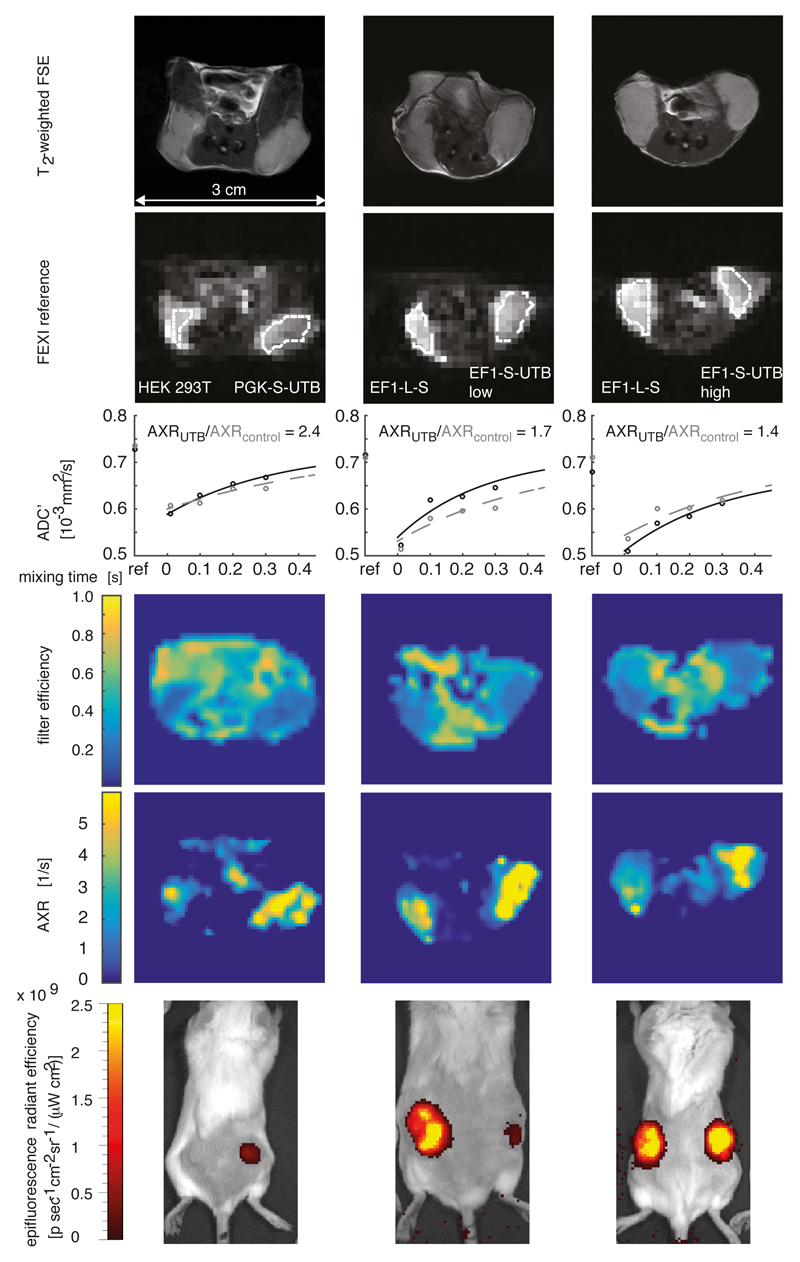
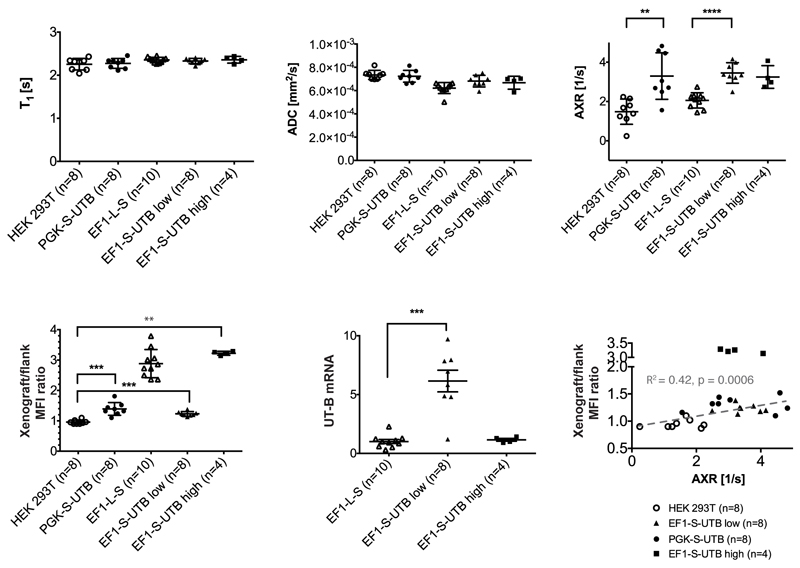
FEXI images were acquired from 8 female mice implanted with EF1-L-S cells in one flank and EF1-S-UTB low cells on the contralateral side. A second group of 4 female mice was implanted with EF1-L-S cells in one flank and EF1-S-UTB high cells on the contralateral side. In two of these animals the EF1-L-S cells did not form xenografts. A third group of 8 female mice were implanted with HEK 293T cells in one flank and PGK-S-UTB cells on the contralateral side. AXR maps of three representative animals from each group are shown in Fig. 3 and in Supplementary Fig. S4. Mean AXR values were calculated from regions-of-interest drawn manually around the xenografts (Fig. 3 a i) – a iii); b i) – b iii)). Mean AXR values in the UT-B-expressing xenografts were significantly higher (p < 0.005, unpaired Mann-Whitney-Wilcoxon test), with a mean AXR of 3.45 ± 0.52 s-1 for EF1-S-UTB low (n=8), 3.25 ± 0.58 s-1 for EF1-S-UTB high (n=4), 3.30 ± 1.18 s-1 for PGK-S-UTB (n=8), 1.48 ± 0.64 s-1 for HEK 293T (n=8), and 2.05 ± 0.39 s-1 for the EF1-L-S (n=10) xenografts (representative fits from each group are shown in Fig. 3 c i) – c iii). Individual AXR and mean values are shown in Fig. 4 c. Filter efficiency (FE) maps (Fig. 3 d i) – d iii)) showed reduced FE in all xenografts, when compared to surrounding tissue. AXR maps (Fig. 3 e i) – e iii)) identified UT-B expressing cells as areas with high transmembrane water exchange, which allowed non-invasive imaging of reporter gene expression with a spatial resolution of 0.93 mm x 0.93 mm x 3 mm. Fluorescence imaging confirmed expression of the UT-B transgene, which was co-expressed with the mStrawberry transgene. Xenograft/flank mean fluorescence intensity ratios were highest in the xenografts grown from cells that had been sorted for the highest levels of red fluorescence; EF1-L-S (2.88 ± 0.46) and EF1-S-UTB high (3.22 ± 0.07), whereas lower fluorescence intensity ratios were observed for PGK-S-UTB (1.31 ± 0.14) and EF1-S-UTB low xenografts (1.23 ± 0.08) as compared to the background from HEK 293T cells (0.95 ± 0.08, Fig. 3 f i) – f iii)). The similar fluorescence intensity ratios for PGK-S-UTB and EF1-S-UTB low xenografts was consistent with their similar AXR values. T1 and ADC values did not show significant differences between UT-B expressing and control xenografts (Fig. 4 a,b), whereas AXR and fluorescence measurements consistently detected transgene expression (Fig. 4 c,d). Remarkably, MRI measurements of AXR had a similar sensitivity to the fluorescence measurements for detecting transgene expression (Fig. 4 c,d), with the caveat that red fluorescence is typically detected from penetration depths up to a few millimetres14, whereas the MRI measurements were collected from a 3 mm thick slice. Moreover, the contrast was higher for the MRI measurements (AXR was 2.33 x higher in EF1-S-UTB low as compared to HEK 293T xenografts, whereas the ratio of the xenograft/flank mean fluorescence intensity ratios was only 1.30).
