Abstract
Purpose
Vertical cup-disc ratio (VCDR) is used as a clinical assessment measure to identify and monitor glaucomatous damage to the optic nerve. Previous genetic studies conducted in European and Asian populations have identified many loci associated with VCDR. The genetic factors in other ethnic populations, such as Latino, influencing VCDR remain to be determined. Here, we describe the first genome-wide association study (GWAS) on VCDR in Latino individuals.
Methods
We conducted this GWAS on VCDR using 4537 Latino individuals who were genotyped by using either the Illumina OmniExpress BeadChip (∼730K markers) or the Illumina Hispanic/SOL BeadChip (∼2.5 million markers). Study subjects were 40 years of age and older. Linear regression, adjusting for age, sex, and principal components of genetic ancestry, was conducted to assess the associations between single nucleotide polymorphisms (SNPs) and VCDR. We imputed SNPs from the 1000 Genomes Project to integrate additional SNPs not directly genotyped.
Results
We replicated two previously reported SNPs that reached GWAS significance, rs1900005 and rs7916697, in the ATOH7-PBLD region, as well as identified two suggestive associations in the CDC7-TGFBR3 region on chromosome 1p22.1 and in the ZNF770-DPH6 region on chromosome 15q14. We discovered a novel SNP, rs56238729 (P = 1.22 × 10−13), in the ATOH7-PBLD region that is significantly associated with VCDR in Latino individuals. We replicated eight previously reported regions, including COL8A1, CDKN2B-CDKN2BAS, BMP2, and CHEK2 (P < 2.17 × 10−3).
Conclusions
Our results discovered a novel SNP that is significantly associated with VCDR in Latino individuals and confirmed previously reported loci, providing further insight into the genetic architecture of VCDR.
Keywords: GWAS, VCDR, Latino Individuals
The optic disc, situated at the back of the eye, is the location where the retinal ganglion cell axons exit the eye to form the optic nerve. Inspection of the optic disc is conducted during routine eye examinations to assess the optic nerve. In particular, the ratio of the vertical diameter of the optic cup to the vertical diameter of the optic disc, called the vertical cup-disc ratio (VCDR), is often calculated to identify and monitor possible glaucomatous damage to the optic nerve. As an important endophenotype for primary open-angle glaucoma (POAG), VCDR characterizes the extent of cupping of the optic nerve, and subsequently, the loss of retinal ganglion cell axons. In numerous epidemiologic studies investigating the prevalence of POAG, VCDR is often used as a clinical diagnostic criterion to identify either suspects or cases of POAG.1–5 Due to the complex etiology of POAG, identifying the factors that influence individual quantitative traits, such as VCDR, may better elucidate the biological mechanisms underlying this disease.
Previous studies have shown substantial differences in VCDR among various ethnic populations. Overall, individuals of African descent reported having the highest VCDR and individuals of European descent exhibiting the lowest, with Chinese and Hispanic individuals displaying VCDR measurements between African and European individuals.6,7 Furthermore, heritability studies have provided additional evidence of a genetic component for VCDR determination. These studies estimate the heritability of this trait to be 48% to 66%.8,9 En masse, these results suggest genetic factors have a major role in VCDR determination and specific genetic variants may influence this trait in different ethnic populations.
There have been a limited number of genome-wide association studies (GWAS) conducted to identify genetic variants associated with VCDR. A previous meta-analysis of GWAS conducted in individuals of European descent identified nine loci associated with VCDR, including CDKN2B, SIX1, SCYL1, CHEK2, ATOH7-PBLD, DCLK1, BCAS3, RERE, and ARID3A.10 Additionally, a meta-analysis of European and Asian individuals identified several novel loci associated with VCDR, such as CDC7-TGFBR3, TMTC2, RPAP3, and BMP2.11 Although these previous studies have identified a number of loci associated with VCDR, these studies were conducted in individuals of European or Asian descent. Because specific genetic variants may be more enriched in one ethnic population compared with another, identifying genetic factors that are both common and unique to different ethnic populations will aid in elucidating racial differences in VCDR, as well as potentially uncovering the pathogenesis of various ocular diseases affecting the optic nerve. To date, there have been no reported GWAS of VCDR in Latino individuals.
Latino individuals are traditionally an underrepresented population in genetic studies, despite being the largest minority group in the United States. We hypothesize conducting a GWAS of VCDR in Latino individuals will replicate previously reported loci that are common to various ethnic populations and, potentially, identify novel loci specific to Latino individuals. Moreover, identifying genetic variants associated with VCDR in Latino individuals may help to elucidate the observed racial differences for this ocular parameter. As such, the purpose of this study was to conduct a GWAS of VCDR in Latino individuals. This study represents the first GWAS of VCDR in a Latino population.
Materials and Methods
Ethics Statement
The institutional review boards at the University of Illinois at Chicago, the University of Southern California Health Sciences Campus, and the Los Angeles Biomedical Research Institute at Harbor-University of California, Los Angeles (UCLA) approved this research and all clinical investigation was conducted according to the principles outlined in the Declaration of Helsinki.
