Abstract
Nonhuman primates are commonly used for cognitive neuroscience research and often surgically implanted with cephalic recording chambers for electrophysiological recording. Aerobic bacterial cultures from 25 macaques identified 72 bacterial isolates, including 15 Enterococcus faecalis isolates. The E. faecalis isolates displayed multi-drug resistant phenotypes, with resistance to ciprofloxacin, enrofloxacin, trimethoprim-sulfamethoxazole, tetracycline, chloramphenicol, bacitracin, and erythromycin, as well as high-level aminoglycoside resistance. Multi-locus sequence typing showed that most belonged to two E. faecalis sequence types (ST): ST 4 and ST 55. The genomes of three representative isolates were sequenced to identify genes encoding antimicrobial resistances and other traits. Antimicrobial resistance genes identified included aac(6’)-aph(2”), aph(3’)-III, str, ant(6)-Ia, tetM, tetS, tetL, ermB, bcrABR, cat, and dfrG, and polymorphisms in parC (S80I) and gyrA (S83I) were observed. These isolates also harbored virulence factors including the cytolysin toxin genes in ST 4 isolates, as well as multiple biofilm-associated genes (esp, agg, ace, SrtA, gelE, ebpABC), hyaluronidases (hylA, hylB), and other survival genes (ElrA, tpx). Crystal violet biofilm assays confirmed that ST 4 isolates produced more biofilm than ST 55 isolates. The abundance of antimicrobial resistance and virulence factor genes in the ST 4 isolates likely relates to the loss of CRISPR-cas. This macaque colony represents a unique model for studying E. faecalis infection associated with indwelling devices, and provides an opportunity to understand the basis of persistence of this pathogen in a healthcare setting.
Introduction
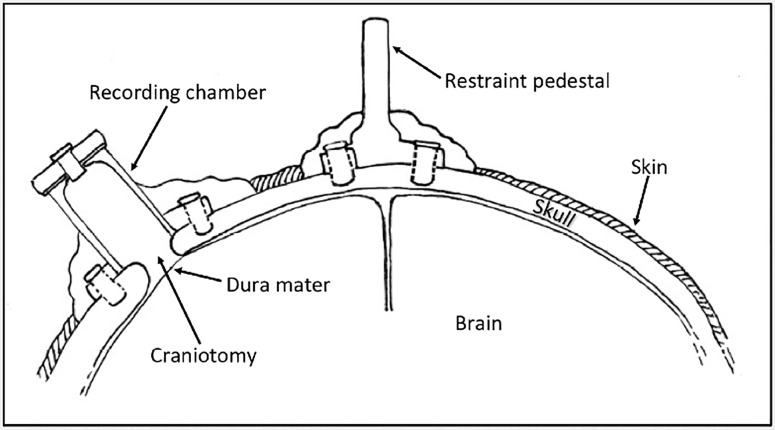
Nonhuman primates (NHP) are an important animal model in cognitive neuroscience research, with the macaque (Macaca spp.) being the most commonly utilized species [1]. The anatomical and functional similarities between the human and macaque brain have been well characterized, and features such as a highly developed cerebral cortex, binocular color vision and front-facing eyes allow comparisons to humans that are impossible in rodent models [1]. Implanted cephalic chambers allow placement of microelectrodes into specific regions of the brain to monitor the activity of individual neurons (Fig 1). Materials commonly used in cephalic recording chamber implants include CILUX plastic (CRIST Instruments), titanium, stainless steel, thermoplastic polyetherimide Ultem (Gray Matter Research), polymethyl methacrylate acrylic and combinations of these materials. Due to the long-term nature of neurophysiology studies, surgically implanted macaques can be maintained for 10 years or longer. A major complication of chronic cephalic implantation is bacterial infection of the recording chambers, with occasional subclinical and clinical cases of meningitis and rare cases of cephalic abscessation [2–5]. Previous publications have reported Staphylococcus aureus, Corynebacterium ulcerans, Candida sp. and Trichosporon beigelii as common bacterial and fungal contaminants of recording chambers and S. aureus, Streptococcus spp., Corynebacterium spp., and Enterococcus spp. as common bacterial isolates from the skin-cranial implant margin [4–6]. Previously, C. ulcerans isolated from the skin-cranial implant margin was found to be sensitive to commonly used antimicrobials, however limited information is available on antimicrobial sensitivities of bacterial species isolated from within cephalic recording chambers. The first goal of this study was to identify microbes colonizing cephalic recording chambers in our macaque colony, and evaluate antimicrobial resistance profiles that may be important for intervention. We hypothesized that antimicrobial resistant microbes would be likely, due to intermittent topical and systemic antimicrobial administration. We identified multi-drug resistant Enterococcus faecalis as the second most prevalent species in recording chambers. E. faecalis is an important nosocomial pathogen of humans, and its ability to form biofilm contributes to catheter-associated blood and urinary tract infections [7, 8]. Recording chambers offer an interface for biofilm formation in cranially-implanted macaques, and we hypothesized that E. faecalis isolated from recording chambers would have similar sequence types, virulence factors, and biofilm genes as human isolates. We highlight the value of implanted macaques as a model of human E. faecalis infection associated with long term indwelling devices.
Fig 1. Illustration of Cephalic Implants Used for Neuroscience Research.
Cephalic recording chambers and restraint pedestals are commonly affixed to the skull using a combination of screws and polymethyl methacrylate acrylic or other polymers.
Materials and Methods
Animals
Twenty-five macaques (19 male, 6 female) with chronic cephalic recording chambers were sampled in August, 2011. The population consisted of 24 rhesus macaques (Macaca mulatta) and 1 cynomolgus macaque (Macaca fasicularis), with a mean and standard deviation age of 10.7 ± 2.2 years (range 6–15 years). All animals were sourced from USDA-approved vendors. Select population characteristics are shown in Table 1. Animals were housed in accordance with The Guide for Care and Use of Laboratory Animals in an AAALAC International-accredited facility. All animal protocols were approved by the Committee on Animal Care at the Massachusetts Institute for Technology. Routine husbandry parameters at the institution include a 12:12 light/dark cycle, and provision of a commercially formulated primate diet (Purina 5038) supplemented with a foraging mixture of fruit, vegetables, nuts and cereal. Water is provided ad libitum except when water regulation is required under approved protocols. Environmental enrichment includes positive interaction with care staff, treats, food puzzles, toys, mirrors, and videos. Macaques are pair-housed, except when precluded by established behavioral incompatibility or for medical reasons, as determined by a veterinarian. The macaques sampled in this study were under investigation by four different cognitive neuroscience research laboratories (A-D; Table 1). For this study, 44 cephalic recording chambers with craniotomies, and 1 without a craniotomy, were sampled. The majority of macaques (11/25) had one cephalic recording chamber; 8/25 had two, and the remaining 6 macaques had three cephalic recording chambers (Table 1). Most (26) of the recording chambers had been in place between one and three years, 12 had been implanted between three and five years, and 7 had been in place less than one year. To minimize discomfort and stress, sampling of cephalic chambers was performed under ketamine anesthesia (10mg/kg, intramuscular injection) during routine semi-annual physical examination. All animals remained on IACUC-approved protocols for cognitive neuroscience at the conclusion of sampling and no animals were euthanized for reasons related to this study.
Table 1. Population Characteristics of Sampled Research Macaques.
