Abstract
The transcriptional regulatory protein Rgg coordinates amino acid catabolism and virulence factor expression in Streptococcus pyogenes. We used a proteomic approach to compare cytoplasmic proteins isolated from S. pyogenes wild-type strain NZ131 (serotype M49) to proteins isolated from an rgg mutant strain during the exponential and stationary phases of growth. Proteins were separated by two-dimensional gel electrophoresis, and 125 protein spots of interest were identified by tandem mass spectrometry. Comparative analysis of proteins isolated from the isogenic strains revealed that growth phase-associated regulation of enzymes involved in the metabolism of arginine (ArcABC), histidine (HutI), and serine (SdhA) was abrogated in the rgg mutant strain, which synthesized the proteins in the exponential phase of growth. In contrast, the enzymes were detected only among wild-type proteins isolated from organisms in the stationary phase of growth. The differences in protein composition were correlated with previously described metabolic changes. In addition, proteins associated with thermal and oxidative stress responses, including ClpE and ClpL, were present in samples isolated from the rgg mutant strain but not in samples isolated from the wild-type strain. The rgg mutant strain was more tolerant to elevated temperature and puromycin than the wild-type strain; however, the mutant was less tolerant to paraquat. We concluded that Rgg is a global regulatory factor that contributes to growth phase-dependent synthesis of proteins associated with secondary metabolism and oxidative and thermal stress responses.
Streptococcus pyogenes is a human-specific pathogen that can cause a variety of diseases, which range in severity from asymptomatic colonization and uncomplicated pharyngitis to invasive diseases such as streptococcal toxic shock syndrome and necrotizing fasciitis. Infection may also induce autoimmune diseases, such as rheumatic fever, acute glomerulonephritis, and various neurological disorders (10). Approximately 20 years ago, the virulence associated with S. pyogenes increased worldwide. Changes in the predominate serotypes in natural populations and in the repertoire of genes encoding exoproteins, including superantigens, are thought to have contributed to the increase in virulence (10, 27, 30). Streptococcal exoproteins, including both cell-associated and soluble proteins, interact directly with host macromolecules to influence host-pathogen interactions.
The regulation of exoprotein synthesis is typically growth phase dependent (22). For example, adhesins and proteins that inhibit complement activation, such as the M protein and C5a peptidase, are primarily expressed in the exponential phase of growth (26). In contrast, hydrolytic enzymes, such as the SPE B protease and nucleases, are primarily expressed during the stationary phase of growth (5, 8, 38). Expression is controlled by a variety of factors, including two-component regulators, such as CsrRS/CovRS (13, 16-18) and FasBCA (21), by so called stand-alone response regulators, such as Mga (10), by RofA-like proteins (1, 34), and by LuxS-mediated quorum sensing (25). In many other species, growth phase-associated regulation also involves alternate sigma factors. However, the chromosome of S. pyogenes does not encode a typical stationary-phase sigma factor, such as RpoS or SigB. Moreover, only a single alternate sigma factor, designated SigX, has been described in S. pyogenes, and this factor has limited activity due to ClpP-dependent degradation (31, 32). Thus, coordinate control of exoprotein synthesis in S. pyogenes probably relies extensively on interactions or cross talk among various regulatory proteins.
Rgg and Rgg-like proteins comprise a family of transcriptional regulatory proteins encoded in the genomes of some low-G+C-content gram-positive bacteria, including Streptococcus, Listeria, Lactobacillus, and Lactococcus; orthologues have not been identified in gram-negative bacteria. The Rgg polypeptide of S. pyogenes consists of 280 amino acids and has a predicted molecular weight of 33,246. Rgg has a helix-turn-helix motif in the amino terminus of the polypeptide, and purified recombinant Rgg proteins from Streptococcus gordonii (40) and S. pyogenes strain HSC5 (known as RopB) (29) bind to DNA in the promoter regions of Rgg-regulated loci. Inactivation of the rgg gene in S. pyogenes strain NZ131 alters the expression of several known or putative virulence factors localized to the cell wall and extracellular milieu. These include the caseinolytic SPE B protease, M protein, C5a peptidase, the SLO cytolysin, and streptokinase, among others (3, 7, 8, 24). Inactivation also affects transcript levels of genes encoding regulatory factors, such as mga, csrRS/covRS, fasBCA, and sagA (7). Finally, an rgg mutant strain ferments arginine and serine in the exponential phase of growth, in contrast to the wild-type strain, which preferentially ferments glucose (6). Thus, Rgg affects both metabolism and virulence factor synthesis in S. pyogenes.
We used a proteomic approach to identify additional Rgg-regulated proteins in S. pyogenes. Cellular proteins isolated from exponential- and stationary-phase cultures of wild-type strain NZ131 were compared to cellular proteins isolated from an isogenic rgg mutant strain. Significant differences in protein composition were discovered between the strains, and proteins of interest were identified by mass spectrometry (MS). The results indicate that Rgg is a global regulatory protein that affects the synthesis or stability of growth phase-regulated proteins associated with amino acid metabolism and oxidative and thermal stress responses.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and protein preparation.
S. pyogenes strain NZ131 (serotype M49) and an isogenic mutant designated NZ131 rgg− were described previously (3, 4). S. pyogenes was grown at 37°C in 50 ml of Todd-Hewitt broth containing 0.2% (wt/vol) yeast extract (THY broth) (Difco Laboratories, Detroit, Mich.) without agitation. After 4, 24, and 48 h of incubation, cultures were centrifuged for 15 min at 13,679 × g at 4°C to collect the bacteria. The bacterial pellets were suspended in 500 μl of lysis buffer, which contained 7 M urea, 2 M thiourea, 4% (wt/vol) 3-[3-(cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 1.0% (vol/vol) immobilized pH gradient (IPG) buffer [Amersham Biosciences (SF) Corp., San Francisco, Calif.], and 75 mM dithiothreitol, and then they were frozen and stored at −80°C. Bacteria were subsequently lysed with a FastPrep protein isolation kit by using a Fastprep instrument (Qbiogene, Carlsbad, Calif.), as described by the manufacturer. A PlusOne 2-D Clean-up kit (Amersham) was used to remove nonprotein contaminants, and protein determinations were done with a PlusOne 2-D Quant kit (Amersham), as described by the manufacturer.