Figure 3. Magnetic resonance imaging (MRI) of UT-B reporter gene expression in xenografts.
a i) - a iii), T2-weighted fast spin echo images of HEK 293T and PGK-S-UT-B xenografts (i), EF1-L-S and EF1-S-UT-B low xenografts (ii) and EF1-L-S and EF1-S-UT-B high xenografts (iii). b i) – b iii), Reference FEXI image (no diffusion-filter). The xenografts are outlined with a broken white line. c i) – c iii), Fits of ADC’ to Equation 1 (see Methods section). The solid black line shows the fit for a region-of-interest (ROI) containing UT-B expressing cells and the dotted grey line shows the fit for an ROI from control cells. The ratio of the AXR values AXRUTB/AXRcontrol is shown as a measure of image contrast. d i) – d iii), FE maps highlight xenograft regions as regions with decreased filter efficiency. e i) – e iii), AXR maps show regions of high transmembrane water exchange in UT-B expressing xenografts. Both the FE and AXR maps were calculated on a pixel-by-pixel basis and were zero-filled by a factor of two to give a 64 x 64 matrix. f i) – f iii), Background-corrected mean fluorescence intensity shows mStrawberry-E2A-UT-B expression on the right flanks, confirming expression of the UT-B transgene in vivo. No red fluorescence was detected in control cells (left flank in f i)). Red fluorescence was also detected in the control EF1-L-S xenografts (left flanks in f ii) and f iii)).
Figure 4. FEXI measurements of UT-B expression in vivo.
a – c, Effect of UT-B expression on T1, ADC and AXR values. d Xenograft/flank mStrawberry mean fluorescence intensity (MFI) ratio. Significance was tested using a Mann-Whitney-Wilcoxon test (significance levels: ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001). e, UT-B mRNA levels (relative to EF1-L-S control cells) in the indicated xenografts. f, Correlation plot between individual xenograft/flank MFI ratios and AXR in the indicated xenografts. With the exception of the EF1-S-UTB high xenografts, there was a linear correlation between AXR and fluorescence intensity ratio (Spearman r = 0.64, p = 0.0008; linear regression with R2 = 0.42 and slope significantly different from zero, p = 0.0006).
The xenograft/flank mean fluorescence intensity ratio correlated with the AXR values (Spearman r = 0.64, p = 0.0008; linear regression with R2 = 0.42 and slope significantly different from zero, p = 0.0006, Fig. 4 f), demonstrating that AXR can be used as a quantitative readout of UT-B gene expression. However, the negative selection pressure imparted by the lower growth rate of the EF1-S-UTB high cells (Fig. 2 b) was manifest in the EF1-S-UTB high xenografts as a loss of UT-B mRNA expression (Fig. 4 e) and an AXR value that was lower than in the EF1-S-UTB low xenografts (Fig. 4 c). Immunostaining confirmed that mStrawberry expression was higher in the EF1-L-S and EF1-S-UTB high xenografts than in the EF1-S-UTB low xenograft and H&E staining showed that this had no discernible effect on cell viability in the xenografts (Supplementary Fig. S6).