| Animal ID | Sex | Age (years) | Laboratory ID | # of Recording Chambers | E. faecalis Isolates |
|---|---|---|---|---|---|
| 1 | F | 15 | A | 1 | Isolate 1 |
| 2 | F | 14 | A | 1 | |
| 3 | F | 12 | A | 1 | |
| 4 | M | 11 | B | 2 | Isolate 2 |
| 5 | M | 10 | B | 3 | Isolates 3, 4, 5 |
| 6 | M | 13 | B | 2 | Isolate 11 |
| 7 | M | 12 | B | 3 | |
| 8 | M | 13 | B | 3 | Isolate 12 |
| 9 | M | 13 | B | 3 | Isolate 13 |
| 10 | M | 10 | B | 11 | |
| 11 | M | 10 | B | 2 | Isolates 14, 15 |
| 12 | M | 12 | C | 2 | |
| 13 | M | 12 | C | 3 | |
| 14 | M | 11 | C | 2 | |
| 15 | M | 8 | C | 1 | Isolate 6 |
| 16 | F | 10 | C | 2 | Isolates 7, 8 |
| 17 | F | 11 | C | 3 | |
| 182 | M | 10 | C | 2 | Isolate 9 |
| 19 | M | 10 | C | 2 | Isolate 10 |
| 20 | F | 11 | D | 1 | |
| 21 | M | 8 | D | 1 | |
| 22 | M | 7 | D | 1 | |
| 23 | M | 7 | D | 1 | |
| 24 | M | 11 | D | 1 | |
| 25 | M | 6 | D | 1 |
1This macaque had a cephalic recording chamber without a craniotomy
2Cynomolgus macaque
Recording Chamber Maintenance and Condition
Primary caregivers were surveyed for the frequency of cephalic recording chamber cleaning, gross appearance of recording chamber discharge, and usage of topical antimicrobial agents and packing material in the chamber (S1 Table). Overall, cephalic recording chambers were typically disinfected daily during periods of recording for most animals, and were cleaned between three times a week to every 3–4 weeks for those not actively being studied. Materials and antimicrobials used in the chambers by different investigative teams varied, and included sterile non-woven sponge balls (Super Stoppers, Metropacifica LLC, Nashville, TN), sterile petroleum jelly, gentamicin sulfate 0.3% ophthalmic solution (Gentak, Akorn Pharmaceuticals, Lake Forest, IL), bacitracin zinc 400U/g-neomycin sulfate 5mg/g-polymyxin B 10,000U/g ophthalmic ointment (Akorn Pharmaceuticals, Lake Forest, IL), 0.5% oxytetracyline with polymixin B (Terramycin, Zoetis, Florham Park, NJ) and a 1:20 dilution of injectable 22.7 mg/ml enrofloxacin (Bayer, Shawnee Mission, Kansas).
Bacterial Culture
Sterile culture swabs (CultureSwab MaxV(+), BD, Franklin Lakes, NJ) were used to sample the interior of cephalic recording chambers, including any discharge present. The swabs were plated onto chocolate agar, trypticase soy agar with 5% sheep blood and MacConkey agar plates and incubated at 37°C in 5% CO2 for 24 h. The swabs, themselves, were then incubated in thioglycollate broth at 37°C for 24 h and re-plated onto the media listed above. Microbial growth was streaked onto blood agar to obtain isolated colonies, which were then identified using the Analytical Profile Index identification system (API 20 E and API 20 Strep, bioMérieux, Durham, NC).
Antimicrobial Susceptibility Testing
Initial antibiotic susceptibility profiles were determined by disk diffusion using the Clinical and Laboratory Standards Institute break points (M2-A10) [9]. Antimicrobial agents tested included ampicillin, amoxicillin/clavulanic acid, bacitracin, cephalothin, erythromycin, gentamicin, oxacillin, trimethoprim-sulfamethoxazole, enrofloxacin, tetracycline, oxytetracycline, ceftriaxone, doxycycline, neomycin, cefazolin, polymyxin B and vancomycin. Chloramphenicol, gentamicin, streptomycin, and vancomycin disks were purchased from BD (BBL Sensi-Disc, Franklin Lakes, New Jersey); the remainder of the antimicrobial disks were obtained from Oxoid (Basingstoke, United Kingdom). Minimum inhibitory concentrations for key antibiotics were determined by broth microdilution in CAMHB (Cation-Adjusted Muëller Hinton Broth) as recommended [10]. For daptomycin MIC determination, the CAMHB was supplemented with calcium to a final concentration of 50 μg/ml. Amoxicillin-clavulanic acid and trimethoprim-sulfamethoxazole combinations were tested for MIC using Etest strips according to the manufacturer’s instructions (Etest, bioMérieux, Durham, NC). The MIC was read at the lowest concentration where the ellipse of inhibited growth intersected the testing strip. All antibiotics used for broth microdilution were purchased from Sigma-Aldrich Chemical Company (St Louis, MO). Control strains included vancomycin-susceptible ATCC E. faecalis 29212 and vancomycin-resistant ATCC E. faecalis 51299.
DNA Extraction
DNA was extracted from overnight broth cultures of E. faecalis using a commercially available kit (Qiagen DNeasy Blood and Tissue Kit, USA). Manufacturer instructions were modified by the addition of 50 μl of lysozyme (50 mg/ml) and 10 μl mutanolysin (2500 U/ml, Sigma-Aldrich) during a 30 minute incubation at 37°C before the addition of proteinase K and buffer.
PCR and MLST
Polymerase chain reaction (PCR) amplification of the D-alanine:D-alanine ligase gene (ddl), 16S rRNA gene, and MLST genes were performed using the listed primers (Table 2), with amplification conditions based on previously published protocols [11, 12]. PCR products were separated by electrophoresis through a 1% agarose gel at 100-120V for 30–40 minutes, prior to ethidium bromide staining and visualization with UV light. Prior to sequencing, PCR products were purified using a commercial kit according to manufacturer instructions (Qiagen, USA), or purified prior to sequencing by a commercial laboratory (QuintaraBio, Cambridge, MA). Purified PCR products underwent Sanger sequencing at the DNA Core Facility at the Massachusetts General Hospital Center for Computational and Integrative Biology, or at a commercial laboratory (QuintaraBio, Cambridge, MA). Sequence types were identified using the Enterococcus faecalis MLST website (http://pubmlst.org/efaecalis) at the University of Oxford [13].
Table 2. Primers Used for the Identification and Characterization of E. faecalis.
| Primer Set | Sequence (5'-3') | Amplicon Size (bp) | Reference |
|---|---|---|---|
| E. faecalis ddl gene | CAGAAGTGAAGAGCACGATG | 647 | In-house design |
| AGGTAAAGTCGTACGGACAT | |||
| 16S universal primer | AGAGTTTGATCCTGGCTGAG | 1550 | Coenye, T., et al.11 |
| AAGGAGGTGATCCAGCCGCA | |||
| pstS—MLST | CGGAACAGGACTTTCGC | 583 | Ruiz-Garbajosa, P., et al.12 |
| ATTTACATCACGTTCTACTTGC | |||
| aroE—MLST | TGGAAAACTTTACGGAGACAGC | 459 | Ruiz-Garbajosa, P., et al.12 |
| GTCCTGTCCATTGTTCAAAAGC | |||
| gdh—MLST | GGCGCACTAAAAGATATGGT | 530 | Ruiz-Garbajosa, P., et al.12 |
| CCAAGATTGGGCAACTTCGTCCCA | |||
| gyd—MLST | CAAACTGCTTAGCTCCAATGGC | 395 | Ruiz-Garbajosa, P., et al.12 |
| CATTTCGTTGTCATACCAAGC | |||
| gki—MLST | GATTTTGTGGGAATTGGTATGG | 438 | Ruiz-Garbajosa, P., et al.12 |
| ACCATTAAAGCAAAATGATCGC | |||
| xpt—MLST | AAAATGATGGCCGTGTATTAGG | 456 | Ruiz-Garbajosa, P., et al.12 |
| AACGTCACCGTTCCTTCACTTA | |||
| yiqL—MLST | CAGCTTAAGTCAAGTAAGTGCCG | 436 | Ruiz-Garbajosa, P., et al.12 |
| GAATATCCCTTCTGCTTGTGCT |
Whole Genome Sequencing, Antimicrobial Resistance Genes and Virulence Factor Identification
E. faecalis isolates #1, #12, and #13, from macaques 1, 7, and 8, respectively, were each sequenced on a single SMRT cell on a Pacific Biosciences RS2 at the University of Massachusetts Deep Sequencing Core Facility. DNA libraries were prepared for sequencing with the SMRTbell Template Prep Kit 1.0 and the DNA/Polymerase Binding Kit P6 v2, according to manufacturer instructions (Pacific Biosciences, Menlo Park, CA). A total of 87,196, 93,299 and 85,195 reads were obtained for genomes from isolates 1, 12 and 13, respectively; resulting in 3 polished contigs for isolates 1 and 12, and 2 polished contigs for isolate 13. N50 read lengths were 24,838, 24,224 and 23,305 bases, and average reference coverage was 327.48, 338.83 and 293.2 for isolates 1, 12, and 13, respectively. Filtered subreads were assembled de novo using the Hierarchical Genome Assembly Process (HGAP 3.0) workflow, with the Celera assembler and assembly polishing by Quiver [14]. Quality trimming was performed during the preassembly stage of HGAP. This whole genome sequencing project has been deposited at DDBJ/ENA/GenBank under the accession numbers MCFU00000000, MCFV00000000 and MCFW00000000 for isolates #1, #12 and #13, respectively. The genome assemblies described in this paper are versions MCFU01000000, MCFV01000000, and MCFW01000000. Assembled genomes were annotated using the Pathosystems Resource Integration Center (PATRIC) annotation service, and the proteomes were compared with vancomycin-susceptible ATCC E. faecalis 29212 [15]. Assembled genomes were analyzed using the PubMLST, ResFinder, VirulenceFinder and PATRIC databases to confirm sequence type and identify genes of interest [13, 15–17]. The ATCC 29212 genome used for comparison was retrieved from GenBank under the accession number CP00816 [18]. Identification thresholds were set at 98% identity over a minimum length of 60% for ResFinder, and 95% identity over a length of 60% for VirulenceFinder. DNA gyrase (gyrA) and topoisomerase IV subunits A and B (ParC and ParE) FASTA protein sequences were compared between macaque isolates and E. faecalis reference strain ATCC 29212 using the multiple sequence alignment tool on PATRIC to identify amino acid polymorphisms contributing to fluoroquinolone resistance.
