Primary embryonic fibroblast cells isolated from NLK-deficient mice proliferate faster and have a shorter cell cycle than wild-type cells. Nemo-like kinase and HDAC1 together negatively regulate Wnt signaling via Tcf/Lef transcription repression and prevent aberrant proliferation of fibroblast cells.
Abstract
The Wnt signaling pathway is essential in regulating various cellular processes. Different mechanisms of inhibition for Wnt signaling have been proposed. Besides β-catenin degradation through the proteasome, nemo-like kinase (NLK) is another molecule that is known to negatively regulate Wnt signaling. However, the mechanism by which NLK mediates the inhibition of Wnt signaling was not known. In the present study, we used primary embryonic fibroblast cells isolated from NLK-deficient mice and showed that these cells proliferate faster and have a shorter cell cycle than wild-type cells. In NLK-knockout cells, we observed sustained interaction between Lef1 and β-catenin, leading to elevated luciferase reporter of β-catenin/Lef1–mediated transcriptional activation. The mechanism for the reduced β-catenin/Lef1 promoter activation was explained by phosphorylation of HDAC1 at serine 421 via NLK. The phosphorylation of HDAC1 was achieved only in the presence of wild-type NLK because a catalytically inactive mutant of NLK was unable to phosphorylate HDAC1 and reduced the luciferase reporter of β-catenin/Lef1–mediated transcriptional activation. This result suggests that NLK and HDAC1 together negatively regulate Wnt signaling, which is vital in preventing aberrant proliferation of nontransformed primary fibroblast cells.
INTRODUCTION
The Wnt pathway is involved in different cellular processes, such as cell fate decisions, cell survival, cell growth, and differentiation, that are responsible for homeostasis of various organs in mammals. Most knockout animals of Wnt-regulated signaling molecules harbor severe phenotypes, including dying during embryogenesis or directly after birth (Aoki and Taketo, 2008). Activation of Wnt receptors leads to downstream signaling consisting of the translocation of β-catenin from the cytosol into the nucleus and further direct binding to the transcription factors T-cell factor (Tcf) and lymphoid-enhancing factor (Lef). This signaling cascade leads to the transcription and expression of Wnt target genes. Depending on the tissue and cell specificity, as well as the type of stimuli, Wnt target genes can regulate the outcome and response of the cell. Two well-known target genes of the Wnt signaling pathway regulating proliferation of the cell are c-Myc and cyclin D1 (Kikuchi, 2000, 2006).
Inactivation of the Wnt signaling pathway is achieved at multiple levels. Degradation of β-catenin through proteasome ubiquitination of β-catenin is the well-established system for antagonizing Wnt signaling. In addition to this system, nemo-like kinase (NLK), which belongs to the atypical mitogen-activated protein kinases, can negatively regulate Wnt signaling. After phosphorylation-mediated activation, NLK, which is a serine threonine kinase, can phosphorylate the substrates involved in different signaling pathways, including Wnt/β-catenin (Ishitani et al., 1999, 2003; Meneghini et al., 1999; Smit et al., 2004), Notch (Rottinger et al., 2006; Ishitani et al., 2010), Smad (Zeng et al., 2007), and NF-κB (Yasuda et al., 2004; Li et al., 2014).
Previously we generated NLK-deficient mice, which were lethal due to the lung phenotype (Ke et al., 2016). These mice are born but die within 24–36 h of birth (Ke et al., 2016), therefore limiting analyses of the function of NLK in vivo. Inactivation of Wnt signaling by NLK is achieved via phosphorylation of the complex Tcf/Lef, which further prevents the interaction of Tcf/Lef with DNA (Ishitani et al., 2003). Further, phosphorylation of histone methyltransferase SET domain bifurcated 1 (SETDB1) by NLK leads to disruption of the PPAR-γ function, which is vital for lineage decisions of mesenchymal stem cells (Takada et al., 2007). Other identified targets for NLK include c-Myb (Kanei-Ishii et al., 2004, 2008), STAT3 (Ohkawara et al., 2004; Kojima et al., 2005), FOXO1 (Kim et al., 2010), and myocyte enhancer factor 2A (Satoh et al., 2007). In the nervous system, phosphorylation of microtubule-associated protein-1B (MAP1B) and paxillin affected cytoskeletal organization and cell adhesion (Ishitani et al., 2009). The interaction between NLK and heat shock protein 27 (HSP27) was shown to protect cancer cells from apoptosis (Shaw-Hallgren et al., 2014).
Overexpression or down-regulation of NLK in both cancerous and noncancerous cell lines showed that NLK can either induce cell death or promote cell proliferation. In the present study, we used NLK−/− mouse embryonic fibroblasts (MEFs) to characterize the role of NLK in these primary cells. Surprisingly, we found that the NLK antagonism of Wnt signaling is not via Lef1 phosphorylation but instead occurs through HDAC1 phosphorylation, which further facilitates binding between HDAC1 and Lef1, preventing Tcf/Lef-mediated promoter activation.
RESULTS
Deletion of NLK does not affect the survival of MEFs
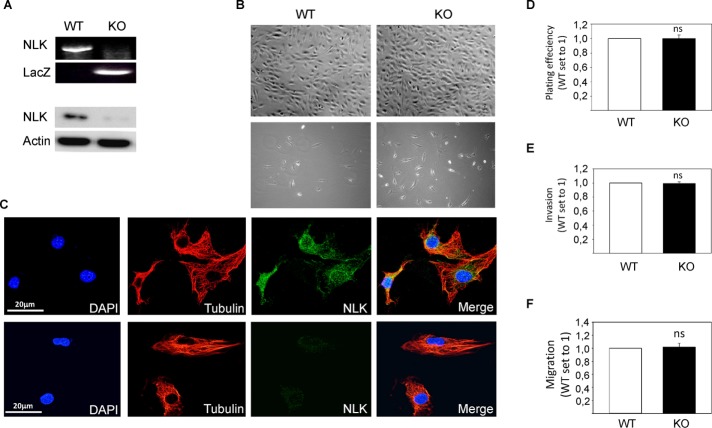
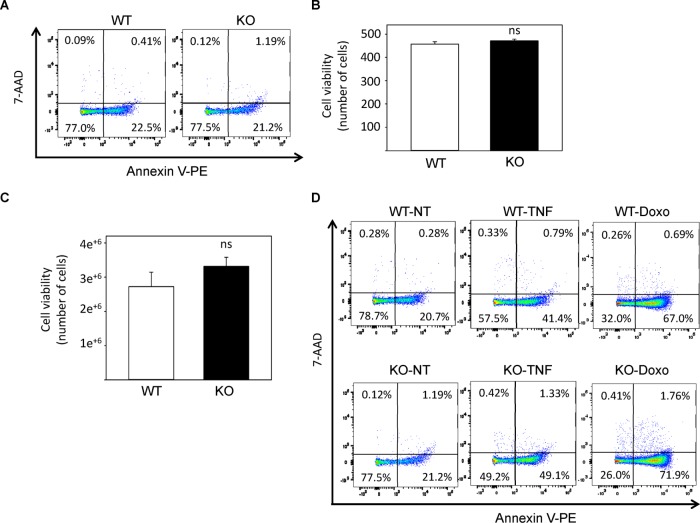
To study the role of NLK in vivo, we isolated NLK−/− (knockout [KO]) MEFs and compared their behavior with NLK+/+ (wild-type [WT]) MEFs obtained from littermate embryos (Figure 1A). NLK+/+ and NLK−/− MEFs were found to exhibit the same morphology, independent of high- and low-density cell culturing (Figure 1B). NLK has been suggested to be localized in the cytoplasm or nuclei, depending on cell type and stimuli (Brott et al., 1998; Ishitani et al., 2011; Shaw-Hallgren et al., 2014). In wild-type MEF cells, endogenous NLK showed strong expression in the cytoplasm and moderate expression in the nucleus (Figure 1C and Supplemental Figure S1). Further, analyzing adhesion, migration, and invasion of cells in vitro showed no differences between NLK−/− and NLK+/+ MEFs (Figure 1, D–F). Previous studies suggested the role of NLK in regulating apoptosis (Yasuda et al., 2003; Emami et al., 2009; Li et al., 2012; Huang et al., 2013). In our system, no significant differences were observed in the number of apoptotic or necrotic cells in MEF-NLK+/+ compared with NLK−/− cells over a period of 12–72 h by using three different methods under noninduced or serum starvation conditions (Figure 2, A–C). In addition, treatment with apoptosis-inducing agents such as tumor necrosis factor α (TNF-α) and doxorubicin did not affect the number of viable cells between NLK+/+ and NLK−/− (Figure 2D and Table 1).