Study Sample
This study was conducted using 4537 samples collected from the Los Angeles Latino Eye Study (LALES), a population-based epidemiologic study examining the prevalence, incidence, and impact of visual impairment and ocular disease in 6357 Latino individuals living in the city of La Puente, Los Angeles County, California, United States.12 All study subjects were 40 years or older and written informed consent was obtained from all study participants.
Measurements of VCDR
Stereoscopic optic disc photography was conducted using the Topcon TRC 50EX Retinal Camera (Topcon Corp. of America, Paramus, NJ, USA) with Ektachrome 100 film (Kodak, Rochester, NY, USA) and was evaluated using a stereoscopic viewer (Asahi viewer; Pentax, Englewood, CO, USA). VCDR measurements were determined by a board-certified ophthalmologist. We used the average VCDR between the left and right eyes as the final VCDR measurement for downstream analysis. If estimates were available for only one eye, the VCDR measurement of that eye was used as a proxy for the final measurement. To convert these data to a standard normal distribution, we applied an inverse normal transformation.
Genotyping and Quality Control
Through LALES and the Mexican American Glaucoma Genetic Study, we genotyped 4996 Latino individuals using either the Illumina OmniExpress BeadChip Kit (730, 525 markers; Illumina, Inc., San Diego, CA, USA; n = 4278) or the Illumina Hispanic/SOL BeadChip (∼2.5 million markers; Illumina, Inc.; n = 718). The Genotyping Laboratory of the Institute for Translational Genomics and Population Sciences at the Los Angeles Biomedical Research Institute at Harbor-UCLA performed the genotyping for this study. The software Illumina GenomeStudio (v2011.1; Illumina, Inc.) was used to call single nucleotide polymorphisms (SNPs). A genotyping call rate less than 97% was used to exclude study participants from further analysis. Duplicates, study participants with sex inconsistency between reported and genetically inferred sex, and missing VCDR measurements were further excluded. Implementation of these exclusion criteria resulted in 4537 study participants remaining for downstream analysis, including 3596 unrelated individuals used as a discovery set (stage 1), and 941 first-degree relatives (417 families) used as a replication set (stage 2, independent of stage 1). Quality control of the genotyped data for the study participants was conducted using the program PLINK (v1.90; available in the public domain at https://www.cog-genomics.org/plink2).13 Overlapping SNPs between the two genotyping chips were retained for analysis and were further omitted if the minor allele frequency (MAF) was less than 1%, the call rate was less than 95%, or the Hardy-Weinberg equilibrium P values were less than 10−6. These exclusion criteria resulted in 576,798 SNPs remaining for downstream analysis. To facilitate the imputation process, SNPs were coded on the forward strand.
Genotype Imputation
To identify additional SNPs associated with VCDR that were not directly genotyped, we performed genotype imputation using Shapeit2,14 Minimac3 (available in the public domain at http://genome.sph.umich.edu/wiki/Minimac3#Download), and the 1000 Genomes Project (1KGP) reference panels. These haplotype reference panels contain approximately 39.7 million variants, substantially increasing the number of SNPs to be tested through the use of linkage disequilibrium (LD). We used the AMR+CEU+YRI reference panels (a combination of Mexican, Colombian, Puerto Rican, CEPH, and Yoruba haplotypes) for imputation because of the unique admixture of different ancestral populations in Latino individuals. This panel has previously been shown to result in the highest genotype imputation accuracy for Latino individuals.15
Using the phased genotypes from Shapeit2, genotype imputation on the phased data was conducted using Minimac3. Imputed genotypes were coded as allelic dosages (estimated counts ranging from 0–2). Low-quality imputed SNPs (i.e., Rsq < 0.80 and MAF < 1%) were filtered out. After applying these quality control parameters, 6,844,888 imputed SNPs were retained for downstream analysis.