Two-dimensional electrophoresis (2-DE).
Isoelectric focusing was done with an IPGphor isoelectric focusing system by using Immobiline dry strips (11 cm) with a linear pH range of 4 to 7, as described by the manufacturer (Amersham). IPG strips were hydrated with 70 μg of protein in 350 μl of sample buffer for 12 h at 20°C. Isoelectric focusing was done at 500 V for 500 V · h, at 1,000 V for 1,000 V · h, and at 8,000 V for 16,000 V · h at 20°C. The strips were incubated in sodium dodecyl sulfate (SDS) equilibration buffer (50 mM Tris-Cl [pH 8.8], 6 M urea, 30% [vol/vol] glycerol, 2% SDS, bromophenol blue) for 10 min. SDS-polyacrylamide gel electrophoresis separation was performed by using a DALT II six electrophoresis apparatus (Amersham) and 10% acrylamide resolving gels (0.1 by 23.4 by 19.5 cm) containing 1% SDS (Duracryl; Genomic Solutions, Ann Arbor, Mich.). The running buffer consisted of 0.25 M Tris, 1.92 M glycine, and 1% (wt/vol) SDS, and electrophoresis was done for approximately 18 h at 75 V. Proteins were stained with Sypro Ruby (Molecular Probes, Eugene, Oreg.), and digital images were acquired with a charge-coupled device camera. Analysis of the gels, including protein spot detection and quantitation, was done with PDQuest software (Bio-Rad, Hercules, Calif.). Protein spots were quantified by adding the values for the pixels comprising each protein spot. Gels were normalized based on the sum of all protein spots detected in each sample.
Protein identification.
Proteins of interest were excised from the SDS-polyacrylamide gel electrophoresis gels with a robotic spot cutter (Bio-Rad). The gel plugs were washed three times for 15 min in 400 μl of 25 mM NH4HCO3-50% acetonitrile (ACN) (Aldrich, Milwaukee, Wis.), incubated in 100% ACN for 5 min, and lyophilized in a SpeedVac for 30 min. The dried gel plugs were hydrated with 25 mM NH4HCO3 containing 10 μg of sequencing-grade trypsin (Sigma Chemical Co., St. Louis, Mo.) per ml. Following incubation at 37°C for approximately 16 h, the trypsin solution was aspirated into a microcentrifuge tube, and additional peptides were recovered from the gel plugs by extracting them twice with 50% ACN-5% trifluroacetic acid for 1 h each time. The extracted peptides were lyophilized in a SpeedVac and suspended in 20 μl of 1% formic acid-2% ACN. A portion (5 to 10 μl) of the peptide mixture was loaded onto a C18 reverse-phase column (13 cm by 25 μm; LC Packings, Sunnyvale, Calif.). Peptides were eluted directly into a Micromass electrospray ionization quadrupole-orthogonal time of flight mass hybrid spectrometer with a 3 to 40% gradient of ACN-0.1% formic acid over 40 min and a flow rate of approximately 20 nl per min. Spectra were obtained in the positive ion mode, deconvoluted, and analyzed with the MassLynx 4.0 software (Micromass). The Protein Lynx Global Server (version 1.2; Micromass) was used to search databases consisting of S. pyogenes genome sequences and the National Center for Biotechnology Information nonredundant genomic databases. Proteins were identified by matching MS-MS spectra from at least two tryptic peptides or by de novo peptide sequence determination when only one MS-MS match was identified.
Thermal and oxidative stress.
Wild-type and rgg mutant strains were grown in THY broth to the mid-exponential phase. Aliquots of the appropriate size were diluted to an A600 of 0.08 with approximately 10 ml of fresh THY broth. Puromycin and paraquat were added at final concentrations of 30 μg/ml and 50 mM, respectively. Stressed and sham-treated cultures were incubated for 24 h at 37°C in 5% CO2. Subsequently, the number of CFU per millimeter was determined by dilution plating on THY agar plates. The detection limit of the assays was 0.6 CFU per ml.
RESULTS
Protein isolation, resolution, and identification.
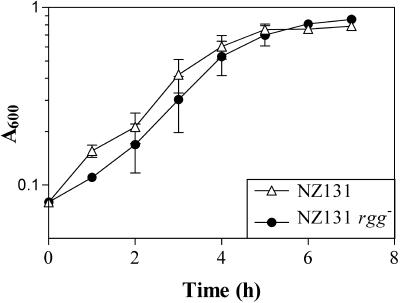
S. pyogenes preferentially ferments glucose when it is available, and depletion of glucose correlates with entry into the stationary phase of growth (5, 6). However, an rgg mutant strain ferments arginine and serine during the exponential phase of growth, even in the presence of glucose (6). To identify changes in protein composition potentially associated with altered metabolism and to characterize the Rgg regulon, we compared proteins isolated from wild-type S. pyogenes after 4, 24, and 48 h of culture to similarly prepared samples isolated from an isogenic rgg mutant strain. The strains grew at similar rates (Fig. 1); however, the growth yield of the mutant strain after 24 and 48 h of culture was greater than that of the wild-type strain (6). The culture conditions were identical to those previously used to measure metabolic precursors and end products (6). Because the previous analysis identified changes in the formation of metabolic end products between 24 and 48 h of growth, it was of interest to determine if the changes were associated with changes in protein composition. Soluble proteins were extracted from whole-cell lysates and separated by 2-DE. The initial results indicated that the majority of soluble proteins had isoelectric points (pI) between 4 and 6 (data not shown). Subsequently, immobilized gradient strips with a separation range of pI 4 to 7 were used. For each strain and time, independent protein isolation was done at least twice, and representative 2-DE gels are shown in Fig. 2 to 4. A comparison of proteins isolated from wild-type and rgg mutant cultures revealed differences in both protein composition and protein abundance (Table 1). A total of 125 protein spots were identified by tandem mass spectrometry, which corresponded to 78 loci. Protein identification was considered to be conclusive if MS-MS spectra from two or more peptides matched a single open reading frame in the databases. For samples in which MS-MS spectra from only a single tryptic peptide matched an open reading frame, the amino acid sequence of the peptide was determined and used to identify the corresponding gene by BLAST searches. In samples prepared from the wild-type strain, enolase (SPy0731; based on serotype M1 genome annotations [14]) was the most abundant protein after 4 and 24 h of culture; glyceraldehye-3-phosphate dehydrogenase (SPy0274) was the most abundant protein after 48 h of culture (see Table S1 at http://www.usd.edu/biomed/biomedfaculty/chaussee/index.shtml). In samples obtained from the rgg mutant strain, enolase (SPy0731) was the most abundant protein at each time (see Table S1 at http://www.usd.edu/biomed/biomedfaculty/chaussee/index.shtml). Few regulatory proteins were identified, probably due to their low levels in the cytoplasm compared to metabolic enzymes.