FIGURE 1:
Deletion of NLK in primary MEFs. (A) Reverse transcription PCR (RT-PCR) of total RNA from MEFs isolated from embryonic day 13.5 wild-type (WT) and NLK knockout (KO) mice. Bottom, protein lysates were prepared from WT and KO MEF cells and analyzed by Western blot. (B) The morphology of the MEF cells at high and low density. (C) WT and KO cells were stained with specific antibodies directed against tubulin (red), NLK (green), and DAPI (blue). (D) Cells were plated into culture dishes and left to attach for 1 h before washing once with PBS and trypsinization for manual counting. (E) Cells were plated on top of Boyden chambers in serum-free medium, whereas serum was placed in the well below. Cells that had been migrating through the Boyden chambers were stained with DAPI and counted. (F) Migration distances of WT and KO cells were compared in a wound-healing assay.
FIGURE 2:
Loss of NLK does not affect the survival of MEF cells. (A) Dot plots showing flow cytometric analysis of apoptosis for NLK WT and KO MEF cells. Cells were harvested, labeled with Annexin V-PE and 7-AAD, and analyzed by flow cytometry. All data are representative of three independent experiments with similar results. (B) The effect of NLK on apoptosis was assessed in both WT and KO MEF cells stained with propidium iodide. A total of 200 live and dead cells were counted under a microscope. Data (±SEM, n = 3) represent the amount of viable cells. (C) WT and KO MEF cells were stained with acridine orange, which stains the entire cell population, and DAPI for staining nonviable cells. Cell viability was measured by using a Nucleocounter NC-3000. Data (±SEM, n = 3) represent the amount of viable cells. (D) Flow cytometric analysis of apoptosis after the treatment of MEF cells with apoptosis-inducing agents, including TNF and doxorubicin (Doxo), as well as of nontreated (NT) cells. Numbers in the quadrants represent the percentage of cells in each quadrant. Viable cells that are negative for Annexin V-PE or 7-AAD are neither apoptotic nor necrotic and are in the lower left quadrant; Apoptotic cells stained for Annexin V-PE but not for 7-AAD are in the lower right quadrant; late apoptotic cells stained for both Annexin V-PE and 7-AAD are in the upper right quadrant; and necrotic cells stained positive for 7-AAD but not for Annexin V-PE are in the upper left quadrant. All data are representative of three independent experiments with similar results.
TABLE 1:
Flow cytometric analysis of apoptosis in NLK-WT and -KO MEF cells.
| Viable cells (%) | Early apoptotic cells (%) | Late apoptotic and necrotic cells (%) | ||
|---|---|---|---|---|
| Nontreated | WT | 73.9 ± 4.2 | 25.6 ± 4.2 | 0.48 ± 0.11 |
| KO | 76.5 ± 4.5 | 22.9 ± 4.6 | 0.67 ± 0.10 | |
| Tumor necrosis factor α | WT | 64.1 ± 14.0 | 34.4 ± 13.5 | 1.4 ± 0.62 |
| KO | 60.0 ± 10.6 | 37.6 ± 11.0 | 2.22 ± 0.71 | |
| Doxorubicin | WT | 52.8 ± 7.4 | 46.0 ± 7.9 | 1.33 ± 0.35 |
| KO | 41.0 ± 8.1 | 55.7 ± 9.12 | 3.3 ± 0.69 |
Results are expressed as the mean ± SD of three different experiments.
Increased proliferation rate of NLK knockout compared with wild-type cells
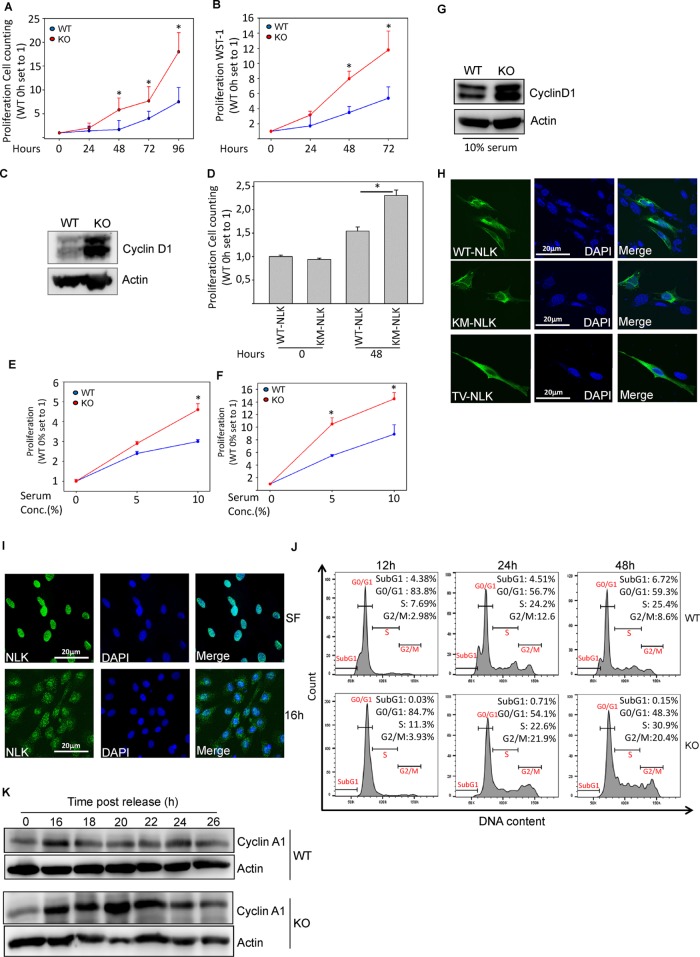
Next we used cell counting (Figure 3A), the WST-1 cell proliferation assay (Figure 3B), and assessment of the levels of cyclin D1 expression (Figure 3C) to monitor the growth rate of MEFs over a period of 5 d and found that NLK-deficient cells grew significantly faster than wild-type cells. To investigate whether NLK activity is necessary for reducing the proliferation rate of cells, we transfected knockout cells with NLK expression plasmid. Overexpression of full-length (WT-NLK) but not the catalytically inactive mutant (KM-NLK) of NLK resulted in a reduced proliferation rate of knockout compared with wild-type cells. Further, overexpression of full-length NLK in knockout cells reduced the levels of cyclin D1 expression (Supplemental Figure S2). These results suggest that activation of NLK is necessary for the limitation of cell growth (Figure 3D). Because we observed an increased proliferation rate of NLK-deficient MEFs compared with wild-type MEF cells in the presence of complete medium supplemented with serum, we tested whether this effect depended on the concentration of serum. In the absence of serum, no significant differences in the proliferation rates after 48 h (Figure 3E) and 72 h (Figure 3F) were observed between the cells, whereas readdition of 5 or 10% serum caused significant differences in proliferation rate after 48 h (Figure 3E) and 72 h (Figure 3F), as well as cyclin D1 expression (10% serum for 48 h; Figure 3G), between wild-type and knockout cells. In the presence of 10% serum, endogenous NLK (Figure 1C) and overexpressed full-length NLK in knockout cells (Figure 3H and Supplemental Figure S3) were exclusively localized in the cytoplasm. To investigate whether the activation of NLK can promote relocalization of NLK in MEF cells, we overexpressed two catalytically inactive NLK mutants in knockout cells. Both mutants were exclusively localized in the cytoplasm in cells cultured with 10% serum (Figure 3H and Supplemental Figure S3). Serum starvation of wild-type MEF cells for 24 h led to accumulation of NLK in the nucleus (Figure 3I, top), and readdition of serum facilitated cytoplasmic localization of NLK after 16 h (Figure 3I, bottom). Consistent with the difference in proliferation rate, we analyzed cell cycle progression after serum starvation of synchronized cells. Twelve hours postrelease of synchronized cells by the addition of serum did not show any significant differences in cell cycle progression between NLK+/+ and NLK−/− cells (Figure 3J). However, after 24 h, the percentage of NLK−/− cells in the G2/M phase (21.9%) was almost twice that of NLK+/+ cells (12.6%). In addition, the rapid progression of NLK−/− (20.4%) compared with NLK+/+ (8.6%) MEF cells entering G2/M phase was observed again after 48 h. In accordance with this finding, synchronized wild-type cells showed a transient increase in the levels of cyclin A1 expression after 16 h, which further declined to basal levels 18 h after release of the cells. In knockout cells, a sustained high level of cyclin A1 expression was observed from 16 until 22 h after release of the cells (Figure 3K). These results suggest that the deletion of NLK promotes a faster cell cycle transition between the S and G2/M phases.
