Abstract
The ygfZ gene product of Escherichia coli represents a large protein family conserved in bacteria to eukaryotes. The members of this family are uncharacterized proteins with marginal sequence similarity to the T-protein (aminomethyltransferase) of the glycine cleavage system. To assist with the functional assignment of the YgfZ family, the crystal structure of the E. coli protein was determined by multiwavelength anomalous diffraction. The protein molecule has a three-domain architecture with a central hydrophobic channel. The structure is very similar to that of bacterial dimethylglycine oxidase, an enzyme of the glycine betaine pathway and a homolog of the T-protein. Based on structural superposition, a folate-binding site was identified in the central channel of YgfZ, and the ability of YgfZ to bind folate derivatives was confirmed experimentally. However, in contrast to dimethylglycine oxidase and T-protein, the YgfZ family lacks amino acid conservation at the folate site, which implies that YgfZ is not an aminomethyltransferase but is likely a folate-dependent regulatory protein involved in one-carbon metabolism.
One-carbon (C1) metabolism mediated by folate coenzymes is essential for major cellular processes, including biosynthesis of nucleotides, vitamins, and amino acids (17). C1 units are derived from catabolism of three donor molecules, serine, glycine, and formate, and are activated by attachment to tetrahydrofolate (THF). In most organisms, serine is the principal contributor to the pool of 5,10-CH2-THF through the action of pyridoxal-5′-phosphate-dependent serine hydroxymethyltransferase (SHMT) (32). Alternatively, C1 units may be obtained from glycine via the glycine cleavage system (GCS) (12) or from formate via 10-HCO-THF synthetase (C1-THF synthase in eukaryotes) (30). The GCS is a multienzyme complex composed of the P-, H-, T- and L-proteins. P-protein (glycine dehydrogenase) catalyzes the pyridoxal-5′-phosphate-dependent decarboxylation of glycine and transfer of the remaining aminomethyl moiety to the lipoyl prosthetic group of H-protein. T-protein (aminomethyltransferase) catalyzes the release of ammonia from the intermediate attached to H-protein and transfer of the methyl group to THF (27). L-protein (lipoamide dehydrogenase) catalyzes the NADH-dependent reoxidation of the dihydrolipoyl residue of H-protein. The GCS is believed to balance the cell requirements for glycine, C1 units, and nitrogen (31). The pool of 5,10-CH2-THF produced by SHMT and GCS and the pool of 10-HCO-THF produced by C1-THF synthase are interconverted by the latter multifunctional enzyme (18).
The regulation of this dynamic metabolic system is complex and not completely understood. Since the GCS provides C1 units for the synthesis of amino acids like serine, methionine, and formylmethionine, it is subject to regulation by the general amino acid control system (34). In Escherichia coli, the components of the GCS are encoded by the genes of the gcvTHP operon, which is regulated by the global regulatory proteins Lrp, PurR, and cAMP receptor protein and by the gcv-specific transcriptional regulators GcvA and GcvR (28, 33). Together, these proteins modulate transcription of the operon in response to the levels of glycine and purines. The repressor capability of GcvR is realized through complex formation with GcvA, whereas the other proteins bind directly to the gcv control region (7).
Closely related to the GCS is the glycine betaine pathway that involves three demethylation steps catalyzed consecutively by betaine-homocysteine methyltransferase, dimethylglycine dehydrogenase (oxidase) (DMGO), and sarcosine dehydrogenase (oxidase) to produce glycine and C1 units (16). Oxidation of the methyl groups of betaine by these dehydrogenases generates formaldehyde, which is efficiently channeled into the folate-C1 pathways by the formation of 5,10-CH2-THF in the presence of THF. In some bacteria, such as Arthrobacter and Corynebacterium spp., the genes encoding SHMT (glyA), heterotetrameric sarcosine dehydrogenase (oxidase) (soxBDAG), 10-CHO-THF hydrolase (purU), DMGO (dmg), formiminotransferase/cyclodeaminase (fic), and 5,10-CH2-THF dehydrogenase (folD) form an operon and are subject to regulation by unidentified proteins (20). At the same time some of these enzymes may also play a regulatory role. For instance, 10-CHO-THF hydrolase functions to balance the pools of 10-CHO-THF and THF in response to the changes in the methionine/glycine ratio (24).
Identification of the regulators of C1 metabolism is important for understanding various cellular processes and ultimately may have implications for drug discovery. A number of human diseases and disorders, including neural tube defects, epithelial cancers, and cardiovascular disease, are associated with folate deficiency and disturbances in folate-mediated reactions (2, 15).