FIG. 1.
Growth of S. pyogenes NZ131 and NZ131 rgg−. The isogenic strains were cultured in THY broth, and the absorbance at 600 nm was determined at different times. The results are the means and standard errors of the means for six independent experiments.
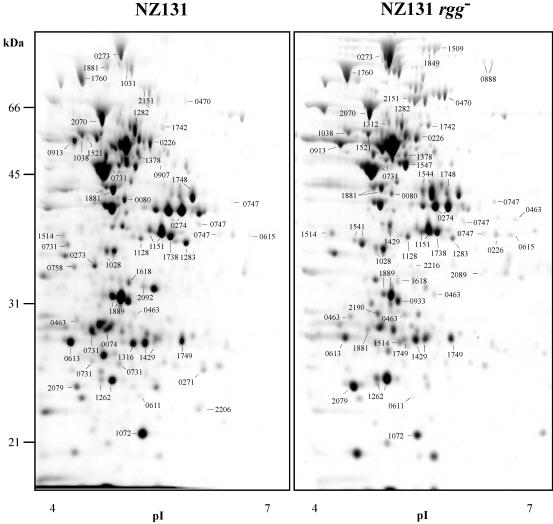
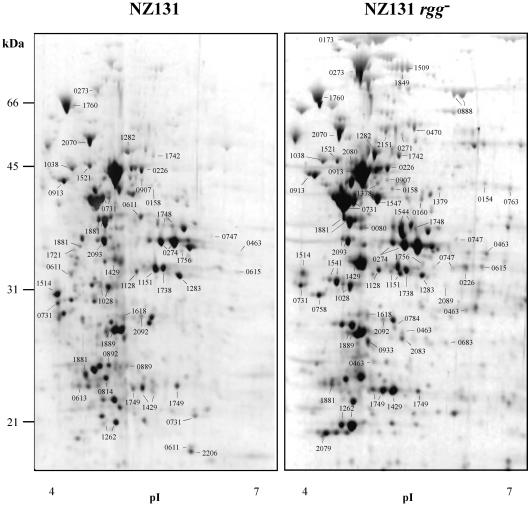
FIG. 2.
Protein compositions of S. pyogenes wild-type and rgg mutant strains after 4 h. Proteins were isolated from the wild-type and rgg mutant strains after 4 h of culture, separated by 2-DE, and stained with Sypro Ruby. The migration positions of molecular weight standards and the orientation of the immobilized pH gradients are indicated. The numbers on the gel correspond to the SPy numbers in the serotype M1 genome sequence annotation (14) and designate proteins identified by tandem mass spectrometry. The original gel size was 25 by 21 by 0.1 cm.
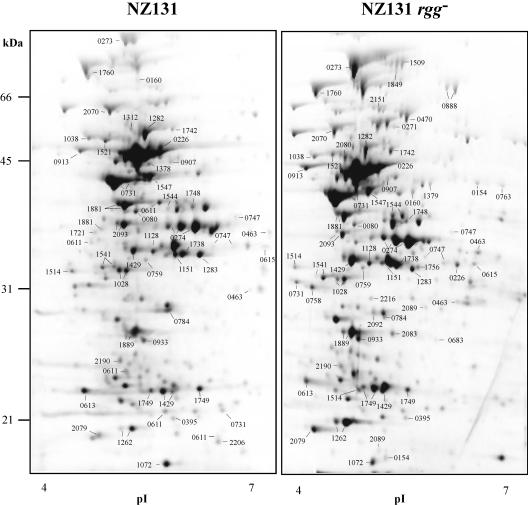
FIG. 4.
Protein compositions of S. pyogenes wild-type and rgg mutant strains after 48 h. Proteins were isolated from wild-type and rgg mutant strains after 48 h of culture, separated by 2-DE, and stained with Sypro Ruby. The migration positions of molecular weight standards and the orientation of the immobilized pH gradients are indicated. The numbers on the gel correspond to the Spy numbers in the serotype M1 genome sequence annotation (14) and designate proteins identified by tandem mass spectrometry. The original gel size was 25 by 21 by 0.1 cm.
TABLE 1.
Number of protein differences between NZ131 and NZ131 rgg− during growtha
| Parameter | No. of spots after:
|
||
|---|---|---|---|
| 4 h | 24 h | 48 h | |
| Interstrain differences in protein spot abundanceb | 38 | 27 | 41 |
| Present only in wild-type samples | 20 | 17 | 22 |
| Present only in rgg mutant samples | 30 | 33 | 36 |
Proteins were isolated from NZ131 and NZ131 rgg− after 4, 24, and 48 h of culture and separated by 2-DE. The results from at least two independent experiments were analyzed with the PDQuest software to identify differences in protein composition and protein abundance between the strains. Protein spots were quantified by adding the pixel values for a spot.