FIGURE 3:
Increased proliferation rate of primary NLK−/− compared with NLK+/+ MEFs. (A) Cell proliferation was determined by cell counting after 24–96 h. The experiment was performed in triplicate. Data are presented as mean ± SEM (*p < 0.05). (B) MEF cells were cultured for 24–96 h before being subjected to WST-1 assay. Data represent the amount of viable cells. The value for each time point was normalized to the value on day 0. Data are presented as mean ± SEM (*p < 0.05). (C) The protein lysates from MEF cells cultured overnight under serum-free condition, readdition of serum for 24 h, and analysis for cyclin D1 expression by Western blotting. (D) NLK−/− cells were transfected with Flag-tagged wild-type NLK (WT-NLK) or the kinase-dead mutant of NLK (K155M, KM-NLK) plasmids for 4 h. After 24 h, cell proliferation was assessed by hemocytometer cell counting. Data represent the amount of viable cells. The value for each data point was normalized to the value of WT-NLK at 0 h. Values are means ± SEM (n = 3, p = 0.00104). (E) WT and KO MEF cells were cultured for 48 h, of which 24 h were in the presence of 0, 5, or 10% serum. Cell proliferation was determined by cell counting. The experiment was performed in triplicate. Values are means ± SEM (5%: p = 0.0000648; 10%: p = 0.00433). (F) WT and KO MEF cells were cultured for 72 h, of which 48 h were in the presence of 0, 5, or 10% serum. Cell proliferation was determined by cell counting. The experiment was performed in triplicate. Values are means ± SEM (5%: p = 0.0000180; 10%: p = 0.000314). (G) The protein lysates from MEF cells were analyzed for cyclin D1 expression by Western blotting. (H) MEF cells were transfected with Flag-tagged wild-type (WT-NLK) or Flag-tagged kinase-dead mutants (K155M; KM-NLK or T286V; TV-NLK) of NLK expression plasmids. The subcellular distribution of NLK was analyzed by using immunofluorescence staining and confocal microscopy. (I) Confocal analysis of MEF cells in the absence of nutrients (starvation; SF) or SF and readdition of complete medium in the presence of serum for 16 h. (J) Effects of NLK knockdown on cell-cycle distribution of MEF cells. Both NLK wild-type and knockout MEF cells were synchronized by serum starvation for 48 h and induced to reenter the cell cycle on serum addition. MEF cells were harvested 12, 24, and 48 h postrelease for PI staining, and the cell-cycle profile was by fluorescence-activated cell sorting (FACS). FACS plots and data are representative of at least two separate experiments. (K) MEFs cells were serum starved for 24 h, trypsinized, and collected after refeeding with FBS for the indicated time points. Cell lysates were analyzed by Western blot.
NLK does not affect the phosphorylation of Lef1
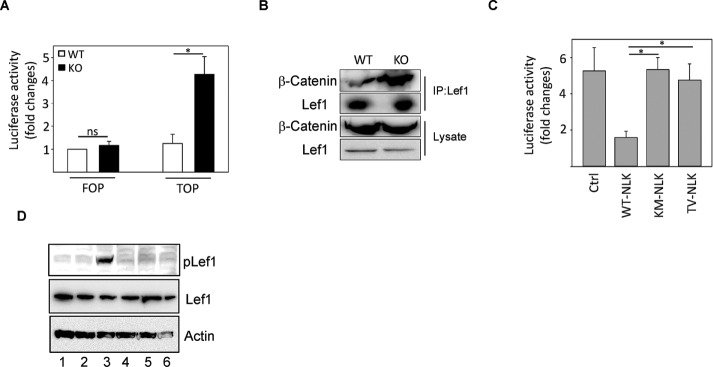
To investigate the signaling pathway by which NLK reduces the proliferation rate of MEF cells, we analyzed previously established Wnt and NF-κB signaling. Deletion of NLK in MEF cells did not affect the NF-κB signaling pathway because no differences in degradation or phosphorylation of IκB-α compared with NLK+/+ cells could be seen (Supplemental Figure S4). Instead, NLK-knockout cells have elevated activation of Tcf/Lef reporter assay TOPFlash (Figure 4A). Further, a significant difference in the interaction between Lef1 and β-catenin was seen when wild-type cells were compared with knockout cells (Figure 4B). Overexpression of wild-type but not mutant NLK constructs reduced Tcf/Lef reporter activity in NLK-deficient cells (Figure 4C). One mechanism by which NLK can reduce Tcf/Lef reporter activation is via the phosphorylation of Lef1 (Ishitani et al., 2003). An analysis of the levels of Lef1 showed that there are no differences in the total levels or phosphorylation of Lef1 in wild-type and knockout cells (Figure 4D, lanes 1 and 2). As a control, we used lung epithelial cells isolated from NLK wild-type mice, which were previously shown to contain high levels of p-Lef (Figure 4D, lane 3; Ke et al., 2016). To determine whether exogenous expression of full-length NLK can affect the phosphorylation of Lef1, we transiently transfected NLK-knockout cells with full-length NLK expression plasmid. It was evident that overexpression of NLK does not alter the phosphorylation status of Lef1 (Figure 4D, lanes 4–6).
FIGURE 4:
Lef1 does not undergo phosphorylation in wild-type MEF cells. (A) Expression plasmids for FOPFlash and TOPFlash were cotransfected into MEFs cells as indicated. Cells were harvested 24 h after transfection, and relative luciferase activity was measured. The luciferase activity was normalized to the Renilla results. The cell sample transfected with FOPFlash alone transfected into MEF-WT served as the control, and its value (relative luciferase activity) was set as 1. The relative luciferase activities of the other samples are normalized to this control. Data are means ± SD (n = 3, p = 0.016). (B) Endogenous immunoprecipitation was performed with an anti-Lef1 antibody and analyzed with an anti–β-catenin or anti-Lef1 antibody. (C) Relative FOP/TOP-driven luciferase activity in MEFs cells cotransfected with Flag-tagged wild-type (WT-NLK) or Flag-tagged kinase-dead mutants (K155M; KM-NLK or T286V; TV-NLK) of NLK expression. Values are means ± SEM (n = 3, WT-NLK and KM-NLK: p = 0.00768; WT-NLK and TV-NLK: p = 0.031). (D) Endogenous total levels or phosphorylation of Lef1 in wild-type (lane 1) and knockout (lane 2) cell lysates, lung epithelial cell lysate isolated from NLK wild-type mice (lane 3), cell lysate from NLK knockout cells transiently transfected with full-length NLK (lane 4), and two catalytically inactivate mutants of NLK (K155M, lane 5; T286V, lane 6) expression plasmids.