Static Biofilm Assay
Biofilm formation was assayed by measuring crystal violet binding, according to previously published protocols with slight modification [19, 20]. Macaque E. faecalis isolates, and positive control ATCC E. faecalis 29212, were plated on tryptic soy agar containing 5% sheep blood, and incubated at 37°C in 5% CO2 for 24 h. Sterile 1 μl disposable loops were used to inoculate isolates into 5 ml of tryptic soy broth supplemented with 1% glucose (w/v). Following overnight incubation at 37°C, the optical density at 600 nm was recorded using a microtiter plate spectrophotometer (Epoch, Biotek Instruments, Inc., Winooski, VT). For each isolate, 2 μl of overnight culture was diluted into 198 μl of tryptic soy broth supplemented with 1% glucose, in triplicate, in 96 well polystyrene plates. For comparison, 4 negative controls containing glucose-supplemented tryptic soy broth without bacterial inoculum were included. Microtiter plates were incubated at 37°C, shaking at 100 rpm and biofilm formation was evaluated at 24 h. Following aspiration of medium and planktonic cells, wells were washed three times with 200 μl of phosphate-buffered saline, inverted, and air-dried for 45 min. The remaining biofilm was fixed with 200 μl of methanol for 20 min, and plates were inverted and air-dried for an additional 45 min. Biofilm was stained with 150 μl of 1% crystal violet for 20 min. Excess stain was removed via aspiration, followed by rinsing under running tap water. After again air-drying, biofilm-bound crystal violet was solubilized via the addition of 150 μl ethanol for 25 min. The absorbance of the extracted dye was measured at 570 nm using a microtiter plate spectrophotometer, and adjusted for the absorbance of the negative control. Biofilm optical density was normalized to the initial bacterial cell mass by dividing the absorbance of the extracted dye by the OD600 of the initial inoculum. Each biofilm test was run in triplicate, and biological replicates were repeated in triplicate on a separate day, independently, to confirm results. Beeswarm plots for each isolate did not qualitatively show systematic bias due to batch effects between experiments, thus data was pooled for analysis (S1 Fig). Beeswarm plots were generated using the Python programming language (Python 3.5.2, matplotlib 1.5.1, seaborn 0.8.0), and Bayesian analysis was done using PyMC3 (ver 3.0 rc2) (S1 Fig). Data was analyzed using a Kruskal-Wallis test with Dunn's multiple correction in GraphPad Prism (GraphPad Software, Inc., La Jolla, CA) with P <0.05 considered significant.
Biofilm Assessment under Flow
A flow cell system was assembled and sterilized as previously described [21, 22], and biofilm formation was assessed using BM2 medium [62 mM potassium phosphate buffer (pH 7), 7 Mm (NH4)2SO4, 2 mM MgSO4, 10 μM FeSO4, 0.4% (wt/vol) glucose]. The chambers were inoculated with overnight cultures of E. faecalis isolates #1, #5, #12 and #13, and bacteria were allowed to attach to the surface of the flow cell chambers for 7 h 40 min. Bacteria were then grown for 43 h under a flow of 2.4 ml/min, after which viable bacteria were stained with SYTO-9 and subsequently visualized using a confocal laser scanning microscope (Zeiss LSM 700). Three-dimensional reconstructions were generated using the Imaris software package (Bitplane AG).
Results
Aerobic Culture and Sensitivity
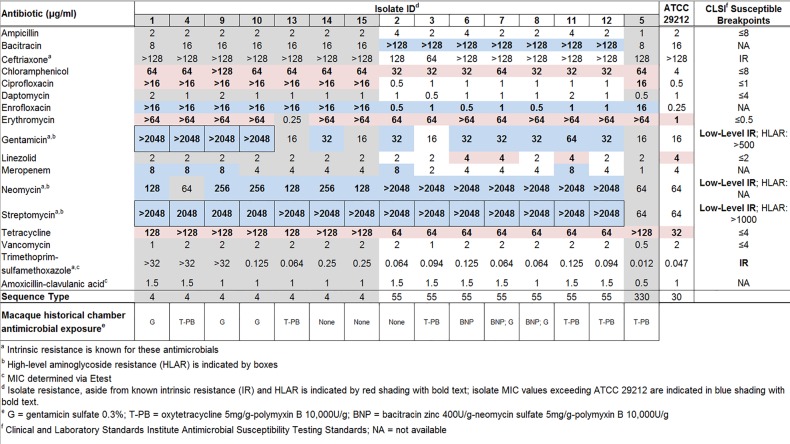
From the 45 cephalic recording chambers sampled, 72 aerobic bacterial isolates were examined, with the most common species being Staphylococcus aureus (n = 20), Enterococcus faecalis (n = 15) and Proteus mirabilis (n = 6) (Table 3). The vast majority of cephalic recording chambers grew polymicrobial cultures with a mean and standard deviation of 2.8 ± 1.5 different species (Table 4). Kirby-Bauer testing revealed that while S. aureus isolates were susceptible to the majority of antimicrobials tested (data not shown), E. faecalis and Proteus spp. isolates were multi-drug resistant (Table 5, S2 Table). Due to the prevalence of E. faecalis in our colony, we further investigated the antimicrobial resistance characteristics of these isolates. MIC determination for the 15 E. faecalis isolates confirmed high-level resistance to aminoglycosides, as well as resistance to bacitracin, chloramphenicol, fluoroquinolones, erythromycin, tetracycline, and trimethoprim-sulfamethoxazole (Fig 2). Aminoglycoside resistance was especially prevalent, with 14/15 isolates displaying high-level resistance to streptomycin, 7/15 isolates resistant to neomycin, and 4/15 isolates with high-level resistance to gentamicin. No resistance to vancomycin, linezolid or daptomycin was identified among isolates. Five isolates (ST 4 isolates #1, #4, #9 and ST 55 isolates #2 and #11) displayed an elevated meropenem MIC (8 μg/ml) compared to ATCC 29212 (4 μg/ml). No CSLI susceptibility breakpoint for meropenem has been established for E. faecalis; however, 8 μg/ml is within the previously reported MIC90 [23, 24]. Isolates #6, #7, #11, and ATCC 29212, possessed MICs to linezolid (4 μg/ml) above the CLSI breakpoint of ≤2 μg/ml. Previous literature suggests these isolates fall within the MIC90 range for the enterococci, and should be classified as intermediate resistant [25, 26].
Table 3. Aerobic Bacterial Culture Results from 45 Chambers1of Research Macaques (n = 25).
| Bacterial Isolate | Number (%) of Isolates |
|---|---|
| Staphylococcus aureus | 20 (27.8) |
| Enterococcus faecalis | 15 (20.8) |
| Proteus mirabilis | 6 (8.3) |
| Group C β-Streptococcus dysgalactiae | 5 (6.9) |
| Proteus vulgaris | 4 (5.6) |
| Staphylococcus intermedius | 4 (5.6) |
| Enterococcus avium | 3 (4.2) |
| Escherichia coli | 2 (2.8) |
| Group A β-Streptococcus pyogenes | 2 (2.8) |
| Leuconostoc spp. | 2 (2.8) |
| Streptococcus uberis | 2 (2.8) |
| Aerococcus viridans 2 | 1 (1.4) |
| Enterococcus hirae | 1 (1.4) |
| Group F β-Streptococcus constellatus | 1 (1.4) |
| Proteus penneri | 1 (1.4) |
| Proteus sp. | 1 (1.4) |
| Staphylococcus epidermidis | 1 (1.4) |
| Staphylococcus xylosus | 1 (1.4) |
| Total | 72 (100) |
1Cultures were pooled for animals with multiple recording chambers
Table 4. The Majority of Macaque Chambers Display Polymicrobial Colonization.