Differences in protein abundance between the isogenic strains at P < 0.05.
Growth phase-associated changes in protein abundance.
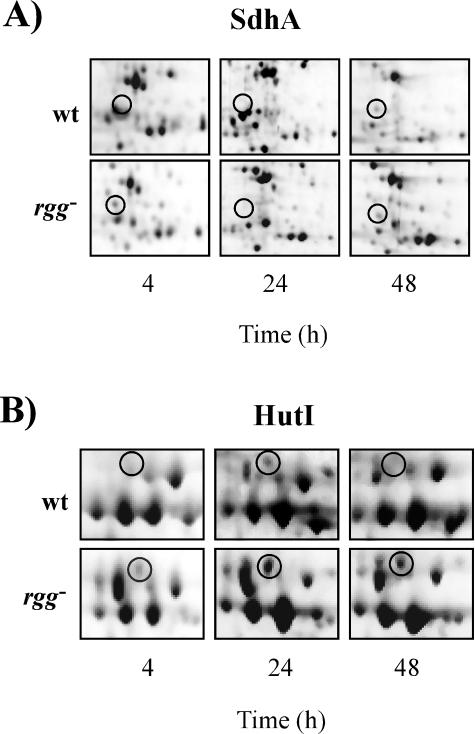
Many of the differences between the strains were differences among growth phase-regulated proteins, as determined by analyzing proteins extracted from wild-type strain NZ131 during the exponential and stationary phases of growth. Serine dehydratase is a heterodimeric enzyme that catalyzes the transformation of l-serine to pyruvate and NH3; pyruvate can subsequently be degraded further to generate ATP. The α-subunit of serine dehydratase (SdhA; SPy2190) was detected in rgg mutant samples isolated after 4 and 24 h of culture but not in the corresponding samples isolated from the wild-type strain. After 48 h of culture, SdhA was detected in samples obtained from both strains (Fig. 5A; Table 2). The results are consistent with the previous finding that the mutant strain ferments serine in the exponential phase of growth (6).
FIG. 5.
Identification of SdhA (A) and HutI (B) in wild-type and rgg mutant samples isolated after 4, 24, and 48 h of culture. The protein positions are indicated by circles. wt, wild-type strain; rgg−, rgg mutant
TABLE 2.
Migration and abundance of selected proteins associated with amino acid metabolism isolated from wild-type and rgg mutant strains
| SPy no.a | Gene product | Observed pI/Mr | Predicted pI/Mr | Mean amt (SEM) after growth forb:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| 4 h
|
24 h
|
48 h
|
|||||||
| Wild type | rgg mutant | Wild type | rgg mutant | Wild type | rgg mutant | ||||
| 1541 | ArcC | 4.7/32.7 | 4.8/33.1 | NDc | 5,434 (170) | 1,786 (397) | 8,673 (381) | 2,624 (1,176) | 3,934 (1,134) |
| 1541 | ArcC | 4.6/33.0 | 4.8/33.1 | ND | 1,048 (1,048) | 199 (199) | 763 (763) | ND | 339 (90) |
| 1544 | ArcB | 5.4/37.9 | 5.3/37.9 | ND | 5,202 (803) | ND | 2,373 (1,114) | 644 (302) | 1,485 (228) |
| 1547 | ArcA/SagP | 5.1/47.7 | 5.0/46.3 | ND | 4,425 (381) | 1,338 (1,338) | 3,606 (299) | 2,173 (539) | 9,542 (6,031) |
| 2081 | HutI | 5.6/41.7 | 5.5/46.8 | ND | 2,726 (2,726) | 779 (482) | 1,769 (1,769) | ND | 4,355 (1,799) |
| 2190 | SdhA | 4.8/27.7 | 4.7/30.0 | ND | 3,073 (83) | ND | 685 (685) | 2,795 (496) | 2,595 (219) |
SPy numbers are based on annotation of the serotype M1 genome sequence.
Protein quantities were determined with the PDQuest software by adding the values for the pixels comprising a protein spot.
ND, not detected.
HutI (SPy2081) hydrolyzes imidazolone 5-propionate to form N-formimino-l-glutamate, which can be further degraded to l-glutamate by HutG (SPy2090). HutI was detected in stationary-phase samples obtained from the wild-type strain but not in exponential-phase samples (Fig. 5B; Table 2). In contrast, HutI was detected in the rgg mutant strain in both the exponential and stationary phases of growth, indicating that rgg inactivation altered growth phase-associated regulation of HutI.
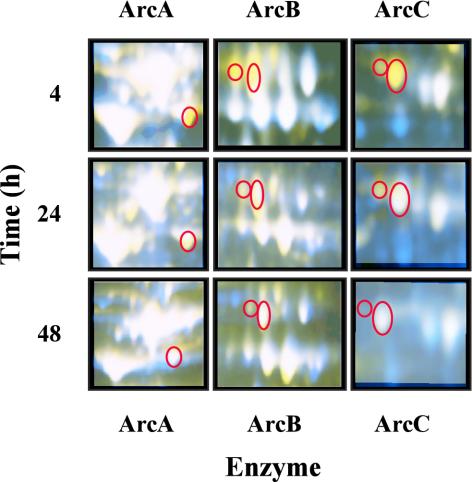
S. pyogenes ferments arginine via the arginine deiminase (ADI) pathway. The first enzyme in the pathway is arginine deiminase (ArcA/SagP; SPy1547), which catalyzes the deamination of arginine to NH3 and citrulline. Ornithine carbamoyltransferase (ArcB; SPy1544) then converts citrulline to CO2, ornithine, and carbamoylphosphate. ATP is generated by carbamate kinase (ArcC; SPy1541)-mediated conversion of carbamoylphosphate to NH3 and CO2. Each enzyme of the ADI pathway (ArcABC) was synthesized by the rgg mutant strain during both the exponential and stationary phases of growth. In contrast, the wild-type strain synthesized the enzymes only during the stationary phase of growth (Fig. 6; Table 2). In addition, the enzymes were more abundant in the rgg mutant strain (Fig. 6; Table 2). Thus, exponential-phase repression of the ADI pathway is dependent on Rgg.