NLK-mediated phosphorylation of HDAC1 at serine 421
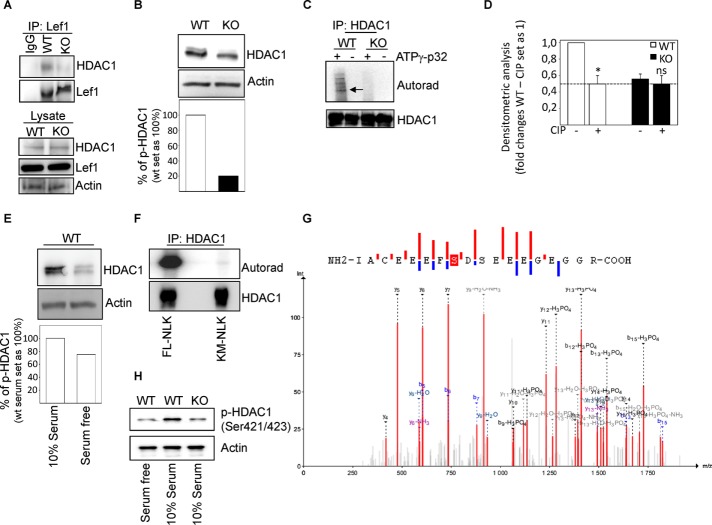
Previously it was demonstrated that the interaction between Lef1 and HDAC1 regulates Lef1 gene transcription (Billin et al., 2000) and that HDAC1 undergoes phosphorylation to initiate its deacetylating and transcriptional repression activities (Pflum et al., 2001). Endogenous immunoprecipitation of Lef1 using cell lysate showed the precipitation of HDAC1 in wild-type but not in knockout cells (Figure 5A), without any differences in the total levels of HDAC1 (Figure 5A, lysate). Further, a slow-migrating band of HDAC1 was observed in wild-type but not in knockout cell lysate (Figure 5B). We confirmed HDAC1 phosphorylation in wild-type cells by using an in vitro kinase assay (Figure 5C), and the treatment of cell lysate with calf intestinal alkaline phosphatase reduced the levels of HDAC1 phosphorylation in wild-type cells (Figure 5D). In addition, endogenous phosphorylation of HDAC1 in wild-type cells was elevated in the presence of serum compared with cells cultured in the serum-free medium for 24 h (Figure 5E). We also found an increase in the total levels of HDAC1 on readdition of serum (Figure 5E). This result was confirmed by an in vitro kinase assay in which HDAC1 phosphorylation was elevated in the presence of full-length but not in the presence of the catalytically inactive mutant of NLK (Figure 5F). Mass spectrometry (MS) analysis using in vitro phosphorylation of HDAC1 in the presence of NLK was performed to identify the specific phosphorylation site(s). The only phosphorylated peptide detected was IACEEEFSDSEEEGEGGR, with one serine phosphorylated, corresponding to phosphorylated serine at position 421 or 423. Several MS/MS spectra were acquired for this peptide, with the lowest expected value at 3.3E-6; the most probable phosphorylation site for all but one of the matched spectra was 421 (Figure 5G), which had a site probability of >92% according to Mascot. However, there was also one occasion of serine 423 as the most probable site, suggested by Mascot and other search engines, albeit with a low Mascot site probability of 72% (Supplemental Figure S5A). The phosphorylated peptide was also detected many times in a longer version with one missed tryptic cleavage and with serine 421 indicated as the most probable site (highest site probability 92%), further suggesting this site as the primary one (Supplemental Figure S5, B–D). Because the peptide (with no missed cleavage) was eluting as one peak in the MS chromatogram, we suggest that phosphorylation at site 421 is predominant and possibly exclusive. We confirmed these data by showing reduced levels of phosphorylated HDAC1 (serine 421 or 423) in serum-free–treated wild-type and knockout compared with wild-type cells cultured in the presence of 10% serum (Figure 5H). Further, a small fraction of p-HDAC1 was relocalized into the cytosol in 10% serum-treated compared with serum free–treated wild-type cells (Supplemental Figure S6).
FIGURE 5:
Phosphorylation of HDAC1 by NLK at serine 421. (A) MEF-WT and MEF-KO cells were immunoprecipitated by using antibodies against HDAC1 and probed with Lef1 or HDAC1 antibodies. (B) Whole-cell lysates were prepared from MEF-WT or MEF-KO cells and subjected to Western blot toward HDCA1 and actin. Levels of phosphorylated and nonphosphorylated protein were quantitated by densitometry and normalized to total protein levels. (C) Top, autoradiographic image from an in vitro kinase assay. Bottom, membrane was immunoblotted with anti-HDAC1 antibody. (D) MEF cells were treated with calf intestinal phosphatase (CIP) and visualized by immunoblotting. Values are means ± SEM (n = 3, WT not CIP-treated and WT CIP-treated: p = 0.000978). (E) Whole-cell lysates were prepared from MEF-WT cells in the absence or presence of serum and subjected to Western blot toward HDAC1 and actin. Levels of phosphorylated and nonphosphorylated protein were quantitated by densitometry and normalized to total protein levels. (F) MEF-KO cells were transfected with Flag-tagged wild-type (WT-NLK) or Flag-tagged kinase-dead mutants (K155M; KM-NLK or T286V; TV-NLK) of NLK expression plasmids, and lysates were immunoprecipitated with antibodies against HDAC1. Immunoprecipitates were subjected to kinase assay. (G) MEF cells transfected with FLAG-tagged WT-, K155M-, or T286V-NLK plasmids were harvested for immunoprecipitation 24 h posttransfection by using FLAG antibody. The protein lysates were separated by SDS–PAGE, and the protein bands were cut and processed for matrix-assisted laser desorption/ionization time-of-flight analysis. Annotated MS/MS spectrum with phosphorylated S421 and peptide sequence with detected fragments. Ions discriminating between S421 and S423 are indicated in the sequence. Asterisk indicates loss of ammonia and water; double asterisks indicate loss of H3PO4. (H) Whole-cell lysates were prepared from WT and KO MEF cells in the absence or presence of serum and subjected to Western blot toward pHDAC1 (Ser-421/423) and actin.
The phosphorylated HDAC1 interacts with Lef1 and prevents Tcf-Lef promoter activation
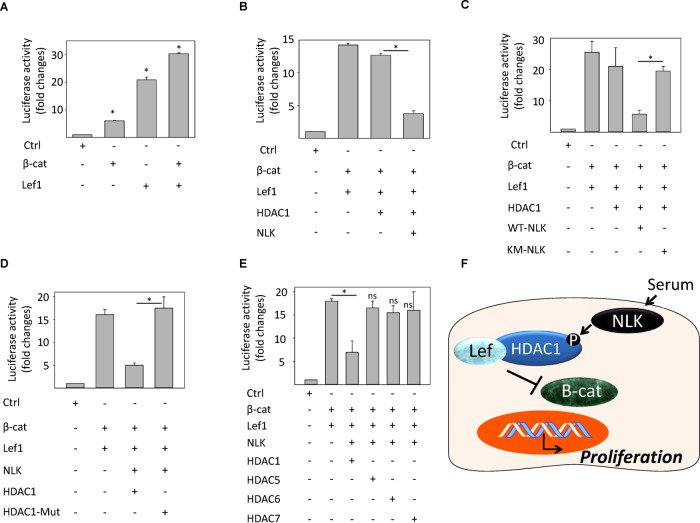
Next we analyzed whether inhibition of the Wnt signaling pathway by NLK is mediated through HDAC1. To test this hypothesis, we transfected NLK-deficient MEF cells with a combination of Lef1, β-catenin, HDAC1, and/or NLK expression plasmids, investigating Tcf/Lef reporter activity. Initially, β-catenin and Lef1 cotransfection showed higher luciferase activation than β-catenin or Lef1-transfected cells (Figure 6A), and overexpression of NLK and HDAC1, but not HDAC1 alone, inhibited Tcf/Lef reporter activity (Figure 6B). This effect was mediated via the activation of NLK because the catalytic mutant of NLK was unable to reduce luciferase activation (Figure 6C). In addition, the HDAC1 deletion mutant was unable to repress Tcf/Lef reporter activity, suggesting that HDAC1 activity is necessary for downstream signaling pathway (Figure 6D). To test the specificity of other HDACs for NLK signaling, we transfected knockout cells with expression plasmids of HDAC1, HDAC5, HDAC6, and HDAC7. We found that HDAC1 was the only HDAC to cause a repression of reporter activity (Figure 6E). These results suggest that the phosphorylation of HDAC1 by NLK leads to repression of Lef1 transcription activity, which is vital for limiting cell proliferation (Figure 6F).