| Number of Isolates | # (Percentage) of Macaques |
|---|---|
| 0 | 1 (4%) |
| 1 | 4 (16%) |
| 2 | 7 (28%) |
| 3 | 5 (20%) |
| 4 | 4 (16%) |
| 5 | 3 (12%) |
| 6 | 1 (4%) |
Table 5. Kirby Bauer Disk Diffusion Results for 15 Macaque Chamber E. faecalis Isolates.
| Antibiotic | # Isolates tested | # Sensitive Isolates (%) | # Intermediate Isolates (%) | # Resistant isolates (%) |
|---|---|---|---|---|
| AMP | 15 | 11 (73%) | 0 | 4 (27%) |
| AMC | 15 | 11 (73%) | 0 | 4 (27%) |
| B | 15 | 2 (13%) | 5 (33%) | 8 (53%) |
| CR | 15 | 0 | 0 | 15 (100%) |
| E | 15 | 0 | 1 (7%) | 14 (93%) |
| GM | 15 | 5 (33%) | 2 (13%) | 8 (53%) |
| OX | 15 | 0 | 0 | 15 (100%) |
| SXT | 15 | 1 (7%) | 0 | 14 (93%) |
| ENO | 15 | 0 | 4 (27%) | 11 (73%) |
| TE | 15 | 0 | 0 | 15 (100%) |
| T | 15 | 0 | 0 | 15 (100%) |
| CRO | 15 | 0 | 0 | 15 (100%) |
| D | 5 | 1 (20%) | 3 (60%) | 1 (20%) |
| N | 15 | 0 | 1 (7%) | 14 (93%) |
| CZ | 15 | 0 | 1 (7%) | 14 (93%) |
| PB | 15 | 0 | 0 | 15 (100%) |
| VA | 13 | 1 (8%) | 5 (38%) | 7 (54%) |
Sensitive, Intermediate and Resistant designations were determined by measuring the zone of inhibition (mm) and comparing to CLSI guidelines9. AMP–Ampicillin, AMC–Amoxicillin/Clavulanic Acid, B–Bacitracin, CR–Cephalothin, E–Erythromycin, GM–Gentamicin, OX–Oxacillin, SXT–Trimethoprim-Sulfamethoxazole, ENO–Enrofloxacin, TE- Tetracycline, T- Oxytetracycline, CRO- Ceftriaxone, D- Doxycycline, N- Neomycin, CZ- Cefazolin, PB- Polymixin B, VA- Vancomycin
Fig 2. Minimum Inhibitory Concentration (MIC) via Broth Microdilution and Etest Results for Macaque Chamber E. faecalis Isolates as Compared to ATCC E. faecalis 29212.
Sequence types and historical recording chamber antimicrobial exposure are indicated for each macaque isolate.
Sequencing and Annotation
Multi-locus sequencing typing identified three sequence types present among the 15 isolates (Fig 2). Most were either ST 4 (n = 7) or ST 55 (n = 7), and a single isolate (isolate #5) was identified as ST 330. Proteome predictions identified 260, 221 and 208 unique proteins encoded by isolates #1 (ST 4), #12 (ST 55) and #13 (ST 4), respectively, not included in those encoded by E. faecalis ATCC 29212 (ST 30; S3 Table). The vast majority (359/691) of these genes were annotated as hypothetical and most likely reside on mobile elements. Mobile element related genes identified included transposases, type III restriction-modification system proteins, phage proteins, transcriptional regulator proteins, and others, which were more abundant in the ST 4 isolates than ST 55. An additional distinguishing feature between the ST 4 isolates and ST 55 isolate is the presence in the latter of the CRISPR (clustered regularly interspaced short palindromic repeats)-cas (CRISPR-associated genes) system. While both ST 4 isolates lacked CRISPR-cas genes, a type II-A CRISPR-cas system was identified in ST 55 isolate 12, based on the presence of cas1, cas2, csn1 and csn2 [27].
Antimicrobial Resistance Genes
A variety of acquired antimicrobial resistance genes were identified in the genomes of isolates #1, #12 and #13 using ResFinder and PATRIC (Table 6). Four genes encoding resistance to aminoglycosides were identified, including str in ST 4, aph(3’)-III and ant(6)-Ia in ST 55, and the bifunctional aminoglycoside-modifying enzyme aac(6’)-aph(2”) in ST 4 isolate #1. Other genes noted among the three isolates included macrolide resistance genes lsa(A) and erm(B), tetracycline resistance genes, tet(M), tet(S), and tet(L), and the chloramphenicol resistance gene cat. Increased trimethoprim-sulfamethoxazole resistance was identified in ST 4 isolate #1 encoded by dfrG. The bacitracin resistance genes bcrA (ATP binding domain of ATP transporter), bcrB (membrane-bound permease of ABC transporter) and bcrR (regulatory protein of the bcrABD operon) were identified in ST 55 isolate #12 using the specialty gene finder in PATRIC [28]. Multiple sequence alignment of the genes encoding topoisomerase IV subunit A (parC) and DNA gyrase subunit A (gyrA) revealed single amino acid polymorphisms in parC codon 80 (Ser to Ile) in gyrA codon 83 (Ser to Ile) in both ST 4 fluoroquinolone-resistant isolates as compared to the less fluoroquinolone-resistant ST 55 isolate #12 and ATCC 29212. No single amino acid polymorphisms were detected in topoisomerase IV subunit B (parE).
Table 6. Selected Antimicrobial Resistance MIC Results and Resistance Genes Identified from Whole Genome Sequence Data.
| Antibiotic (μg/ml) | Isolate Id c | Relevant Resistance Genes Identifiedd | |||
|---|---|---|---|---|---|
| 1 | 13 | 12 | ATCC 29212 | ||
| Bacitracin | 8 | 16 | >128 | 16 | bcrA, bcrB, bcrR (isolate 12) |
| Chloramphenicol | 64 | 64 | 32 | 4 | cat (all isolates) |
| Ciprofloxacin | >16 | >16 | 1 | 0.5 | parC (S80I) and gyrA (S83I) mutations (isolates 1 and 13) |
| Enrofloxacin | >16 | >16 | 1 | 0.25 | parC (S80I) and gyrA (S83I) mutations (isolates 1 and 13) |
| Erythromycin | >64 | 0.25 | >64 | 1 | ermB (isolates 1 and 12), lsa(A) (all isolates and 29212) |
| Gentamicina | >2048 | 16 | 32 | 16 | aac(6’)-aph(2”) (isolate 1) |
| Neomycina | 128 | 128 | >2048 | 64 | aph(3’)-III (isolate 12) |
| Streptomycina | >2048 | >2048 | >2048 | 64 | str (isolates 1 and 13), aph(3’)-III and ant(6)-Ia (isolate 12) |
| Tetracycline | 128 | 128 | 64 | 32 | tetM (all isolates and 29212), tetS (isolates 1 and 13), tetL (isolate 12) |
| Trimethoprim-sulfamethoxazoleb | >32 | 0.064 | 0.094 | 0.047 | dfrG (isolate 1) |
| Sequence Type | 4 | 4 | 55 | 30 | |
a High-level aminoglycoside resistance (HLAR) is indicated by boxes
b MIC determined by Etest
c Isolate resistance, aside from known intrinsic resistance and HLAR, is indicated by red shading with bold text; isolate MIC values exceeding ATCC 29212 are indicated in blue shading with bold text.
d Assembled genomes were uploaded to ResFinder with a 98% threshold for gene identification and a minimum length of 60%. The PATRIC specialty gene finder tool was used to identify bcrABR genes. Multiple sequence alignment was performed in PATRIC to identify amino acid polymorphisms conferring fluoroquinolone resistance.