Statistical Analysis
We inferred principal components of genetic ancestry using the program EIGENSOFT.16 To make comparisons with reference populations of known ancestry, we included reference panels of unrelated Northern Europeans (CEU, n = 87) and West Africans (YRI, n = 88) from the 1000 Genomes Project,17 and Native Americans (n = 105).18 We retained and included the first four principal components as covariates in downstream association analyses. To assess control of population stratification, the genomic control inflation factor19 was calculated and a quantile-quantile (Q-Q) plot was generated to visualize the distribution of the test statistics. Linear regression, adjusting for age, sex, and principal components of genetic ancestry, was conducted to assess the associations between SNPs and VCDR among study participants in the discovery set using PLINK (v1.90).13 Additionally, an additive genetic effects model was assumed. To account for relatedness among individuals in the replication set, we used a linear mixed-effects model (Proc Mixed procedure of SAS v9.4; SAS Institute, Cary, NC, USA) to test the associations between SNPs and VCDR, adjusting for age, sex, and principal components of genetic ancestry. The empirical “sandwich” estimator and compound symmetry covariance structure were used during linear mixed-effects modeling. The software EMMAX (Efficient Mixed-Model Association Expedited; available in the public domain at http://csg.sph.umich.edu/kang/emmax/download/)20 was used to analyze genotyped and imputed SNPs for the full study sample, the discovery and replication sets combined, using linear mixed-effects modeling to account for population stratification and relatedness, adjusting for age, sex, kinship, and principal components of genetic ancestry. Allelic dosage was used to account for genotype imputation uncertainty for imputed SNPs in EMMAX. SNPs with a P < 1 × 10−6 in the discovery set were retained and analyzed in the replication set. SNPs reaching the genome-wide significance threshold (P < 5 × 10−8) were declared significant and SNPs reaching P < 1 × 10−6 were declared suggestive during the full study sample analysis. The program simpleM21–23 (available in the public domain at http://simplem.sourceforge.net) was used to identify the effective number of independent tests as a multiple testing correction method for the replication of previously published loci. Conditional association analysis was performed by including the lead SNP as a covariate in the regression model. Graphing was performed using R24 (available in the public domain at https://www.r-project.org) and LocusZoom25 (available in the public domain at http://csg.sph.umich.edu/locuszoom) (hg19/1KGP 2014 AMR).
Pathway Analysis
We performed pathway analysis using SKAT-O26 (available in the public domain at https://www.hsph.harvard.edu/skat/download/) and QIAGEN's Ingenuity Pathway Analysis (IPA)27 (QIAGEN, Redwood City, CA, USA) on the maximum number of unrelated individuals from both the discovery and replication sets to identify canonical pathways influencing VCDR. Based on the genomic positions of the GRCh37/hg19 assembly, directly genotyped SNPs were mapped to autosomal genes and a ±50 kb gene boundary was used to capture proximal regulatory and other functional elements influencing gene regulation. Gene-set associations were conducted using SKAT-O,26 adjusting for age, sex, and principal components of genetic ancestry. The commercial software IPA analyzed genes associated with VCDR for the enrichment of canonical pathways.27 Significantly enriched canonical pathways, P < 0.05 after Benjamini-Hochberg multiple testing adjustment,28 were reported.
Results
Study Sample
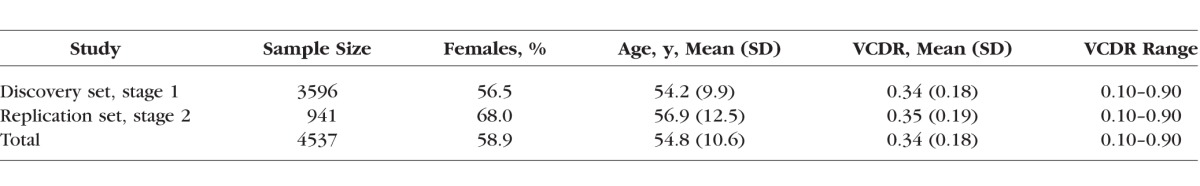
Table 1 presents the descriptive statistics for the overall study population, as well as the discovery (stage 1) and replication (stage 2) sets separately. For the entire study sample, the mean (SD) age was 54.8 (10.6) years, with the mean age of the discovery and replication sets as 54.2 (9.9) years and 56.9 (12.5) years, respectively. The proportion of females in the entire study was 58.9%: 56.5% in the discovery set and 68.0% in the replication set. Together, the average VCDR (SD) was 0.34 (0.18; range, 0.10–0.90), with the average of the discovery and replication sets as 0.34 (0.18; range, 0.10–0.90) and 0.35 (0.19; range, 0.10–0.90), respectively.
Table 1.
Descriptive Statistics of the Study Sample
Genome-Wide Association Results
The genomic control inflation factor19 was moderate, λ = 1.03. Supplementary Figure S1 displays the Q-Q plot of the observed P values versus the expected P values. As seen in the plot, the observed P values do not deviate from the null, except at the extreme tail. Both the genomic control inflation factor and the Q-Q plot indicate proper control of population stratification in this sample of Latino individuals.
