Abstract
The bacterial SOS regulon is strongly induced in response to DNA damage from exogenous agents such as UV radiation and nalidixic acid. However, certain mutants with defects in DNA replication, recombination, or repair exhibit a partially constitutive SOS response. These mutants presumably suffer frequent replication fork failure, or perhaps they have difficulty rescuing forks that failed due to endogenous sources of DNA damage. In an effort to understand more clearly the endogenous sources of DNA damage and the nature of replication fork failure and rescue, we undertook a systematic screen for Escherichia coli mutants that constitutively express the SOS regulon. We identified mutant strains with transposon insertions in 42 genes that caused increased expression from a dinD1::lacZ reporter construct. Most of these also displayed significant increases in basal levels of RecA protein, confirming an effect on the SOS system. As expected, this collection includes genes, such as lexA, dam, rep, xerCD, recG, and polA, which have previously been shown to cause an SOS constitutive phenotype when inactivated. The collection also includes 28 genes or open reading frames that were not previously identified as SOS constitutive, including dcd, ftsE, ftsX, purF, tdcE, and tynA. Further study of these SOS constitutive mutants should be useful in understanding the multiple causes of endogenous DNA damage. This study also provides a quantitative comparison of the extent of SOS expression caused by inactivation of many different genes in a common genetic background.
Most bacteria, including Escherichia coli, elicit the SOS response following DNA damage (reviewed in references 32 and 106; see also reference 23). This response involves the transcriptional induction of a regulon with more than 30 genes, many involved in DNA damage repair, bypass, and tolerance mechanisms (e.g., recA, lexA, umuDC, polB, sulA, etc.). Expression of the SOS regulon is controlled by the RecA and LexA proteins. In the uninduced state, LexA protein represses SOS genes by binding to SOS boxes upstream of each gene (53). Following DNA damage, RecA protein becomes activated in the presence of single-stranded DNA and a nucleoside triphosphate. Activated RecA protein functions as a coprotease, mediating cleavage of LexA repressor and thus activating transcription of SOS regulon genes. As the cell recovers from the treatment, the inducing signal (single-stranded DNA) diminishes, and RecA protein returns to its unactivated state. Continued synthesis of LexA protein restores repression of the SOS genes and thereby returns the cell to the uninduced state.
The SOS response is generally studied by inducing DNA damage with exogenous agents. Two of the best-studied inducers are UV irradiation and nalidixic acid. Treatment of E. coli with UV directly damages DNA by causing the formation of photoproducts, including pyrimidine dimers (32). The presence of these lesions is not sufficient to cause SOS induction, but rather the SOS inducing signal is generated when the cell attempts to replicate the damaged DNA (91, 93). The primary target for nalidixic acid is DNA gyrase, a type II topoisomerase that alters DNA topology by creating a transient double-strand break in DNA (64, 88, 107). Nalidixic acid inhibits gyrase by stabilizing a normally transient reaction intermediate called the cleavage complex, which is necessary but not sufficient for induction of the SOS response (10, 26). There is conflicting evidence concerning the involvement of DNA replication in SOS induction by nalidixic acid (37, 93).
In general, DNA lesions caused by UV irradiation, topoisomerase poisons, cross-linking agents, and alkylating agents all block the progress of replication forks (42, 45, 68, 101). Blocked replication forks are implicated in generating the SOS-inducing signal in bacteria (see above) and the intra-S-phase checkpoint in eukaryotes (25). Blocked forks can also lead to mutations and genome rearrangements, and this genomic instability is relevant to cancer progression in mammals (13, 70).
Although cells must deal with the stress of exogenous DNA-damaging agents at times, they must always cope with basal levels of endogenous DNA damage from sources such as reactive oxygen species (19, 69). Endogenous DNA damage and perhaps subtle defects in the replication machinery can lead to replication fork failure, and it is now believed that bacterial recombination systems evolved specifically from the need to restart stalled or blocked replication forks (19, 20). Support for this conclusion comes from the observation of chronic SOS induction in E. coli mutants with defects in replication, recombination, and/or repair genes, including dam, dnaQ, lig, polA, priA, recG, recN, rep, and uvrD (see Discussion and references in Table 3). For example, PriA plays a central role in rescuing stalled replication forks, and priA strains presumably elicit a constitutive SOS phenotype because they are deficient in rescuing stalled forks resulting from endogenous DNA damage (60).
TABLE 3.
Summary of constitutive mutants
| Functional group | Straina | No. of insertionsb | Assay resultsc
|
Functional or putative functiond | Functional reference | Constitutive reference | |
|---|---|---|---|---|---|---|---|
| I | II | ||||||
| Replication, recombination, and repair | dam mutant | 3 | • | • | DNA adenine methylase | 61 | 83, 84, 89 |
| dnaQ mutant | 2 | ○ | • | Epsilon subunit of DNA polymerase III (3′-5′ exonuclease) | 50 | 89, 100 | |
| polA mutant | 1 | • | • | DNA polymerase I | 50 | 5, 89 | |
| recG mutant | 4 | • | • | Branch-specific helicase or fork reversal | 67 | 2, 56 | |
| recN mutant | 1 | • | Recombination and repair gene | 85 | 14, 27, 99 | ||
| rep mutant | 7 | • | Helicase | 58 | 81, 105 | ||
| ruvA mutant | 3 | • | ○ | Holiday junction helicase (branch migration) | 108 | 2 | |
| ruvB mutant | 2 | • | ○ | Holiday junction helicase (branch migration) | 108 | 2e | |
| ruvC mutant | 2 | • | ○ | Holiday junction endonuclease | 108 | 2, 55 | |
| uvrD mutant | 2 | • | • | DNA-dependent helicase II | 32 | 6, 81, 89 | |
| Chromosome dimer resolution | ftsK mutant | 14 | • | • | Cell division protein or SpoIIIa-like function | 24, 28 | 54 |
| xerC mutant | 8 | • | • | Site-specific recombinase | 44 | 40 | |
| xerD mutant | 7 | • | • | Site-specific recombinase | 44 | 40 | |
| yncD yddW mutantf | 1 | • | ○ | Putative outer membrane receptor for iron transport or putative lipoprotein | 9 | This work | |
| Transcription and transcription regulation | lexA mutant | 2 | • | • | Global regulator (repressor) for SOS regulon | 32 | 73 |
| rpoZ mutant | 1 | • | • | RNA-polymerase subunit omega (polar on recG?) | 12 | This work | |
| spoT mutant | 6 | • | • | ppGpp synthetase (polar on recG?) | 12 | This work | |
| Nucleoside metabolism | dcd mutant | 1 | • | • | 2′-Deoxycytosine deaminase | 75 | This work |
| folA mutant | 1 | • | Dihydrofolate reductase | 36 | This work | ||
| folK mutant | 2 | ○ | • | 6-Hydroxymethyl-7,8-dihydropterin pyrophosphokinase | 36 | This work | |
| purA mutant | 1 | ○ | Adenylosuccinate synthase | 110 | This work | ||
| purE mutant | 1 | ○ | Phosphoribosylaminoimidazole carboxylase | 110 | This work | ||
| purF mutant | 2 | • | • | Glutamine phosphoribosylpyrophosphate amidotransferase | 110 | This work | |
| purL mutant | 1 | ○ | Phosphoribosylformylglycinamide synthase | 110 | This work | ||
| thyA mutant | 1 | • | Thymidylate synthase | 75 | 1g | ||
| Membrane structure/function | acrB mutant | 14 | ○ | Drug efflux pump | 76 | This work | |
| cvpA mutant | 1 | • | ○ | Colicin V production protein (polar on purF?) | 29, 30 | This work | |
| dsbB mutant | 1 | ○ | Disulfide oxidoreductase | 46 | This work | ||
| ompA mutant | 1 | ○ | Outer membrane protein A | 76 | This work | ||
| tolC mutant | 9 | ○ | Channel-tunnel spanning the outer membrane and periplasm | 76 | This work | ||
| Miscellaneous | arpB mutant | 1 | ○ | ○ | Putative regulatory protein with ankyrin repeats | 9 | This work |
| damX mutant | 4 | • | • | Overproduction interferes with cell division (polar on dam?) | 57, 59 | This work | |
| ftsE mutant | 1 | ○ | ○ | ABC transporter? Salt transport? | 22, 34 | This work | |
| ftsX mutant | 3 | • | ○ | ABC transporter? Salt transport? | 22, 34 | This work | |
| rluD ecfD mutanth | 1 | ○ | Pseudouridylate synthase; null mutants grow poorly or putative lipoprotein regulated by sigma E | 9, 21, 79, 87 | This work | ||
| This work | |||||||
| tdcE mutant | 2 | • | • | Threonine dehydratase | 41 | This work | |
| tynA mutant | 1 | • | • | Tyramine oxidase | 74 | This work | |
| yebC mutant | 1 | ○ | • | Putative membrane protein (polar on ruvC?) | 4, 9 | This work | |
| yehB mutant | 1 | ○ | ○ | Putative outer membrane export usher protein | 9 | This work | |
| yejM mutant | 1 | ○ | Putative sulfatase | 9 | This work | ||
| yfgM mutant | 1 | ○ | Putative N-terminal transmembrane domain | 9 | This work | ||
| yhcB mutant | 1 | ○ | ○ | Conserved bacterial protein of unknown function | 9 | This work | |
Alternate gene names: arpB, b1720/b1721; ecfD, b2595; envC, yibP; rluD, sfhB; yfgL, b2512; yfgM, b2513; yncD, b1451; yddW, b1491.