FIG. 6.
Pseudocolored proteins after 4, 24, and 48 h of culture (as indicated on the left). ArcA, ArcB, and ArcC are circled. Yellow indicates that the protein is more abundant in the rgg mutant samples, white indicates that the levels in the two strains are similar, and blue indicates that the protein is more abundant in samples from the wild-type cultures.
In addition, growth phase-associated regulation of a putative virulence factor known as the 67-kDa myosin cross-reactive protein (SPy0470 (14) was abrogated in the rgg mutant strain; the mutant strain synthesized the protein during both the exponential and stationary phases of growth, in contrast to the wild-type strain, which synthesized the protein only in the exponential phase of growth (see Table S1 at http://www.usd.edu/biomed/biomedfaculty/chaussee/index.shtml).
Metabolic enzymes.
With the exception of phosphoglycerate mutase, glycolytic enzymes were more abundant in samples obtained from the wild-type strain (see Table S1 at http://www.usd.edu/biomed/biomedfaculty/chaussee/index.shtml). Among glycolytic enzymes, relatively little change in abundance was observed in wild-type samples isolated during different growth phases. Thus, Rgg significantly affects the synthesis of alternate pathways of catabolism but not the abundance of glycolytic enzymes.
S. pyogenes must synthesize acetyl coenzyme A for biosynthesis from pyruvate via (i) pyruvate-formate lyase, (ii) the pyruvate dehydrogenase complex, and (iii) dismutation of pyruvate via pyruvate oxidase and lactate oxidase, although, pyruvate oxidase does not appear to be encoded in the S. pyogenes genome (M. A. Chaussee, unpublished data). Trace amounts of formate are produced by the rgg mutant strain during the stationary phase of growth but not in the exponential phase (6). In contrast, essentially no formate is produced during growth of the wild-type strain (6). Consistent with this finding, pyruvate-formate lyase (Pfl; SPy1849) was detected in protein samples isolated from the rgg mutant strain after 4 and 24 h of culture; however, the enzyme was not present in samples prepared from wild-type cultures. In contrast, subunits of the pyruvate dehydrogenase complex were more abundant in samples obtained from the wild-type strain. The complex consists of three subunits (E1, E2, and E3) encoded by four adjacent genes (SPy1026, SPy1028, SPy1029, and SPy1031). AcoB (a subunit of E1; SPy1028) and the E3 subunit, dihydrolipoamide dehydrogenase (AcoL; SPy1031), were more abundant in samples obtained from the wild-type strain after 24 and 48 h of culture than in similarly prepared samples from the rgg mutant strain.
Stress-associated proteins.
Inactivation of rgg also affected the abundance of proteins associated with tolerance to oxidative and thermal stresses (Table 3). ClpL (SPy0888) is a class III chaperone that in other streptococci is responsive to thermal stress (9). We identified two ClpL isoforms, which migrated near the predicted position (Table 3), in samples obtained from the rgg mutant strain during both the exponential and stationary phases of growth; cognate proteins were not detected in samples obtained from the wild-type strain. Similarly, ClpE (SPy1509) was detected in samples obtained from the rgg mutant strain during the exponential and stationary phases of growth, but it was not among the proteins isolated from the wild-type strain (Table 3). In addition, ClpP (SPy0395) was more abundant in samples obtained from the mutant strain after 48 h of culture; however, the levels were similar after 4 and 24 h of culture (Table 3). Together, the results show that rgg inactivation is associated with increased synthesis or stability of ClpL, ClpP, and ClpE.
TABLE 3.
Migration and abundance of stress-associated proteins isolated from wild-type and rgg mutant strains
| SPy no.a | Gene product | Observed pI/Mr | Predicted pI/Mr | Mean amt (SEM) after growth forb:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| 4 h
|
24 h
|
48 h
|
|||||||
| Wild type | rgg mutant | Wild type | rgg mutant | Wild type | rgg mutant | ||||
| 0158 | Anion resistance | 5.3/48.5 | 5.3/46.3 | NDc | ND | ND | 478 (243) | ND | 1,338 (125) |
| 0395 | ClpP | 5.7/22.1 | 5.5/21.6 | ND | ND | ND | ND | 411 (102) | 815 (183) |
| 0888 | ClpL | 6.1/75.3 | 5.8/77.3 | ND | 266 (170) | ND | 1,622 (145) | ND | 1,107 (135) |
| 0888 | ClpL | 6.2/74.4 | 5.8/77.3 | ND | 289 (210) | ND | 2,567 (489) | ND | 1,806 (519) |
| 1509 | ClpE | 5.5/82.3 | 5.3/84.7 | ND | 1,494 (153) | ND | 2,783 (650) | ND | ND |
| 1760 | DnaK | 4.5/69.2 | 4.6/64.9 | 8,365 (1,969) | 9,378 (223) | 15,600 (8,845) | 19,695 (1,350) | 10,123 (2,557) | 7,555 (1,951) |
| 2070 | GroEL | 4.7/57.3 | 4.8/57.1 | 16,449 (3,138) | 11,585 (2,186) | 5,975 (2,468) | 12,918 (1,640) | 6,700 (1,408) | 7,196 (421) |
| 2079 | Ahp | 4.6/20.6 | 4.7/20.5 | 2,785 (78) | 10,939 (1,945) | 674 (674) | 3,190 (3,190) | 4,103 (72) | 10,243 (1,526) |
| 2080 | NoxI | 4.8/53.8 | 4.9/54.8 | ND | ND | ND | 751 (751) | ND | 2,795 (2,795) |
| 2216 | HtrA | 5.3/31.0 | 7.6/42.7 | ND | 423 (170) | ND | 651 (27) | ND | 564 (139) |
SPy numbers are based on annotation of the serotype M1 genome sequence.