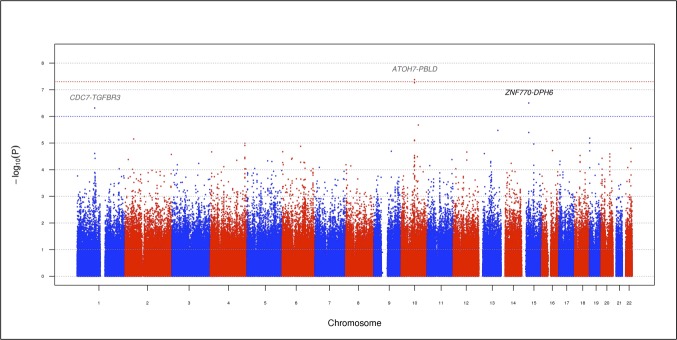
Figure 1 displays a Manhattan plot of the genome-wide P values from the discovery set association analysis. The results for the top SNPs (P < 1 × 10−6) are summarized in Table 2. One SNP, rs1900005 (P = 4.17 × 10−8, GRCh37/hg19 position 69,998,055), on chromosome 10q21.3 reached the GWAS significance level P < 5 × 10−8. The minor allele A (MAF = 0.36) was associated with a reduction in VCDR with β (SE) = −0.13 (0.02). This SNP is located 6.2 kb upstream of the ATOH7 (atonal bHLH transcription factor 7) gene and 44 kb downstream of the PBLD (phenazine biosynthesis-like protein domain containing) gene. The second most significant SNP, rs7916697 (P = 5.44 × 10−8, GRCh37/hg19 position 69,991,853), is located 6.2 kb upstream of rs1900005 and was borderline GWAS significant. This SNP is situated in the 5′ untranslated region (5′UTR) of ATOH7. The minor allele A of rs7916697 (MAF = 0.38) is associated with a decrease in VCDR with β (SE) = −0.13 (0.02). The third most significant SNP is rs16960773 (P = 3.15 × 10−7, GRCh37/hg19 position 35,604,502), located on chromosome 15q14 and situated 324 kb upstream of ZNF770 (zinc finger protein 770) and 53.2 kb downstream of DPH6 (diphthamine biosynthesis 6). The minor allele G (MAF = 0.08) is associated with a reduction in VCDR with β (SE) = −0.23 (0.04). The last most significant SNP is rs1192419 (P = 4.85 × 10−7, GRCh37/hg19 position 92,080,059), located on chromosome 1p22.1 and positioned 88.7 kb downstream of CDC7 (cell division cycle 7) and 65.8 kb downstream of TGFBR3 (transforming growth factor, beta receptor III). The minor allele A (MAF = 0.29) of rs1192419 is associated with an increase in VCDR with β (SE) = 0.13 (0.03).
Figure 1.
Manhattan plot displaying the –log10 (P values) for the association between VCDR and the 576,798 SNPs in the discovery set (stage 1). The red and blue horizontal dotted lines indicate genome-wide significant associations (P = 5 × 10−8) and suggestive associations (P = 1 × 10−6), respectively. Previously reported and novel loci associated with VCDR are shown in dark gray (CDC7-TGFBR3 and ATOH7-PBLD) and black (ZNF770-DPH6), respectively. Single nucleotide polymorphisms are plotted by genomic position.
Table 2.
Summary Results for the Top-Ranking Genotyped SNPs Associated With VCDR in Latino Individuals
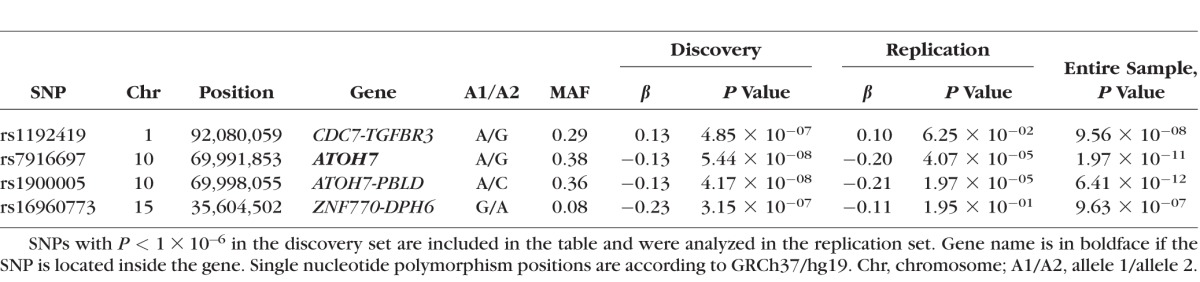
We then analyzed these top SNPs in the replication set using linear mixed-effects models. As displayed in Table 2, the direction of effect for the top four SNPs are consistent with the directions observed in the discovery set. The associations for three SNPs (rs1192419, rs7916697, and rs1900005) were strengthened when the full study sample was analyzed. In addition to rs1900005 remaining significant, rs7916697 also became genome-wide significant (P = 1.97 × 10−11) and rs1192419 became borderline genome-wide significant (P = 9.56 × 10−8) after analyzing the discovery and replication sets together. The SNPs rs7916697 and rs1900005 have previously been reported to be associated with VCDR in European and Asian individuals and the results reported in this study confirm these associations in a Latino population.