FIGURE 6:
HDAC1 in the presence of NLK represses Lef1 transcription activity. (A) TOPFlash/FOPFlash luciferase activity assay in MEFs cells transfected with β-catenin and Tcf expression plasmids. Values are means ± SEM (n = 3, control and β-catenin: p = 0.0000199; control and Lef1: p = 0.00000343; Control and Lef1/β-catenin: p = 0.0000000102). (B) TOPFlash/FOPFlash luciferase activity assay in MEFs cells transfected with β-catenin, Tcf, HDAC1, and/or NLK expression plasmids. Values are means ± SEM (n = 3, p = 0.0000101). (C) TOPFlash luciferase activity assay in MEFs cells transfected with β-catenin, Tcf, HDAC1, Flag-tagged wild-type (WT-NLK), and/or Flag-tagged, kinase-dead, catalytically inactive mutant (NLK-KM) of NLK expression plasmids. Values are means ± SEM (n = 3, p = 0.000134). (D) TOPFlash luciferase activity assay in MEFs cells transfected with β-catenin, Tcf, full-length HDAC1, or catalytically inactive mutant of HDAC1 (HDAC1-Mut). Values are means ± SEM (n = 3, p = 0.000513). (E) TOPFlash/FOPFlash luciferase activity assay in MEFs cells transfected with β-catenin, Tcf, HDAC1, HDAC5, HDAC6, and/or HDAC7. Values are means ± SEM (n = 3, p = 0.000860). (F) Model of how NLK activation leads to phosphorylation of HDAC1, which, in turn, binds to Lef1 and represses transcriptional activation of Wnt-target genes.
DISCUSSION
The Wnt signaling pathway plays an important role in development, and it is essential that this system is specifically regulated for homeostasis. Dysregulation in this signaling leads to the development of human diseases. Downstream target proteins in this signaling pathway consist of the transcription factors Tcf and Lef, which can mediate receptor activation by initiating gene transcription and expression. Besides NLK, Wnt signaling can be limited via other signaling molecules, including ICAT, Groucho, and β-catenin degradation in mammalian cells (Cadigan and Peifer, 2009).
To analyze whether the role of NLK in the inhibition of Wnt signaling can be overridden by other known negative regulators of this signaling, we generated NLK-deficient mice. The deletion of NLK in animals leads to a lethal phenotype, suggesting that negative regulation of the Wnt signaling pathway cannot be compensated by other molecules (Ke et al., 2016). Because of the lethal phenotype of NLK-deficient mice, we used MEFs isolated from NLK-deficient mice to identify the mechanism of NLK-mediated inhibition of Wnt signaling. NLK-deletion MEF cells have no differences in adhesion, migration, invasion, or plate efficiency from wild-type cells. Further, neither survival nor apoptosis was found to be affected when wild-type were compared with NLK−/− cells in the absence or presence of cell death–inducing agents such as TNF-α and doxorubicin. Instead, we NLK-deficient primary cells grow much faster than wild-type cells. This effect depended on the concentration of serum in which the cells were cultured, and the removal of serum prevented cell growth. More specifically, the rapid proliferation of NLK-deficient cells was achieved via a faster S transition into the G2/M phase. Indeed, the peak of Wnt signaling during the G2/M phase was previously reported (Olmeda et al., 2003; Davidson et al., 2009). In this scenario, NLK could function as a target protein for limiting Wnt signaling in the S-phase transition into the G2/M phase of the cell cycle.
In NLK-knockout cells, there is a constitutive interaction between Lef1 and β-catenin, which could explain the elevated Tcf/Lef promoter activity in these cells. Further, overexpression of full-length but not the catalytically inactive mutant of NLK in knockout cells reduced Tcf/Lef promoter activity. Previously it was shown that Lef1 phosphorylation by NLK prevents the binding of Lef1 to the promoter/DNA (Ishitani et al., 2003). Surprisingly, in our system, reduced binding of Tcf and β-catenin and Tcf/Lef promoter activation were not dependent on the phosphorylation status of Lef1. To identify the mechanism causing reduced Tcf/Lef promoter activity, we investigated the involvement of HDAC1. The role of histone acetylation in the transcription of Wnt target genes was previously described (Wohrle et al., 2007), and HDAC1 was identified as a repressor for the Wnt-induced transcription (Billin et al., 2000). This is achieved via direct interaction between HDAC1 and Tcf/Lef proteins, causing a repression in transcription (Daniels and Weis, 2005). However, the mechanisms that regulate the activation or inactivation of HDACs are largely unknown. Similarly, the mechanism that regulates the interaction of HDAC1 with Tcf and upstream factors that regulate this interaction has not yet been explained.
HDAC1 belongs to class I HDACs; it has been shown that HDAC1 can regulate the expression of key cell cycle–regulated genes. Different posttranslational modifications (PTMs) of HDACs were shown to modify the function and regulate the activation of these enzymes. Phosphorylation is one of the known PTMs that can affect the activity of HDACs, and serine 421 and 423 phosphorylation promotes the activation of HDAC1, whereas mutation of these sites blocks the deacetylation activity of HDAC1 (Pflum et al., 2001). The kinases responsible for phosphorylation of HDAC1 include not only casein kinase II but also, to some extent, protein kinase A (Cai et al., 2001). Additional tyrosine phosphorylation (Tyr-221) was discovered through MS analysis, but the role of this phosphorylation is not yet known (Rush et al., 2005). During hypoxia (Pluemsampant et al., 2008) and virus infection (Poon et al., 2003), HDAC1 phosphorylation correlated with increased HDAC enzymatic activity, suggesting that phosphorylation is vital for signaling factors of different pathways to carry on different biological responses. In our model, in wild-type cells, HDAC1 is associated with Lef1, whereas this interaction is lost in knockout cells. Further, NLK promotes the phosphorylation of HDAC1, mainly at serine 421 in the presence of serum. The phosphorylation of HDAC1 was achieved only in the presence of full-length NLK; a catalytically inactive mutant of NLK was unable to phosphorylate HDAC1. In addition, the phosphorylation of HDAC1 limited Tcf-Lef promoter activation in MEF cells. Because the deacetylase mutant of HDAC1 could not block the downstream signaling, this suggests that HDAC1 activation is necessary for reducing Tcf/Lef gene transcription. HDAC isoform specificity provides evidence that Tcf-Lef signaling can be limited only by HDAC1 and not with the other HDACs, including HDAC5, HDAC6, and HDAC7 in MEF cells.
In the present study, we showed that NLK activation phosphorylates HDAC1 at serine 421, which further causes Tcf/Lef transcription repression and a reduced proliferation rate of wild-type cells. NLK-deficient cells harbor constitutive interactions between β-catenin and Lef, as well as elevated promoter activation, which, in turn, facilitate a shorter S into the G2/M phase of the cell cycle.
MATERIALS AND METHODS
Cell culture
NLK KO (NLK−/−) mice were previously generated by inserting a cassette containing lacZ reporter gene upstream of exon 2 of the NLK gene (Ke et al., 2016). All mice were maintained in specific pathogen-free housing at the Clinical Research Centre in Malmö, and the animal experiments were performed according to the national and international guidelines of the European Union. Further, all experimental protocols were approved by the Swedish regional (Malmö-Lund) ethical committee (application M336-12). MEFs were isolated from embryos on day 13.5 in WT and NLK-deficient (KO) mice. The cells were cultured in DMEM that was supplemented with 4500 mg/l l-glucose, 4 mM/l l-glutamine, and sodium pyruvate (HyClone, Thermo Scientific), 20% fetal bovine serum (FBS), and 100 IU/ml penicillin and streptomycin mixture (Life Technologies).