The protein described in this report, which is proposed to have a role in C1 metabolism, has emerged as a target in a structural genomics project aimed at the functional assignment of proteins through three-dimensional structure determination (5). The ygfZ gene of E. coli encodes an uncharacterized protein with a molecular mass of 36 kDa. Homologs of this protein are present in most bacteria and eukaryotes, but not in archaea. Based on marginal (15%) sequence identity to the T-protein of the GCS, the YgfZ protein has been annotated as a putative aminomethyltransferase. The crystal structure of YgfZ revealed a three-domain ring-like protein molecule with a deep hydrophobic cavity and no apparent active site. When the structure of DMGO from Arthrobacter globiformis (14) became available, it appeared that the two structures are strikingly similar. DMGO is an enzyme of the betaine pathway and a homolog of the T-protein (26% sequence identity). Based on structural superposition, a folate-binding site was identified in the central cavity of YgfZ, and the ability of YgfZ to bind folate derivatives was confirmed experimentally. However, in contrast to DMGO and T-protein, the YgfZ family lacks amino acid conservation at the folate site, which implies that YgfZ is probably not an aminomethyltransferase and may be not an enzyme at all but rather is a folate-dependent regulatory protein involved in C1 metabolism.
MATERIALS AND METHODS
Cloning, expression, and purification.
The ygfZ gene was PCR amplified from E. coli ATCC 700926 genomic DNA and subcloned into a pET100/D-TOPO plasmid by using the TOPO cloning technology (Invitrogen). The construct contained a sequence coding for a peptide with the thrombin-cleavable His tag attached to the N-terminal methionine. The selenomethionine (SeMet)-containing protein was produced in E. coli strain B834(DE3) that was transformed with the plasmid. Cells were grown on minimal medium containing 50 mg of seleno-l-methionine per ml and 100 mg of ampicillin per ml to an A600 of 0.5 and induced with 1 mM isopropyl-β-d-thiogalactoside for 3 h at 37°C. The protein was purified by column chromatography in four steps. After the initial Ni-nitrilotriacetic acid column, the His tag was cleaved with thrombin. The material was then applied to a benzamidine column (to remove thrombin) and again to a Ni-nitrilotriacetic acid column (to remove the nonhydrolyzed His-tagged protein). The final step included gel filtration on a Sephacryl S100 HR column.
Crystallization and structure determination.
YgfZ crystals were grown at room temperature by vapor diffusion in hanging drops by mixing 1.5 μl of a 15-mg/ml protein solution with 1.5 μl of a reservoir solution containing 30% saturated ammonium sulfate and 0.1 M sodium acetate (pH 4.5). The crystals reached the maximum size (0.2 mm) in few days. YgfZ crystals belong to space group P3121 with the unit cell parameters a = b = 151.0 Å and c = 68.0 Å. There is one protein molecule in the asymmetric unit, which gives a specific volume of 6.2 Å3/Da. Crystals were frozen in the mother liquor supplemented with 50% saturated lithium formate. X-ray data were collected at 100 K at the IMCA-CAT beamline 17-BM at the Advanced Photon Source (Argonne, Ill.) equipped with a Mar CCD (165-mm) detector, and they were processed with HKL2000 (29).
The structure was solved by the multiwavelength anomalous diffraction method by using one SeMet protein crystal. X-ray data at 2.8-Å resolution were measured at four wavelengths (Table 1). Six selenium sites were located by the shake-and-bake method (38) and were used for phasing with MLPHARE/DM (3). The atomic model was built by using O (11) and was refined with REFMAC (22) by using the low-energy data set (Table 1). The final model contains 325 amino acid residues. The remains of the His tag (three residues at the N terminus) and the C-terminal Glu326 are not visible in the electron density map. Pro157 is in the cis conformation. The atomic B-factors are high (64 Å2 on average), and the value corresponds to the mean B-factor calculated from the Wilson plot (Table 1). The polypeptide tracing is unambiguous, but the positional errors for many side chains may be quite high due to the low resolution of the data.
TABLE 1.