Results From Imputed SNPs
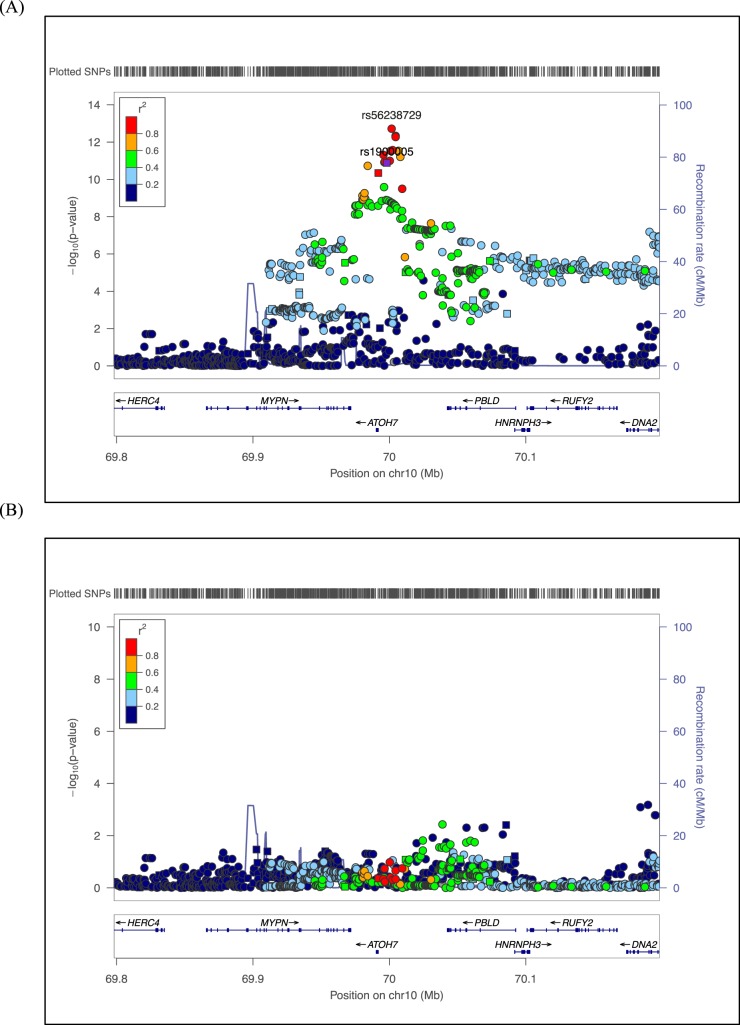
To interrogate additional SNPs not directly genotyped, we performed genotype imputation on the full study sample. After retaining SNPs of high quality (Rsq ≥ 0.80), no additional genomic regions reached GWAS significance. Within the ATOH7-PBLD region, numerous imputed SNPs reached genome-wide significance, including several SNPs more significant than those directly genotyped (Supplementary Table S1). The most significant SNP in this region is rs56238729 (P = 1.22 × 10−13, Rsq = 0.98), located 9.8 kb downstream of rs7916697. This SNP represents a novel association with VCDR. The regional SNP association plot for the ATOH7-PBLD region is presented in Figure 2A. Genotyped SNPs are plotted as squares, and imputed SNPs as circles.
Figure 2.
Regional SNP association plots for the ATOH7-PBLD region. (A) The most significant directly genotyped SNP, rs1900005, using the entire study sample is plotted in purple. Genotyped and imputed SNPs (Rsq ≥ 0.80) are plotted as squares and circles, respectively. Genes are shown below the SNPs and the arrows indicate the strand orientation for each gene. The color coding in the plot represents the level of LD with rs1900005. After imputation, rs56238729 is the most significant SNP in the ATOH7-PBLD region. (B) Regional SNP association plot conditioning on rs56238729, the most significant SNP in the ATOH7-PBLD region.
Conditional Analysis
To determine whether additional SNPs contribute to the VCDR association, we conducted conditional analysis in the ATOH7-PBLD region. As shown in Figure 2B, conditioning on the most significant SNP, rs56238729, by including this SNP as a covariate into linear regression models resulted in all immediate SNP associations to be reduced toward the null with no other SNP remaining significant. These data suggest rs56238729 is the leading SNP of the VCDR association and further nullifies the associations of surrounding SNPs in the ATOH7-PBLD region, including SNPs that have previously been reported.
Analysis of Previously Reported Loci for VCDR
We investigated previously reported VCDR loci identified in populations of European and Asian descent to determine whether these associations are consistent in a Latino population. Table 3 summarizes these results. Of the previously reported SNPs, 25 SNPs exhibited high imputation quality (Rsq ≥ 0.80) and when further analyzed, we observed 32% (8/25) having a P < 0.05 in our Latino population. Furthermore, we observed consistent directions of associations with all previous SNPs, except one (rs301801). To account for multiple testing and to avoid penalties associated with a traditional Bonferroni correction, we used simpleM21–23 to calculate the effective number of independent tests. This method identified 23 independent tests, resulting in a Bonferroni correction significance level of P = 2.17 × 10−3. We were able to replicate five index SNPs in three regions (CDC7-TGFBR3, CDKN2B-CDKN2BAS, and ATOH7-PBLD) after multiple testing correction (shown in boldface in Table 3). To replicate genomic regions associated with VCDR, we extracted the most significant directly genotyped SNP within ±100 kb of the previously reported SNPs. This approach identified an additional five SNPs in five regions surviving the multiple testing correction, replicating the associations between VCDR and the COL8A1, HSF2, RPAP3, TMTC2, and BMP2 regions.