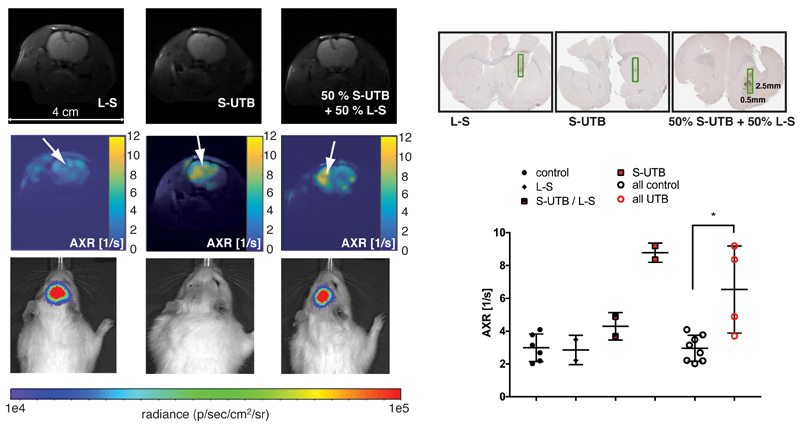
Rat brain cells were transduced in vivo by direct intracranial injection of lentiviruses encoding either mStrawberry and UT-B (S-UTB), luciferase and mStrawberry (L-S) or a 50:50 mixture of the two lentiviruses. There was a ~ 2-fold increase in AXR at the site of injection of lentivirus encoding S-UTB when compared with controls that received only the L-S virus (n = 6 animals, n = 2 per injection, Fig. 5 a,b). Viral transduction was confirmed in the animals that received the L-S virus by bioluminescence imaging of luciferase expression (Fig. 5 c) and by histological staining for mStrawberry in all rats (Fig. 5 d) (Supplementary Fig. S7). ROI analysis of the needle track, observed in the T2-weighted images (Fig. 5 a), and an ROI on the contralateral side of the brain confirmed an increase in AXR in brain regions injected with virus encoding for UT-B (Fig. 5 e). The AXR was 2.95 ± 0.79 s-1 (n = 8) from all non UT-B expressing regions (L-S and control) and 6.54 ± 2.66 s-1 (n = 4) from UT-B expressing regions (mean ± SD; p ≤ 0.05, Mann-Whitney-Wilcoxon test). The mStrawberry positive cells in the ROIs highlighted in (Fig. 5 d i)-iii)) constituted 22.4 ± 12.3 % of all cells.
Figure 5. Magnetic resonance imaging (MRI) of UT-B reporter gene expression in rat brain.
a i)-iii), T2-weighted images showing the injection site. b i)-iii), AXR maps overlaid on the corresponding reference images showing increased AXR at the site of S-UTB virus injection (virus injection sites are marked with white arrows). Images were acquired at 21 days after virus injection. c i)-iii), Bioluminescence imaging confirming luciferase activity. d i)-iii), Histological slices confirming mStrawberry expression. e, AXR values from ROIs at the injection site of the indicated virus and from control regions on the contralateral side (n = 6 control ROIs from normal brain, n = 2 for L-S, n = 2 for S-UTB / L-S, n = 2 for S-UTB; mean ± SD; *p ≤ 0.05, Mann-Whitney-Wilcoxon test). All control indicates L-S and control.
The 200% higher AXR values in both UT-B expressing xenografts (Fig. 4 c) and in transduced brain cells in vivo (Fig. 5e) mean that UT-B is the most sensitive endogenous contrast mechanism MRI gene reporter to date. For example, lysine-rich protein8 gave only a 4.7 % enhancement, tyrosinase15 a 35% increase in contrast in T2-weighted images and ferritin16 a 12 % increase in R2 (Supplementary Table S1). Detection of UT-B expression using AXR measurements is similar conceptually to CEST measurements of lysine-rich protein expression. However, in contrast to these measurements, in which rapid exchange of protons between micro- to millimolar concentrations of lysine-rich protein17 (Supplementary Material S2) and intracellular bulk water is measured (the proton exchange rate for poly-L-lysine is 403 s-1 18) FEXI measures exchange between a much larger pool of protons in intracellular water (> 40 M) and the extracellular water pool, albeit at a much slower exchange rate (~ 4 – 6 s-1AXR). The much larger pool size of intracellular water, when compared to the water exchangeable protons bound to lysine-rich protein, compensates for the slower transmembrane exchange measured by FEXI. CEST detection of LRP might be improved by using more sophisticated methods for measuring CEST contrast19. Nevertheless, comparison of the contrast-to-noise ratio (CNR)20 and CNR efficiency of the in vivo FEXI data presented here with CEST data from a recent study9 shows that FEXI detection of UT-B is more sensitive (CNR of 4.3 versus 1.6 and CNR efficiency of 1.1 versus 0.5) (Supplementary Table S2). FEXI has a second advantage compared with CEST in that contrast can only arise from intact cells.