Protein quantities were determined with the PDQuest software by adding the values for the pixels comprising a protein spot.
ND, not detected.
The oxidoreductases AhpC (SPy2079) and Nox1 (SPy2080) were more abundant in samples from the rgg mutant stain (Table 3). The loci are adjacent to each other in the S. pyogenes genome and are separated by 20 bp, which suggests that they are cotranscribed; AhpC is proximal to the putative promoter. AhpC was 3.9, 4.7, and 2.5 times more abundant in protein samples isolated from the rgg mutant strain than in protein samples isolated from the wild-type strain after 4, 24, and 48 h of growth, respectively. Nox1 was detected in samples from the mutant strain after 24 and 48 h of culture but not in corresponding samples prepared from the wild-type strain (Table 3). Increased expression of oxidoreductases in the rgg mutant strain is functionally consistent with derepression of enzymes associated with the cellular response to oxidative stress.
HtrA (SPy2216), also known as DegP, is a serine protease that degrades misfolded proteins targeted for secretion. In samples isolated after 4 and 24 h of culture, HtrA was detected only in rgg mutant samples (Table 3); however, after 48 h, HtrA was detected in samples from both strains but was more abundant in the rgg mutant strain (Table 3).
Finally, a putative toxic anion resistance protein (SPy0158) was detected in samples from the rgg mutant strain after 24 and 48 h of culture but not in samples from the wild-type strain (Table 3). Although little information is available regarding the function of this protein, tellurite toxicity may result from the oxidation of cellular macromolecules. Taken together, the results show that inactivation of rgg affects the abundance of proteins associated with tolerance to thermal and oxidative stresses.
Responses to thermal and oxidative stresses.
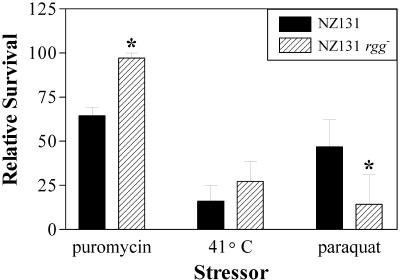
To determine if differences in stress-associated proteins affected the ability to survive thermal and oxidative stresses, the viabilities of the isogenic strains were determined following exposure to puromycin, an elevated temperature, and paraquat. Puromycin is an aminoacyl-tRNA analogue that inhibits protein synthesis and elicits a heat shock-like response in gram-negative and gram-positive bacteria (15, 36, 39). The rgg mutant strain was more tolerant to puromycin and incubation at 41°C than the wild-type strain (Fig. 7). To assess changes in the tolerance to oxidative stress, the strains were similarly exposed to paraquat, which generates hydroxyl ion radicals. In contrast to results obtained with thermal stresses, the rgg mutant strain was more susceptible to paraquat (Fig. 7). Taken together, the results show that the difference in the abundance of stress-related proteins is associated with an altered phenotypic response to thermal and oxidative stresses.
FIG. 7.
Bacterial survival following exposure to thermal stress and oxidative stress. Wild-type and rgg mutant cultures were exposed to puromycin, 41°C, or paraquat for 24 h; sham-treated cultures were used as controls. Viability was determined by dilution plating and is expressed as the CFU per milliliter for cultures exposed to stress divided by the CFU per milliliter for control cultures. The values are the means ± standard errors of the means obtained in five independent experiments. An asterisk indicates that the difference in viability between the strains was significant (P < 0.05).
DISCUSSION
Rgg is a global regulatory protein that coordinates metabolic activity (6) and the expression of virulence factors localized to the cell wall and extracellular environment (3, 7, 8, 24). We compared proteins isolated from wild-type S. pyogenes to proteins isolated from an isogenic rgg mutant strain to identify additional Rgg-regulated proteins. The results show that growth phase-associated regulation of proteins involved in amino acid catabolism and tolerance to thermal and oxidative stresses is influenced, directly or indirectly, by Rgg. In addition, the results provide insight into growth phase-associated changes in protein composition.
The culture conditions used to isolate proteins from the wild-type and rgg mutant strains of S. pyogenes were identical to those previously used to quantitate metabolic precursors and end products during growth (6). A previous analysis showed that the rgg mutant strain ferments arginine and serine in the exponential phase of growth, even in the presence of glucose (6). Consistent with these results, enzymes associated with arginine (ArcABC) and serine (SdhA) catabolism were detected in exponential-phase samples obtained from the rgg mutant strain but not in exponential-phase samples obtained from the wild-type strain, which synthesized ArcABC and SdhA only in the stationary phase of growth. The results demonstrate that growth phase-dependent synthesis or stability of proteins involved in the catabolism of arginine and serine is regulated by an Rgg-dependent mechanism.
In general, the absence of growth phase-associated regulation in the rgg mutant strain is similar to results obtained following the inactivation of alternate sigma factors. For example, SigB coordinates cellular responses to stress in several gram-positive bacteria (2, 23). Moreover, sigB deletion affects the expression of secreted virulence factors, including caseinolytic proteases (20, 28, 33, 41). Similarly, the alternate sigma factor RpoS of gram-negative bacteria controls the expression of virulence-associated exoproteins, including caseinolytic proteases (19), stress-responsive proteins, and enzymes mediating amino acid catabolism in the stationary phase of growth (43). Neither a SigB orthologue nor an RpoS orthologue is encoded in the genome of S. pyogenes (14). Moreover, the S. pyogenes genome encodes only a single alternate sigma factor, designated SigX, which has limited activity due to low levels of transcription and ClpP-dependent proteolysis (31, 32). Thus, in S. pyogenes, genome-wide changes in expression during the transition to the stationary phase of growth are probably mediated by interactions among transcriptional regulatory proteins, including Rgg.