Table 3.
Comparison With Previously Reported SNPs Associated With VCDR in Latino Individuals
Pathway Analysis
To determine whether canonical pathways were enriched with genes associated with VCDR, we performed pathway analysis using IPA. After adjusting for multiple testing, the only pathway significantly associated with VCDR was the “pathogenesis of multiple sclerosis (MS)” pathway (P = 7.41 × 10−3). Of the nine genes comprising this pathway, five genes from our dataset overlapped with these genes, including CCL3, CCL4, CXCL9, CXCL10, and CXCL11.
Discussion
This study represents the first GWAS conducted on VCDR in Latino individuals. We identified two genome-wide significant SNPs, rs1900005 and rs7916697, associated with VCDR, confirming the involvement of the ATOH7-PBLD region. We also identified suggestive associations in the CDC7-TGFBR3 and ZNF770-DPH6 regions. We discovered a novel SNP, rs56238729, in the ATOH7-PBLD region to be significantly associated with VCDR in Latino individuals after genotype imputation from the 1KGP reference panels. Moreover, we were able to replicate genomic regions previously associated with VCDR, including COL8A1, HSF2, RPAP3, TMTC2, and BMP2. Results from our pathway analysis identified one canonical pathway associated with VCDR.
The most significant SNPs in our study reside in the ATOH7-PBLD region. Previous GWAS studies, including Ramdas et al.10 and Springelkamp et al.,11 identified many SNPs in the ATOH7-PBLD region to be associated with VCDR. In addition to VCDR, earlier GWAS have also associated this region with optic disc area,10,29–31 cup area,29,30 and POAG.10 Both rs1900005 and rs7916697, the most and second most significant SNPs in our study, respectively, have previously been associated with VCDR.11 In particular, rs7916697 resides in the 5′UTR region of ATOH7, a single exon gene that plays a role in retinal ganglion cell development.32 Moreover, rs7916697 has been associated with a reduction in optic disc area31 and was identified to have a significant interactive effect with rs1063192 in an Afro-Caribbean population, resulting in a reduction in POAG risk.33 Taken together, our results are consistent with previous studies of these SNPs being strongly associated with glaucoma-related quantitative traits and may have a biological role in the pathogenesis of POAG.
The third and fourth most significant SNPs indicate suggestive associations in the CDC7-TGFBR3 and ZNF770-DPH6 regions, respectively. Similar to the previous region, the CDC7-TGFBR3 region has been reported to be associated with VCDR,11 optic disc area,10,31 and POAG.34 Moreover, expression of both CDC7 and TGFBR3 have been observed in numerous human ocular tissues, most notably the optic disc and optic nerve.34 The SNP rs1192419 has specifically been associated with VCDR11 and an increase in disc area.29 Additionally, common variants within the genomic region on chromosome 15q14, in which ZNF770-DPH6 resides, has previously been associated with refractive error and myopia.35 This study independently confirms the associations of rs1900005 and rs7916697 in the ATOH7-PBLD region with VCDR in a sample of Latino individuals and suggests additional loci in the CDC7-TGFBR3 and ZNF770-DPH6 regions.
Although ophthalmologists routinely assess the VCDR to diagnose and monitor the progression of POAG, the cupping of the optic nerve may not solely be a result of glaucoma and may result from other conditions, such as optic neuritis.36 Our pathway results implicate an association between the pathogenesis of MS and VCDR. Multiple sclerosis is a demyelinating disease of the central nervous system that commonly affects vision. Patients with MS were found to have a higher VCDR compared with healthy controls, suggesting enlarging of the optic cup due to the thinning of the retinal nerve fiber layer may be explained by the predilection of the disease to afflict the optic nerves.37 Moreover, several of the genes included in this pathway code for chemokines that were shown to be at higher concentrations in the aqueous humor of glaucomatous eyes compared with cataract controls.38 Collectively, our pathway results suggest the biological mechanisms influencing VCDR and MS may share common genetic constituents.