Western blot
Proteins were electrophoretically separated on 7.5% gels and transferred to polyvinylidene difluoride membranes (Millipore). Membranes were blocked by incubation with phosphate-buffered saline (PBS) containing 5% nonfat milk. Primary antibodies used were anti-NLK, anti–cyclin D1 (Abcam), anti–cyclin A, anti-HDAC1, anti-IκBα, anti-pIκBα (Santa Cruz Biotechnology), anti-actin (MP Biomedicals), anti-tubulin (Sigma-Aldrich), anti-Lef1 (Thermo Scientific), and anti-pLef1 (Millipore). Secondary horseradish peroxidase–labeled antibodies used were from GE Healthcare and Dako. For the chemiluminescence reaction, Supersignal Substrate (Thermo Scientific) was used according to the manufacturer’s instructions. Chemiluminescence was detected with an LAS-1000 charge-coupled device camera (Fujifilm) and Image Reader LAS-1000 Pro version 2.6 software (Fujifilm).
Adhesion, migration, and invasion assay
For the adhesion assay, MEF cells were seeded at a density of 15 × 103 cells/ml on six-well plates. After 1 h of incubation at 37°C in 5% CO2, cells were trypsinized and counted by using a Bürker chamber (Hirschmann).
For the scratch wound–healing migration assay, confluent MEF cells (>90% confluence) were rinsed with PBS by using a sterile 200-μl pipette tip. Three separate wounds (scratches) through the cells were made. After 6 h, a phase-contrast photograph was taken above and just below each line, and migration was quantified as area units.
For the Boyden chamber invasion assay, MEF cells were placed on the upper layer of a cell-permeable membrane and incubated for 6–8 h. The cells that migrated through the membrane were stained and counted.
Transfections
MEFs were transfected with a Targefect kit (Targeting Systems) according to the manufacturer’s instructions. In brief, cells were seeded at ∼70% confluence. Transfections were initiated after 24 h of seeding with 2 µg of DNA and 6 µl of Targefect-MEF transfection reagent (Targeting Systems) in DMEM containing 10% FBS.
Luciferase assay
MEFs were transfected with 2 µg of TOPFlash or FOPFlash reporter plasmids for 24 h. Cells were washed once with PBS and then lysed for 30 min at room temperature. The lysates were clarified by centrifugation at 14,000 rpm for 10 min, and 30 µl of each lysate was used to measure luciferase reporter gene expression (Dual-Luciferase Reporter Assay; Promega). The luciferase activity was normalized to Renilla luciferase activity. All experiments were performed in duplicate at least three times.
Analysis of cell growth
MEFs were seeded in 100-µl medium to a density of 2000 cells/well in 96-well culture plates. The amount of viable cells was assessed by incubating the cells with the tetrazolium salt 4-[3-(4-Iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1, 3-benzen disulfonate (WST-1; Roche) for 4 h to a water-soluble formazan dye. Absorbance (450, 690 nm) was measured in an Antons 2020 enzyme-linked immunosorbent assay plate reader (Antons Labtech Instruments).
Cell proliferation in the presence of different serum concentrations
For cell counting, NLK+/+ and NLK−/− MEF cells were seeded in triplicate with 70,000 cells/well in six-well plates. Cells seeded overnight were washed twice with PBS, and fresh medium containing 0, 5, or 10% FBS was added. Cells on the plate were washed once with PBS and harvested by using trypsin followed by centrifugation (1400 rpm, 5 min) and resuspended in medium. Cells were diluted in trypan blue and manually counted by using a hemocytometer. For the transfection studies, NLK−/− cells were seeded in duplicate on 24-well plates with 1.5 × 104 cells/well. After 24 h in culture, cells were transfected for 4 h with 0.8 µg of DNA, 1.5 µl of Targefect-MEF transfection reagent, and 2.5 µl of Virofect (Targeting Systems) in DMEM containing 10% FBS. At 0 and 24 h after transfection, cells were stained with trypan blue and counted manually by using a hemocytometer.
Immunofluorescence and confocal microscopy
MEFs were fixed with 4% paraformaldehyde (PFA) after removal of medium, followed by permeabilization and blocking with 5% normal goat serum and 0.3% Triton X-100 (Sigma-Aldrich) in PBS for 30 min. Primary antibodies toward NLK (Abcam), Flag (Sigma-Aldrich), p-HDAC1–Ser-421/423 (GeneTex), and α-tubulin (Sigma-Aldrich) were diluted in PBS, added to the cells, and incubated for 1 h at room temperature. Cells were washed four times with PBS and incubated with Alexa Fluor 488– or Alexa Fluor 546–conjugated antibodies (Molecular Probes). Coverslips were mounted on object slides in Vectashield with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories). Images were captured by using a 40× oil objective and Zeiss LSM 710 confocal system.
In vitro kinase assay
Wild-type and knockout MEF cells were transfected with full-length wild-type NLK (Flag-NLKWT) or catalytically inactive mutant of NLK (Flag-NLKKM), lysed, and immunoprecipitated with anti-HDAC1 antibody. Immunoprecipitates were purified by washing five times with lysis buffer, once with lysis buffer containing high salt (0.5 M NaCl and 1 mM dithiothreitol [DTT]), and once with 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) and 1 mM DTT. The pellet was incubated with 1 mM ATP in 40 µl of kinase buffer (10 mM MgCl2, 10 mM HEPES, pH 7.4, and 1 mM DTT) for 30 min at 30°C.
Kinase assay
Cells were transfected with Flag-tagged wild-type (WT-NLK) or Flag-tagged kinase-dead mutants (K155M; KM-NLK) of NLK expression plasmids followed by immunoprecipitation using anti-Flag antibody. Immunoprecipitates were purified by washing twice with lysis buffer containing high salt (0.625 M NaCl), three times with PBS containing high salt (0.625 M NaCl), and twice with PBS. The pellet was incubated with 5 μCi of [γ-32P]ATP in 40 µl of kinase buffer (10 mM MgCl2, 10 mM HEPES, pH 7.4, and 1 mM DTT) for 30 min at 30°C. Samples were separated by SDS–PAGE and visualized by autoradiography.
Cell viability
For propidium iodide (PI) staining, the same number of cells were serum starved for 48 h, fixed in 4% PFA, and washed twice with PBS before being incubated with a solution containing 3.5 µM Tris-HCl, pH 7.6, 10 mM NaCl, 50 µg/ml PI, 20 µg/ml RNase, and 0.1% Igepal CA-630 for 20 min in the dark. After incubation, coverslips were washed four times with PBS and mounted on object slides with PVA-DABCO (9.6% polyvinyl alcohol, 24% glycerol, and 2.5% 1,4-diazabicyclo (2.2.2) octane in 67 mM Tris-HCl, pH 8.0). Viable and nonviable cells were counted through visualization of PI.
MEFs were seeded in triplicate in 60-mm plates containing 5 × 105 cells/plate and serum starved for 48 h. Cells were collected and stained according to the manufacturer’s protocol (Nucleocounter NC-3000; Chemometec). Briefly, the medium was transferred to Falcon tubes, and after washing of cells with PBS, this was also transferred to Falcon tubes. Cells were trypsinized, and the cell suspension was transferred to the Falcon tubes before being spun down at 1400 rpm for 5 min. The cell pellet was washed once in PBS and finally resuspended in PBS. A representative volume of cell suspension was transferred to an Eppendorf tube, and one volume of solution 13 (final dilution 1:20) was added and mixed well. Then 10 µl of the cell/solution 13 suspension was loaded onto an NC-Slide A8, and viability analysis was run on the Nucleocounter NC-3000.
Annexin V-PE apoptosis assay
To determine viable and apoptotic cells, MEF cells were stained with Annexin V-PE along with 7-aminoactinomycin D (7-AAD) by following the manufacturer’s protocol supplied with the PE Annexin V apoptosis detection kit I (BD PharMingen). Briefly, cells seeded in a 10-cm Petri dish at 2 × 105 cell density were allowed to reach 70–80% confluency before treatment. After TNF-α (10 ng/ml) or doxorubicin (1 μM) treatment, the supernatant together with the respective adhered cells was harvested and washed twice with ice-cold PBS before being suspended in 100 μl of binding buffer. Next Annexin V-PE (5 µl) and 7-AAD (5 µl) were added, and the mixture was incubated for 15 min in the dark. Finally, 400 μl of binding buffer was added to the cells and analyzed by using a flow cytometer (BD FACSVerse). The data analysis was done by using FlowJo version 10 software.