X-ray data and refinement statistics
| Data set | Low-E | Peak | Inflection | High-E |
|---|---|---|---|---|
| Wavelength (Å) | 1.000 | 0.9793 | 0.9797 | 0.9724 |
| Resolution (Å) | 2.8 | 2.8 | 2.8 | 2.8 |
| No. of unique reflections | 21,998 (2,412)a | 42,083b | 41,425b | 42,346b |
| Completeness (%) | 98.5 (97.1) | 98.8 | 96.6 | 99.4 |
| Redundancy | 4.9 (4.1) | 3.0 | 5.3 | 5.0 |
| Rsym (ΣI-<I>|)/ΣI) | 0.055 (0.44) | 0.058 | 0.083 | 0.065 |
| <I/σ> | 22.4 (2.1) | 13.6 | 17.3 | 20.9 |
| Rcryst (Σ‖Fo|-|Fc‖)/(Σ|Fo|) | 0.193 | |||
| Rfree (3% data) | 0.252 | |||
| No. of protein atoms | 2,536 | |||
| No. of water molecules | 215 | |||
| RMSD in bonds (Å) | 0.013 | |||
| RMSD in angles (°) | 1.6 | |||
| Mean B-factor, model (Å2) | 64.4 | |||
| Mean B-factor, Wilson plot (Å2) | 71.3 |
The statistics for the high-resolution shell (2.9 to 2.8 Å) are in parentheses.
Anomalous pairs not merged.
The solvent-accessible surface area was calculated with the program AREAIMOL from the CCP4 package (3) as the area of the locus of the center of the probe. This should be distinguished from the contact area, which is the area of the atom surface accessible to the probe. The probe radius was 1.4 Å.
Fluorescence measurements.
Fluorescence intensity was measured at 25°C by using a Fluoromax-2 spectrofluorometer (Jobin Yvon) in a 200-μl quartz cell. Tryptophan residues in the protein were excited at 280 nm, and emission was monitored at 335 nm with a slit width of 5 nm. The reaction mixture contained 1 ml of a 100 nM protein solution in 20 mM Tris-HCl (pH 7.5)-100 mM NaCl. It was titrated with increasing amounts of each ligand by sequential addition of 1 to 2 μl of stock solutions, so that the total dilution of the initial solution did not exceed 2.5%. Because both folic acid and THF display significant fluorescence with the maximum emission spectra at 355 nm, their final concentrations were limited to 3 and 0.3 μM, respectively, to maintain the ligand contribution to the measured fluorescence below 25%. All readings were corrected for background emission of the buffer and free ligand solutions (folic acid in water and THF in 0.1% mercaptoethanol [to prevent oxidation]).
The dissociation constant (Kd) was determined by using the following equation: ΔF = ΔFmax[L]/([L] + [Kd]), where [L] is the free ligand concentration. The fraction of the protein containing a bound ligand molecule (ΔF/ΔFmax) was defined as the fraction of the total quenchable tryptophan fluorescence that was quenched at each point of the titration. ΔFmax was estimated by extrapolation at high [L] values of a plot of 1/ΔF versus 1/[L].
Accession code of the structure.
The atomic coordinates and structure factors of the YgfZ protein have been deposited in the Protein Data Bank under accession code 1NRK.
RESULTS
Amino acid conservation pattern.
The sequence similarity search with YgfZ from E. coli performed with PSI-BLAST (1) resulted in a large number of statistically significant hits with Z scores below 10−3. Alignment of these sequences by using CLUSTALW (35) revealed a conserved pattern, K-G-C-Y/F-X-G-Q-E spanning residues 226 to 233 of the E. coli protein. This pattern was used at the PROSITE server (6) to pick up additional members of the protein family. This search resulted in more than 100 proteins considered to constitute a family on the grounds of the overall sequence similarity and the conservation pattern. The PROSITE search produced no false positives. Therefore, the sequence motif K-G-C-Y/F-X-G-Q-E can serve as a fingerprint of the protein family.
The YgfZ homologs are widely represented in bacteria and in eukaryotes, including fungi, plants, insects, and mammals, but not in archaea. Besides the fingerprint motif, very few residues are conserved throughout the entire family (Fig. 1). These residues include the basic amino acids Arg and Lys in positions 68, 237, and 245 that must have functional importance and several glycine residues that typically play a structural role.
FIG. 1.
Structure-based sequence alignment of the protein families represented by YgfZ from E. coli (YGFZ), glycine cleavage system T-protein from E. coli (GCST), and dimethylglycine oxidase from A. globiformis (DMGO). Residues strictly conserved in each protein family are in black boxes, and residues conserved in 90% family members are in open boxes. The catalytic Asp in T-protein and DMGO is indicated by a star, and the folate-anchoring Glu is indicated by a triangle. The alignment was prepared with ESPRIPT (8).