This is the first GWAS of VCDR in Latino individuals and several limitations exist. First, the Latino population is historically understudied. As far as we know, our dataset is currently the only Latino genetic dataset with ophthalmic phenotypes. Furthermore, the three-way admixture of Latino individuals makes it even more challenging in genetics research (Supplementary Fig. S2).15,39,40 Using STRUCTURE (available in the public domain at http://pritchardlab.stanford.edu/structure.html),41,42 we estimated that our Latino subjects on average had 3%, 53%, and 44% African, European, and Native American ancestries, respectively (Supplementary Fig. S3). We performed a fixed-effects meta-analysis for the discovery and replication sets on the top genotyped SNPs using METAL43 (available in the public domain at http://csg.sph.umich.edu/abecasis/Metal/download/) and obtained similar results as EMMAX (Supplementary Table S2). These results suggest population stratification and genetic relatedness were properly controlled for in our analysis, despite Latino individuals being a three-way admixed population. However, we emphasize the need for replication in an independent Latino cohort. Second, we did not conduct secondary analyses adjusting for disc area, an ocular parameter known to be correlated with VCDR.44 Unfortunately, at the time of the VCDR data collection, disc area was not collected. Given a previously reported reduction in significance for VCDR-associated variants after adjusting for disc area,11 a similar trend may be observed for our results.
In conclusion, in the first GWAS of VCDR in Latino individuals, we discovered a novel SNP that is significantly associated with VCDR in Latino individuals. In addition, two SNPs reached genome-wide significance, replicating associations in the ATOH7-PBLD region. We were also able to replicate associations with several previously reported genomic regions for VCDR in this population. Our pathway results identified a novel association between pathogenesis of MS and VCDR, suggesting potential shared genetic factors influencing both VCDR and MS. The findings from this study suggest that many genetic factors influencing VCDR are shared among ethnic populations.
Supplementary Material
Acknowledgments
The authors thank the study participants from LALES and the staff who aided in data collection and processing.
Supported in part by National Institutes of Health (NIH; Bethesda, MD, USA) Grants R01EY022651 (XG), U10EY011753 (RV), and P30EY001792 (departmental core grant). The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences Grant UL1TR000124 and National Institute of Diabetes and Digestive and Kidney Diseases Diabetes Research Center Grant DK063491 to the Southern California Diabetes Research Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH.
Disclosure: D.R. Nannini, None; M. Torres, None; Y.-D.I. Chen, None; K.D. Taylor, None; J.I. Rotter, None; R. Varma, None; X. Gao, None
References
- 1. Varma R,, Ying-Lai M,, Francis BA,, et al. Prevalence of open-angle glaucoma and ocular hypertension in Latinos: the Los Angeles Latino Eye Study. Ophthalmology. 2004; 111: 1439–1448. [DOI] [PubMed] [Google Scholar]
- 2. Mitchell P,, Smith W,, Attebo K,, Healey PR. Prevalence of open-angle glaucoma in Australia. The Blue Mountains Eye Study. Ophthalmology. 1996; 103: 1661–1669. [DOI] [PubMed] [Google Scholar]
- 3. Quigley HA,, West SK,, Rodriguez J,, Munoz B,, Klein R,, Snyder R. The prevalence of glaucoma in a population-based study of Hispanic subjects: Proyecto VER. Arch Ophthalmol. 2001; 119: 1819–1826. [DOI] [PubMed] [Google Scholar]
- 4. Leske MC,, Connell AM,, Schachat AP,, Hyman L. The Barbados Eye Study. Prevalence of open angle glaucoma. Arch Ophthalmol. 1994; 112: 821–829. [DOI] [PubMed] [Google Scholar]
- 5. Klein BE,, Klein R,, Sponsel WE,, et al. Prevalence of glaucoma. The Beaver Dam Eye Study. Ophthalmology. 1992; 99: 1499–1504. [DOI] [PubMed] [Google Scholar]
- 6. Knight OJ,, Girkin CA,, Budenz DL,, Durbin MK,, Feuer WJ,, Cirrus OCTNDSG. Effect of race, age, and axial length on optic nerve head parameters and retinal nerve fiber layer thickness measured by Cirrus HD-OCT. Arch Ophthalmol. 2012; 130: 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Varma R,, Tielsch JM,, Quigley HA,, et al. Race-, age-, gender-, and refractive error-related differences in the normal optic disc. Arch Ophthalmol. 1994; 112: 1068–1076. [DOI] [PubMed] [Google Scholar]
- 8. Klein BE,, Klein R,, Lee KE. Heritability of risk factors for primary open-angle glaucoma: the Beaver Dam Eye Study. Invest Ophthalmol Vis Sci. 2004; 45: 59–62. [DOI] [PubMed] [Google Scholar]
- 9. Charlesworth J,, Kramer PL,, Dyer T,, et al. The path to open-angle glaucoma gene discovery: endophenotypic status of intraocular pressure, cup-to-disc ratio, and central corneal thickness. Invest Ophthalmol Vis Sci. 2010; 51: 3509–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ramdas WD,, van Koolwijk LM,, Ikram MK,, et al. A genome-wide association study of optic disc parameters. PLoS Genet. 2010; 6: e1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Springelkamp H,, Hohn R,, Mishra A,, et al. Meta-analysis of genome-wide association studies identifies novel loci that influence cupping and the glaucomatous process. Nat Commun. 2014; 5: 4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Varma R,, Paz SH,, Azen SP,, et al. The Los Angeles Latino Eye Study: design, methods, and baseline data. Ophthalmology. 2004; 111: 1121–1131. [DOI] [PubMed] [Google Scholar]