Inactivation of rgg affects the transcript levels of several regulatory genes, including mga, fasBCA, sagA, and csrRS/covRS (7). Inactivation of csrR/covR also affects transcription of rgg (16). Thus, Rgg interacts with other regulons to coordinate gene expression. Based on this interpretation, at least some of the differences in protein abundance associated with rgg inactivation are likely to be due to perturbations of other regulatory circuits. For example, levels of transcripts encoding a variety of stress-associated proteins are affected by CsrR/CovR (16). Similarly, inactivation of csrS/covS affects the pathogen's ability to tolerate pH, thermal, and osmotic stresses (11). In addition, dysregulation of amino acid catabolism could affect the NADH/NAD+ ratio in the cell, which is likely to perturb regulatory circuits that are responsive to oxidative stresses. In this regard, regulatory circuits that control amino acid catabolism are often responsive to stress. For example, ADI expression is responsive to pH stress in Streptococcus mutans and Lactobacillus sakei (37) and to heat shock and low O2 tension in Streptococcus suis (42) and Pseudomonas aeruginosa, respectively (12, 35). Clearly, more information regarding the hierarchal relationships among regulatory circuits is necessary to distinguish between loci directly affected by Rgg and loci affected by perturbations of other regulatory circuits. To maintain infection in the human host, S. pyogenes must adapt to changes in the availability of essential metabolic precursors. Host proteins, peptides, and free amino acids are particularly important since the pathogen is auxotrophic for most amino acids. In addition, amino acid fermentation is likely to contribute to the viability of the pathogen during infection of carbohydrate-limited anatomical sites. Rgg-regulated synthesis of secondary catabolic operons, hydrolytic exoenzymes, and proteins associated with the tolerance to stress may promote dissemination of the pathogen to more favorable ecological niches. For example, a decrease in or cessation of growth due to the depletion of primary metabolic substrates could signal an Rgg-dependent regulatory response that would relieve the repression of alternate catabolic operons and promote the synthesis of hydrolytic exoproteins, such as the SPE B protease and nucleases. The resulting degradation of host tissues could facilitate dissemination and liberate metabolic substrates, including amino acids and peptides. Simultaneous induction of stress-related proteins may protect the pathogen from oxidative stresses associated with inflammation and thermal stress due to fever. Importantly, severe diseases, such as necrotizing fasciitis, are characterized by massive tissue destruction during the invasion of subcutaneous tissues.
In summary, our results show that the global transcriptional regulatory protein Rgg coordinates the expression of virulence factor synthesis and growth phase-associated expression of metabolic enzymes and stress-responsive gene products. These results are an important step towards defining the Rgg regulon and mapping interactions among regulatory circuits that control virulence factor expression in S. pyogenes.
FIG. 3.
Protein compositions of S. pyogenes wild-type and rgg mutant strains after 24 h. Proteins were isolated from wild-type and rgg mutant strains after 24 h of culture, separated by 2-DE, and stained with Sypro Ruby. The migration positions of molecular weight standards and the orientation of the immobilized pH gradients are indicated. The numbers on the gel correspond to the SPy numbers in the serotype M1 genome sequence annotation (14) and designate proteins identified by tandem mass spectrometry. The original gel size was 25 by 21 by 0.1 cm.
Acknowledgments
We thank I. Biswas and K. Weaver for reviewing the manuscript and Emily McDowell for technical assistance.
The work was supported by NIH grant P20 RR16479-02 from the National Center for Research Resources and by Public Health Service grant RO1401507 from the National Institutes of Health to M.S.C.
REFERENCES
- 1.Beckert, S., B. Kreikemeyer, and A. Podbielski. 2001. Group A streptococcal rofA gene is involved in the control of several virulence genes and eukaryotic cell attachment and internalization. Infect. Immun. 69:534-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan, P. F., S. J. Foster, E. Ingham, and M. O. Clements. 1998. The Staphylococcus aureus alternative sigma factor σB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J. Bacteriol. 180:6082-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaussee, M. S., D. Ajdic, and J. J. Ferretti. 1999. The rgg gene of Streptococcus pyogenes NZ131 positively influences extracellular SPE B production. Infect. Immun. 67:1715-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaussee, M. S., D. Gerlach, C.-E. Yu, and J. J. Ferretti. 1993. Inactivation of the streptococcal erythrogenic toxin B gene (speB) in Streptococcus pyogenes. Infect. Immun. 61:3719-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaussee, M. S., E. R. Phillips, and J. J. Ferretti. 1997. Temporal production of streptococcal erythrogenic toxin B (streptococcal cysteine proteinase) in response to nutrient depletion. Infect. Immun. 65:1956-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaussee, M. S., G. A. Somerville, L. Reitzer, and J. M. Musser. 2003. Rgg coordinates virulence factor synthesis and metabolism in Streptococcus pyogenes. J. Bacteriol. 185:6016-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaussee, M. S., G. L. Sylva, D. E. Sturdevant, L. M. Smoot, M. R. Graham, R. O. Watson, and J. M. Musser. 2002. Rgg influences the expression of multiple regulatory loci to coregulate virulence factor expression in Streptococcus pyogenes. Infect. Immun. 70:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaussee, M. S., R. O. Watson, J. C. Smoot, and J. M. Musser. 2001. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. Infect. Immun. 69:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, I. H., J. H. Shim, S. W. Kim, S. N. Kim, S. N. Pyo, and D. K. Rhee. 1999. Limited stress response in Streptococcus pneumoniae. Microbiol. Immunol. 43:807-812. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalton, T. L., and J. R. Scott. 2004. CovS inactivates CovR and is required for growth under conditions of general stress in Streptococcus pyogenes. J. Bacteriol. 186:3928-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Angelis, M., L. Mariotti, J. Rossi, M. Servili, P. F. Fox, G. Rollan, and M. Gobbetti. 2002. Arginine catabolism by sourdough lactic acid bacteria: purification and characterization of the arginine deiminase pathway enzymes from Lactobacillus sanfranciscensis CB1. Appl. Environ. Microbiol. 68:6193-6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Federle, M. J., K. S. McIver, and J. R. Scott. 1999. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J. Bacteriol. 181:3649-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frees, D., and H. Ingmer. 1999. ClpP participates in the degradation of misfolded protein in Lactococcus lactis. Mol. Microbiol. 31:79-87. [DOI] [PubMed] [Google Scholar]