The T-proteins of the GCS were picked up in the second round of the PSI-BLAST search. The sequence similarity between YgfZ and T-protein is quite low; e.g., the E. coli proteins have only 15% identical residues. Moreover, with the exception of glycine residues, none of the residues conserved among T-proteins is conserved in the YgfZ family, nor does the YgfZ conservation pattern match the T-protein sequence. This casts serious doubt on the putative function of YgfZ as an aminomethyltransferase, at least regarding the T-protein chemistry. We consider YgfZ a separate protein family different from the T-protein of the GCS and its homologs, such as DMGO.
Description of the structure.
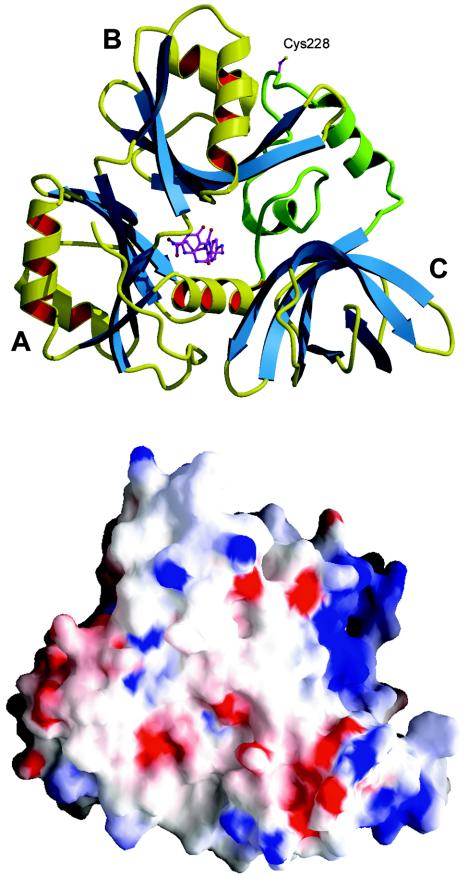
The crystal structure of YgfZ was determined by the multiwavelength anomalous diffraction method by using a selenomethionine protein. The protein molecule has a globular shape, and the dimensions are 60 by 50 by 30 Å. The molecule consists of three domains arranged in a ring-like structure with a narrow central channel (Fig. 2). Domain A includes residues 1 to 26 and 114 to 196. Domain B includes residues 27 to 113 and may be considered an insertion in domain A. Both of these domains have a ferredoxin-like fold (i.e., an α+β sandwich with an antiparallel β-sheet and two α-helices on one side of the sheet). In a deviation from a canonical four-strand β-sheet, domain B has an additional β-strand, and domain A has two additional β-strands. The ferredoxin fold is common in many functionally unrelated and nonhomologous proteins, such as metallochaperones, protease propeptides, and RNA-binding domains of various enzymes (23). Despite the topological similarity of domains A and B, no sequence homology between them was detected.
FIG. 2.
Ribbon diagram (upper diagram) and electrostatic surface potential diagram (lower diagram) (blue, positive; red, negative) of the YgfZ molecule. The linker between domains A and C is shown in green. Cys228 indicates the location of the fingerprint sequence K226GCYTGQE233. The THF molecule (not observed in the crystal structure) is the folate-binding site. The diagrams were produced with MOLSCRIPT (13), RASTER3D (19), and GRASP (25).
Domain C comprises 80 C-terminal residues. It has the topology of a six-strand antiparallel β-barrel with a Greek key connection pattern. The topologically identical domains are present in the C-terminal regions of elongation factors Tu and eEF-1α and in the gamma subunit of the initiation factor eIF2. These domains are involved in binding of the TΨC loop of tRNA (26).
A 50-residue segment comprising residues 197 to 245 cannot be attributed to either domain. It is sandwiched between domains B and C and, with the exception of two short α-helices, lacks secondary structure elements.
The interfaces between the domains are extensive. The surface areas of domains A, B, and C buried in these interfaces are 1,400, 2,000, and 1,000 Å2, respectively. When these areas are compared to the total surface area of each domain (about 6,000 Å2), the values indicate the tight domain packing in the protein molecule. On the other hand, the three-domain structure provides sufficient conformational flexibility that may be utilized for, e.g., altering the shape of the central channel. Domain C is probably the least restricted part of the structure; it has the smallest interface and a significantly higher average B-factor (96 Å2) than domains A (45 Å2) and B (57 Å2). High B-factor values reflect loose packing of the protein in crystals that contain 80% solvent.