Virulence Factor and Biofilm Formation-Associated Genes
Genes encoding virulence factors from isolates #1, #12 and #13 were identified using VirulenceFinder and the specialty feature tool in PATRIC (Table 7). Genes associated with the cytolysin toxin (cylA, cylB, cylL and cylM) were identified in both ST 4 isolates. Many genes associated with biofilm formation were identified, including aggregation substance (agg), enterococcal surface protein (espfs), endocarditis and biofilm-associated pili genes (ebpA, ebpB, ebpC), collagen adhesion precursor (ace), gelatinase toxin (gelE) and sortase (SrtA). The ST 4 isolates possessed more biofilm-associated factors compared to ST 55 isolate 12, which lacked aggregation substance and gelatinase. Because of this finding, we further hypothesized that ST 4 isolates would produce more biofilm than ST 55 isolates and performed biofilm assays to evaluate this hypothesis. Other virulence factors identified included sex pheromone-associated genes (cad, cCF10, camE, cOB1), the cell wall adhesion expressed in serum gene (efaAfs), the enterococcal Rgg-like regulator gene associated with macrophage persistence (ElrA), hyaluronidases (hylA, hylB), and the thiol peroxidase gene to protect against oxidative stress (tpx) (Table 7)[29–36]
Table 7. Acquired Virulence Factor Genes Identified Using VirulenceFindera and PATRICb.
| ST 4 | ST 55 | ST 30 | |||
|---|---|---|---|---|---|
| Virulence Factor Functionc | Gene | Isolate 1 | Isolate 13 | Isolate 12 | ATCC 29212d |
| Collagen adhesin precursor | ace | X | X | X | X |
| Aggregation substance | agg | X | X | X | |
| Endocarditis and biofilm-associated pili genes | ebpA | X | X | X | X |
| ebpB | X | X | X | X | |
| ebpC | X | X | X | X | |
| Cell wall adhesin expressed in serum | efaAfs | X | X | X | X |
| Enterococcal surface protein | esp | X | X | X | |
| Gelatinase toxin (metalloendoprotease) | gelE | X | X | X | |
| Sortase A | SrtA | X | X | X | X |
| Cytolysin (hemolysin-bacteriocin) | cylL | X | X | X | |
| Post-translational cytolysin modification | cylM | X | X | X | |
| Transport of cytolysin | cylB | X | X | X | |
| Activation of cytolysin | cylA | X | X | X | |
| Sex pheremone | cad | X | X | X | X |
| Sex pheremone cAM373 precursor | camE | X | X | X | X |
| Sex pheremone | cCF10 | X | X | X | X |
| Sex pheremone | cOB1 | X | X | X | X |
| Enterococcal Rgg-like regulator | ElrA | X | X | X | X |
| Hyaluronidase | hylA | X | X | X | X |
| hylB | X | ||||
| Thiol peroxidase (oxidative stress resistance) | tpx | X | X | X | X |
a Assembled genomes were uploaded to ResFinder with a 95% threshold for gene identification and a minimum length of 60%. https://cge.cbs.dtu.dk/services/VirulenceFinder/
b The PATRIC Specialty Gene Finder tool was used to confirm virulence factors following annotation.
c Genes associated with biofilm production are designated with blue shading and bolded text and genes associated with cytolysin toxin production are designated with red shading and bolded text.
d Genome was obtained from GenBank accession CP008816.
Biofilm Production
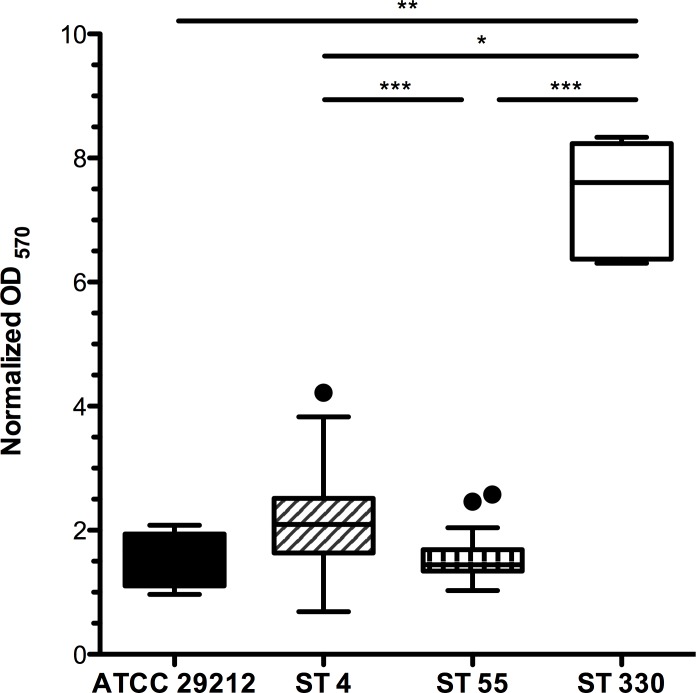
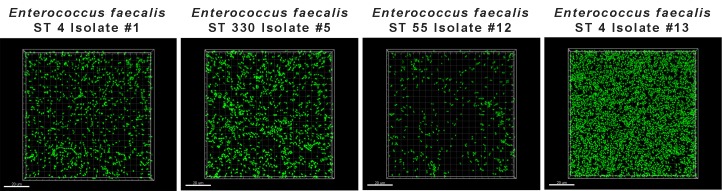
Significant differences in biofilm production, as measured by crystal violet binding, were noted among the E. faecalis isolates (Fig 3, P < 0.0001, Kruskal-Wallis with Dunn’s Multiple Comparison). The ST 330 isolate was a robust biofilm former, producing more biofilm than the ST 4, ST 55 isolates, or control ATCC E. faecalis 29212 (Fig 3, S4 Table). Flow cell assays also revealed that ST 55 isolate #12 exhibited a biofilm-deficient phenotype when compared to ST 4 isolates #1, and #13 and ST 330 isolate #5 (Fig 4). On the other hand, ST 4 isolate #13 showed a hyper-biofilm phenotype compared to all other isolates tested (Fig 4).
Fig 3. 24 hour Biofilm Production for 15 E. faecalis Isolates Assessed by Crystal Violet Staining.
ST 4 and ST 330 isolates produce significantly more biofilm than ST 55 isolates (see also S4 Table). Mean normalized OD570 for pooled data were 2.016 ±.0.016 for ST 4 isolates, 1.500 ± 0.2942 for ST 55 isolates, 8.191 ± 0.1489 for the ST 330 isolate and 1.894 ± 0.1833 for the ATCC 29212 control strain.
Fig 4. Biofilm Growth of E. faecalis in Flow Cell Chambers.
Attached cells grown in flow cell chambers for 43 h were stained green with SYTO-9, and visualized at 63X magnification. Images are representative for each isolate.
Discussion
Enterococcus faecalis is an important cause of healthcare-associated infections, and is the third most common cause of central line-associated bloodstream infections, the fifth most common cause of catheter-associated urinary tract infections, and the sixth most common cause of healthcare-associated infections overall [37]. The enterococci possess mechanisms of both acquired and intrinsic resistance to multiple antimicrobial agents, making nosocomial infection especially difficult to treat successfully. In this study, our E. faecalis isolates were identified from chronic cephalic implants, and are likely associated with biofilm. Most isolates belonged to two multi-drug resistant sequence types and we identified a variety of resistance genes and virulence genes from representative sequenced whole genomes. Minimum inhibitory concentration testing via broth dilution was elected to both confirm initial Kirby Bauer disk diffusion results and quantify the magnitude of resistance. There were some discrepancies between disk diffusion and MIC results; specifically, disk diffusion suggested some isolates displayed resistance to ampicillin, amoxicillin-clavulanic acid and vancomycin. MIC testing found all isolates to be below the resistant breakpoint for ampicillin (8 μg/ml) and vancomycin (4 μg/ml). While no CLSI resistance breakpoint is noted for amoxicillin-clavulanic acid, all isolates and the ATCC control strain 29212 had an MIC between 0.5–1.5 μg/ml. Discrepancies between disk diffusion classification and MIC susceptibilities may be attributed to not recording disk diffusion results at exactly 24 hours, or error in agar inhibitory zone measurement.
Antimicrobial resistance profiles obtained by MIC and Etest related well to antimicrobial resistance genes identified from representative whole genome sequence data (Table 6). Gentamicin MIC profiles also matched well, but not perfectly, with a history of gentamicin exposure. Specifically, E. faecalis isolates #1, #7, #8, #9 and #10 (5/15 isolates; 33%) were isolated from macaques 1, 16, 18 and 19 with a history of gentamicin sulfate administration into their recording chambers. While ST 4 isolates #1, #9 and #10 display high-level gentamicin resistance (HLGR), ST 55 isolates 7 and 8 from macaque 16 lacked HLGR. The aac(6’)-aph(2”) gene has previously been established to confer HLGR, which is supported by the differences in MIC between HLGR ST 4 isolate #1 (>2048 μg/ml) and non-HLGR isolate #13 (16 μg/ml) [38]. The presence of the aac(6’)-aph(2”) gene was confirmed to be in close proximity to genes encoding plasmid recombination enzymes in ST 4 isolate #1. The acquisition of such mobile-element-derived genes may be inhibited by an intact CRISPR-cas system, which was identified in ST 55 isolate #12 [39]. Additionally, both ST 4 and ST 55 isolates display high-level streptomycin resistance, and ST 55 isolates also demonstrate increased resistance to neomycin. The presence of high-level aminoglycoside resistance is significant because it abolishes synergistic treatment with the combination of an aminoglycoside and cell-wall inhibitor [38, 40].