Cell dilution experiments in vitro (Supplementary Fig. S8) and our lentivirus experiments in vivo (Fig. 5) indicate a UT-B detection sensitivity of 104-105 cells/voxel (Supplementary Material S3), which is similar to the limit for 19F NMR detection of cells labelled with fluorinated tracers3, 21. Modeling experiments presented in Supplementary Fig. S9 predict that detection of UT-B at lower cell densities might be possible.
The sensitivity of AXR measurements depends on the extent to which the diffusion filter suppresses extracellular signal, and on the background water exchange rate. Background AXR might change owing to other factors, such as inflammation 22, which has the potential to confound measurements of UT-B expression. One drawback of FEXI is the limited spatial resolution (~0.94 x 0.94 x 3 mm3 in the current study) because of the loss of signal resulting from the use of two diffusion modules. 3D-FEXI implementations using retrospective gating and slab-selective diffusion filtering could be used to improve both signal-to-noise ratio and spatial resolution, especially if combined with parallel imaging. Sensitivity might also be improved by increasing UT-B expression, although when highly expressed (greater then 700 μM, see Supplementary Material S2) it can inhibit cell growth in vitro (Fig. 2 b) and result in loss of expression in proliferating cells in vivo (Fig. 4 e). An inducible system for UT-B gene expression might be helpful, or other water channels, such as the aquaporins could be adopted because they transport water at similar rates to UT-B23 but have a different mechanism and therefore might not inhibit cell growth upon over-expression. Even though low levels of UT-B expression did not affect cell growth in vitro, the long-term effects of UT-B expression on cell function will need to be examined.
The FEXI experiment described here could be used in the clinic. Although sensitivity would decrease at the lower magnetic field strengths used in the clinic (~3-fold at 3 T) magnetic field homogeneity would be improved, making the EPI readout used in the FEXI experiment less prone to artefacts24.
In summary, we report that UT-B can be used as a contrast agent-free, gene reporter for MRI that relies on endogenously generated contrast, and whose expression is detected by increases in transmembrane water exchange. The UT-B reporter could be applied to track cells non-invasively, to analyse cellular differentiation states in vivo or to monitor the delivery and expression of gene therapy vectors.
Online Methods
Chemicals
Unless stated otherwise, all chemicals were purchased from Sigma-Aldrich (St. Louis, USA).
Sucrose Phantom
For diffusion measurements and gradient calibrations, a phantom containing different sucrose concentrations was prepared1,2. Five microtainer tubes (Beckton, Dickinson and Company, Franklin Lakes, USA) containing between 10 %-50 % w/v sucrose (in 10 % increments) were placed in a 50 mL cylindrical tube filled with water (Corning Incorporated, Corning, USA), giving final sucrose concentrations of 0.3-1.5 M.
Cell Culture
Human embryonic kidney cells (HEK 293T) were obtained from the Cancer Research UK Cambridge Institute Biorepository. The cells were authenticated using short-tandem repeat genetic profiling (STR) yielding a 100% match to the HEK 293T cell line published on ATCC database. Mycoplasma tests were negative. Cells were grown in Dulbecco’s modified Eagle’s medium, with 10 % fetal bovine serum, and 5 % L-glutamine, in 5 % CO2 at 37°C. Cells were passaged when in exponential growth phase at 80 % confluency. Cell viability and total cell count were assessed using an automated Trypan blue dye exclusion assay (ViCell, Beckman Coulter, High Wycombe, UK). MRI experiments in vitro used ca. 1.6 x 108 cells, which were washed with phosphate-buffered saline (PBS), concentrated to give a dense cell suspension (800 g, 5 min, 4°C), transferred with a 1 ml pipette into 0.6 ml Eppendorf tubes and then centrifuged again at moderate speed to remove air bubbles (20 g, 5 min, 4°C). UT-B activity was inhibited by incubating the cells with 170 mM N-N’-Dimethylthiourea (10 min, 37°C) before they were pelleted, as above, and imaged.
Generation of virally transduced cell lines
An EF1 promoter was used to drive transcription of the red fluorescent protein, mStrawberry, and the urea transporter (Mm_UTB, NCBI reference sequence NM_001171010.1) or firefly luciferase. The mStrawberry coding sequence was separated from the UT-B or luciferase coding sequences by an E2A sequence (EF1-S-UTB or EF1-L-S), which resulted in the expression of mStrawberry and UT-B or luciferase in equimolar concentrations from a single mRNA transcript. The generation of a PGK-S-UTB plasmid, where expression was driven by a PGK promoter, has been described previously3. The transgenes were assembled into the lentiviral vector, pBOBI4 (a gift from the Verma laboratory, Salk Institute, La Jolla, USA). Lentiviruses were produced by co-transfecting HEK 293T cells with EF1-S-UTB, EF1-L-S or PGK-S-UTB plasmids and the packaging plasmids. Supernatants containing lentiviruses were collected 72 h after transfection, mixed with polybrene (8 μg/ml) and used to infect HEK 293T cells. After 72 h cells displaying similar levels of red fluorescence were sorted, using a BD FACSAria cell sorter.