Structural similarity to DMGO.
The structure of YgfZ is strikingly similar to that of DMGO from A. globiformis (14), an enzyme of the betaine catabolism pathway. DMGO is formed by fusion of two subunits that catalyze separate half-reactions, the flavin adenine dinucleotide-dependent amine oxidation of dimethylglycine and the THF-dependent conversion of the iminium intermediate to sarcosine. The two active sites are connected by an internal cavity that enables sequestration of the reactive iminium intermediate and avoids formation of toxic formaldehyde. YgfZ matches the THF-binding subunit of DMGO, which resides in the C-terminal region of the sequence.
A DALI (9) similarity search yielded DMGO as a top hit, with a Z score of 23.6, which corresponds to the root-mean-square deviation (RMSD) of 3.0 Å for 296 common Cα atoms. The overall superposition of YgfZ and DMGO shows that the three domains composing the protein molecule have slightly different relative orientations in these structures. The domains can be individually superimposed with RMSD of 2.2, 1.2, and 1.6 Å for domains A, B, and C, respectively. The somewhat higher value for domain A is due to the shift of the helical segments.
Folate-binding site.
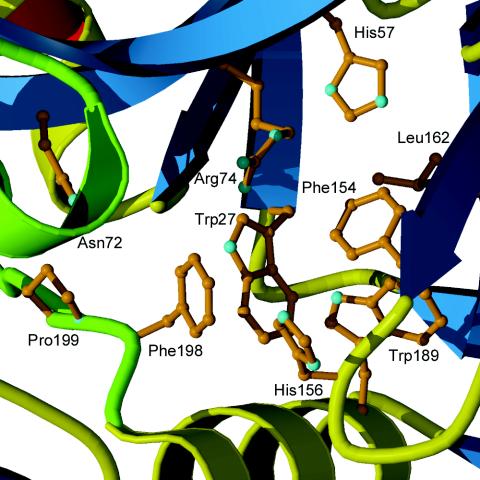
The crystal complexes of DMGO with folic acid and 5-CHO-THF (folinic acid) indicate that the binding site of the cofactor is in the central channel of the ring structure (14). Folate is bound in a kinked conformation, with the pterin group deeply imbedded in the protein (Fig. 2). The binding pocket is predominantly hydrophobic, and the interacting residues come from domains A and B and a central portion of the linker. The carboxyl group of Glu658 forms a bidentate hydrogen bond with the pteroyl amino groups N2 and N3. From a comparison of the DMGO structures with different ligands it was noted that binding of THF is accompanied by conformational changes in the enzyme, most of which result in improved hydrophobic packing with the folate ring system (14).
Based on the structural similarity to DMGO, it was possible to locate the putative folate-binding site in YgfZ and to model the cofactor binding. Overall, the binding pocket is hydrophobic and has a large fraction of aromatic amino acids, which corresponds to the nature of interactions observed in DMGO (Fig. 3). Leu162 occupies a position equivalent to Glu658 in DMGO, implying that the hydrogen bonds to N2 and N3 of pterin are not preserved in YgfZ. Although some adjustments of the surrounding residues are needed to accommodate a folate molecule, the modeling study suggests that YgfZ is capable of binding folate derivatives.
FIG. 3.
Central channel entrance in YgfZ. Leu162 occupies the position of the folate anchor Glu658 in DMGO, and Asn72 is present in place of the catalytic Asp552 residue.
Binding of folic acid and THF by YgfZ.
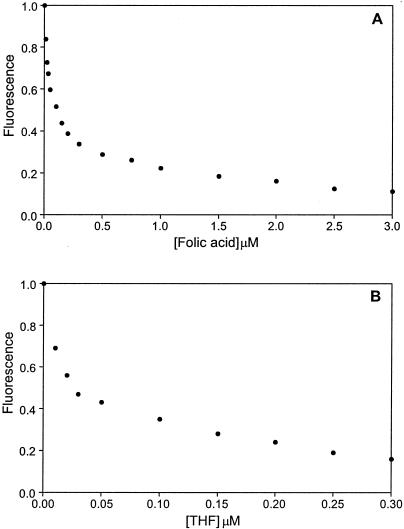
Binding of folic acid and THF by YgfZ was assessed by measuring the fluorescence of tryptophan residues. There are seven tryptophan residues in the YgfZ molecule. Two of these residues, Trp27 and Trp189, are located in the central cavity and may directly interact with the ligand according to the model. The observed dramatic quenching of the Trp fluorescence upon addition of THF or folic acid to the protein sample (Fig. 4) confirmed the ability of YgfZ to bind these ligands. Titration of the ligands at a fixed protein concentration (100 nM) produced a hyperbolic curve that gave dissociation constants of about 100 nM for folic acid and 20 nM for THF.