- 16.Graham, M. R., L. M. Smoot, C. A. Migliaccio, K. Virtaneva, D. E. Sturdevant, S. F. Porcella, M. J. Federle, G. J. Adams, J. R. Scott, and J. M. Musser. 2002. Virulence control in group A streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. USA 99:13855-13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heath, A., V. J. DiRita, N. L. Barg, and N. C. Engleberg. 1999. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect. Immun. 67:5298-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heath, A., A. Miller, V. J. DiRita, and C. N. Engleberg. 2001. Identification of a major, CsrRS-regulated secreted protein of group A streptococcus. Microb. Pathog. 31:81-89. [DOI] [PubMed] [Google Scholar]
- 19.Hulsmann, A., T. M. Rosche, I.-S. Kong, H. M. Hassan, D. M. Beam, and J. D. Oliver. 2003. RpoS-dependent stress response and exoenzyme production in Vibrio vulnificus. Appl. Environ. Microbiol. 69:6114-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreikemeyer, B., M. D. Boyle, B. A. Buttaro, M. Heinemann, and A. Podbielski. 2001. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol. Microbiol. 39:392-406. [DOI] [PubMed] [Google Scholar]
- 22.Kreikemeyer, B., K. S. McIver, and A. Podbielski. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224-232. [DOI] [PubMed] [Google Scholar]
- 23.Kullik, I. I., and P. Giachino. 1997. The alternative sigma factor sigmaB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch. Microbiol. 167:151-159. [DOI] [PubMed] [Google Scholar]
- 24.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyon, W. R., J. C. Madden, J. C. Levin, J. L. Stein, and M. G. Caparon. 2001. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 42:145-157. [DOI] [PubMed] [Google Scholar]
- 26.McIver, K. S., and J. R. Scott. 1997. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J. Bacteriol. 179:5178-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musser, J. M., and R. M. Krause. 1998. The revival of group A streptococcal diseases, with a commentary on staphylococcal toxic shock syndrome, p. 185-218. In R. M. Krause (ed.), Emerging infections. Academic Press, New York, N.Y.
- 28.Nadon, C. A., B. M. Bowen, M. Wiedmann, and K. J. Boor. 2002. Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70:3948-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neely, M. N., W. R. Lyon, D. L. Runft, and M. Caparon. 2003. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J. Bacteriol. 185:5166-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson, K., P. M. Schlievert, R. K. Selander, and J. M. Musser. 1991. Characterization and clonal distribution of four alleles of the speA gene encoding pyrogenic exotoxin A (scarlet fever toxin) in Streptococcus pyogenes. J. Exp. Med. 174:1271-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Opdyke, J. A., J. R. Scott, and C. P. Moran, Jr. 2003. Expression of the secondary sigma factor σX in Streptococcus pyogenes is restricted at two levels. J. Bacteriol. 185:4291-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Opdyke, J. A., J. R. Scott, and C. P. Moran, Jr. 2001. A secondary RNA polymerase sigma factor from Streptococcus pyogenes. Mol. Microbiol. 42:495-502. [DOI] [PubMed] [Google Scholar]
- 33.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Podbielski, A., M. Woischnik, B. A. Leonard, and K. H. Schmidt. 1999. Characterization of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 31:1051-1064. [DOI] [PubMed] [Google Scholar]
- 35.Sriskandan, S., D. Moyes, L. K. Buttery, J. Wilkinson, T. J. Evans, J. Polak, and J. Cohen. 1997. The role of nitric oxide in experimental murine sepsis due to pyrogenic exotoxin A-producing Streptococcus pyogenes. Infect. Immun. 65:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steiner, K., and H. Malke. 2001. relA-independent amino acid starvation response network of Streptococcus pyogenes. J. Bacteriol. 183:7354-7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi, N., and T. Yamada. 1999. Acid-induced acid tolerance and acidogenicity of non-mutans streptococci. Oral Microbiol. Immunol. 14:43-48. [DOI] [PubMed] [Google Scholar]
- 38.Unnikrishnan, M., J. Cohen, and S. Sriskandan. 1999. Growth-phase-dependent expression of virulence factors in an M1T1 clinical isolate of Streptococcus pyogenes. Infect. Immun. 67:5495-5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.VanBogelen, R. A., and F. C. Neidhardt. 1990. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:5589-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vickerman, M. M., M. Wang, and L. J. Baker. 2003. An amino acid change near the carboxyl terminus of the Streptococcus gordonii regulatory protein Rgg affects its abilities to bind DNA and influence expression of the glucosyltransferase gene gtfG. Microbiology 149:399-406. [DOI] [PubMed] [Google Scholar]
- 41.Wiedmann, M., T. J. Arvik, R. J. Hurley, and K. J. Boor. 1998. General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180:3650-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winterhoff, N., R. Goethe, P. Gruening, M. Rohde, H. Kalisz, H. E. Smith, and P. Valentin-Weigand. 2002. Identification and characterization of two temperature-induced surface-associated proteins of Streptococcus suis with high homologies to members of the arginine deiminase system of Streptococcus pyogenes. J. Bacteriol. 184:6768-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zinser, E. R., and R. Kolter. 1999. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J. Bacteriol. 181:5800-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]