The plasmid-derived bcrABD operon with an upstream regulator bcrR has previously been identified to confer bacitracin resistance in E. faecalis [28]. We identified bcrA, bcrB and bcrR in ST 55 isolate #12 with 89%, 89% and 94% homology, respectively, to previously reported bacitracin resistance genes [28]. Previous mutagenesis experiments have established that the bcrD gene encoding undecaprenol kinase is not required for high-level bacitracin resistance [28]. While macaque 8 (ST 55 isolate #12) does not have a history of bacitracin exposure, macaques 15 and 16 (ST 55 isolates 6, 7 and 8) did have chamber exposure to triple-antibiotic ointment containing bacitracin and are our hypothesized source of selective pressure in our colony.
Multiple genes conferring resistance to tetracyclines were identified, including both ribosomal protection genes and genes encoding efflux pumps [41]. Macaques 5, 6, 8 and 9, from which E. faecalis isolates #3, #4, #5, #11, #12 and #13 were identified, had a history of exposure to combination oxytetracycline-polymyxin B ointment inside recording chambers (Fig 2, S1 Table). Isolates #4, #5 and #13 displayed a higher MIC (≥ 128 μg/ml) than isolates #3, #11 and #12, suggesting that the tetS gene may be responsible for conferring increased tetracycline resistance as compared to tetL and tetM.
Two single amino acid polymorphisms previously identified to confer fluoroquinolone resistance were observed in genes encoding DNA gyrase subunit A (gyrA) and topoisomerase IV subunit A (parC) in both ST 4 isolates sequenced [42, 43]. Enrofloxacin is the most commonly used fluoroquinolone in our colony and has been administered systemically both perioperatively, and therapeutically for clinical cases of wounds or suspected meningitis. Macaque 3 had chamber exposure to dilute enrofloxacin but E. faecalis was not isolated from the chamber of this individual.
The ability of the enterococci to form biofilms contributes to the pathogenicity of implant-associated infections, as mature biofilms allow E. faecalis to withstand antimicrobial agents at 10-1000-fold greater concentrations than those required to control planktonic bacteria [44]. We identified virulence factor genes associated with biofilm formation including aggregation substance (agg), enterococcal surface protein (esp), adhesion of collagen from E. faecalis (ace), gelatinase (gelE), endocarditis and biofilm-associated pili genes (Ebp) and sortase A (SrtA) [35, 45–48]. ST 4 isolates produced significantly more biofilm than ST 55 isolates. Interestingly, one ST 330 isolate formed a robust static biofilm, and additional genetic analysis will be needed to relate this phenotype to genotype (Fig 3). Whole genome sequencing of representative strains showed that ST 55 isolate 12 lacked both agg and gelE, which were present in ST 4 isolates. Nevertheless, we cannot attribute increased biofilm production by ST 4 to the presence of agg and gelE, as the agg- and gelE-positive ATCC 29212 E. faecalis strain showed no significant differences in biofilm-forming ability compared to either ST 4 or ST 55 isolates. It is probable that increased biofilm production by ST 4 and ST 330 isolates, compared to ST 55 isolates, is polygenic in nature, as multiple genes including esp, agg, gelE, and srtA have been individually shown to contribute to biofilm formation [49–52]. As well as genes associated with biofilm formation, we identified the presence of genes encoding the cytolysin toxin in both ST 4 isolates sequenced. The cytolysin toxin has been shown to increase the lethality of infection in multiple species, including mice, rabbits and humans, and can act synergistically with aggregation substance to increase lethality of infection in a rabbit endocarditis model [53, 54]. Due to the rarity of confirmed Enterococcus-associated implant complications we cannot definitively assess how ST 4 isolates bearing the cytolysin toxin contribute to pathogenicity in macaques with chronic implants.
Our macaque colony models an environment with many similarities to humans possessing indwelling devices in a long-term care facility. Macaques are housed in a high-density environment, with approximately 12 animals per housing room. Human intensive care units (ICUs) vary in size, but recent reports have suggested a median ICU bed density in the range of 12–30 beds [55, 56]. Similar to a mixed-population ICU, macaques are housed in a mixed population, and surgically naïve individuals and individuals with cephalic implants of varying duration can be pair-housed or housed in close proximity within the same room. Besides topical antimicrobial exposure inside recording chambers, macaques in our vivarium are exposed to systemic antimicrobial therapy during the perioperative period and when clinically indicated, such as prophylactic treatment for wounds resulting from an altercation with a conspecific. This chronic intermittent antimicrobial exposure provides a selective pressure for resistant isolates to emerge, as well as provide a niche for intestinal overgrowth and permit breakdown of colonization resistance [57]. MIC testing suggests that our E. faecalis isolates remain susceptible to ampicillin, amoxicillin-clavulanic acid, vancomycin, daptomycin and meropenem. Vancomycin, daptomycin and meropenem are not used in our facility and will continue to be excluded from clinical use while isolates remain susceptible to second-generation penicillins, such as ampicillin.
Finally, chronic cephalic implants serve as a nidus for biofilm formation and persistent infection. A variety of bacterial species, including Staphylococcus, Enterococcus, and Corynebacterium spp. have been previously cultured from the skin margins of macaque cephalic implants [4, 5]. It is unknown whether chamber colonization results from translocation of skin flora or from fecal contamination. Because of the parallels between the macaque colony under study and a human hospital environment, it is not surprising to see similarities in identified pathogens. Specifically, the main two E. faecalis sequence types, ST 4 and ST 55, detected in the macaque population have been previously identified as agents of human infection [58–61].While the majority of chamber infections are subclinical in the macaque colony, the potential importance of meningitis, cephalic abscess, and bacteremia/septicemia to animal health maintenance necessitates a thorough understanding of the bacterial species colonizing implants, their antimicrobial resistance profiles and the appropriate use of antimicrobials, when warranted. The samples analyzed here were recovered from aerobic cultures, and so it remains possible, perhaps likely, that additional anaerobic microbes are present in the mixed microbial biofilms, which remain to be identified.
With improved understanding of multi-drug resistant bacteria colonizing research macaques, veterinary protocols have been updated to address judicious use of antimicrobials in our facility. Topical instillation of antimicrobial agents by investigators is now prohibited and only allowed under veterinary supervision. The veterinary staff directs regular disinfection of cephalic recording chambers, with frequency of disinfection dependent on culture results, as well as the amount and appearance of discharge. Disinfection protocols for cephalic recording chambers have been standardized to 1–2% povodone iodine solution and saline under strict aseptic technique. Perioperative trimethoprim-sulfamethoxazole and enrofloxacin have been replaced by cefazolin, and post-operative ceftriaxone use is restricted to surgical procedures involving a craniotomy. Animals with suspected meningitis undergo thorough diagnostic work-up, including culture and sensitivity testing of isolates in cerebrospinal fluid to guide choices in antimicrobial therapy and minimize further spread of bacterial resistance genes within the macaque colony.
Conclusions
Chronic cephalic implants in macaques used for cognitive neuroscience research can serve as a nidus of multi-drug resistant bacterial pathogens. We have identified polymicrobial colonization as a common feature, and noted that S. aureus, Proteus spp. and E. faecalis are the most prevalent organisms associated with these implants. Multi-locus sequence typing revealed that two major E. faecalis sequence types associated with human infection are present in our colony. Cephalic implants also serve as a minimally invasive access location for studying biofilm formation on medical-grade biomaterials to provide better insight on the mechanisms of enterococcal persistence in a healthcare setting. Further investigation is needed to understand how S. aureus, Proteus and other bacteria contribute to chronic polymicrobial chamber infections. Future studies will investigate E. faecalis colonization of implanted devices, feces and other sites at multiple time points to evaluate sources and reservoirs for these hospital pathogens. Additional investigation using in vitro biofilm models will help elucidate the roles of varying combinations of pathogens and the effectiveness of different disinfection and treatment regimens in controlling implant colonization.
Supporting Information
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We acknowledge Ellen Buckley and Carolyn Madden for their diagnostic microbiology expertise, and Dr. Robert Marini, Dr. Mary Patterson and Monica Siddalls for their veterinary support. We acknowledge Eric J Ma for his assistance with beeswarm plot generation. The contributions of SEW, MTL, ET, and JDF were supported by NIH grants T32 OD010978 and P30 ES02109 (to JGF). The contributions of FL and MSG were supported by PHS grant AI072360, AI108710, and the Harvard-wide Program on Antibiotic Resistance AI083214. CDLF-N holds a postdoctoral scholarship from Fundación Ramón Areces (Spain).