RNA extraction, reverse transcription and qPCR
Total RNA was isolated from cells using an RNeasy kit (Qiagen). For xenograft material, RNA was extracted using RLT Reagent (Qiagen, Venlo, Netherlands) using the manufacturer's protocol. Total RNA (1 μg) was used for first strand cDNA synthesis with SuperScript II Reverse Transcriptase and oligo dT primers (Invitrogen, Carlsbad, USA). Real time PCR was performed with SYBR® Green PCR Master Mix (Applied Biosystems, Waltham, USA) using Quantitect primers (Qiagen: Mm_SLC14a1 QT00117502; Hs_ACTB QT00095431; Hs_B2M QT00088935) in a QuantStudio™ 6 Flex Real-Time PCR System (Applied Biosystems, Waltham, USA). All reactions were performed in triplicate. The relative amount of all mRNAs was calculated using the comparative CT method after normalization to the Geomean of β-actin and β2-microglobulin (B2M).
Functional Assay for UT-B expression
UT-B activity was assessed by placing cells in a hypoosmotic solution (0.225 % NaCl) and assessing their viability. In this hypoosmotic medium UT-B expressing cells, with faster transmembrane water transport, were expected to increase in volume at a greater rate than non-expressing cells and therefore show lower viability. Cells were grown to 60 % confluence and then resuspended in 0.9 % and 0.225 % NaCl solutions, incubated on ice for 5 min, and then their viability and total cell count assessed using ViCell. For each experiment 50 images were analysed and each experiment was repeated four times. Cell viability was defined as the percentage of unstained viable cells at 0.225 % NaCl compared to the number at physiological salt concentration (0.9 % NaCl). The quoted error is the standard deviation of the mean value of the percentage of unstained viable cells in four independent experiments.
Cell Implantation
Severe combined immune-deficient (SCID) mice, 6–8-weeks old, (Charles River Ltd. Margate, UK) were implanted subcutaneously with either 1 x 107 control HEK 293T cells, or HEK 293T cells transfected with PGK-S-UTB, or EF1-S-UTB or EF1-L-S lentiviral vectors. The cells were suspended in PBS before injection. UT-B expressing cells were injected on the left dorsal flank and control cells on the contralateral flank. Prior to implantation, cells were counted and their viability assessed (ViCell). Imaging was performed using mice with similar xenograft burden. Xenograft volume was determined using the ellipsoidal volume formula: 1/2 x length x width2. Experiments were conducted in compliance with project and personal licenses issued under the Animals (Scientific Procedures) Act of 1986 and were designed according to the UK Co-ordinating Committee on Cancer Research guidelines for the welfare of animals in experimental neoplasia. The Cancer Research UK Cambridge Institute Animal Welfare and Ethical Review Body approved the work.
Viral Transfection in vivo
Lentiviral particles were produced as described previously5 and titred using a MACSQuantVYB benchtop flow cytometer. Six week-old (150-190g) female CD IGS rats (Charles River UK Limited, Kent) were anaesthetized by inhalation of 1-2% isoflurane (Isoflo, Abbotts Laboratories Ltd., UK) in air/O2 (flow rate 2L min-1). Analgesia was administered subcutaneously (Vetergesic (Alstoe, York, UK) containing 0.3 mg/ml buprenorphine hydrochloride and 0.135% w/v chlorocresol diluted 1:10 in 0.9% sodium chloride, and 1 ml/kg of subcutaneous Rimadyl LA (Pfizer, New York, US) containing carpofen 5 mg/kg diluted 1: 10 in 0.9% sodium chloride). Respiratory rate was monitored at maintained at approximately 60 breaths min-1 with core body temperature maintained via a heat pad. Animals were transferred to a stereotactic surgical frame (Kopf, Tujunga, US) and the head underwent three point fixation using ear bars and a metal bit. The scalp was shaved and washed with aqueous Videne (Ecolab, US). A midline incision was performed and pericranium stripped. A 1 mm burr hole was drilled freehand at 2 mm anterior and 3 mm lateral to the bregma (right-side). A 23-gauge full displacement syringe (SGE Analytical Science, Melbourne, Australia) was filled with 5 μl of virus mixture at 1.7 x 1010 viral particles μL-1 before being inserted 6 mm intracranially. Upon withdrawal by 1 mm, the contents of the syringe were injected into the right caudate nucleus at 2 μl min-1. The syringe was removed over 30 seconds and the burr hole filled with bone wax (Ethicon, Norderstedt, Germany). Interrupted 6/0 undyed absorbable vicryl rapide sutures (Ethicon, Norderstedt, Germany) were used to close the wound, which was then covered using tissue glue (GLUture, US) after which the animal was recovered in a hot box. Rimadyl LA (1:10 0.9% saline) was given for 48 hours post operatively as analgesia.
MRI
Magnetic Resonance Imaging was performed using a 9.4 T horizontal bore small animal MRI scanner (Agilent, Palo Alto, USA) using a transmit-receive 4 cm inner diameter Millipede™ coil (Agilent, Palo Alto, CA). After placement of the animal in the scanner, scout images were acquired using 3D gradient echo and coronal fast spin echo sequences for animal positioning. First order shimming was performed using an automated non-selective FID-based procedure and was then further optimized using slice-selective manual shimming using a PRESS sequence. Line widths for 3 mm axial slices were ca. 90 Hz in vivo and 30 Hz in vitro. Mice were anesthetized by inhalation of 1.5 – 2.5 % Isoflurane (Isoflo, Abbotts Laboratories, Maidenhead, UK) in 75 air / 25 % O2 (2 L min-1) delivered via a facemask. Body temperature was maintained by blowing warm air through the magnet bore. Breathing rate (30-40 bpm) and body temperature (37 °C) were monitored during experiments (Biotrig, Small Animal Instruments, Stony Brook, USA). Respiratory triggering was used for all measurements such that images were acquired during the expiration phase. An axial slice of 3 mm thickness was positioned in the center of the two xenografts and a reference image was acquired using a fast spin echo sequence (repetition time ≥ 2 s, matrix size: 256 x 256, FOV: 3 cm x 3 cm, TE = 20 ms, turbo factor = 8). RF pulse power calibration was performed using a slice at the same spatial position but 5 mm thick. MRI of rat brains was performed with the same protocol as described above but on a 7 T horizontal bore scanner (Agilent, Palo Alto, USA) using a 72 mm transmit coil with a rat head quadrature receive coil (Rapid Biomedical, Rimpar, Germany) using a FOV of 4 cm x 4 cm.
