In consenting to publish an extravagantly long report of the most tedious kind—a genome analysis (10)—this journal has shown what appears to be extraordinary indulgence. The genome is that of P1, a bacteriophage that, unlike certain of its cousins, neither raises hopes nor instills fear. P1 phage particles are not the “great hope of universal therapy and prophylaxis,” as Felix d'Hérelle believed bacteriophages that target pathogens might be (quoted in reference 14). In the more than 50 years since the discovery of P1, the phage has not been drafted for a combat role in therapy (11). As for the P1 prophage, it does not confer toxigenicity on a harmless bacterium, as do the prophages that have given us, among other troubles, diphtheria, cholera, toxic shock syndrome, and botulism (4), nor does P1 normally subvert antibiotic treatments, despite its capacity to acquire and transfer multidrug resistance genes while sacrificing nothing more than some of its extensive terminal redundancy. P1, if not a phage “of interest,” is nevertheless an interesting phage. It has long been the workhorse of gene transduction, and its packaging and recombination systems are much in use for the genetic engineering of eukaryotes. What sets P1 apart from the majority of characterized temperate phages is the autonomy of the prophage. P1 lysogenizes its hosts as a plasmid of low copy number and, because it has the viral option, can be manipulated more conveniently than other large plasmids, most of which are horizontally transmitted by conjugation without extensive amplification. But is P1 such a very important phage as to merit the royal treatment accorded the analysis of its genome? Yes, it is. P1 opens new perspectives on familiar biological topics, to one of which I shall return shortly. It also boasts a distinguished history.
P1 has played a prominent role in the development of molecular biology. The experiments of Werner Arber and Daisy Dussoix (beginning with reference 1), showing P1 to be capable of conferring a phenotype on DNA, can be said to have ushered in the new biology in which restriction-modification (R-M) plays such a key role. The several contributions of P1 are widely known, and the reader can refresh his memory of them by reading the introductory section of the accompanying report (10). That gene-by-gene survey of P1 has the virtue of bringing together scattered information to give a broad picture of the phage and is the most ambitious overview of P1 to be published since 1988 (15). Although not conceived as a review of the literature, the report seeks to provide a meaningful context for the new sequence data and so offers the reader, as an alternative to casual table-hopping, the opportunity to make a more intimate acquaintance with the subject.
Much more attention has been bestowed on T4 and on lambda than on P1. The only recent P1 review (9) is modest in scope. T4 is the subject of a comprehensive compendium, revised in 1994 (8), and a major recent review (12). Lambda is honored by two sequential volumes, known respectively among “lambdologists” as the “lambda bible” (7) and, with unmerited pessimism, “lambda's tombstone” (6). A separate monograph that is largely about the lambda immunity switch is a movie script awaiting a Fellini (13). Simulators are finding the so-called “genetic switch” irresistible (2), and a movie of the simulacra, if not by a famous director, is sure to come. For the record, lambda has no genetic switch; the one so named is epigenetic. P1, on the other hand, does possess a genetic switch, and it is used for exchanging specificity elements on delicate appendages that laboratory strains of lambda appear to have lost, tail fibers.
Knowledge of P1 is not as extensive as knowledge of T4 or lambda, but in some respects, it is not far behind. The R-M system of P1, although overshadowed by the subsequently discovered type II R-M systems, has been thoroughly studied. Genes required for plasmid maintenance, including the complex immunity system of P1, have been analyzed in detail, and the critical DNA regions were sequenced several years ago. On the other hand, large regions of P1 remained unsequenced until now. Relatively little is known about morphogenesis of the virion beyond the evidence obtained from early genetic and electron micrographic studies. The present analysis fills a number of obvious gaps, albeit with several tentative and imprecise assignments of genes to structural, regulatory, and enzymatic functions.
Probably the most useful function of the genome analysis is to make a larger public aware that our present understanding of P1 raises questions of general interest, some that deserve revival and some that could not have been asked before. The absence, until now, of any broad review on P1 in a widely circulated journal means that much that is known about the phage has not been given the attention deserved. There are exceptions. Notice is being taken of how efficiently P1 breaks free from the confines of its host, and P1 partitioning genes are finally creating a flurry of interest as the curtain rises on the blurry ballet (with glowing tutus courtesy of Aequorea victoria). Other topics, unaided by the appeal of liberation or of false color, have fallen into benign neglect; the P1 immunity circuitry is an obvious case in point.