Data Availability
All bacterial sequence files are available from the DDBJ/ENA/GenBank database (accession number(s) MCFU01000001-MCFU01000003; MCFV01000001-MCFV01000003; and MCFW01000001-MCFW01000002).
Funding Statement
This research was supported by National Institutes of Health (www.nih.gov) grants T32 OD010978 and P30 ES02109 (to JGF) and 5P01AI083214-07 (to MSG). The contributions of authors FL and MSG were supported by United States Public Health Service (www.usphs.gov) grants AI072360, AI108710, and the Harvard-wide Program on Antibiotic Resistance AI083214. CDLF-N holds a postdoctoral scholarship from Fundación Ramón Areces (Spain). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Perretta G. Non-human primate models in neuroscience research. Scandinavian Journal of Laboratory Animal Sciences. 2009. [Google Scholar]
- 2.Fremont JJ, Marini RP, Fox JG, Rogers AB. Acute respiratory distress syndrome in two rhesus macaques (Macaca mulatta). J Am Assoc Lab Anim Sci. 2008;47(5):61–6. PubMed Central PMCID: PMCPMC2691537. [PMC free article] [PubMed] [Google Scholar]
- 3.Leblanc M, Berry K, McCort H, Reuter JD. Brain Abscess in a Rhesus Macaque (Macaca mulatta) with a Cephalic Implant. Comp Med. 2013;63(4):367–72. [PMC free article] [PubMed] [Google Scholar]
- 4.Bergin IL, Chien CC, Marini RP, Fox JG. Isolation and characterization of Corynebacterium ulcerans from cephalic implants in macaques. Comp Med. 2000;50(5):530–5. [PubMed] [Google Scholar]
- 5.Lee G, Danneman P, Rufo R, Kalesnykas R, Eng V. Use of Chlorine Dioxide for Antimicrobial Prophylactic Maintenance of Cephalic Recording Devices in Rhesus Macaques (Macaca mulatta). Contemporary topics in laboratory animal science / American Association for Laboratory Animal Science. 1998;37(2):59–63. Epub 2002/11/29. [PubMed] [Google Scholar]
- 6.Venezia J, Cassiday PK, Marini RP, Shen Z, Buckley EM, Peters Y, et al. Characterization of Corynebacterium species in macaques. Journal of medical microbiology. 2012;61(Pt 10):1401–8. Epub 2012/06/23. PubMed Central PMCID: PMCPmc3541768. 10.1099/jmm.0.045377-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kafil HS, Mobarez AM. Spread of Enterococcal Surface Protein in Antibiotic Resistant Entero-coccus faecium and Enterococcus faecalis isolates from Urinary Tract Infections. The open microbiology journal. 2015;9:14–7. Epub 2015/07/15. PubMed Central PMCID: PMCPmc4493631. 10.2174/1874285801509010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paganelli FL, Willems RJ, Leavis HL. Optimizing future treatment of enterococcal infections: attacking the biofilm? Trends in microbiology. 2012;20(1):40–9. Epub 2011/12/16. 10.1016/j.tim.2011.11.001 [DOI] [PubMed] [Google Scholar]
- 9.Performance standards for antimicrobial disk susceptibility tests; Approved Standard, M02-A10— 10th ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2009.
- 10.Methods for dilution antimicrobial susceptibility tests of bacteria that grow aerobically; Approved Standard, M07-A9— 9th ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2012.
- 11.Coenye T, Falsen E, Vancanneyt M, Hoste B, Govan JR, Kersters K, et al. Classification of Alcaligenes faecalis-like isolates from the environment and human clinical samples as Ralstonia gilardii sp. nov. International Journal of Systematic Bacteriology. 1999;49 Pt 2:405–13. [DOI] [PubMed] [Google Scholar]
- 12.Ruiz-Garbajosa P, Bonten MJ, Robinson DA, Top J, Nallapareddy SR, Torres C, et al. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J Clin Microbiol. 2006;44(6):2220–8. PubMed Central PMCID: PMCPMC1489431. 10.1128/JCM.02596-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595 Epub 2010/12/15. PubMed Central PMCID: PMCPmc3004885. 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nature methods. 2013;10(6):563–9. Epub 2013/05/07. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- 15.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42:D581–91. PubMed Central PMCID: PMCPMC3965095. 10.1093/nar/gkt1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–4. Epub 2012/07/12. PubMed Central PMCID: PMCPmc3468078. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, et al. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol. 2014;52(5):1501–10. Epub 2014/02/28. PubMed Central PMCID: PMCPmc3993690. 10.1128/JCM.03617-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minogue TD, Daligault HE, Davenport KW, Broomall SM, Bruce DC, Chain PS, et al. Complete Genome Assembly of Enterococcus faecalis 29212, a Laboratory Reference Strain. Genome announcements. 2014;2(5). Epub 2014/10/08. PubMed Central PMCID: PMCPmc4175211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tendolkar PM, Baghdayan AS, Gilmore MS, Shankar N. Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis. Infect Immun. 2004;72(10):6032–9. Epub 2004/09/24. PubMed Central PMCID: PMCPmc517584. 10.1128/IAI.72.10.6032-6039.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stepanović S, Vuković D, Hola V, Bonaventura G. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS: acta pathologica, microbiologica, et immunologica Scandinavica. 2007. [DOI] [PubMed] [Google Scholar]
- 21.de la Fuente-Nunez C, Reffuveille F, Mansour SC, Reckseidler-Zenteno SL, Hernandez D, Brackman G, et al. D-enantiomeric peptides that eradicate wild-type and multidrug-resistant biofilms and protect against lethal Pseudomonas aeruginosa infections. Chemistry & biology. 2015;22(2):196–205. Epub 2015/02/24. PubMed Central PMCID: PMCPmc4362967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reffuveille F, de la Fuente-Nunez C, Mansour S, Hancock RE. A broad-spectrum antibiofilm peptide enhances antibiotic action against bacterial biofilms. Antimicrob Agents Chemother. 2014;58(9):5363–71. Epub 2014/07/02. PubMed Central PMCID: PMCPmc4135845. 10.1128/AAC.03163-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King A, Boothman C, Phillips I. Comparative in-vitro activity of meropenem on clinical isolates from the United Kingdom. J Antimicrob Chemother. 1989;24 Suppl A:31–45. [DOI] [PubMed] [Google Scholar]
- 24.Schouten MA, Voss A, Hoogkamp-Korstanje JA. Antimicrobial susceptibility patterns of enterococci causing infections in Europe. The European VRE Study Group. Antimicrob Agents Chemother. 1999;43(10):2542–6. PubMed Central PMCID: PMCPMC89517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gemmell CG. Susceptibility of a variety of clinical isolates to linezolid: a European inter-country comparison. J Antimicrob Chemother. 2001;48(1):47–52. [DOI] [PubMed] [Google Scholar]
- 26.Yu ZJ, Chen Z, Cheng H, Zheng JX, Pan WG, Yang WZ, et al. Recurrent linezolid-resistant Enterococcus faecalis infection in a patient with pneumonia. Int J Infect Dis. 2015;30:49–51. 10.1016/j.ijid.2014.06.017 [DOI] [PubMed] [Google Scholar]
- 27.Chylinski K, Makarova KS, Charpentier E, Koonin EV. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42(10):6091–105. PubMed Central PMCID: PMCPMC4041416. 10.1093/nar/gku241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manson JM, Keis S, Smith JM, Cook GM. Acquired bacitracin resistance in Enterococcus faecalis is mediated by an ABC transporter and a novel regulatory protein, BcrR. Antimicrob Agents Chemother. 2004;48(10):3743–8. PubMed Central PMCID: PMCPMC521867. 10.1128/AAC.48.10.3743-3748.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.An FY, Clewell DB. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J Bacteriol. 2002;184(7):1880–7. PubMed Central PMCID: PMCPMC134937. 10.1128/JB.184.7.1880-1887.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antiporta MH, Dunny GM. ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J Bacteriol. 2002;184(4):1155–62. PubMed Central PMCID: PMCPMC134800. 10.1128/jb.184.4.1155-1162.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clewell DB, An FY, White BA, Gawron-Burke C. Streptococcus faecalis sex pheromone (cAM373) also produced by Staphylococcus aureus and identification of a conjugative transposon (Tn918). J Bacteriol. 1985;162(3):1212–20. PubMed Central PMCID: PMCPMC215906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakayama J, Abe Y, Ono Y, Isogai A, Suzuki A. Isolation and structure of the Enterococcus faecalis sex pheromone, cOB1, that induces conjugal transfer of the hemolysin/bacteriocin plasmids, pOB1 and pYI1. Biosci Biotechnol Biochem. 1995;59(4):703–5. [DOI] [PubMed] [Google Scholar]