Filter-Exchange Imaging and Data Analysis
A filter-exchange imaging (FEXI) sequence was optimized using a phantom containing different compartments with sucrose concentrations in the range from 0.3 to 1.5 M. Sucrose-water solutions serve as a simple two-compartment model system, consisting of a fast-diffusing free water and slow diffusing bound water. The principles of the FEXI sequence are illustrated in Fig. 1 a in the main text. Application of the diffusion filter suppresses signal from the apparently fast diffusing water (in this case in the extracellular space) leading to a reduction in the measured ADC, denoted ADC’(tm), compared to the ADC at equilibrium ADCref. The reduction in ADC’ compared to ADCref directly after application of the diffusion filter is quantified by the filter efficiency σ, where σ = 1 − ADC′(tm = 0)/ADCref. With increasing mixing time, tm, between the filter and detection modules, exchange of water between the intra- and extracellular compartments leads to relaxation of ADC’ back to its equilibrium value, which is determined by the apparent exchange rate, AXR (Fig. 1 b). Assuming negligible effects from longitudinal and transverse relaxation and an exchange that is slow when compared to the timescale of the pulsed field gradient spin echo blocks, this equilibration process can be described by6:
| (1) |
The storage and recovery pulses (pulse 3 and 4) in the FEXI sequence (Fig. 1 a) were phase cycled, resulting in a first order correction for the effect of T1 during the storage module. Spoiler gradients before, during, and after the storage module removed unwanted coherence pathways. Diffusion gradients were applied along three gradient axes using two field strengths (1.5; 7) G/cm in the readout module and 10.8 G/cm in the filter module. Taking into account the diffusion weighting of the slice selection gradients, the corresponding b-values were (39; 628) s/mm2 in the readout module and bf = 1496 s/mm2 in the filter module. Only two diffusion gradient strengths were used in the readout module (1.5; 7) G/cm, which were similar to those used previously6, in order to acquire ADC’ values at different mixing times as quickly as possible. These two gradient strengths were chosen to be sufficient to achieve flow suppression at the low b-value and significant diffusion-dependent attenuation at the high b-value. Mixing times were acquired in interleaved order tm = (0.01; 0.3; 0.2; 0.1) s. Both diffusion time and diffusion gradient durations were kept the same in the filter and readout modules: diffusion time Δ= Δϕ = 0.012 ms, diffusion gradient duration δ = δϕ = 0.08 ms. Images were acquired using a single-shot EPI readout; matrix size, 32 x 32; slice thickness, 3 mm; FOV, 3 cm x 3 cm. The sequence was triggered with a respiration sensor leading to a TR ≥ 3 s. Images were acquired with 8 transients for bf = 0 and 16 for all other acquisitions. Two reference scans with no phase encoding and positive and negative readout polarity were acquired for each EPI dataset. For each b-value, two fully phase encoded scans with positive and negative readout polarities were combined in order to eliminate Nyquist (N/2) ghosts. The total acquisition time was approximately 15 min. For the sucrose phantom experiments, a multi-shot (n = 16) EPI-readout was used to reduce chemical shift artefacts. Imaging parameters were the same as for the in vivo experiments with the following exceptions: encoding matrix, 64 x 64; slice thickness, 10 mm; FOV, 4 cm x 4 cm; 8 averages; TR = 5 s, tm = (0.01; 0.3; 0.1; 0.4; 0.2; 0.5) s. For the cell experiments, a dual-shot EPI-readout was used. Imaging parameters were the same as for the in vivo experiments with the following exceptions: slice thickness, 5 mm; FOV, 2.5 cm x 2.5 cm; 4 averages; TR = 4 s; tm = (0.01; 0.2; 0.05; 0.25; 0.1; 0.3; 0.15; 0.35) s.
T1-mapping and DWI
For T1-mapping, an inversion recovery Turbo-FLASH sequence was used, which employed a non-slice-selective adiabatic inversion pulse with 15 inversion times (TI), which were acquired interleaved, between 0.1 s and 10 s (TI = [0.1; 0.4; 0.9; 1.8; 4; 0.2; 0.5; 1.2; 2.3; 6; 0.3; 0.7; 1.5; 2.8; 10] s). Images were acquired with an encoding matrix of 128 x 64; slice thickness, 3 mm; FOV, 3 cm x 3 cm; time between inversion pulses = 12 s. Total acquisition time was 3 min. Diffusion-weighted spin echo EPI was used for ADC measurements, with six b-values (1228; 984; 740; 496; 250; 1; s/mm2). Images were acquired with an encoding matrix size of 64 x 64; slice thickness, 3 mm; FOV, 3 cm x 3 cm; Δ = 0.012 ms; diffusion gradient duration δ = 0.08 ms. The sequence was triggered with a respiration sensor leading to a TR ≥ 3 s. Four averages were acquired in a segmented EPI readout (n = 2). Total acquisition time was approximately 4 min.