Number of independent insertions isolated in each gene. Single hits were confirmed by P1 transduction.
Performance of transposon insertion mutant strains in quantitative assays. I, β-galactosidase assay; II, RecA Western blot assay. Open circles indicate those strains whose assay results exceeded that of the wild type by more than 1 standard deviation; closed circles indicate those strains whose assay results were at least twice as high as wild-type levels.
Functions according to the NCBI website (http://www.ncbi.gov).
Although ruvB mutants were not directly assayed in the cited study, we assume they are SOS constitutive since RuvAB acts as a complex.
This insertion mutant has two transposons (see text).
thyA mutants have never been assayed for the SOS constitutive phenotype; however, this review suggests that they are SOS constitutive based on their phenotypes.
The transposon insertion in this strain is located between the two genes.
Surprisingly, a systematic search for SOS constitutive mutants has never been reported. The identification of additional SOS constitutive strains should aid our understanding of processes that lead to endogenous DNA damage and may also shed light on the interplay between replication, recombination, and repair. We therefore undertook such a screen and identified transposon insertions in 42 genes that cause some level of constitutive SOS expression. In addition to many genes that were previously known to cause an SOS constitutive phenotype, we identified a variety of new genes, including some with quite high levels of SOS expression. This study also provides a direct comparison of the levels of constitutive SOS expression of many different knockout mutations in the same genetic background.
MATERIALS AND METHODS
Materials and media.
Kanamycin, nalidixic acid, and o-nitrophenyl-β-d-galactopyranoside (ONPG) were purchased from Sigma, while 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was purchased from Gold Biotechnology, Inc. Oligonucleotides were purchased from Sigma Genosys. L broth (LB) contained Bacto Tryptone (10 g/liter), yeast extract (5 g/liter), and sodium chloride (10 g/liter), while LB plates had the same composition plus Bacto Agar (14 g/liter).
Transposon mutagenesis and primary nalidixic acid screening.
SOS constitutive mutants were isolated during two different screens of transposon libraries in E. coli strain JH39 [F− sfiA11 thr-1 leu-6 hisG4 argE3 ilv(Ts) galK2 srl(?) rpsL31 lacΔU169 dinD1::MudI(Apr lac)] (38, 39, 80). The EZ::TN <KAN-2> Tnp Transposome kit from Epicentre (Madison, Wis.) was used, according to the manufacturers directions, to generate the libraries. The first set of mutants was identified during a screen for mutants with altered SOS induction in response to nalidixic acid (unpublished data). Approximately 19,000 insertion strains were selected based on their resistance to kanamycin. The colonies were lifted onto nitrocellulose filters (Schleicher & Schuell), which were then transferred to LB plates containing nalidixic acid (50 μg/ml) and X-Gal (60 μg/ml). The lifted colonies were incubated overnight at 37°C, and the plates were checked for color development at approximately 4 and 12 h. A small fraction of the colonies displayed unusual color intensities, including mutants with reduced color relative to that of JH39 (unpublished data) and dark-blue mutants with increased color intensity. The second screen for SOS constitutive mutants was similar to the first, except that approximately 15,000 transposon insertion mutants were plated directly on LB plates containing X-Gal (60 μg/ml) plus kanamycin (60 μg/ml), allowing direct screening of mutants that constitutively express the dinD1::lacZ fusion.
DNA techniques and sequencing.
Genomic DNA was purified using a MasterPure DNA purification kit from Epicentre by the protocol described by the manufacturer, except that phenol-chloroform extraction and ethanol precipitation were performed after the RNase treatment. The resulting DNA pellet was resuspended in 30 μl of T5E1 buffer (5 mM Tris-HCl [pH 7.8], 1 mM EDTA). The DNA samples were sequenced by the Duke University Cancer Center DNA Analysis Facility by use of a modification of the automated sequencing protocol (60 cycles, 60°C annealing temperature). The sequencing primer was KAN-2 FP-1 (5′-ACCTACAACAAAGCTCTCATCAACC-3′), which is the forward primer from the EZ::TN <KAN-2> Tnp Transposome kit.
Phage P1 transductions.
Transductions were performed by the method of Silhavy et al. (98), with selection for the kanamycin resistance gene of the transposon.
Liquid β-galactosidase assay.
Overnight cultures were diluted 100-fold and grown for 2 h unless otherwise indicated. Two 1.0-ml samples were removed, and the cells were pelleted in a microcentrifuge. One of the pellets was used immediately in the liquid β-galactosidase assay, while the other was frozen for future analysis by Western blotting (see below). β-Galactosidase assays were performed essentially as described by Miller (71). Briefly, the pellets were resuspended in Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 10 mM dithiothreitol). Two drops of chloroform and 1 drop of 0.1% sodium dodecyl sulfate (SDS) were added to the cell suspension, which was then vortexed vigorously for 10 s. Appropriate amounts of the lysate were mixed with Z-buffer to a final volume of 1.0 ml. To start the reactions, 200 μl of ONPG (4 mM) was added, and the reaction mixtures were incubated at 30°C until a moderate yellow color was observed. The reactions were stopped with 0.5 ml of 1 M sodium bicarbonate, and the cellular debris was pelleted. The optical density was recorded with an ANTHOS 2001 plate reader with a 405-nm filter. Miller units were calculated as follows: units = 1,000[(OD405/(t × v × OD600)], where OD405 is the optical density at 405 nm of the reaction product, OD600 reflects the cell density at 600 nm, t is the reaction time in minutes, and v is the volume of culture used in the assay.
Western blots.
After a brief thaw on ice, the cell pellets were resuspended in 100 μl of cracking buffer (60 mM Tris-HCl, 1% [wt/vol] SDS, 1% [vol/vol] β-mercaptoethanol, 10% [vol/vol] glycerol, 0.01% [wt/vol] bromophenol blue). The cells were lysed by boiling the suspension for 10 min. Appropriate dilutions of the extracts were separated on SDS-10% polyacrylamide gels (Criterion gels; Bio-Rad) and blotted onto a polyvinylidene difluoride membrane (Pall Corporation) at 400 mA for 45 min in Western transfer buffer (25 mM glycine, 25 mM Tris-HCl, 10% methanol) by use of a Genie blotter (Idea Scientific Co.). Blots were probed with a monoclonal RecA-specific antibody (ARM-321; StressGen Biotech) at a 1:5,000 dilution and a secondary antibody linked to horseradish peroxidase (GAM-HRP; Bio-Rad) at a 1:30,000 dilution. Bands were visualized using ECL-Plus detection reagent (Amersham). The chemifluorescent signal was imaged by use of the blue fluorescence mode on a STORM Phosphorimager and quantitated using ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.), while the chemiluminescent signal was imaged with film.
In addition to sample extracts, all blots contained a serial dilution of purified RecA protein (New England Biolabs) that was used to generate a standard curve. The amount of RecA protein in each sample was calculated by use of the RecA standard curve and corrected for cell density and loading dilution.
RESULTS
Isolation of transposon mutants with constitutive expression from the dinD promoter.
To identify SOS constitutive mutants, we searched for transposon insertions that cause constitutive expression from the dinD promoter. We used the EZ::TN <KAN-2> Tnp Transposome kit (Epicentre) to create a library of transposon mutants with the JH39 strain of E. coli. This strain has a dinD1::lacZ fusion that provides a convenient reporter for the SOS response, allowing constitutive mutants to be identified by a simple color screen (39). The strain also has a sfiA (sulA) mutation, which reduces filamentation and inhibition of cell division upon SOS induction (32).
One set of potential SOS constitutive mutants was identified during a screen for transposon mutants with altered SOS induction in response to nalidixic acid (unpublished data). In that screen, approximately 19,000 colonies with random transposon insertions were selected based on kanamycin resistance. The colonies were lifted onto nitrocellulose filters, which were then transferred to plates containing nalidixic acid and X-Gal. After overnight incubation, 66 of these colonies were scored as dark-blue mutants with increased reporter gene expression relative to that of JH39.
There were two obvious reasons for this dark-blue phenotype. Some of these colonies presumably had mutations in drug permeability or efflux mechanisms, leading to a higher effective drug concentration inside the cells (and thus greater induction of the reporter construct). Alternatively, some of the colonies were likely SOS constitutive mutants. The latter group of mutants was identified by streaking the 66 mutants on plates containing X-Gal without nalidixic acid. We found that 45 of the mutants displayed a darker-blue color than the JH39 control and were therefore classified as putative SOS constitutive mutants.
These 45 mutants were obtained from a screen with a different purpose by a somewhat circuitous route. In particular, we were concerned that the presence of nalidixic acid had affected the screen (e.g., we might have missed weakly constitutive mutants because the response to nalidixic acid was so strong). To overcome this limitation, we pursued a second screen for transposon mutants that constitutively express the reporter construct. Approximately 15,000 colonies from a transposon library of JH39 were screened directly by including X-Gal in the selection plates. We thereby obtained an additional 128 transposon insertion mutants that appeared to express more β-galactosidase than the parental control. After being restreaked on X-Gal plates, 20 of these mutants looked indistinguishable from the parental JH39 and were therefore discarded as false positives. Thus, from the two different primary screens, we obtained a total of 153 putative SOS constitutive mutants that passed the secondary screen.