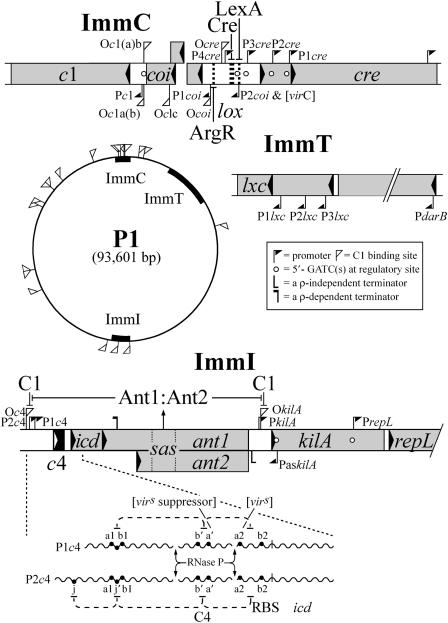
The relevant genes are located in three separate regions: ImmC, ImmI, and ImmT (Fig. 1). In ImmC is the c1 master repressor gene (an analog of the unrelated cI of lambda) and the gene for a C1 inhibitor, coi. In ImmI is a gene that codes for an immunity-specific translational repressor RNA (C4). It blocks a ribosome binding site on a downstream portion of the RNA of which C4 is a part. The resulting failure to synthesize an inhibitor of cell division, Icd, makes the message susceptible to termination by the bacterial Rho protein at a site early in the cotranslated Ant1 message. In this indirect way, C4 blocks the synthesis of the message for a pair of immunity-specific antirepressor (Ant) proteins. Their action, preferentially in cis, is also indirect, requiring formation of a 1:1 complex and activation at a specific site, sas, within the ant DNA. ImmT, the tertiary component of the circuitry, encodes a corepressor, Lxc, that enhances the affinity of C1 for operators to which it is already bound. It lowers the expression of c1 through an autoregulatory loop and, dependent upon the relative affinity of each particular operator for C1, may directly decrease or indirectly increase the transcription it regulates.
FIG. 1.
The tripartite immunity circuitry of P1. The diagram is based largely on work reviewed in reference 5. Additional relevant citations can be found in the accompanying report (10), which the reader is urged to consult. Gray boxes representing genes are drawn approximately to scale. The central circle represents the P1 genome. O, C1 binding sites; P, promoters. The capacity of certain gene products to bind to a target is indicated by a line from the ligand to its target (at the blunt end). Target sites, except for C1 binding sites (see the boxed key), are indicated by thick dashed vertical lines. The site to which Cre binds is a locus of site-specific recombination (lox). The binding of ArgR to a putative binding site nearby is presumed to assist in constraining the directionality of the recombination reaction towards dimer resolution. ArgR, LexA, and RNase P are bacterial proteins. The arrow from sas (site of ant specificity) to Ant1-Ant2 (the complex of the two Ant proteins) signifies that the complex is activated by binding to sas, from which site it can alleviate repression at flanking operator sites in cis. The alternative transcripts that code for C4 RNA, Icd and the Ant proteins are indicated by wiggly lines. Regions of these transcripts marked with a prime are indicated by dashed connecting lines as susceptible of interaction with similarly named, complementary regions that are unprimed. Note that a point mutation in region a2 that confers virulence (constitutive Ant synthesis) can be suppressed by a complementary mutation in region a′. The complementarity of region j with region j′ was pointed out by Jochen Heinrich (personal communication). RBS (overlapping region b2), ribosome binding site used for the expression of icd and ant1.