FIG. 4.
Quenching of tryptophan fluorescence upon addition of folic acid (A) and THF (B). The protein concentration was 100 nM. The graphs were prepared with PSI-Plot (Poly Software).
DISCUSSION
Asp552 was implicated to serve as a general base in the THF methylation reaction in DMGO (14). In the crystal structure this amino acid forms a hydrogen bond to the N10 amino group of folinic acid. An acidic residue is conserved in this position in all T-protein-like enzymes, which ensures their catalytic ability. In the absence of a conserved amino acid in either this or any other position in the central cavity, it is hard to see how YgfZ would catalyze a reaction that involves folate.
It should be noted that an aspartic acid occupies the position equivalent to Asp552 (Asp72 in E. coli YgfZ numbering) in all eukaryotic family members and in the representatives of the alpha division of the Proteobacteria. In all other members of the YgfZ family, neither Asp nor Glu is present at this position. Thus, it is possible that the family includes two subfamilies, one with a potential for enzymatic activity and the other without such a potential. The phylogenetic distribution among the YgfZ homologs implies that this division may be related to the evolution of this protein family.
The only fragment that bears residues conserved in the entire YgfZ family is the octapeptide fingerprint motif located in the interdomain linker on the surface of the protein. All residues of the motif except Tyr229 are exposed to solvent and are not in contact with each other or with the rest of the molecule. The strict conservation of these residues suggests their functional importance. As a theoretical possibility, one might consider a structural rearrangement that would bring the fingerprint loop to the folate-binding site. Although such a conformational transition cannot be ruled out, it seems to be unlikely given its massive scale.
As an alternative hypothesis, we suggest that YgfZ may function as a signal transducer by sensing certain folate derivatives (e.g., THF) for which it has a high affinity. Folate binding in the central cavity would trigger the conformational changes in the protein, such as the relative movement of the domains. The domain mobility in YgfZ can be predicted from the difference in the average B-factors and from superposition on the DMGO structure, as discussed above. Some adjustment of the hydrophobic pocket was required to accommodate even structurally similar ligands in DMGO (14), suggesting that the transfer from the free state to the bound state may cause significant conformational changes. The interdomain linker must be sensitive to the domain movements, and hence it would be an ideal location for the interaction with the target molecule. The conserved sequence motif would ensure specific target recognition. The presence of two glycine residues in the motif indicates the conformational flexibility of the polypeptide fragment that may be important for signal transduction.
The central residue of the motif, Cys228, bears a reactive thiol group. In the present structure, Cys228 forms an unusual intermolecular disulfide bridge with the equivalent residue from another YgfZ molecule. The protein is thus a symmetric dimer sitting on the crystallographic twofold axis. The interface is not particularly extensive and covers about 7% of the monomer surface. There are primarily nonspecific van der Waals interactions between the two molecules. With the exception of Cys228, none of the invariant residues is involved in the interaction, suggesting that the dimer is not functionally relevant. If a possible recognition role of the fingerprint motif is considered, Cys228 may be a key anchor in the molecular interaction.
Next to the putative recognition site is a concave positively charged surface between domains B and C (Fig. 2). Importantly, the basic character of the residues covering the surface is conserved in the YgfZ family. The charge and the shape of the surface suggest that it may bind a nucleic acid. The tRNA-binding role of the topologically similar domains in translation factors supports this suggestion (26). It is tempting to speculate that YgfZ may act as a transcriptional regulator. One such uncharacterized protein has recently been shown to recognize a conserved CATCN7CTTCTT motif present in the promoter regions of the yeast gcv genes (10). The formation of the complex is responsive to THF, indicating that glycine-specific control may be mediated via folate derivatives. The CATCN7CTTCTT motif is also found in the promoter of the DFR1 gene encoding dihydrofolate reductase, which catalyzes de novo synthesis of THF.