Identification of transposon locations by DNA sequencing.
We used the MasterPure DNA purification kit (Epicentre) to purify genomic DNA from each mutant strain and the forward primer from the EZ::TN transposon kit to sequence from one end of the transposon in each strain. Using a modified automated sequencing protocol, we were able to obtain at least 100 bp of unambiguous sequence from each mutant. With those sequences, the E. coli K-12 genome was searched by using BLAST to determine which gene had been interrupted by the transposon. The precise locations of transposon insertions for the final mutant collection are available online (see Table S1 in the supplemental material).
The collection of 153 mutants included transposon insertions in 65 different genes. Two of these genes (trpB and phage Mu gam gene) are located within the fusion construct (11); these insertions presumably affected the expression of the lacZ gene independent of any SOS response and were therefore discarded. Twenty-two genes from this collection had two or more independent hits, results that strongly argued that the transposon was causative in these cases. The other 41 genes were hit just once, and so we were initially uncertain if the transposon was causative. To resolve this uncertainty, we performed P1 transductions to move each transposon insertion mutation into a clean JH39 background, using the kanamycin resistance marker. Upon transduction, 25 of the insertion mutants displayed constitutive expression of the reporter construct, verifying that the transposon insertion was causative in each case. Thirteen of the insertion mutants were found not to be causative (data not shown) and were therefore discarded from the collection.
We were unable to transduce three of the mutants (the gmhA, sppA, and yigG mutants) because they were resistant to P1 phage and to another transducing phage (T4). We initially included these three in subsequent experiments, but all three failed two quantitative assays (described below) and were therefore eliminated from the collection. Thus, after a secondary screen, DNA sequencing, and phage P1 transductions, our collection contained potential SOS constitutive mutants with insertions in 47 different genes (see Table S1 in the supplemental material).
Analysis of constitutive expression from the reporter construct.
A qualitative estimate of constitutive expression from the reporter construct was made by carefully comparing colonies of the transposon mutants on plates containing X-Gal (data not shown). Mutants such as the lexA, rep, and ruvA mutants were much bluer than JH39, while others such as the acrA, purA, and yfgL mutants were slightly bluer than JH39. We were concerned that the results of the plate assay could be misleading for any of a variety of reasons: (i) the signal accumulates over a long period of time (overnight growth) and might therefore saturate, (ii) colony morphology might alter the appearance of the blue signal, and (iii) mutations that affect membrane integrity or transport (i.e., in acrA, acrB, or tolC) might affect uptake or export of the X-Gal indicator and thus result in false positives. To address these potential problems, we quantitated expression of the reporter construct by use of liquid β-galactosidase assays.
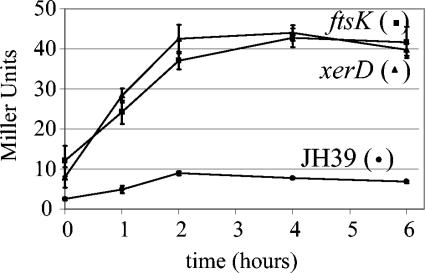
We first performed a time course assay during growth of the wild-type strain, JH39, and two mutant strains, the ftsK and xerD mutants. The zero time point was established when the cultures reached mid-log phase (OD600, ∼0.4 to 0.6), and 1-ml samples were removed and analyzed at 0, 1, 2, 4, and 6 h. The wild-type strain displayed low levels of β-galactosidase throughout the experiment, starting at 2.5 Miller units and reaching a maximum of 9.0 Miller units at the 2-h time point (Fig. 1; note that Miller units are corrected for cell density). The ftsK and xerD mutant strains exhibited three to five times the β-galactosidase activity of the wild-type strain throughout the time course, reaching a maximum of approximately 40 Miller units.
FIG. 1.
Time course of β-galactosidase expression in JH39 and the xerD and ftsK mutants. Overnight cultures of each strain were diluted and grown in LB at 37°C until the OD600 reached approximately 0.5 (zero time point). At the indicated time points, 1-ml samples were removed and assayed for β-galactosidase activity. Values represent the average of results of three independent experiments, with standard deviations indicated by error bars.
The ftsK and xerD mutants reached maximal β-galactosidase activities approximately 2 h after mid-log phase. We therefore used this 2-h time point for assays of the collection of transposon insertion mutants (Table 1). The wild type displayed 7.8 ± 1.4 Miller units (average ± standard deviation; n = 24 experiments), similar to that reported previously for a strain carrying this reporter construct (48). Not surprisingly, the lexA strain displayed the most activity, 26-fold more than that of the wild type. Many of the other strains also showed increased levels of reporter gene expression compared to the wild type, including the dam (7.1-fold), dcd (3.5-fold), and dnaQ (1.7-fold) strains. However, the β-galactosidase levels in 13 of the mutant strains were not significantly different from that of the wild type (Table 1).
TABLE 1.
β-Galactosidase expression levels
| Straina | β-Galactosidase expressionb
|
|
|---|---|---|
| Miller unitsc | Relative unitsd | |
| JH39 | 7.8 (±1.4) | 1.00 |
| lexA mutant | 202.0 (±84.7) | 26.00 |
| dam mutant | 55.1 (±15.8) | 7.09 |
| xerC mutant | 46.9 (±3.2) | 6.04 |
| purF mutant | 43.1 (±1.1) | 5.55 |
| xerD mutant | 43.1 (±3.0) | 5.54 |
| ftsK mutant | 37.9 (±4.6) | 4.87 |
| polA mutant | 33.0 (±4.2) | 4.24 |
| tynA mutant | 30.1 (±1.3) | 3.88 |
| spoT mutant | 29.8 (±4.8) | 3.83 |
| tdcE mutant | 28.7 (±1.2) | 3.70 |
| damX mutant | 28.3 (±0.7) | 3.64 |
| dcd mutant | 26.9 (±0.8) | 3.46 |
| rpoZ mutant | 25.8 (±0.3) | 3.32 |
| recG mutant | 23.1 (±2.2) | 2.97 |
| uvrD mutant | 22.5 (±1.3) | 2.90 |
| ftsX mutant | 22.2 (±12.8) | 2.86 |
| ruvC mutant | 21.3 (±6.2) | 2.73 |
| rep mutant | 20.4 (±3.7) | 2.63 |
| ruvB mutant | 18.3 (±1.7) | 2.35 |
| cvpA mutant | 17.6 (±1.4) | 2.26 |
| yncD yddW mutante | 17.4 (±4.7) | 2.23 |
| thyA mutant | 17.2 (±0.7) | 2.21 |
| ruvA mutant | 17.0 (±1.8) | 2.19 |
| folA mutant | 16.7 (±0.8) | 2.15 |
| recN mutant | 15.5 (±1.1) | 2.00 |
| yebC mutant | 15.0 (±0.2) | 1.93 |
| ftsE mutant | 14.5 (±4.9) | 1.87 |
| dnaQ mutant | 13.1 (±4.0) | 1.69 |
| yhcB mutant | 11.7 (±0.7) | 1.50 |
| purL mutant | 10.6 (±0.9) | 1.36 |
| purE mutant | 10.4 (±3.0) | 1.34 |
| folK mutant | 10.2 (±3.9) | 1.31 |
| yehB mutant | 9.9 (±0.2) | 1.27 |
| arpB mutant | 9.6 (±0.6) | 1.24 |
| envC mutant | 8.3 (±0.7) | 1.07 |
| tolC mutant | 8.0 (±1.3) | 1.03 |
| yfgL mutant | 7.7 (±0.2) | 0.99 |
| rluD ecfD mutantf | 7.7 (±0.5) | 0.99 |
| yejM mutant | 7.6 (±1.4) | 0.98 |
| yfgM mutant | 7.5 (±0.2) | 0.97 |
| acrB mutant | 7.4 (±0.6) | 0.96 |
| acrA mutant | 7.4 (±1.2) | 0.95 |
| ompA mutant | 7.2 (±1.1) | 0.93 |
| htrA mutant | 7.1 (±0.8) | 0.92 |
| purA mutant | 6.4 (±0.1) | 0.82 |
| surA mutant | 6.3 (±0.7) | 0.81 |
| dsbB mutant | 6.1 (±0.2) | 0.79 |
| recA mutantg | 4.1 (±0.1) | 0.53 |
Alternate gene names: arpB, b1720/b1721; ecfD, b2595; envC, yibP; rluD, sfhB; yfgL, b2512; yfgM, b2513; yncD, b1451; yddW, b1491.
A line indicates the levels of expression that are equivalent to the wild-type level plus 1 standard deviation.
Number of β-galactosidase units expressed after 2 h of growth (post-log phase). The values shown are the average of results for at least three independent experiments. Standard deviations are shown in parentheses.
Relative units of β-galactosidase compared to that of the wild type (JH39).
This insertion mutant has two transposons (see text).
The transposon insertion in this strain is located between the two genes.
A recA control strain (see legend to Fig. 2 for information on this strain).
Many of the strains with expression levels not significantly different from that of the wild type have insertions that disrupt genes that affect membrane structure or function, including acrA, acrB, htrA, ompA, and tolC. As stated above, these mutants might give false readings in the plate assay. On the other hand, it is possible that the plate assay might be more sensitive than the liquid assay because the signal can accumulate during overnight growth.