The design of this elaborate system permits immunity to be rapidly established, uninterruptedly maintained, and, should conditions become unfavorable, promptly and irreversibly lifted. An important contribution to the bistability of the switch comes from the mutually antagonistic actions of C1 and Coi: Coi binds to and inactivates C1; C1 represses the transcription of coi at one of its two promoters. Moreover, in vitro studies indicate that the ImmC region operators (in particular those that control c1 and coi transcription) are prone to looping through C1 bridges that Lxc significantly strengthens, although the net effect on repression is not large. In the ternary complex of Lxc with operator-bound C1, the repressor is protected from binding to Coi. Probably Lxc is of most importance during the establishment or lifting of immunity, but the benefits to the phage of possessing this little protein are not obvious. An additional control on Coi, more properly on coi expression, most likely comes from the bacterial host protein LexA, a sensor of DNA damage. The location of a LexA binding site in the P1 DNA at P2coi suggests that LexA represses coi transcription at this promoter but releases repression in response to an SOS signal. Lysogenic induction mediated by DNA damage apparently occurs without cleavage of the master repressor. A mutation to constitutive expression of coi at P2coi results in virulence (virC). The LexA binding site is close to lox (the locus of Cre-mediated recombination), and both sites are located in the path of opposing coi and cre transcripts. The region appears to be the nexus of several interconnected regulatory elements. What the consequences of their interconnections may be is a knotty problem.
Of the operator sites that bind C1, most are asymmetric and bind C1 as a monomer. The operator at which the C1 protein exerts autorepression (Oc1ab) is exceptional; it is an incomplete inverted repeat and binds two C1 molecules. Why a bivalent operator? Perhaps the robustness of the regulatory circuit is thereby enhanced.
The multiplicity of operator sites scattered over the P1 genome (Fig. 1) precludes the possibility that immunity specificity might be changed by mutation in the c1 gene. Too many operator sites would have to be changed as well. At ImmI, however, immunity specificity can be altered, and with relative ease. The heteroimmunity of the related phages P1 and P7 (their ability to plate on lysogens of the other) can be attributed to only four base changes in ImmI. These have been mapped to complementary regions in C4 and its target. P1 and P7 exhibit a further specificity difference (as regards their capacity to induce a related prophage on superinfection). It has been identified with a small region, sas, within otherwise nearly identical ant genes of the two phages (J. Heinrich and H. Schuster, unpublished data). Alterations in the separate immunity specificity conferred by sas affect the Ant proteins and their target simultaneously. How many changes are required to change specificity at this site is unclear, but a new specificity did arise, and through changes in a relatively small region. It is tempting to speculate that the capacity to alter the specificity of immunity, like the capacity to alter host range through recombinational switching of tail fiber genes, confers a selective advantage. ImmI may offer a flexibility that is excluded at ImmC. Could the Byzantine complexity of the controls at ImmI be the outcome, not of successive host-parasite accommodations, but of competition among related phages?
The transcription of c4 and the ant gene from two promoters, one under C1 control (P2c4) and the other not (P1c4), suggests that P2c4 is involved in establishing immunity and P1c4 is involved primarily in its maintenance. Establishment is a complex kinetic process in which the separate or overlapping stages are sensitive to the state of the cell. What are those stages and how are they influenced? How, and under what circumstances, do the noxious proteins in and adjacent to the c4 operon—the inhibitor of cell division, Icd, and KilA (a lethal paralog of the Ant proteins)—contribute to the well-being of P1? Might they function during lysogenization to influence cell cycle parameters? Finally, what are the role and mode of action of sas? Does the antirepressor complex Ant1-Ant2 move along the DNA or loop from sas to nearby operator sites such as OkilA, which controls lytic replication? Might the capacity of P1 or P7 to plate on a heteroimmune lysogen without prophage induction mean that appreciable replication of the superinfecting phage precedes a complete lifting of immunity that is presumably required for phage maturation? A delay in the lifting of immunity until the replication of the superinfecting phage had switched from theta to rolling circle replication would allow transcription from the supercoil-sensitive antisense promoter, PaskilA, with the result that the lifting of immunity would not result in new initiations of replication.
Note also that both the ImmC and ImmI genes contain sets of GATC sequences in strategic locations. These sequences are potential targets of DNA adenine methylation, a reaction that enzymes of both E. coli and P1 can catalyze. This much has been known for some time, but the genome analysis has revealed, in addition, that the P1 DNA adenine methylase has an N-terminal extension with putative cytosine methylase activity. Methylation is known to play a critical role in the packaging of P1 DNA, but what is its role at each of the several potential methylation sites in the ImmC and ImmI regions and elsewhere in the genome, and what functions do the possible dual activities of the P1 enzyme have?