Cell cycle analysis
For cell-cycle analysis, MEFs were seeded overnight and synchronized in serum-free medium for 48 h. The cells were then collected over the indicated time points after being released from serum starvation and washed with PBS before the pellets were fixed with 70% ice-cold ethanol at −20°C. The fixed cells were first washed and then suspended in PI staining solution (20 μg/ml PI, 200 μg/ml RNase A in PBS containing 0.1% Triton X-100), and finally incubated at 37°C for 30 min. For each condition, 10,000 cells were analyzed by a flow cytometer (BD FACSVerse), and the cell-cycle profile was analyzed by using FlowJo version 10 software.
Mass spectrometry analysis
Protein samples were separated by SDS–PAGE, and selected gel bands were excised for analysis. After reduction and alkylation using DTT and IAA, respectively, in-gel digestion was performed by using porcine trypsin (Promega) as endoprotease. After extraction, peptides were phosphoenriched by using TiO2 Mag Sepharose beads (GE Healthcare Life Sciences). Dry peptides were resuspended in 0.1% formic acid and separated by using an Eksigent NanoLC 2D system (Eksigent, Dublin, CA) that was coupled to an LTQ-Orbitrap XL ETD mass spectrometer (Thermo Fisher Scientific, Germany) operated in data-dependent acquisition mode. The peptides were loaded and washed for 15 min on a precolumn (Acclaim PepMap 100, C18, 3-μm particle size, 50-μm diameter; Thermo Fisher Scientific, Germany) at a constant flow of 5 μl/min solvent B (0.1% formic acid [FA] in acetonitrile [ACM]) before separation on an analytical column (10-μm fused silica emitter, 75-μm × 16-cm Pico Tip Emitter; New Objective) packed in-house with C18 material ReproSil-Pur. A linear 60-min gradient in 0.1% FA buffer from 3–90% ACN at a flow rate of 300 nl/min was used. The MS/MS analysis was performed by using multistage activation and top 10 collision-induced dissociation and detection in the linear ion trap, rejecting molecules with an unassigned charge state or a charged state. MS/MS data were converted into Mascot Generic Format (MGF) by using ProteoWizard (www.nature.com/nbt/journal/v30/n10/full/nbt.2377.html) and searched against the SwissProt part of the UniProt database. Mascot 2.4.1 (www.matrixscience.com) searches were performed from Proteios Software Environment (http://pubs.acs.org/doi/abs/10.1021/pr900189c). Complementary searches in X!Tandem and MS-GF+ (www.nature.com/ncomms/2014/141031/ncomms6277/full/ncomms6277.html) were performed by using SearchGUI (www.ncbi.nlm.nih.gov/pubmed/21337703), and annotated spectra were visualized in PeptideShaker (www.nature.com/nbt/journal/v33/n1/full/nbt.3109.html). For all searches, 10-ppm precursor tolerance and 0.5-Da fragment tolerance, fixed carbamidomethylation of C, variable oxidation of M and variable phosphorylation of ST, and up to one missed cleavage were used as search settings.
Statistical analyses
Statistical analyses were performed by using Sigma Plot 12.5 Software. Results are expressed as mean ± SEM, mean ± SD, or percentages. p < 0.05 was deemed statistically significant. Comparisons between groups were performed by using the Mann–Whitney U test. A correlation analysis was performed by determining the Pearson product moment correlation coefficient.
Supplementary Material
Acknowledgments
We are grateful to Kunihiro Matsumoto for WT-NLK and NLK-K155M expression plasmids. All other expression plasmids were obtained from Addgene (Cambridge, MA). This work was supported by the Swedish Cancer Foundation, the Magnus Bergvall Foundation, the Gunnar Nilsson Cancer Foundation, and the Gyllenstiernska Krapperup Foundation and by funding from the European Research Council under the European Union’s Seventh Framework Program for Research and Technology Development, Grant Agreement 260460, awarded to R.M.
Abbreviations used:
- HDAC1
histone deacetylase 1
- HSP27
heat shock protein 27
- KO
knockout
- Lef
lymphoid-enhancing factor
- MAP1B
microtubule-associated protein1B
- MEF
mouse embryonic fibroblast
- MS
mass spectrometry
- NLK
nemo-like kinase
- PTM
posttranslational modification
- RT-PCR
reverse transcription PCR
- SETDB1
set domain bifurcated 1
- Tcf
transcription factors T-cell factor
- TNF-α
tumor necrosis factor α
- WT
wild type
Footnotes
The authors declare no competing financial interests.
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-07-0547) on November 30, 2016.
REFERENCES
- Aoki K, Taketo MM. Tissue-specific transgenic, conditional knockout and knock-in mice of genes in the canonical Wnt signaling pathway. Methods Mol Biol. 2008;468:307–331. doi: 10.1007/978-1-59745-249-6_24. [DOI] [PubMed] [Google Scholar]
- Billin AN, Thirlwell H, Ayer DE. Beta-catenin-histone deacetylase interactions regulate the transition of LEF1 from a transcriptional repressor to an activator. Mol Cell Biol. 2000;20:6882–6890. doi: 10.1128/mcb.20.18.6882-6890.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brott BK, Pinsky BA, Erikson RL. Nlk is a murine protein kinase related to Erk/MAP kinases and localized in the nucleus. Proc Natl Acad Sci USA. 1998;95:963–968. doi: 10.1073/pnas.95.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan KM, Peifer M. Wnt signaling from development to disease: insights from model systems. Cold Spring Harb Perspect Biol. 2009;1:a002881. doi: 10.1101/cshperspect.a002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai R, Kwon P, Yan-Neale Y, Sambuccetti L, Fischer D, Cohen D. Mammalian histone deacetylase 1 protein is posttranslationally modified by phosphorylation. Biochem Biophys Res Commun. 2001;283:445–453. doi: 10.1006/bbrc.2001.4786. [DOI] [PubMed] [Google Scholar]
- Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol. 2005;12:364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- Davidson G, Shen J, Huang YL, Su Y, Karaulanov E, Bartscherer K, Hassler C, Stannek P, Boutros M, Niehrs C. Cell cycle control of wnt receptor activation. Dev Cell. 2009;17:788–799. doi: 10.1016/j.devcel.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Emami KH, Brown LG, Pitts TE, Sun X, Vessella RL, Corey E. Nemo-like kinase induces apoptosis and inhibits androgen receptor signaling in prostate cancer cells. Prostate. 2009;69:1481–1492. doi: 10.1002/pros.20998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Jiang Y, Lu W, Zhang Y. Nemo-like kinase associated with proliferation and apoptosis by c-Myb degradation in breast cancer. PLoS One. 2013;8:e69148. doi: 10.1371/journal.pone.0069148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani S, Inaba K, Matsumoto K, Ishitani T. Homodimerization of Nemo-like kinase is essential for activation and nuclear localization. Mol Biol Cell. 2011;22:266–277. doi: 10.1091/mbc.E10-07-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani T, Hirao T, Suzuki M, Isoda M, Ishitani S, Harigaya K, Kitagawa M, Matsumoto K, Itoh M. Nemo-like kinase suppresses Notch signalling by interfering with formation of the Notch active transcriptional complex. Nat Cell Biol. 2010;12:278–285. doi: 10.1038/ncb2028. [DOI] [PubMed] [Google Scholar]