Quantitative Western blots of RecA protein levels.
The dinD1::lacZ fusion has been used as a convenient reporter for the SOS response (38, 39, 48). However, it seemed possible that some of the transposon insertion mutants might increase expression of the reporter construct in an SOS-independent manner. Also, the sequence of the dinD SOS box is similar to SOS operators that bind LexA tightly and therefore are expressed efficiently only in response to higher levels of damage (31, 52, 53). Thus, dinD might not be responsive to weak SOS signals. Because of these concerns, a second independent measure of SOS was performed. RecA protein itself is upregulated during the SOS response, and so we measured RecA levels by use of quantitative Western blot analyses with the same 2-h time point samples used in the β-galactosidase assays.
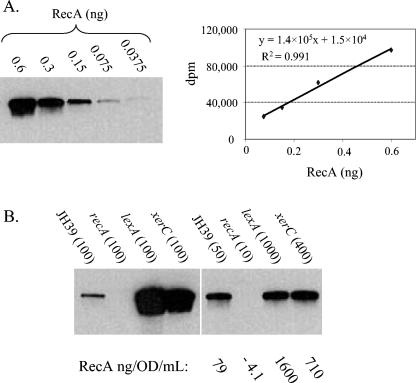
We first established a standard curve for RecA and found good linearity within an eightfold range of protein amount (Fig. 2A) (0.075 to 0.6 ng). For each of the mutant lysates, we developed an appropriate dilution to fall within this linear range. For example, 100-fold dilutions of the lexA and xerC lysates were out of the linear range of the curve (Fig. 2B). We used these test dilutions as a guide and found that 1,000-and 400-fold dilutions of the lexA and xerC lysates, respectively, allowed accurate quantitation (Fig. 2B). As expected, an extract from a control recA insertion mutant showed no detectable signal, even when loaded at levels 10-fold higher than that of the wild-type extract.
FIG. 2.
Quantitative Western blots for RecA protein in parental JH39 and insertion mutant strains. (A) The indicated amounts (in nanograms) of purified RecA protein were subjected to polyacrylamide gel electrophoresis and visualized by Western blotting. A regression of the RecA standard curve is shown. (B) Extracts of the indicated strains were loaded in equal dilutions (left) or in dilutions that fit within the RecA standard curve (right). Values in parentheses indicate dilutions (n-fold). Quantitation of the dilutions that fit within the standard curve is shown below the gel. The recA insertion mutant used as a control is a JH39 derivative with the EZ-TN <KAN-2> transposon located 99 bp downstream of the recA start codon.
The parental JH39 strain had 80.8 ± 22.5 (n = 15 experiments) ng of RecA/OD unit/ml (Table 2), which translates to roughly 1,600 ± 400 molecules of RecA per cell. This is within the range previously observed for other strains (between 1,000 and 10,000 molecules [47, 90, 93]). As in the liquid β-galactosidase assays, the lexA strain displayed the highest constitutive levels, with a 22-fold increase in RecA levels over that of the parental wild-type strain (Table 2). Results from the two assays were generally similar, especially for the stronger constitutive mutants. Thirty-six of the strains showed a significant increase in RecA levels over that of the parental control (Table 2), and 19 showed more than twice as much RecA as the control (e.g., dam mutant, 5-fold; dcd mutant, 3.1-fold; and dnaQ mutant, 2.1-fold). For five insertion mutants, the expression levels fell below the level of significance in both quantitative assays (the acrA, envC, htrA, surA, and yfgL mutants), and these mutants were therefore eliminated from our collection of constitutive mutants.
TABLE 2.
RecA expression levels
| Straina | RecA levelb,c | Relative levelc,d |
|---|---|---|
| JH39 | 80.8 | 1.00 |
| lexA mutant | 1,765.1 | 21.85 |
| ftsK mutant | 619.5 | 7.67 |
| xerC mutant | 462.7 | 5.73 |
| tynA mutant | 430.2 | 5.32 |
| purF mutant | 404.9 | 5.01 |
| dam mutant | 403.1 | 4.99 |
| xerD mutant | 399.7 | 4.95 |
| spoT mutant | 351.1 | 4.35 |
| recG mutant | 309.3 | 3.83 |
| polA mutant | 294.7 | 3.65 |
| damX mutant | 257.3 | 3.18 |
| folK mutant | 251.1 | 3.11 |
| dcd mutant | 248.9 | 3.08 |
| rpoZ mutant | 243.5 | 3.01 |
| tdcE mutant | 219.9 | 2.72 |
| uvrD mutant | 179.8 | 2.23 |
| dnaQ mutant | 165.8 | 2.05 |
| yncD yddW mutante | 165.3 | 2.05 |
| yebC mutant | 165.2 | 2.04 |
| yejM mutant | 159.7 | 1.98 |
| yhcB mutant | 155.5 | 1.92 |
| yehB mutant | 149.4 | 1.85 |
| ftsE mutant | 148.8 | 1.84 |
| arpB mutant | 142.9 | 1.77 |
| purA mutant | 142.4 | 1.76 |
| rluD ecfD mutantf | 128.1 | 1.59 |
| ruvA mutant | 123.7 | 1.53 |
| ftsX mutant | 120.8 | 1.50 |
| dsbB mutant | 120.1 | 1.49 |
| ruvC mutant | 119.6 | 1.48 |
| yfgM mutant | 116.0 | 1.44 |
| cvpA mutant | 115.0 | 1.42 |
| tolC mutant | 110.2 | 1.36 |
| acrB mutant | 107.0 | 1.32 |
| ruvB mutant | 105.1 | 1.30 |
| ompA mutant | 104.9 | 1.30 |
| yfgL mutant | 102.4 | 1.27 |
| recN mutant | 102.2 | 1.26 |
| rep mutant | 101.6 | 1.26 |
| envC mutant | 96.2 | 1.19 |
| purE mutant | 92.8 | 1.15 |
| thyA mutant | 90.1 | 1.12 |
| acrA mutant | 87.1 | 1.08 |
| purL mutant | 83.2 | 1.03 |
| htrA mutant | 76.4 | 0.95 |
| surA mutant | 73.0 | 0.90 |
| folA mutant | 71.5 | 0.88 |
| recA mutantg | 0.0 | 0.00 |
Alternate gene names: arpB, b1720/b1721; ecfD, b2595; envC, yibP; rluD, sfhB; yfgL, b2512; yfgM, b2513; yncD; b1451; yddW, b1491.
Level expressed in nanograms per OD unit per milliliter after 2 h of growth (post-log phase). The value shown for JH39 represents the average for 15 independent experiments, and the standard deviation was ±22.5. All other values shown are the average for two independent experiments. The values from the data in Fig. 2 are not included in these averages.
A line indicates the levels of expression that are equivalent to the wild-type level plus 1 standard deviation.
Amount of RecA protein relative to that of the wild type (JH39).
This insertion mutant has two transposons (see text).
The transposon insertion in this strain is located between the two genes.
A recA control strain (see legend to Fig. 2 for information on this strain).
Characterization of an unstable mutant.
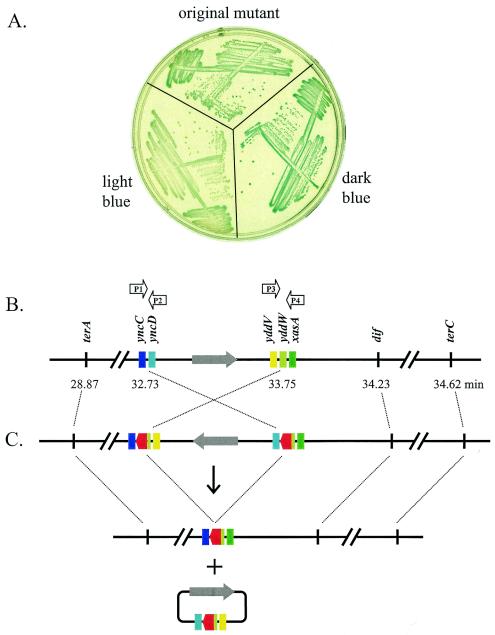
We obtained an unusual transposon insertion mutant that is strikingly unstable, with about 70 to 90% of the colonies appearing dark blue and the rest appearing light blue (Fig. 3A). When a dark-blue colony was picked from the original streak, the phenotype remained unstable, with a mix of dark-blue and light-blue colonies (Fig. 3A, dark-blue streak). However, when a light-blue colony was picked, the phenotype was stable, with all resulting colonies being light blue (Fig. 3A, light-blue streak, color similar to that of the JH39 parental strain). Because this insertion mutant was isolated only once, we performed P1 transductions using the kanamycin resistance marker and found that the phenotypes described above were transducible: P1 lysates made from dark-blue colonies resulted in both dark-blue and light-blue transductant colonies, while lysates made from light-blue colonies resulted in only light-blue transductant colonies.
FIG. 3.