Transcription factors typically bind DNA with a pair of helix-turn-helix motifs that interact with nucleotide bases in the major groove (21). However, the concave surface in YgfZ seems to favor unspecific DNA binding and does not reveal any pattern for recognizing a particular nucleotide sequence. On the other hand, the presence of two recognition surfaces, one for targeting a protein and the other for nucleic acid binding, suggests that YgfZ may be part of a nucleoprotein complex. Targeting a transcription factor is a definite possibility.
Taking into account the sensitivity to folates, one may hypothesize that YgfZ is a folate-dependent regulatory protein that may affect the expression of the proteins in response to changes in the environment. Consistent with this hypothesis is the fact that the yeast homolog of YgfZ, CAF17 (CCR4-associated factor), is a component of the transcriptional regulatory complex. The data were obtained from yeast two-hybrid whole-genome screening (36). The CCR4 regulator is an evolutionarily conserved transcriptional regulator involved in controlling mRNA initiation, elongation, and degradation (4).
Another observation related to the putative DNA-binding function of YgfZ is particularly interesting because it involves a THF-dependent enzyme. A single-stranded DNA-binding activity has been reported for C1-THF synthase from different organisms (37). This protein does not bind double-stranded DNA or RNA but does bind single-stranded DNA with high affinity and in a sequence-independent fashion. Among various possibilities, we believe that C1-THF synthase might regulate gene expression of one of the enzymes involved in C1 metabolism.
Analysis of the genomic context indicates that ygfZ does not belong to any particular gene string, although in some enterobacteria, including E. coli, Yersinia pestis, and Salmonella enterica serovar Typhimurium, the ygfZ gene is located upstream of the gcv operon coding for the GCS proteins. This proximity may reflect involvement of YgfZ in the regulation of C1 pools related to the GCS. Further experiments should identify the molecular partners of YgfZ and reveal its role in C1 metabolism. The three-dimensional structure of YgfZ helps to establish a new protein family widely represented in bacteria and eukaryotes whose members are topologically similar to, but functionally different from, the T-protein-like enzymes.
Acknowledgments
We are grateful to Alexey Murzin for pointing out the structural similarity between YgfZ and DMGO and to the reviewer who drew our attention to the conservation of the active site Asp in some members of the YgfZ family. The use of the IMCA-CAT beamline is acknowledged.
This work was supported by National Institutes of Health grant P01-GM57890. Use of the APS was supported by the U.S. Department of Energy Basic Energy Sciences Office of Science under contract W-31-109-Eng-38.
Certain commercial materials, instruments, and equipment are identified in this paper in order to specify the experimental procedure as completely as possible. In no case does such identification imply a recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials, instruments, or equipment identified are necessarily the best available for the purpose.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, L. B. (ed.). 1995. Folate in health and disease. Marcel Dekker, New York, N.Y.
- 3.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 4.Denis, C. L., and J. Chen. 2003. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 73:221-250. [DOI] [PubMed] [Google Scholar]
- 5.Eisenstein, E., G. L. Gilliland, O. Herzberg, J. Moult, J. Orban, R. J. Poljak, L. Banerjei, D. Richardson, and A. J. Howard. 2000. Biological function made crystal clear—annotation of hypothetical proteins via structural genomics. Curr. Opin. Biotechnol. 11:25-30. [DOI] [PubMed] [Google Scholar]
- 6.Falquet, L., M. Pagni, P. Bucher, N. Hulo, C. J. Sigrist, K. Hofmann, and A. Bairoch. 2002. The PROSITE database, its status in 2002. Nucleic Acids Res. 30:235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghrist, A. C., G. Heil, and G. V. Stauffer. 2001. GcvR interacts with GcvA to inhibit activation of the Escherichia coli glycine cleavage operon. Microbiology 147:2215-2221. [DOI] [PubMed] [Google Scholar]
- 8.Gouet, P., E. Courcelle, D. I. Stuart, and F. Metoz. 1999. ESPript: multiple sequence alignments in PostScript. Bioinformatics 15:305-308. [DOI] [PubMed] [Google Scholar]