- 13. Chang CC,, Chow CC,, Tellier LC,, Vattikuti S,, Purcell SM,, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015; 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Delaneau O,, Marchini J; 1000 Genomes Project Consortium. Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat Commun. 2014; 5: 3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao X,, Haritunians T,, Marjoram P,, et al. Genotype imputation for Latinos using the HapMap and 1000 Genomes Project Reference Panels. Front Genet. 2012; 3: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Patterson N,, Price AL,, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006; 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abecasis GR,, Auton A,, et al. ; 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mao X,, Bigham AW,, Mei R,, et al. A genomewide admixture mapping panel for Hispanic/Latino populations. Am J Hum Genet. 2007; 80: 1171–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Devlin B,, Roeder K. Genomic control for association studies. Biometrics. 1999; 55: 997–1004. [DOI] [PubMed] [Google Scholar]
- 20. Kang HM,, Sul JH,, Service SK,, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010; 42: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gao X,, Starmer J,, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008; 32: 361–369. [DOI] [PubMed] [Google Scholar]
- 22. Gao X. Multiple testing corrections for imputed SNPs. Genet Epidemiol. 2011; 35: 154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao X,, Becker LC,, Becker DM,, Starmer JD,, Province MA. Avoiding the high Bonferroni penalty in genome-wide association studies. Genet Epidemiol. 2010; 34: 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 25. Pruim RJ,, Welch RP,, Sanna S,, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010; 26: 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee S,, Emond MJ,, Bamshad MJ,, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012; 91: 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. QIAGEN. Ingenuity Pathway Analysis. Available at: https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/.
- 28. Benjamini Y,, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995; 57: 289–300. [Google Scholar]
- 29. Springelkamp H,, Mishra A,, Hysi PG,, et al. Meta-analysis of genome-wide association studies identifies novel loci associated with optic disc morphology. Genet Epidemiol. 2015; 39: 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Macgregor S,, Hewitt AW,, Hysi PG,, et al. Genome-wide association identifies ATOH7 as a major gene determining human optic disc size. Hum Mol Genet. 2010; 19: 2716–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Khor CC,, Ramdas WD,, Vithana EN,, et al. Genome-wide association studies in Asians confirm the involvement of ATOH7 and TGFBR3, and further identify CARD10 as a novel locus influencing optic disc area. Hum Mol Genet. 2011; 20: 1864–1872. [DOI] [PubMed] [Google Scholar]
- 32. Brown NL,, Dagenais SL,, Chen CM,, Glaser T. Molecular characterization and mapping of ATOH7, a human atonal homolog with a predicted role in retinal ganglion cell development. Mamm Genome. 2002; 13: 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cao D,, Jiao X,, Liu X,, et al. CDKN2B polymorphism is associated with primary open-angle glaucoma (POAG) in the Afro-Caribbean population of Barbados, West Indies. PLoS One. 2012; 7: e39278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li Z,, Allingham RR,, Nakano M,, et al. A common variant near TGFBR3 is associated with primary open angle glaucoma. Hum Mol Genet. 2015; 24: 3880–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Solouki AM,, Verhoeven VJ,, van Duijn CM,, et al. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet. 2010; 42: 897–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rebolleda G,, Noval S,, Contreras I,, Arnalich-Montiel F,, Garcia-Perez JL,, Munoz-Negrete FJ. Optic disc cupping after optic neuritis evaluated with optic coherence tomography. Eye (Lond). 2009; 23: 890–894. [DOI] [PubMed] [Google Scholar]
- 37. Syc SB,, Warner CV,, Saidha S,, et al. Cup to disc ratio by optical coherence tomography is abnormal in multiple sclerosis. J Neurol Sci. 2011; 302: 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chua J,, Vania M,, Cheung CM,, et al. Expression profile of inflammatory cytokines in aqueous from glaucomatous eyes. Mol Vis. 2012; 18: 431–438. [PMC free article] [PubMed] [Google Scholar]
- 39. Gao X,, Gauderman WJ,, Marjoram P,, et al. Native American ancestry is associated with severe diabetic retinopathy in Latinos. Invest Ophthalmol Vis Sci. 2014; 55: 6041–6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nannini D,, Torres M,, Chen YD,, et al. African ancestry is associated with higher intraocular pressure in Latinos. Ophthalmology. 2016; 123: 102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Falush D,, Stephens M,, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003; 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pritchard JK,, Stephens M,, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000; 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Willer CJ,, Li Y,, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010; 26: 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ramdas WD,, Wolfs RC,, Hofman A,, de Jong PT,, Vingerling JR,, Jansonius NM. Heidelberg Retina Tomograph (HRT3) in population-based epidemiology: normative values and criteria for glaucomatous optic neuropathy. Ophthalmic Epidemiol. 2011; 18: 198–210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.