An unstable insertion mutant. (A) The freezer stock of the unstable insertion mutant (the yncD yddW mutant) was streaked onto an LB plate containing X-Gal (60 μg/ml) and kanamycin (60 μg/ml). A dark-blue and a light-blue colony were picked and restreaked onto a second plate of the same composition, along with a new streak from the same freezer stock (original mutant). The plates were incubated overnight at 37°C and photographed the next morning. The light-blue color is very similar to that of parental strain, JH39 (i.e., SOS repressed). (B) The gene organization of the wild-type terminal region is depicted, with emphasis on the sites involved in the transposition events. PCR primers used in the DNA analysis are indicated above the relevant reading frames, which are color coded (shades of blue, green, and yellow). A large grey arrow indicates the orientation of the intervening 47-kb chromosomal segment. (C) The inferred genome arrangement of the inversion mutant, prior to excisive recombination, is depicted above a downward arrow. Homologous recombination between the directly repeated transposon insertions (indicated in red) can excise a circle containing one copy of the transposon and a segment of the terminus region (diagram below +). Loss of the excised circle during cell division is proposed to explain the generation of colonies that are SOS repressed but contain a large deletion of terminus DNA.
Given the unusual characteristics of this mutant, we performed extensive sequencing and PCR analyses with DNA from this mutant. The results imply that the blue colonies contain chromosomal DNA with two directly repeated transposons, flanking an intervening inversion of 47 kb (Fig. 3C). In addition, these cells apparently contain significant amounts of circular DNA resulting from excisive recombination between the directly repeated transposons (Fig. 3C, circular diagram at bottom). The evidence for this interpretation is as follows. DNA from this mutant was difficult to sequence, but when sequencing was successful, the sequence reproducibly read towards yncD with the forward primer of the transposon and towards yddV with the reverse primer (Fig. 3C, circular diagram at bottom). PCR analysis indicated that this mutant contains four junctions between the transposon and E. coli DNA. Primers from all four flanking regions (P1, P2, P3, and P4) (Fig. 3B) were synthesized and tested in every possible combination. PCR products were generated with four primer pairs, P1-P4, P2-P3, P1-P3, and P2-P4, and in each case, the size was that expected for DNA containing an intervening transposon (data not shown). No product was obtained with the “natural” primer pairs P1-P2 or P3-P4, but these did lead to the expected products with DNA from the wild-type parental control (e.g., as in Fig. 3B).
Based on DNA analysis, we conclude that the white colonies are generated by excisive recombination followed by loss of the excised circle. Thus, the DNA sequence from the white colonies read towards yncC with the forward transposon primer and towards xasA with the reverse primer. Furthermore, this DNA gave the PCR product expected for the 47-kb deletion mutant with the P1-P4 primer pair (i.e., as for the chromosomal DNA with a single transposon shown in Fig. 3C).
DISCUSSION
Many cellular processes are directly or indirectly involved in maintaining genome integrity. In an effort to further understand these processes, we undertook a screen for SOS constitutive mutants. Such mutants presumably have difficulty repairing endogenous DNA damage and/or suffer higher levels of endogenous damage. We screened approximately 34,000 colonies and found that transposon insertions in 47 genes led to constitutive expression of the reporter gene construct as judged by plate assays. Strains with insertions into each of these 47 genes were isolated multiple times, or the insertions were shown to be causative by P1 transduction. Since results from the plate assay can be misleading, we also performed quantitative assays for β-galactosidase and RecA protein levels. Five insertion mutants were eliminated because they failed both quantitative assays, leaving 42 mutants in the final collection. These are listed by functional category in Table 3, which also summarizes the performance of each mutant during the quantitative assays. For each mutant listed, we indicate whether the assay results were significantly higher, or twofold higher, than those of the wild type. This study provides the first quantitative comparison of many different SOS constitutive mutants in the same genetic background.
Based on previous observations, we expected to find many of the genes listed in Table 3. The most obvious is lexA, inactivation of which would cause constitutive expression of all SOS genes, including dinD and recA. lexA null mutants can be difficult to obtain due to constitutive expression of the sfiA (sulA) gene (32, 73), but our parental strain (JH39) has a sfiA mutation.
We expected to find priA mutants, which are known SOS constitutive mutants (60, 78). Perhaps we did not obtain these because they grow very poorly on rich media (62). This possibility relates to the biggest limitation of our screen, that we can identify only nonessential genes and might potentially miss semiessential genes due to growth defects. We also point out that some of the constitutive mutants identified (Table 3) could conceivably be false positives. In particular, six of the mutant strains failed one quantitative test and barely passed the other (the purE, purL, acrB, ompA, tolC, and yfgM mutants).
DNA replication, repair, and recombination.
As expected, we isolated numerous strains with transposon insertions in genes involved in DNA replication, recombination, and repair, and many of these were among the most strongly constitutive in the quantitative tests (Table 3). Mutations in all of these genes have previously been identified as causing an SOS constitutive phenotype (see references cited in Table 3). The constitutive nature of strains with these mutations likely results from altered replication fork dynamics, aberrant repair reactions, and/or failed rescue of replication forks.
The dam gene encodes DNA adenine methylase (Dam), an essential component of the mismatch repair system (61). Cells lacking Dam cannot distinguish newly replicated strands from template strands and thereby suffer double-strand breaks via the MutH endonuclease (3). The product of the uvrD gene, helicase II, participates in nucleotide excision repair and methyl-directed mismatch repair (32). However, the constitutive nature of uvrD strains is apparently caused by a requirement for UvrD in removing secondary structures that can lead to gap formation during lagging strand synthesis (89). Similarly, polA-deficient strains are thought to be SOS constitutive due to single-strand gaps on the lagging strand (5, 89).
The ɛ subunit of replicative DNA polymerase III is encoded by dnaQ. Although the molecular details are unclear, mutations in this gene have been shown to cause aberrant DNA replication that leads to excessive single-stranded DNA and/or broken replication forks; these in turn cause an SOS constitutive phenotype and unusually high rates of direct repeat recombination (72, 94, 100). The Rep helicase also plays a role in replication fork progression, probably by removing DNA-bound proteins (15, 63, 109). It has been proposed that chromosomes break more frequently in rep cells due to frequent replication pauses (69, 105).
Although the molecular function of RecN is not clear, it plays a role in the repair of double-strand breaks (85). Presumably, the delayed or reduced repair of endogenous double-strand breaks in the recN mutant causes an increase in the SOS-inducing signal. We found that our recN mutant convincingly passed the β-galactosidase assay (2-fold increase) but not the RecA protein assay (1.26-fold increase). These observations are very consistent with previous studies of recN mutants. Simic et al. (99) found that recN mutants were induced twofold compared to the wild type with a lacZ fusion to the sfiA (sulA) gene, and the strength of the sfiA SOS box is similar to that of dinD (31, 53).On the other hand, Chua et al. (14) found that recN mutants were induced for SOS only 1.26-fold compared to the wild type with a lacZ fusion to the recA promoter on a low-copy number plasmid (14). These results suggest that SOS constitutive mutants can show reproducible and presumably meaningful differences in the two quantitative assays.
ruv null mutants are known to be SOS constitutive, hypersensitive to UV light, and filamentous (2, 55). RuvABC resolves Holliday junction recombination intermediates (reviewed in reference 108) and is also believed to play a role in replication fork reversal during replication restart (67, 70, 95, 96). In addition to RuvABC, another branch-specific helicase, RecG, plays a role in replication fork reversal (66, 67). Mutations in recG have previously been shown to cause an SOS constitutive phenotype (2, 56). The recG gene is within an operon with the gene order gmk-rpoZ-spoT-spoU-recG, with promoters upstream of gmk (P1) and rpoZ (P2) (12, 33, 92). We also isolated SOS constitutive mutants with insertions in rpoZ and spoT (Table 3), and these could be constitutive due to reduced transcription of the downstream recG gene. However, we did not isolate insertions in spoU, which might also be expected if there is a polar effect on recG expression. In addition, the rpoZ and spoT strains appeared darker blue than the recG strain in the plate assays (data not shown), suggesting a more direct involvement of these two gene products in the SOS phenotype. RpoZ and SpoT are both involved in the stringent response, which can impact the survival of E. coli after DNA damage (65).
CDR.
Dimeric chromosomes can be generated by crossover events during recombinational repair of replication forks, and these dimers must be resolved so they can segregate to daughter cells. Chromosome dimer resolution (CDR) requires the XerCD recombinase, the dif site, and the FtsK cell division protein (7, 8, 51, 102). dif, xerC, xerD, and ftsK mutants have previously been shown to be SOS constitutive (40, 51, 54, 104), and SOS induction is blocked in these mutants if cell division is inhibited (40, 54). These and other results led to a model in which unresolved dimer chromosomes are broken during cell division, thus creating the SOS-inducing signal (40). Indeed, Prikryl et al. (86) presented direct evidence that the terminal region of the chromosome is frequently broken and degraded when CDR is blocked and that a mutation in recD can prevent the degradation. Consistent with these past studies, the xerC, xerD, and ftsK insertion mutants identified in our screen were strongly constitutive in both quantititative assays (Table 3).
We also isolated an unusual transposon insertion mutant that apparently contains dual transposons flanking a 47-kb chromosomal inversion in the terminal region of the chromosome, close to the dif site (Fig. 3). Previous work of Perals et al. (82) helps to explain the unusual phenotype of this mutant. These authors showed that inversions of terminal DNA (not including the dif site) can interfere with XerCD-mediated recombination. Two of the inversions that have this effect contain the 47-kb segment inverted in our mutant strain (along with additional adjoining DNA). Their interpretation of this and other results involves oriented DNA sites, quite possibly the Rag motifs (5′-RRRAGGGY-3′), that exist in this region of the chromosome. By this model, the Rag motifs serve as directional markers for DNA translocation by FtsK. This DNA translocation is proposed to bring the dif sites together at the septum for CDR (18). Accordingly, when this DNA translocation is disturbed by incorrectly oriented motifs, one of the dif sites does not end up at the septum. Failure of CDR then leads to breakage of dimeric chromosomes, just as in the CDR-deficient mutants discussed above. This model is sufficient to explain the SOS constitutive phenotype of our inversion mutant.