- Ishitani T, Ishitani S, Matsumoto K, Itoh M. Nemo-like kinase is involved in NGF-induced neurite outgrowth via phosphorylating MAP1B and paxillin. J Neurochem. 2009;111:1104–1118. doi: 10.1111/j.1471-4159.2009.06400.x. [DOI] [PubMed] [Google Scholar]
- Ishitani T, Ninomiya-Tsuji J, Matsumoto K. Regulation of lymphoid enhancer factor 1/T-cell factor by mitogen-activated protein kinase-related Nemo-like kinase-dependent phosphorylation in Wnt/beta-catenin signaling. Mol Cell Biol. 2003;23:1379–1389. doi: 10.1128/MCB.23.4.1379-1389.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani T, Ninomiya-Tsuji J, Nagai S, Nishita M, Meneghini M, Barker N, Waterman M, Bowerman B, Clevers H, Shibuya H, et al. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature. 1999;399:798–802. doi: 10.1038/21674. [DOI] [PubMed] [Google Scholar]
- Kanei-Ishii C, Ninomiya-Tsuji J, Tanikawa J, Nomura T, Ishitani T, Kishida S, Kokura K, Kurahashi T, Ichikawa-Iwata E, Kim Y, et al. Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK. Genes Dev. 2004;18:816–829. doi: 10.1101/gad.1170604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanei-Ishii C, Nomura T, Takagi T, Watanabe N, Nakayama KI, Ishii S. Fbxw7 acts as an E3 ubiquitin ligase that targets c-Myb for nemo-like kinase (NLK)-induced degradation. J Biol Chem. 2008;283:30540–30548. doi: 10.1074/jbc.M804340200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke H, Masoumi KC, Ahlqvist K, Seckl MJ, Rydell-Tormanen K, Massoumi R. Nemo-like kinase regulates the expression of vascular endothelial growth factor (VEGF) in alveolar epithelial cells. Sci Rep. 2016;6:23987. doi: 10.1038/srep23987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi A. Regulation of beta-catenin signaling in the Wnt pathway. Biochem Biophys Res Commun. 2000;268:243–248. doi: 10.1006/bbrc.1999.1860. [DOI] [PubMed] [Google Scholar]
- Kikuchi A, Kishida S, Yamamoto H. Regulation of Wnt signaling by protein-protein interaction and posttranslational modifications. Exp Mol Med. 2006;38:1–10. doi: 10.1038/emm.2006.1. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim Y, Lee J, Chung J. Regulation of FOXO1 by TAK1-Nemo-like kinase pathway. J Biol Chem. 2010;285:8122–8129. doi: 10.1074/jbc.M110.101824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H, Sasaki T, Ishitani T, Iemura S, Zhao H, Kaneko S, Kunimoto H, Natsume T, Matsumoto K, Nakajima K. STAT3 regulates Nemo-like kinase by mediating its interaction with IL-6-stimulated TGFbeta-activated kinase 1 for STAT3 Ser-727 phosphorylation. Proc Natl Acad Sci USA. 2005;102:4524–4529. doi: 10.1073/pnas.0500679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SZ, Zhang HH, Liang JB, Song Y, Jin BX, Xing NN, Fan GC, Du RL, Zhang XD. Nemo-like kinase (NLK) negatively regulates NF-kappa B activity through disrupting the interaction of TAK1 with IKKbeta. Biochim Biophys Acta. 2014;1843:1365–1372. doi: 10.1016/j.bbamcr.2014.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Cui G, Wang J, Yu Z, Zhao L, Lv Z. Nemo-like kinase (NLK) involves in neuronal apoptosis after traumatic brain injury. Cell Mol Neurobiol. 2012;32:381–389. doi: 10.1007/s10571-011-9766-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini MD, Ishitani T, Carter JC, Hisamoto N, Ninomiya-Tsuji J, Thorpe CJ, Hamill DR, Matsumoto K, Bowerman B. MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature. 1999;399:793–797. doi: 10.1038/21666. [DOI] [PubMed] [Google Scholar]
- Ohkawara B, Shirakabe K, Hyodo-Miura J, Matsuo R, Ueno N, Matsumoto K, Shibuya H. Role of the TAK1-NLK-STAT3 pathway in TGF-beta-mediated mesoderm induction. Genes Dev. 2004;18:381–386. doi: 10.1101/gad.1166904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmeda D, Castel S, Vilaro S, Cano A. Beta-catenin regulation during the cell cycle: implications in G2/M and apoptosis. Mol Biol Cell. 2003;14:2844–2860. doi: 10.1091/mbc.E03-01-0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflum MK, Tong JK, Lane WS, Schreiber SL. Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J Biol Chem. 2001;276:47733–47741. doi: 10.1074/jbc.M105590200. [DOI] [PubMed] [Google Scholar]
- Pluemsampant S, Safronova OS, Nakahama K, Morita I. Protein kinase C(PKC)K2 is a key activator of histone deacetylase in hypoxia-associated tumors. Int J Cancer. 2008;122:333–341. doi: 10.1002/ijc.23094. [DOI] [PubMed] [Google Scholar]
- Poon AP, Liang Y, Roizman B. Herpes simplex virus 1 gene expression is accelerated by inhibitors of histone deacetylases in rabbit skin cells infected with a mutant carrying a cDNA copy of the infected-cell protein no. 0. J Virol. 2003;77:12671–12678. doi: 10.1128/JVI.77.23.12671-12678.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottinger E, Croce J, Lhomond G, Besnardeau L, Gache C, Lepage T. Nemo-like kinase (NLK) acts downstream of Notch/Delta signalling to downregulate TCF during mesoderm induction in the sea urchin embryo. Development. 2006;133:4341–4353. doi: 10.1242/dev.02603. [DOI] [PubMed] [Google Scholar]
- Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- Satoh K, Ohnishi J, Sato A, Takeyama M, Iemura S, Natsume T, Shibuya H. Nemo-like kinase-myocyte enhancer factor 2A signaling regulates anterior formation in Xenopus development. Mol Cell Biol. 2007;27:7623–7630. doi: 10.1128/MCB.01481-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw-Hallgren G, Chmielarska Masoumi K, Zarrizi R, Hellman U, Karlsson P, Helou K, Massoumi R. Association of nuclear-localized Nemo-like kinase with heat-shock protein 27 inhibits apoptosis in human breast cancer cells. PLoS One. 2014;9:e96506. doi: 10.1371/journal.pone.0096506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit L, Baas A, Kuipers J, Korswagen H, van de Wetering M, Clevers H. Wnt activates the Tak1/Nemo-like kinase pathway. J Biol Chem. 2004;279:17232–17240. doi: 10.1074/jbc.M307801200. [DOI] [PubMed] [Google Scholar]
- Takada I, Mihara M, Suzawa M, Ohtake F, Kobayashi S, Igarashi M, Youn MY, Takeyama K, Nakamura T, Mezaki Y, et al. A histone lysine methyltransferase activated by noncanonical Wnt signalling suppresses PPAR-gamma transactivation. Nat Cell Biol. 2007;9:1273–1285. doi: 10.1038/ncb1647. [DOI] [PubMed] [Google Scholar]
- Wohrle S, Wallmen B, Hecht A. Differential control of Wnt target genes involves epigenetic mechanisms and selective promoter occupancy by T-cell factors. Mol Cell Biol. 2007;27:8164–8177. doi: 10.1128/MCB.00555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda J, Tsuchiya A, Yamada T, Sakamoto M, Sekiya T, Hirohashi S. Nemo-like kinase induces apoptosis in DLD-1 human colon cancer cells. Biochem Biophys Res Commun. 2003;308:227–233. doi: 10.1016/s0006-291x(03)01343-3. [DOI] [PubMed] [Google Scholar]
- Yasuda J, Yokoo H, Yamada T, Kitabayashi I, Sekiya T, Ichikawa H. Nemo-like kinase suppresses a wide range of transcription factors, including nuclear factor-kappaB. Cancer Sci. 2004;95:52–57. doi: 10.1111/j.1349-7006.2004.tb03170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng YA, Rahnama M, Wang S, Sosu-Sedzorme W, Verheyen EM. Drosophila Nemo antagonizes BMP signaling by phosphorylation of Mad and inhibition of its nuclear accumulation. Development. 2007;134:2061–2071. doi: 10.1242/dev.02853. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.