Abstract
Bacteriophage T4 encodes three ADP-ribosyltransferases, Alt, ModA, and ModB. These enzymes participate in the regulation of the T4 replication cycle by ADP-ribosylating a defined set of host proteins. In order to obtain a better understanding of the phage-host interactions and their consequences for regulating the T4 replication cycle, we studied cloning, overexpression, and characterization of purified ModA and ModB enzymes. Site-directed mutagenesis confirmed that amino acids, as deduced from secondary structure alignments, are indeed decisive for the activity of the enzymes, implying that the transfer reaction follows the Sn1-type reaction scheme proposed for this class of enzymes. In vitro transcription assays performed with Alt- and ModA-modified RNA polymerases demonstrated that the Alt-ribosylated polymerase enhances transcription from T4 early promoters on a T4 DNA template, whereas the transcriptional activity of ModA-modified polymerase, without the participation of T4-encoded auxiliary proteins for middle mode or late transcription, is reduced. The results presented here support the conclusion that ADP-ribosylation of RNA polymerase and of other host proteins allows initial phage-directed mRNA synthesis reactions to escape from host control. In contrast, subsequent modification of the other cellular target proteins limits transcription from phage early genes and participates in redirecting transcription to phage middle and late genes.
Posttranslational ADP-ribosylation of proteins is catalyzed by ADP-ribosyltransferases (ADP-RTs), which have been identified in viral, bacterial, and eukaryotic systems. Transfer of the ADP-ribose moiety from the substrate NAD+ to a specific amino acid residue, frequently histidine or arginine within a target protein, modulates the activity of the acceptor. ADP-ribosylation changes the electrostatic potential of a target protein by introducing two phosphate groups and may affect protein-DNA as well as protein-protein interactions. ADP-RTs were initially discovered as the exotoxins of pathogenic bacteria. Therefore, most of our knowledge concerning these proteins has been gained by biochemical, genetic, and structural studies performed on bacterial toxins, such as those produced by Corynebacterium diphtheriae (15, 30), Bordetella pertussis (34), Vibrio cholerae (46), Pseudomonas aeruginosa (33), and Escherichia coli (45). New putative bacterial toxins were found to be encoded in the genomes of Streptococcus pyogenes and Salmonella enterica serovar Typhi (50).
To date, the family of ADP-RTs comprises more than 40 enzymes, including bacterial exotoxins as well as a variety of other enzymes, such as the eukaryotic mono- and poly-ADP-RTs, which include T-cell differentiation alloantigens like RT6 (9), poly-ADP-RTs (56), the dinitrogenase reductase regulation factor DraT (51, 77), and enzymes encoded by T-even bacteriophages. The bacteriophage T4 gene products Alt (76 kDa), ModA (23 kDa), and ModB (24 kDa) appear to actively regulate gene expression during the transition from host to phage protein synthesis.
The T4 Alt protein initially acts as a structural component of the phage head. At the time of infection, it enters the host cell with phage DNA and immediately displays enzymatic activity. This protein, purified from infected cells, has been found to efficiently ADP-ribosylate one of the two α subunits of host RNA polymerase in the carboxy-terminal domain at Arg265 (19, 31, 32, 49, 52). Isolation and partial characterization of a recombinant Alt protein (GenBank accession number X15811), now available in larger amounts, revealed that host RNA polymerase subunits β, β′, and σ are also ADP-ribosylated, as are a number of other E. coli proteins (36, 37). ADP-ribosylation of RNA polymerase, as catalyzed by Alt, triggers the preferred transcription from T4 early promoters as possibly the first step by which T4 gains control over the metabolism of the host cell (60, 72, 73).
The ModA and ModB proteins are encoded by two neighboring T4 genes (44, 63) under control of the strong early promoter Pe 12.8, and their activities are directed against different cellular target proteins. As first demonstrated by Skorko et al. (57), ModA-catalyzed ADP-ribosylation, in contrast to Alt activity, targets both α subunits of RNA polymerase at residue Arg265. Also in contrast to Alt activity, recombinant ModA has no activity towards the β, β′, and σ subunits. It has been inferred that the differences in the molecular masses of gpAlt (76 kDa) and gpModA (23 kDa) might be responsible for the different approaches to the α subunits. The ModA-induced ADP-ribosylation of α subunits inhibits transcription from T4 early promoters (21). Host polymerase may thus be conditioned to interact with other T4-encoded transcription factors that may be active in the middle mode (14, 27, 28, 68) or late transcription (26, 74, 75, 76).