Mindful that an intriguing question, like a charming Paris bistro, is likely to seem less interesting if brought to one's attention by someone else, I will not persist in depriving the reader of the pleasures of independent thinking. Nor will I make matters worse by suggesting how the questions raised here might be answered. But it may be relevant to point out that some major clues to the structure of the immunity circuitry came from studies of relationships between P1 and its close relative, P7. It is hard to imagine how sas might have been discovered from studies of P1 alone. Crucial to the discovery was the paradoxical ability of mutants constitutive for ant expression to plate on bacteria harboring a prophage repressed by the C1 gene carried by the infecting phage, but without a significant yield of phages of the prophage type appearing in the bursts. The existence of a site of ant specificity (sas) was postulated to reconcile these observations, and a comparison of the sequences of the ant genes of the two phages revealed precisely where sas is located. A major benefit of having the complete sequence of P1 available now and having that of P7 available soon (M. Łobocka, personal communication) will be to enlarge possibilities for comparative and evolutionary research.
Comparative in silico studies are receiving increasing attention with the current flood of sequence information, and they will continue to play a major role in understanding the biology of phages. That attention should not distract us from the recognition, as Tom Bickle has put it, “that there is still a place for wet work” (3). The publication of an analysis of the P1 genome does honor to a phage with a distinguished past, but its primary function is as a resource for future studies, wet, dry, and perhaps even funded.
Acknowledgments
I thank Ryland Young for suggesting that I write this article, David Lane, Ian Molineux, Dhruba Chattoraj and Małgorzata Łobocka for astute constructive criticism, and Jochen Heinrich for unpublished information and advice on the immunity circuitry.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Arber, W., and D. Dussoix. 1962. Host specificity of DNA produced by Escherichia coli. I. Host controlled modification of bacteriophage lambda. J. Mol. Biol. 5:18-36. [DOI] [PubMed] [Google Scholar]
- 2.Arkin, A., J. Ross, and H. H. McAdams. 1998. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics 149:1633-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bickle, T. A. 2004. Restricting restriction. Mol. Microbiol. 51:3-5. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, E. F., B. M. Davis, and B. Hochhut. 2001. Bacteriophage-bacteriophage interactions in the evolution of pathogenic bacteria. Trends Microbiol. 9:137-144. [DOI] [PubMed] [Google Scholar]
- 5.Heinrich, J., M. Velleman, and H. Schuster. 1995. The tripartite immunity system of phages P1 and P7. FEMS Microbiol. Rev. 17:121-126. [DOI] [PubMed] [Google Scholar]
- 6.Hendrix, R. W., J. W. Roberts, F. W. Stahl, and R. A. Weisburg (ed.). 1983. Lambda II. Cold Spring Harbor Laboratory, New York, N.Y.
- 7.Hershey, A. D. (ed.). 1971. The bacteriophage lambda. Cold Spring Harbor Laboratory, New York, N.Y.
- 8.Karam, J. D., J. W. Drake, K. N. Kreuzer, G. Mosig, D. H. Hall, F. A. Eiserling, L. W. Black, E. K. Spicer, E. Kutter, K. Carlson, and E. S. Miller (ed.). 1994. Molecular biology of bacteriophage T4, 2nd ed. ASM Press, Washington, D.C.
- 9.Lehnherr, H. Bacteriophage P1. In R. Calendar (ed.), The bacteriophages, in press. Oxford University Press, New York, N.Y.
- 10.Łobocka, M. B., D. J. Rose, G. Plunkett III, M. Rusin, A. Samojedny, H. Lehnherr, M. B. Yarmolinsky, and F. R. Blattner. 2004. The genome of bacteriophage P1. J. Bacteriol. 186:7032-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merril, C. R., D. Scholl, and S. L. Adhya. 2003. The prospect for bacteriophage therapy in Western medicine. Nat. Rev. Drug Discovery 2:489-497. [DOI] [PubMed] [Google Scholar]
- 12.Miller, E. S., E. Kutter, G. Mosig, F. Arisaka, T. Kunisawa, and W. Rüger. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67:1092-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ptashne, M. 1992. A genetic switch. Phage λ and higher organisms, 2nd ed. Cell Press and Blackwell Science, Cambridge, Mass.
- 14.Stent, G. S. 1963. Molecular biology of bacterial viruses. W. H. Freeman & Co., San Francisco, Calif.
- 15.Yarmolinsky, M. B., and N. Sternberg. 1988. Bacteriophage P1, p. 291-438. In R. Calendar (ed.), The bacteriophages, vol. 1. Plenum Publishing Corporation, New York, N.Y. [Google Scholar]