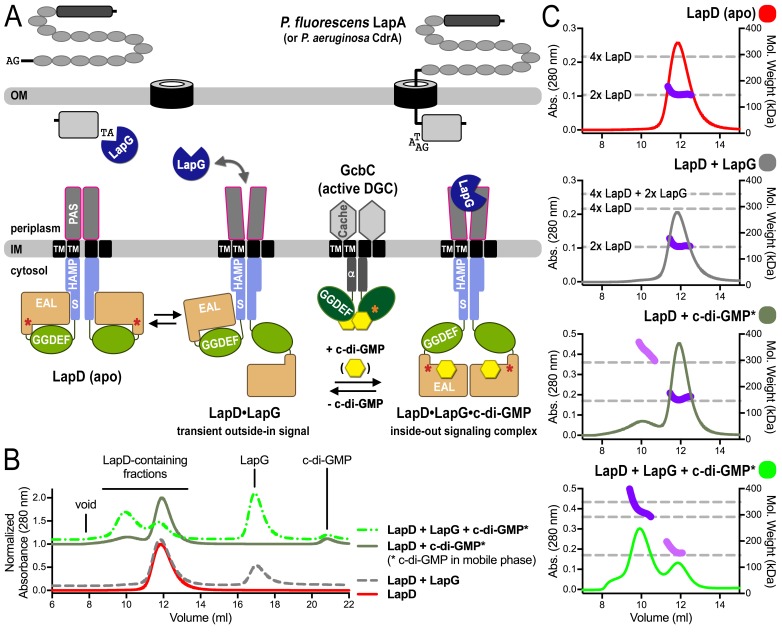
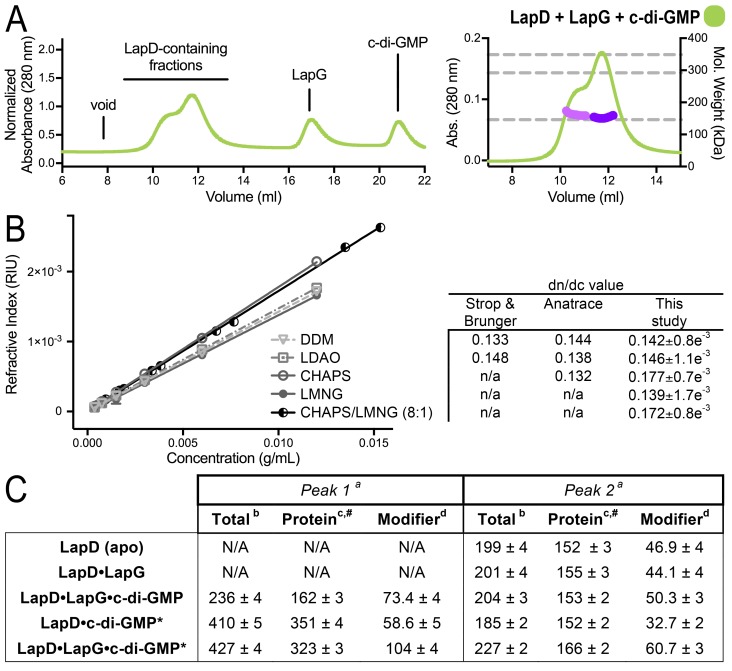
Figure 1. SEC-MALS reveals a switch of LapD dimers to dimer-of-dimers upon ligand binding.
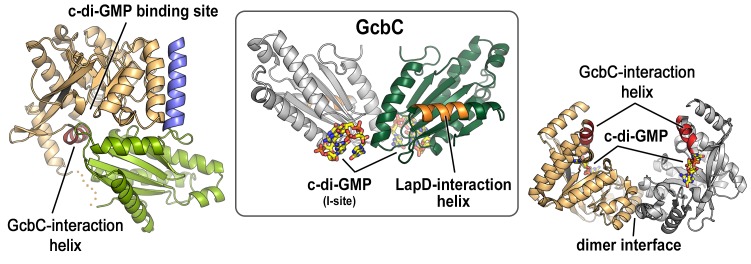
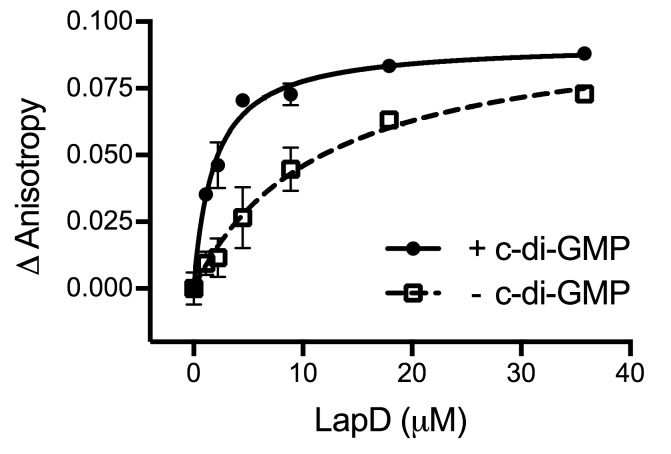
(A) Working model for c-di-GMP-dependent regulation of the periplasmic protease LapG via the inner membrane protein LapD. Concerted conformational changes expose a periplasmic binding site for LapG on LapD, sequestering the protease away from its substrates, the adhesin proteins LapA in P. fluorescens or CdrA in P. aeruginosa. (DGC, diguanylate cyclase; red/orange asterisks indicate interaction helices in LapD/GcbC). (B). Size-exclusion chromatograms for detergent-solubilized LapD in different states. Samples were prepared as described in the Material and Methods. (Asterisks: c-di-GMP was included in the mobile phase). (C) Molecular weight of LapD in solution. Peak fractions were analyzed by in-line SEC-MALS. (Absorbance at 280 nm: Traces colored according to (B); molecular weight determination: Dark and light purple dots; theoretical molecular weights based on sequence: Horizontal dashed lines.) Data are representative of two biological replicates using independent protein preparations.