Fluorescence and Bioluminescence Imaging
At 2–3 weeks post cell implantation and immediately before the MRI measurements, mice were shaved at the site of implantation and the planar red fluorescence emission from transgenic cells imaged using an IVIS-200 camera (Perkin Elmer, Waltham, MA). The animals were anesthetized using 2% Isoflurane and 75 % air / 25 % O2 (flow rate 2 L min-1), and the images analyzed using Living Image software vs. 4.4 (Perkin Elmer). Images were acquired with epi-illumination, small binning, aperture range F2-F8, exposure 1-3s, and the field of view was varied between 13 and 19.4 cm. The excitation and emission pass band ranges were 500-550 nm and 575–650 nm, respectively. A red background pass band correction filter (460–490 nm) was used to suppress autofluorescence. Rats were anesthetized and bioluminescence imaged using the IVIS-200 camera at approximately 5 minutes after luciferin injection (1 ml of 15mg/ml solution i.p.).
Histology and immunohistochemistry
Sections of formalin-fixed paraffin-embedded tissue (10 μm for xenografts and 5μm for brain tissue) were stained with H&E or with an anti-RFP rabbit polyclonal antibody that crossreacts with mStrawberry (1:100 dilution) (Abcam ab34771) and detected using Leica’s Polymer Refine Kit in combination with the Bond automated system (Leica Biosystems DS9800). Slides were scanned at x20 magnification with a resolution of 0.5 μm per pixel on an Aperio AT2 (Leica Biosystems). Images were analyzed using the Aperio Imagescope Nuclear version 9 algorithm to quantify the proportion of cells with positive nuclear staining.
Statistical analyses
All results are expressed as mean ± S.D. Statistical significance was tested using a Mann-Whitney-Wilcoxon test. Sample size estimates were not performed since both mean and S.D. had to be determined in the experiment for each group. Animal MRI experiments were not randomized and not blinded. Real-time PCR analysis of the xenografts was performed double-blinded, both during the experiment and during analysis.
Data availability
Image data and associated metadata can be accessed at: http://dx.doi.org/10.17863/CAM.4374
Supplementary Material
Editors summary.
Urea transporter can be used as a sensitive, contrast agent-free gene reporter for magnetic resonance imaging in cells and whole animals.
Acknowledgements
This work was supported by a Cancer Research UK Programme grant to KMB (17242) and by the CRUK-EPSRC Imaging Centre in Cambridge and Manchester (16465). FS is in receipt of funding from the Alexander von Humboldt Foundation in the form of a Feodor Lynen Research Fellowship. We thank S. Patrick for the PGK-S-UTB cell line and his help with the UT-B functional assay and D. Honess and D. McIntyre for helpful suggestions and support with the MRI experiments. We thank M. Strzelecki for cell sorting (Flow Cytometry Facility, Cancer Research UK CI) , and the Histopathology Facility and the BRU unit (Cancer Research UK CI) for their help.
Footnotes
Author Contributions. F.S., S.R. and K.M.B designed the research; F.S, S.R., D.H., P.D., S.M., R.M., A.W., L.M., A. A. N. performed the research; F.S., S.R and A.A.N. analysed the data; S.R. generated the virally transduced cell lines and lentiviral particles, R.J.M.F contributed the brain model; and F.S. and K.M.B. wrote the paper.
The authors declare no competing financial interests.
References
- 1.Weissleder R, et al. In vivo magnetic resonance imaging of transgene expression. Nat Med. 2000;6:351–355. doi: 10.1038/73219. [DOI] [PubMed] [Google Scholar]
- 2.Kircher MF, Gambhir SS, Grimm J. Noninvasive cell-tracking methods. Nat Rev Clin Oncol. 2011;8:677–688. doi: 10.1038/nrclinonc.2011.141. [DOI] [PubMed] [Google Scholar]
- 3.Ahrens ET, Bulte JW. Tracking immune cells in vivo using magnetic resonance imaging. Nat Rev Immunol. 2013;13:755–763. doi: 10.1038/nri3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vandsburger MH, Radoul M, Cohen B, Neeman M. MRI reporter genes: applications for imaging of cell survival, proliferation, migration and differentiation. NMR Biomed. 2013;26:872–884. doi: 10.1002/nbm.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patrick PS, et al. Dual-modality gene reporter for in vivo imaging. Proc Natl Acad Sci U S A. 2014;111:415–420. doi: 10.1073/pnas.1319000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deans AE, et al. Cellular MRI contrast via coexpression of transferrin receptor and ferritin. Magn Reson Med. 2006;56:51–59. doi: 10.1002/mrm.20914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zurkiya O, Chan AW, Hu X. MagA is sufficient for producing magnetic nanoparticles in mammalian cells, making it an MRI reporter. Magn Reson Med. 2008;59:1225–1231. doi: 10.1002/mrm.21606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilad AA, et al. Artificial reporter gene providing MRI contrast based on proton exchange. Nat Biotechnol. 2007;25:217–219. doi: 10.1038/nbt1277. [DOI] [PubMed] [Google Scholar]