The instability of our insertion mutant can be explained by related data showing that direct repeat recombination is greatly stimulated in the terminal region when CDR is compromised (16, 17). This presumably reflects the stimulatory nature of the DNA breaks induced when dimers are not properly resolved by CDR. In the case of our inversion mutant, direct repeat recombination (followed by excised circle loss as shown in Fig. 3C) could lead to deletion of the intervening inverted segment, and cells with this deletion should be competent for CDR and therefore SOS repressed.
Two interesting issues remain. First, based on the DNA sequencing results, the transposon-chromosome junctions that appear to be most prominent in DNA from our inversion mutant are those from the inferred DNA circle. This finding raises the possibility that this circle can replicate, perhaps via recombination-dependent replication (see references 49 and 77). Second, we do not understand how this unusual transposon insertion or inversion mutant was generated. We have sequenced more than 230 other transposon insertion mutants without difficulty, indicating that none of these other mutants had dual transposons. We note that only one such SOS constitutive mutant was isolated from about 34,000 transposon insertion mutants and so generation of this kind of mutant appears to be an infrequent event.
Nucleotide metabolism and pool depletion.
We found a number of insertions in nucleotide metabolism genes, with the strongest SOS constitutive mutants being the dcd and purF mutants (Table 3). All eight genes in this category are involved in maintaining balanced nucleotide pools (36, 75, 110). We presume that unbalanced pools lead to frequent misincorporation and/or replication fork stalling, which in turn leads to the constitutive SOS phenotype. It is important to note that all of our studies were conducted with rich media, and we have not systematically tested whether the SOS phenotypes of these mutants are altered by provision of exogenous bases or nucleosides.
Our isolation of thyA mutants is not surprising, since thyA mutants have previously been shown to have endogenous DNA damage and many characteristics expected for SOS constitutive mutants (for a recent review, see reference 1). For decades, it has been known that thyA mutants undergo “thymineless death” upon thymine starvation. The mechanism of thymineless death appears to be quite complex and is still not entirely clear, but this phenomenon has some clinical relevance because of its relationship to the cytotoxic mechanism of anticancer and antibacterial agents that interrupt thymidine metabolism (1).
Mutations that inactivate dcd are known to cause a 10-fold increase in dCTP pools and as much as 4-fold decrease in dTTP pools (75), which could obviously pose problems for the replication complex. The PurF enzyme is the first enzyme in the purine biosynthesis pathway (110). The products of purA, purE, and purL are in the same pathway and were also isolated in our screen. It is unclear why insertions in these genes did not cause responses as strong as that seen with the purF mutant.
Membrane structure and function.
We are less certain that the mutant strains identified as belonging to the membrane structure and function category in Table 3 are truly SOS constitutives. As stated above, strains with mutations in these genes could give a misleading readout in the plate assay. In support of this, four original members of this group (the acrA, envC, htrA, and surA mutants) were eliminated because they were not significantly different from the wild type in the two quantitative assays, and three others (the acrB, ompA, and tolC mutants) failed one quantitative test and barely passed the other. The only mutant in this group whose results in both assays were significantly different from those of the wild type was the cvpA mutant (Table 3). A previously analyzed transposon insertion into cvpA had a polar effect on purF (29), and we therefore suspect that the constitutive phenotype is caused by a polar effect on purF (see above).
Miscellaneous mutants.
We isolated strains with constitutive insertions in the ftsX and ftsE genes, which share an operon with ftsY (34). Based on sequence comparisons and the work of de Leeuw et al. (22), FtsE and FtsX likely constitute an ABC transporter, although the substrate and biological role of the complex are unknown. The original point mutations in the ftsX and ftsE genes caused a temperature-sensitive filamentation phenotype, and mutant cells lacking FtsE display a filamentous phenotype and require high salt for viability at any temperature (22). ABC transporters are known to act on a variety of substrates, and some constitute efflux systems in gram-negative bacteria (43). We therefore speculate that the phenotype of ftsE and ftsX mutants might be explained by the accumulation of some genotoxic substance that causes SOS induction, filamentation, and potentially cell death.
The tynA and tdcE insertion mutants were strongly constitutive in both quantitative assays. The tynA gene codes for an aromatic amine oxidase and is upregulated in the presence of monoamine compounds such as tyramine (74). The tdcE gene codes for a threonine dehydratase and shares an operon with six other genes in the order tdcABCDEFG (35, 41). TdcE can convert 2-ketobutyrate to propionyl-coenzyme A and formate but may conceivably act on other substrates. The simplest explanation for the SOS constitutive phenotype of tynA and tdcE mutants is that TynA and TdcE degrade compounds that are otherwise genotoxic. We hope to further explore these interesting metabolic issues by isolating extragenic suppressor mutations of ftsX, ftsE, tynA, and tdcE.
We also isolated strains with insertions in eight genes with uncharacterized functions, with the strongest phenotypes in the damX and yebC insertions (Table 3). These two insertions might cause the SOS phenotype indirectly as they are cotranscribed upstream of dam and ruvC, respectively, (57, 97, 103). Nonetheless, the product of damX may well play a role in the SOS phenotype, since overexpression of this gene has been shown to induce cell filamentation (59).
Concluding remarks.
This study has significantly expanded the number of genes known to cause an SOS constitutive phenotype upon inactivation. In addition, our results provide an isogenic comparison of the extent of fusion gene expression in many different knockout mutants. The collection of 42 genes presumably includes most of the nonessential genes that are required to maintain genomic stability. The SOS constitutive phenotypes likely result from unusually high levels of DNA breaks, gaps, and/or aberrant replication forks. Previously, nearly all the genes known to cause an SOS constitutive phenotype upon inactivation were genes involved directly in DNA replication, recombination, or repair. Many of the mutants we isolated were indeed in this functional group. However, we also isolated a number of strong SOS constitutive mutants with mutations in genes involved in nucleoside metabolism, presumably reflecting aberrant replication fork dynamics due to unbalanced nucleotide pools. In addition, strong SOS constitutive mutants with mutations in the genes ftsX, ftsE, tynA, and tdcE, along with many other weaker constitutive mutants, provide leads that might allow the identification of new genotoxic metabolites.
Supplementary Material
Acknowledgments
We thank Joe Heitman for the JH39 strain and for helpful discussions about using the SOS reporter system. We are also grateful to Christina Marks for her technical assistance. Finally, we are deeply indebted to Katie Newmark for initiating the companion screen and to Jennifer Reineke-Pohlhaus for helpful insights and discussion.
This work was supported by NIH grants GM065206 and CA60836 to K.N.K. E.K.O. was supported in part by Department of Defense research grant DAMD-00-01-0235.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org.