- 9.Holm, L., and C. Sander. 1998. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 26:316-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong, S. P., M. D. Piper, D. A. Sinclair, and I. W. Dawes. 1999. Control of expression of one-carbon metabolism genes of Saccharomyces cerevisiae is mediated by a tetrahydrofolate-responsive protein binding to a glycine regulatory region including a core 5′-CTTCTT-3′ motif. J. Biol. Chem. 274:10523-10532. [DOI] [PubMed] [Google Scholar]
- 11.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 12.Kikuchi, G. 1973. The glycine cleavage system: composition, reaction mechanism, and physiological significance. Mol. Cell. Biochem. 1:169-187. [DOI] [PubMed] [Google Scholar]
- 13.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 14.Leys, D., J. Basran, and N. S. Scrutton. 2003. Channelling and formation of ′active' formaldehyde in dimethylglycine oxidase. EMBO J. 22:4038-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucock, M. 2000. Folic acid: nutritional biochemistry, molecular biology, and role in disease processes. Mol. Genet. Metab. 71:121-138. [DOI] [PubMed] [Google Scholar]
- 16.MacKenzie, C. G., and W. R. Frisell. 1958. The metabolism of dimethylglycine by liver mitochondria. J. Biol. Chem. 232:417-427. [PubMed] [Google Scholar]
- 17.Matthews, M. G. 1996. One-carbon metabolism, p. 600-611. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 18.McKenzie, K. Q., and E. W. Jones. 1977. Mutants of formyltetrahydrofolate interconversion pathway of Saccharomyces cerevisiae. Genetics 86:85-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merritt, E. A., and D. J. Bacon. 1997. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 20.Meskys, R., R. J. Harris, V. Casaite, J. Basran, and N. S. Scrutton. 2001. Organization of the genes involved in dimethylglycine and sarcosine degradation in Arthrobacter spp.: implications for glycine betaine catabolism. Eur. J. Biochem. 268:3390-3398. [DOI] [PubMed] [Google Scholar]
- 21.Muller, C. W. 2001. Transcription factors: global and detailed views. Curr. Opin. Struct. Biol. 11:26-32. [DOI] [PubMed] [Google Scholar]
- 22.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 23.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247:536-540. [DOI] [PubMed] [Google Scholar]
- 24.Nagy, P. L., A. Marolewski, S. J. Benkovic, and H. Zalkin. 1995. Formyltetrahydrofolate hydrolase, a regulatory enzyme that functions to balance pools of tetrahydrofolate and one-carbon tetrahydrofolate adducts in Escherichia coli. J. Bacteriol. 177:1292-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 26.Nissen, P., M. Kjeldgaard, S. Thirup, G. Polekhina, L. Reshetnikova, B. F. Clark, and J. Nyborg. 1995. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270:1464-1472. [DOI] [PubMed] [Google Scholar]
- 27.Okamura-Ikeda, K., K. Fujiwara, and Y. Motokawa. 1982. Purification and characterization of chicken liver T-protein, a component of the glycine cleavage system. J. Biol. Chem. 257:135-139. [PubMed] [Google Scholar]
- 28.Okamura-Ikeda, K., Y. Ohmura, K. Fujiwara, and Y. Motokawa. 1993. Cloning and nucleotide sequence of the gcv operon encoding the Escherichia coli glycine-cleavage system. Eur. J. Biochem. 216:539-548. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 30.Paukert, J. L., and J. C. Rabinowitz. 1980. Formyl-methenyl-methylenetetrahydrofolate synthetase (combined): a multifunctional protein in eukaryotic folate metabolism. Methods Enzymol. 66:616-626. [DOI] [PubMed] [Google Scholar]
- 31.Piper, M. D., S. P. Hong, G. E. Ball, and I. W. Dawes. 2000. Regulation of the balance of one-carbon metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 275:30987-30995. [DOI] [PubMed] [Google Scholar]
- 32.Schirch, L. 1984. Serine hydroxymethyltransferase, p. 399-431. In R. L. Blakley and S. J. Benkovic (ed.), Folates and pterins, vol. 1. John Wiley & Sons, New York, N.Y. [Google Scholar]
- 33.Stauffer, L. T., S. J. Fogarty, and G. V. Stauffer. 1994. Characterization of the Escherichia coli gcv operon. Gene 142:17-22. [DOI] [PubMed] [Google Scholar]
- 34.Stauffer, L. T., and G. V. Stauffer. 1994. Characterization of the gcv control region from Escherichia coli. J. Bacteriol. 176:6159-6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 37.Wahls, W. P., J. M. Song, and G. R. Smith. 1993. Single-stranded DNA binding activity of C1-tetrahydrofolate synthase enzymes. J. Biol. Chem. 268:23792-23798. [PubMed] [Google Scholar]
- 38.Weeks, C. M., and R. Miller. 1999. Optimizing shake-and-bake for proteins. Acta Crystallogr. Sect. D Biol. Crystallogr. 55:492-500. [DOI] [PubMed] [Google Scholar]