- 33.Brinster S, Posteraro B, Bierne H, Alberti A, Makhzami S, Sanguinetti M, et al. Enterococcal leucine-rich repeat-containing protein involved in virulence and host inflammatory response. Infect Immun. 2007;75(9):4463–71. PubMed Central PMCID: PMCPMC1951196. 10.1128/IAI.00279-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155(Pt 6):1749–57. 10.1099/mic.0.026385-0 [DOI] [PubMed] [Google Scholar]
- 35.Nielsen HV, Flores-Mireles AL, Kau AL, Kline KA, Pinkner JS, Neiers F, et al. Pilin and sortase residues critical for endocarditis- and biofilm-associated pilus biogenesis in Enterococcus faecalis. J Bacteriol. 2013;195(19):4484–95. PubMed Central PMCID: PMCPMC3807452. 10.1128/JB.00451-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.La Carbona S, Sauvageot N, Giard JC, Benachour A, Posteraro B, Auffray Y, et al. Comparative study of the physiological roles of three peroxidases (NADH peroxidase, Alkyl hydroperoxide reductase and Thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis. Mol Microbiol. 2007;66(5):1148–63. 10.1111/j.1365-2958.2007.05987.x [DOI] [PubMed] [Google Scholar]
- 37.Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol. 2013;34(1):1–14. 10.1086/668770 [DOI] [PubMed] [Google Scholar]
- 38.Chow JW. Aminoglycoside resistance in enterococci. Clin Infect Dis. 2000;31(2):586–9. 10.1086/313949 [DOI] [PubMed] [Google Scholar]
- 39.Palmer KL, Gilmore MS. Multidrug-resistant enterococci lack CRISPR-cas. MBio. 2010;1(4). PubMed Central PMCID: PMCPMC2975353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bantar CE, Micucci M, Fernandez Canigia L, Smayevsky J, Bianchini HM. Synergy characterization for Enterococcus faecalis strains displaying moderately high-level gentamicin and streptomycin resistance. J Clin Microbiol. 1993;31(7):1921–3. PubMed Central PMCID: PMCPMC265661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller WR, Munita JM, Arias CA. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti Infect Ther. 2014;12(10):1221–36. 10.1586/14787210.2014.956092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petersen A, Jensen LB. Analysis of gyrA and parC mutations in enterococci from environmental samples with reduced susceptibility to ciprofloxacin. FEMS Microbiol Lett. 2004;231(1):73–6. 10.1016/S0378-1097(03)00929-7 [DOI] [PubMed] [Google Scholar]
- 43.Onodera Y, Okuda J, Tanaka M, Sato K. Inhibitory activities of quinolones against DNA gyrase and topoisomerase IV of Enterococcus faecalis. Antimicrob Agents Chemother. 2002;46(6):1800–4. PubMed Central PMCID: PMCPMC127212. 10.1128/AAC.46.6.1800-1804.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohamed JA, Huang DB. Biofilm formation by enterococci. J Med Microbiol. 2007;56(12):1581–8. [DOI] [PubMed] [Google Scholar]
- 45.Sussmuth SD, Muscholl-Silberhorn A, Wirth R, Susa M, Marre R, Rozdzinski E. Aggregation substance promotes adherence, phagocytosis, and intracellular survival of Enterococcus faecalis within human macrophages and suppresses respiratory burst. Infect Immun. 2000;68(9):4900–6. PubMed Central PMCID: PMCPMC101694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shankar V, Baghdayan AS, Huycke MM, Lindahl G, Gilmore MS. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun. 1999;67(1):193–200. PubMed Central PMCID: PMCPMC96296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nallapareddy SR, Singh KV, Sillanpaa J, Zhao M, Murray BE. Relative contributions of Ebp Pili and the collagen adhesin ace to host extracellular matrix protein adherence and experimental urinary tract infection by Enterococcus faecalis OG1RF. Infect Immun. 2011;79(7):2901–10. PubMed Central PMCID: PMCPMC3191960. 10.1128/IAI.00038-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hancock LE, Perego M. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J Bacteriol. 2004;186(17):5629–39. PubMed Central PMCID: PMCPMC516840. 10.1128/JB.186.17.5629-5639.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M, et al. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Applied and environmental microbiology. 2001;67(10):4538–45. Epub 2001/09/26. PubMed Central PMCID: PMCPmc93200. 10.1128/AEM.67.10.4538-4545.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chuang-Smith ON, Wells CL, Henry-Stanley MJ, Dunny GM. Acceleration of Enterococcus faecalis biofilm formation by aggregation substance expression in an ex vivo model of cardiac valve colonization. PloS one. 2010;5(12):e15798 Epub 2011/01/07. PubMed Central PMCID: PMCPmc3012704. 10.1371/journal.pone.0015798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kristich CJ, Li Y-H, Cvitkovitch DG, Dunny GM. Esp-Independent Biofilm Formation by Enterococcus faecalis. Journal of Bacteriology. 2004;186(1):154–63. 10.1128/JB.186.1.154-163.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guiton PS, Hung CS, Kline KA, Roth R, Kau AL, Hayes E, et al. Contribution of autolysin and Sortase a during Enterococcus faecalis DNA-dependent biofilm development. Infect Immun. 2009;77(9):3626–38. PubMed Central PMCID: PMCPMC2738007. 10.1128/IAI.00219-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Tyne D, Martin MJ, Gilmore MS. Structure, Function, and Biology of the Enterococcus faecalis Cytolysin. Toxins. 2013;5(5):895–911. 10.3390/toxins5050895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chow JW, Thal LA, Perri MB, Vazquez JA, Donabedian SM, Clewell DB, et al. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob Agents Chemother. 1993;37(11):2474–7. PubMed Central PMCID: PMCPMC192412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Costa DK, Barg FK, Asch DA, Kahn JM. Facilitators of an Interprofessional Approach to Care in Medical and Mixed Medical/Surgical ICUs: A Multicenter Qualitative Study. Res Nurs Health. 2014;37(4):326–35. 10.1002/nur.21607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hyzy RC, Flanders SA, Pronovost PJ, Berenholtz SM, Watson S, George C, et al. Characteristics of intensive care units in Michigan: Not an open and closed case. J Hosp Med. 2010;5(1):4–9. 10.1002/jhm.567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vollaard EJ, Clasener HA. Colonization resistance. Antimicrob Agents Chemother. 1994;38(3):409–14. PubMed Central PMCID: PMCPMC284472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watanabe S, Kobayashi N, Quinones D, Nagashima S, Uehara N, Watanabe N. Genetic diversity of enterococci harboring the high-level gentamicin resistance gene aac(6')-Ie-aph(2'')-Ia or aph(2'')-Ie in a Japanese hospital. Microb Drug Resist. 2009;15(3):185–94. 10.1089/mdr.2009.0917 [DOI] [PubMed] [Google Scholar]
- 59.Freitas AR, Novais C, Ruiz-Garbajosa P, Coque TM, Peixe L. Clonal expansion within clonal complex 2 and spread of vancomycin-resistant plasmids among different genetic lineages of Enterococcus faecalis from Portugal. Journal of Antimicrobial Chemotherapy. 2009;63(6):1104–11. 10.1093/jac/dkp103 [DOI] [PubMed] [Google Scholar]
- 60.Yang JX, Li T, Ning YZ, Shao DH, Liu J, Wang SQ, et al. Molecular characterization of resistance, virulence and clonality in vancomycin-resistant Enterococcus faecium and Enterococcus faecalis: A hospital-based study in Beijing, China. Infect Genet Evol. 2015;33:253–60. 10.1016/j.meegid.2015.05.012 [DOI] [PubMed] [Google Scholar]
- 61.Sun H, Wang H, Xu Y, Jones RN, Costello AJ, Liu Y, et al. Molecular characterization of vancomycin-resistant Enterococcus spp. clinical isolates recovered from hospitalized patients among several medical institutions in China. Diagn Microbiol Infect Dis. 2012;74(4):399–403. 10.1016/j.diagmicrobio.2012.09.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All bacterial sequence files are available from the DDBJ/ENA/GenBank database (accession number(s) MCFU01000001-MCFU01000003; MCFV01000001-MCFV01000003; and MCFW01000001-MCFW01000002).