- 9.Farrar CT, et al. Establishing the Lysine-rich Protein CEST Reporter Gene as a CEST MR Imaging Detector for Oncolytic Virotherapy. Radiology. 2015;275:746–754. doi: 10.1148/radiol.14140251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patrick PS, et al. Detection of transgene expression using hyperpolarized 13C urea and diffusion-weighted magnetic resonance spectroscopy. Magn Reson Med. 2015;73:1401–1406. doi: 10.1002/mrm.25254. [DOI] [PubMed] [Google Scholar]
- 11.Ogami A, Miyazaki H, Niisato N, Sugimoto T, Marunaka Y. UT-B1 urea transporter plays a noble role as active water transporter in C6 glial cells. Biochem Biophys Res Commun. 2006;351:619–624. doi: 10.1016/j.bbrc.2006.10.097. [DOI] [PubMed] [Google Scholar]
- 12.Smouha E, Neeman M. Compartmentation of intracellular water in multicellular tumor spheroids: diffusion and relaxation NMR. Magn Reson Med. 2001;46:68–77. doi: 10.1002/mrm.1161. [DOI] [PubMed] [Google Scholar]
- 13.Lasic S, Nilsson M, Latt J, Stahlberg F, Topgaard D. Apparent exchange rate mapping with diffusion MRI. Magn Reson Med. 2011;66:356–365. doi: 10.1002/mrm.22782. [DOI] [PubMed] [Google Scholar]
- 14.Deliolanis NC, et al. Performance of the red-shifted fluorescent proteins in deep-tissue molecular imaging applications. J Biomed Opt. 2008;13:044008. doi: 10.1117/1.2967184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weissleder R, et al. MR imaging and scintigraphy of gene expression through melanin induction. Radiology. 1997;204:425–429. doi: 10.1148/radiology.204.2.9240530. [DOI] [PubMed] [Google Scholar]
- 16.Cohen B, Dafni H, Meir G, Harmelin A, Neeman M. Ferritin as an endogenous MRI reporter for noninvasive imaging of gene expression in C6 glioma tumors. Neoplasia. 2005;7:109–117. doi: 10.1593/neo.04436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Zijl PC, Yadav NN. Chemical exchange saturation transfer (CEST): what is in a name and what isn't? Magn Reson Med. 2011;65:927–948. doi: 10.1002/mrm.22761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMahon MT, et al. Quantifying exchange rates in chemical exchange saturation transfer agents using the saturation time and saturation power dependencies of the magnetization transfer effect on the magnetic resonance imaging signal (QUEST and QUESP): Ph calibration for poly-L-lysine and a starburst dendrimer. Magn Reson Med. 2006;55:836–847. doi: 10.1002/mrm.20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaiss M, Schmitt B, Bachert P. Quantitative separation of CEST effect from magnetization transfer and spillover effects by Lorentzian-line-fit analysis of z-spectra. J Magn Reson. 2011;211:149–155. doi: 10.1016/j.jmr.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Welvaert M, Rosseel Y. On the definition of signal-to-noise ratio and contrast-to-noise ratio for FMRI data. PLoS One. 2013;8:e77089. doi: 10.1371/journal.pone.0077089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janjic JM, Ahrens ET. Fluorine-containing nanoemulsions for MRI cell tracking. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2009;1:492–501. doi: 10.1002/wnan.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabolli V, et al. Critical role of aquaporins in interleukin 1beta (IL-1beta)-induced inflammation. J Biol Chem. 2014;289:13937–13947. doi: 10.1074/jbc.M113.534594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang B, Verkman AS. Analysis of double knockout mice lacking aquaporin-1 and urea transporter UT-B. Evidence for UT-B-facilitated water transport in erythrocytes. J Biol Chem. 2002;277:36782–36786. doi: 10.1074/jbc.M206948200. [DOI] [PubMed] [Google Scholar]
- 24.Oh SH, et al. Distortion correction in EPI at ultra-high-field MRI using PSF mapping with optimal combination of shift detection dimension. Magn Reson Med. 2012;68:1239–1246. doi: 10.1002/mrm.23317. [DOI] [PubMed] [Google Scholar]
- 1.Delakis I, Moore EM, Leach MO, Wilde JPD. Developing a quality control protocol for diffusion imaging on a clinical MRI system. Physics in Medicine and Biology. 2004;49:1409–1422. doi: 10.1088/0031-9155/49/8/003. [DOI] [PubMed] [Google Scholar]
- 2.Lavdas I, Behan KC, Papadaki A, McRobbie DW, Aboagye EO. A phantom for diffusion-weighted MRI (DW-MRI) J Magn Reson Imaging. 2013;38:173–179. doi: 10.1002/jmri.23950. [DOI] [PubMed] [Google Scholar]
- 3.Patrick PS, et al. Detection of transgene expression using hyperpolarized 13C urea and diffusion-weighted magnetic resonance spectroscopy. Magn Reson Med. 2015;73:1401–1406. doi: 10.1002/mrm.25254. [DOI] [PubMed] [Google Scholar]
- 4.Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma IM. Development of a self-inactivating lentivirus vector. J Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 6.Nilsson M, et al. Noninvasive mapping of water diffusional exchange in the human brain using filter-exchange imaging. Magn Reson Med. 2013;69:1573–1581. doi: 10.1002/mrm.24395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Image data and associated metadata can be accessed at: http://dx.doi.org/10.17863/CAM.4374