REFERENCES
- 1.Ahmad, S. I., S. H. Kirk, and A. Eisenstark. 1998. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu. Rev. Microbiol. 52:591-625. [DOI] [PubMed] [Google Scholar]
- 2.Asai, T., and T. Kogoma. 1994. Roles of ruvA, ruvC and recG gene functions in normal and DNA damage-inducible replication of the Escherichia coli chromosome. Genetics 137:895-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Au, K. G., K. Welsh, and P. Modrich. 1992. Initiation of methyl-directed mismatch repair. J. Biol. Chem. 267:12142-12148. [PubMed] [Google Scholar]
- 4.Balasubramanian, S., T. Schneider, M. Gerstein, and L. Regan. 2000. Proteomics of Mycoplasma genitalium: identification and characterization of unannotated and atypical proteins in a small model genome. Nucleic Acids Res. 28:3075-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bates, H., S. K. Randall, C. Rayssiguier, B. A. Bridges, M. F. Goodman, and M. Radman. 1989. Spontaneous and UV-induced mutations in Escherichia coli K-12 strains with altered or absent DNA polymerase I. J. Bacteriol. 171:2480-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bierne, H., M. Seigneur, S. D. Ehrlich, and B. Michel. 1997. uvrD mutations enhance tandem repeat deletion in the Escherichia coli chromosome via SOS induction of the RecF recombination pathway. Mol. Microbiol. 26:557-567. [DOI] [PubMed] [Google Scholar]
- 7.Blakely, G., S. Colloms, G. May, M. Burke, and D. Sherratt. 1991. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 3:789-798. [PubMed] [Google Scholar]
- 8.Blakely, G., G. May, R. McCulloch, L. K. Arciszewska, M. Burke, S. T. Lovett, and D. J. Sherratt. 1993. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell 75:351-361. [DOI] [PubMed] [Google Scholar]
- 9.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 10.Burden, D. A., and N. Osheroff. 1998. Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochim. Biophys. Acta 1400:139-154. [DOI] [PubMed] [Google Scholar]
- 11.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 12.Cashel, M., D. R. Gentry, V. J. Hernandez , and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff ., M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 13.Chakraverty, R. K., and I. D. Hickson. 1999. Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. Bioessays 21:286-294. [DOI] [PubMed] [Google Scholar]
- 14.Chua, K. L., Y. K. Mak, and P. Oliver. 1993. Expression of the recA gene in recombination-deficient (rec-) strains of Escherichia coli. Biochimie 75:775-783. [DOI] [PubMed] [Google Scholar]
- 15.Colasanti, J., and D. T. Denhardt. 1987. The Escherichia coli rep mutation. X. Consequences of increased and decreased Rep protein levels. Mol. Gen. Genet. 209:382-390. [DOI] [PubMed] [Google Scholar]
- 16.Corre, J., F. Cornet, J. Patte, and J. M. Louarn. 1997. Unraveling a region-specific hyper-recombination phenomenon: genetic control and modalities of terminal recombination in Escherichia coli. Genetics 147:979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corre, J., J. Patte, and J. M. Louarn. 2000. Prophage lambda induces terminal recombination in Escherichia coli by inhibiting chromosome dimer resolution. An orientation-dependent cis-effect lending support to bipolarization of the terminus. Genetics 154:39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corre, J., and J. M. Louarn. 2002. Evidence from terminal recombination gradients that FtsK uses replichore polarity to control chromosome terminus positioning at division in Escherichia coli. J. Bacteriol. 184:3801-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 20.Cox, M. M. 2001. Historical overview: searching for replication help in all of the rec places. Proc. Natl. Acad. Sci. USA 98:8173-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dartigalongue, C., D. Missiakas, and S. Raina. 2001. Characterization of the Escherichia coli sigma E regulon. J. Biol. Chem. 276:20866-20875. [DOI] [PubMed] [Google Scholar]
- 22.de Leeuw, E., B. Graham, G. J. Phillips, C. M. Hagen-Jongman, B. Oudega, and J. Luirink. 1999. Molecular characterization of Escherichia coli FtsE and FtsX. Mol. Microbiol. 31:983-993. [DOI] [PubMed] [Google Scholar]
- 23.Defais, M., and R. Devoret, R. 2000. SOS response. Nature Encyclopedia of Life Sciences. Nature Publishing Group, London, United Kingdom. [Online.] http://www.els.net/ [doi:10.1038/npg.els. 0000580]
- 24.Donachie, W. D. 2002. FtsK: Maxwell's demon? Mol. Cell. 9:206-207. [DOI] [PubMed] [Google Scholar]
- 25.Donaldson, A. D., and J. J. Blow. 2001. DNA replication: stable driving prevents fatal smashes. Curr. Biol. 11:R979-R982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunman, P. M., L. Ren, M. S. Rahman, V. A. Palejwala, H. S. Murphy, M. R. Volkert, and M. Z. Humayun. 2000. Escherichia coli cells defective for the recN gene display constitutive elevation of mutagenesis at 3,N(4)-ethenocytosine via an SOS-induced mechanism. Mol. Microbiol. 37:680-686. [DOI] [PubMed] [Google Scholar]
- 28.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fath, M. J., H. K. Mahanty, and R. Kolter. 1989. Characterization of a purF operon mutation which affects colicin V production. J. Bacteriol. 171:3158-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fath, M. J., L. H. Zhang, J. Rush, and R. Kolter. 1994. Purification and characterization of colicin V from Escherichia coli culture supernatants. Biochemistry 33:6911-6917. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez de Henestrosa, A. R., T. Ogi, S. Aoyagi, D. Chafin, J. J. Hayes, H. Ohmori, and R. Woodgate. 2000. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 35:1560-1572. [DOI] [PubMed] [Google Scholar]
- 32.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis,p/ 407-464. ASM Press, Washington, D.C.
- 33.Gentry, D. R., and R. R. Burgess. 1986. The cloning and sequence of the gene encoding the omega subunit of Escherichia coli RNA polymerase. Gene 48:33-40. [DOI] [PubMed] [Google Scholar]
- 34.Gill, D. R., G. F. Hatfull, and G. P. Salmond. 1986. A new cell division operon in Escherichia coli. Mol. Gen. Genet. 205:134-145. [DOI] [PubMed] [Google Scholar]
- 35.Goss, T. J., H. P. Schweizer, and P. Datta. 1988. Molecular characterization of the tdc operon of Escherichia coli K-12. J. Bacteriol. 170:5352-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green, J. M., B. P. Nichols, and R. G. Matthews. 1996. Folate biosynthesis, reduction and polyglutamylation, p. 665-673. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin , K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 37.Gudas, L. J., and A. B. Pardee. 1976. DNA synthesis inhibition and the induction of protein X in Escherichia coli. J. Mol. Biol. 101:459-477. [DOI] [PubMed] [Google Scholar]
- 38.Heitman, J., and P. Model. 1987. Site-specific methylases induce the SOS DNA repair response in Escherichia coli. J. Bacteriol. 169:3243-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heitman, J., and P. Model. 1991. SOS induction as an in vivo assay of enzyme-DNA interactions. Gene 103:1-9. [DOI] [PubMed] [Google Scholar]
- 40.Hendricks, E. C., H. Szerlong, T. Hill, and P. Kuempel. 2000. Cell division, guillotining of dimer chromosomes and SOS induction in resolution mutants (dif, xerC and xerD) of Escherichia coli. Mol. Microbiol. 36:973-981. [DOI] [PubMed] [Google Scholar]
- 41.Hesslinger, C., S. A. Fairhurst, and G. Sawers. 1998. Novel keto acid formate-lyase and propionate kinase enzymes are components of an anaerobic pathway in Escherichia coli that degrades l-threonine to propionate. Mol. Microbiol. 27:477-492. [DOI] [PubMed] [Google Scholar]
- 42.Hiasa, H., D. O. Yousef, and K. J. Marians. 1996. DNA strand cleavage is required for replication fork arrest by a frozen topoisomerase-quinolone-DNA ternary complex. J. Biol. Chem. 271:26424-26429. [DOI] [PubMed] [Google Scholar]
- 43.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67-113. [DOI] [PubMed] [Google Scholar]
- 44.Hill, T. M. 1996. Features of the chromosomal terminus, p. 1602-1614. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C C. Lin , K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 45.Hong, G., and K. N. Kreuzer. 2003. Endonuclease cleavage of blocked replication forks: an indirect pathway of DNA damage from antitumor drug-topoisomerase complexes. Proc. Natl. Acad. Sci. USA 100:5046-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kadner, R. J. 1996. Cytoplasmic: membrane, p. 58-87. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin , K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 47.Karu, A. E., and E. D. Belk. 1982. Induction of E. coli recA protein via recBC and alternate pathways: quantitation by enzyme-linked immunosorbent assay (ELISA). Mol. Gen. Genet. 185:275-282. [DOI] [PubMed] [Google Scholar]
- 48.Kenyon, C. J., and G. C. Walker. 1980. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc. Natl. Acad. Sci. USA 77:2819-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kogoma, T. 1997. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61:212-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kornberg, A., and T. A. Baker. 1992. DNA replication. W.H. Freeman, New York, N.Y.
- 51.Kuempel, P. L., J. M. Henson, L. Dircks, M. Tecklenburg, and D. F. Lim. 1991. dif, a recA-independent recombination site in the terminus region of the chromosome of Escherichia coli. New Biol. 3:799-811. [PubMed] [Google Scholar]
- 52.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lewis, L. K., G. R. Harlow, L. A. Gregg-Jolly, and D. W. Mount. 1994. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J. Mol. Biol. 241:507-523. [DOI] [PubMed] [Google Scholar]
- 54.Liu, G., G. C. Draper, and W. D. Donachie. 1998. FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol. Microbiol. 29:893-903. [DOI] [PubMed] [Google Scholar]
- 55.Lloyd, R. G., F. E. Benson, and C. E. Shurvinton. 1984. Effect of ruv mutations on recombination and DNA repair in Escherichia coli K12. Mol. Gen. Genet. 194:303-309. [DOI] [PubMed] [Google Scholar]
- 56.Lloyd, R. G., and C. Buckman. 1991. Genetic analysis of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J. Bacteriol. 173:1004-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lobner-Olesen, A., E. Boye, and M. G. Marinus. 1992. Expression of the Escherichia coli dam gene. Mol. Microbiol. 6:1841-1851. [DOI] [PubMed] [Google Scholar]
- 58.Lohman, T. M., and K. P. Bjornson. 1996. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 65:169-214. [DOI] [PubMed] [Google Scholar]
- 59.Lyngstadaas, A., A. Lobner-Olesen, and E. Boye. 1995. Characterization of three genes in the dam-containing operon of Escherichia coli. Mol. Gen. Genet. 247:546-554. [DOI] [PubMed] [Google Scholar]
- 60.Marians, K. J. 2000. PriA-directed replication fork restart in Escherichia coli. Trends Biochem. Sci. 25:185-189. [DOI] [PubMed] [Google Scholar]
- 61.Marinus, M. G. 1996. Methylation of DNA, p. 782-791. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin , K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 62.Masai, H., T. Asai, Y. Kubota, K. Arai, and T. Kogoma. 1994. Escherichia coli PriA protein is essential for inducible and constitutive stable DNA replication. EMBO J. 13:5338-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matson, S. W., D. W. Bean, and J. W. George. 1994. DNA helicases: enzymes with essential roles in all aspects of DNA metabolism. Bioessays 16:13-22. [DOI] [PubMed] [Google Scholar]
- 64.Maxwell, A., and S. E. Critchlow. 1998. Mode of action, p. 119-166. In J. Kuhlmann, A. Dalhoff, and H. J. Zeiler (ed.), Quinolone antibacterials. Springer, Berlin, Germany.
- 65.McGlynn, P., and R. G. Lloyd. 2000. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell 101:35-45. [DOI] [PubMed] [Google Scholar]
- 66.McGlynn, P., and R. G. Lloyd. 2001. Action of RuvAB at replication fork structures. J. Biol. Chem. 276:41938-41944. [DOI] [PubMed] [Google Scholar]
- 67.McGlynn, P., and R. G. Lloyd. 2002. Genome stability and the processing of damaged replication forks by RecG. Trends Genet. 18:413-419. [DOI] [PubMed] [Google Scholar]
- 68.McHugh, P. J., W. R. Sones, and J. A. Hartley. 2000. Repair of intermediate structures produced at DNA interstrand cross-links in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:3425-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Michel, B., S. D. Ehrlich, and M. Uzest. 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 16:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Michel, B., M. J. Flores, E. Viguera, G. Grompone, M. Seigneur, and V. Bidnenko. 2001. Rescue of arrested replication forks by homologous recombination. Proc. Natl. Acad. Sci. USA 98:8181-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 72.Morag, A. S., C. J. Saveson, and S. T. Lovett. 1999. Expansion of DNA repeats in Escherichia coli: effects of recombination and replication functions. J. Mol. Biol. 289:21-27. [DOI] [PubMed] [Google Scholar]
- 73.Mount, D. W. 1977. A mutant of Escherichia coli showing constitutive expression of the lysogenic induction and error-prone DNA repair pathways. Proc. Natl. Acad. Sci. USA 74:300-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murooka, Y., H. Azakami, and M. Yamashita. 1996. The monoamine regulon including syntheses of arylsulfatase and monoamine oxidase in bacteria. Biosci. Biotechnol. Biochem. 60:935-941. [DOI] [PubMed] [Google Scholar]
- 75.Neuhard, J., and R. Kelln. 1996. Biosynthesis and conversions of pyrimidines, p. 580-599. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin , K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 76.Nikaido, H. 1996. Outer membrane, p. 29-47. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin , K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd. ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 77.Nishitani, H., M. Hidaka, and T. Horiuchi. 1993. Specific chromosomal sites enhancing homologous recombination in Escherichia coli mutants defective in RNase H. Mol. Gen. Genet. 240:307-314. [DOI] [PubMed] [Google Scholar]
- 78.Nurse, P., K. H. Zavitz, and K. J. Marians. 1991. Inactivation of the Escherichia coli priA DNA replication protein induces the SOS response. J. Bacteriol. 173:6686-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ofengand, J., and A. Bakin. 1997. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J. Mol. Biol. 266:246-268. [DOI] [PubMed] [Google Scholar]
- 80.Ohta, T., M. D. Sutton, A. Guzzo, S. Cole, A. E. Ferentz, and G. C. Walker. 1999. Mutations affecting the ability of the Escherichia coli UmuD′ protein to participate in SOS mutagenesis. J. Bacteriol. 181:177-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ossanna, N., and D. W. Mount. 1989. Mutations in uvrD induce the SOS response in Escherichia coli. J. Bacteriol. 171:303-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Perals, K., F. Cornet, Y. Merlet, I. Delon, and J. M. Louarn. 2000. Functional polarization of the Escherichia coli chromosome terminus: the dif site acts in chromosome dimer resolution only when located between long stretches of opposite polarity. Mol. Microbiol. 36:33-43. [DOI] [PubMed] [Google Scholar]
- 83.Peterson, K. R., K. F. Wertman, D. W. Mount, and M. G. Marinus. 1985. Viability of Escherichia coli K-12 DNA adenine methylase (dam) mutants requires increased expression of specific genes in the SOS regulon. Mol. Gen. Genet. 201:14-19. [DOI] [PubMed] [Google Scholar]
- 84.Peterson, K. R., and D. W. Mount. 1993. Analysis of the genetic requirements for viability of Escherichia coli K-12 DNA adenine methylase (dam) mutants. J. Bacteriol. 175:7505-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Picksley, S. M., P. V. Attfield, and R. G. Lloyd. 1984. Repair of DNA double-strand breaks in Escherichia coli K12 requires a functional recN product. Mol. Gen. Genet. 195:267-274. [DOI] [PubMed] [Google Scholar]
- 86.Prikryl, J., E. C. Hendricks, and P. L. Kuempel. 2001. DNA degradation in the terminus region of resolvase mutants of Escherichia coli, and suppression of this degradation and the Dif phenotype by recD. Biochimie 83:171-176. [DOI] [PubMed] [Google Scholar]
- 87.Raychaudhuri, S., J. Conrad, B. G. Hall, and J. Ofengand. 1998. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA 4:1407-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reece, R. J., and A. Maxwell. 1991. DNA gyrase: structure and function. Crit. Rev. Biochem. Mol. Biol. 26:335-375. [DOI] [PubMed] [Google Scholar]
- 89.SaiSree, L., M. Reddy, and J. Gowrishankar. 2000. lon incompatibility associated with mutations causing SOS induction: null uvrD alleles induce an SOS response in Escherichia coli. J. Bacteriol. 182:3151-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Salles, B., and C. Paoletti. 1983. Control of UV induction of recA protein. Proc. Natl. Acad. Sci. USA 80:65-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Salles, B., and M. Defais. 1984. Signal of induction of recA protein in E. coli. Mutat. Res. 131:53-59. [DOI] [PubMed] [Google Scholar]
- 92.Sarubbi, E., K. E. Rudd, H. Xiao, K. Ikehara, M. Kalman, and M. Cashel. 1989. Characterization of the spoT gene of Escherichia coli. J. Biol. Chem. 264:15074-15082. [PubMed] [Google Scholar]
- 93.Sassanfar, M., and J. W. Roberts. 1990. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol. 212:79-96. [DOI] [PubMed] [Google Scholar]
- 94.Saveson, C. J., and S. T. Lovett. 1997. Enhanced deletion formation by aberrant DNA replication in Escherichia coli. Genetics 146:457-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Seigneur, M., V. Bidnenko, S. D. Ehrlich, and B. Michel. 1998. RuvAB acts at arrested replication forks. Cell 95:419-430. [DOI] [PubMed] [Google Scholar]
- 96.Seigneur, M., S. D. Ehrlich, and B. Michel. 2000. RuvABC-dependent double-strand breaks in dnaBts mutants require RecA. Mol. Microbiol. 38:565-574. [DOI] [PubMed] [Google Scholar]
- 97.Sharples, G. J., and R. G. Lloyd. 1991. Resolution of Holliday junctions in Escherichia coli: identification of the ruvC gene product as a 19-kilodalton protein. J. Bacteriol. 173:7711-7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 99.Simic, D., B. Vukovic-Gacic, A. Ajanovic, and J. Knezevic-Vukcevic. 1991. Activation of RecA protein in recombination-deficient strains of Escherichia coli following DNA-damaging treatments. Mutat. Res. 254:255-262. [DOI] [PubMed] [Google Scholar]
- 100.Slater, S. C., M. R. Lifsics, M. O'Donnell, and R. Maurer. 1994. holE, the gene coding for the theta subunit of DNA polymerase III of Escherichia coli: characterization of a holE mutant and comparison with a dnaQ (epsilon-subunit) mutant. J. Bacteriol. 176:815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sonoda, E., M. S. Sasaki, C. Morrison, Y. Yamaguchi-Iwai, M. Takata, and S. Takeda. 1999. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol. 19:5166-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Steiner, W., G. Liu, W. D. Donachie, and P. Kuempel. 1999. The cytoplasmic domain of FtsK protein is required for resolution of chromosome dimers. Mol. Microbiol. 31:579-583. [DOI] [PubMed] [Google Scholar]
- 103.Takahagi, M., H. Iwasaki, A. Nakata, and H. Shinagawa. 1991. Molecular analysis of the Escherichia coli ruvC gene, which encodes a Holliday junction-specific endonuclease. J. Bacteriol. 173:5747-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tecklenburg, M., A. Naumer, O. Nagappan, and P. Kuempel. 1995. The dif resolvase locus of the Escherichia coli chromosome can be replaced by a 33-bp sequence, but function depends on location. Proc. Natl. Acad. Sci. USA 92:1352-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Uzest, M., S. D. Ehrlich, and B. Michel. 1995. Lethality of rep recB and rep recC double mutants of Escherichia coli. Mol. Microbiol. 17:1177-1188. [DOI] [PubMed] [Google Scholar]
- 106.Walker, G. C. 1996. The SOS response of Escherichia coli, p. 1400-1416. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin , K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 107.Wang, J. C. 1996. DNA topoisomerases. Annu. Rev. Biochem. 65:635-692. [DOI] [PubMed] [Google Scholar]
- 108.West, S. C. 1997. Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 31:213-244. [DOI] [PubMed] [Google Scholar]
- 109.Yancey-Wrona, J. E., and S. W. Matson. 1992. Bound Lac repressor protein differentially inhibits the unwinding reactions catalyzed by DNA helicases. Nucleic Acids Res. 20:6713-6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zalkin, H., and P. Nygaard. 1996. Biosynthesis of purine nucleotides, p. 561-579. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin , K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.