The modB gene was detected only recently in the course of sequencing experiments performed to identify the reading frame of the modA gene (GenBank accession number X98695 for ModA and ModB) (44, 63). Partial characterization of the ModB protein has revealed that the ribosomal S1 protein (8, 38) is a target for ModB-catalyzed ADP-ribosylation; however, a number of other proteins involved in translation and cell regulation are also modified (R. Depping, C. Lohaus, H. E. Meyer, and W. Rüger, unpublished data).
Here we describe a new and effective procedure to renature both of the overexpressed T4 Mod proteins to obtain amounts that allow studies of the effects of the individual ribosylation reactions in vitro. Catalytic properties of the T4 ADP-RTs were demonstrated and compared, and transcription assays were performed with ADP-ribosylated host RNA polymerase. For the transcription experiments, the Alt enzyme was used as a control. This study provided additional evidence that the modification catalyzed by the Alt protein enhances transcription from T4 early promoters, whereas ADP-ribosylation catalyzed by the ModA protein hampers early transcription. Mutation analysis of ModA and ModB demonstrated that functional and structural features common to most members of the ADP-RT family are also valid for the T4 ModA and ModB proteins. The data presented here are consistent with ADP-ribosyltransfer following an Sn1-type mechanism to dissociate the nicotinamide portion of NAD+. This reaction generates a positively charged oxocarbenium intermediate stabilized by one or two Glu residues of the consensus pattern (the EXE motif). The ADP-ribose moiety remains in the enzyme binding site until it is transferred to the acceptor protein (5). Our mutation data are consistent with this mechanism, and the active site glutamic acid residues Glu165 (ModA) and Glu173 (ModB), the conserved Arg72 (ModA) and Arg73 (ModB) residues for NAD+ binding, and other residues of the predicted active center are important for enzyme activity.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are shown in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacqZΔM15 Tn10 (Tetr)] | Stratagene (La Jolla, Calif.) |
| E. coli DH5α | F−gyrA96 (Nalr) recA1 relA1 endA1 thi-1 hsdR17 (rK− mK+) glnV44 deoR Δ(lacZYA-argF)U169 [φ80dΔ(lacZ)M15] | Fermentas (St. Leon-Rot, Germany) |
| E. coli BL21(DE3) | F−dcm ompT hsdS(rB− mB−) gal λ(DE3) | Novagen (Bad Soden, Germany) |
| E. coli C41(DE3) | Derivative of E. coli BL21(DE3) allowing overexpression of proteins at an elevated level without a toxic effect | 43 |
| Plasmids | ||
| pBAD/HisB | Expression vector with araBAD promoter and N-terminal His tag | 23 |
| pBADmodA | pBAD/HisB with modA | 63 |
| pBN19 | Vector expressing DnaK (Tct) | 6, 7 |
| pET-11d | Expression vector with T7 promoter and lac operator | Novagen |
| pET-16b(+) | Expression vector with T7 promoter and lac operator that allow N-terminal fusion to a cleavable His tag sequence for rapid affinity purification | Novagen |
| pET-22b(+) | Expression vector with T7 promoter and lac operator | Novagen |
| pET-32b(+) | Expression vector with T7 promoter and lac operator | Novagen |
| pGroESL | Vector expressing GroEL and GroES (Cam) | 22 |
| pT-GroE | Vector expressing GroEL and GroES (Cam) | 12 |
| pTKRI | alt gene plasmid vector | 36 |
| p16modA | pET-16(+)with modA encoding a cleavable His tag sequence at the N terminus | 63 |
| p16modB | pET-16b(+) with modB | 63 |
Overexpression of the ADP-ribosyltransferases ModA and ModB.
For isolation of DNA, transformation, ligation, and sequencing we used standard procedures described by Ausubel et al. (3). The modA or modB gene was cloned downstream from the T7 late promoter into the NcoI-BamHI restriction sites of vector pET-11d. To obtain the transferases as His-tagged polypeptides for purification of the proteins, modA was cloned into the BamHI-BamHI restriction sites and modB was cloned into the NdeI-BamHI restriction sites of pET-16b(+). The vectors carrying modA or modB or the corresponding mutated genes were transformed into E. coli C41(DE3) by CaCl2 treatment, screened for the production of the recombinant enzymes, and grown as described below. E. coli C41(DE3) is a derivative of the widely used strain BL21(DE3). Plausible explanations for the improved performance of strain C41(DE3) for protein overexpression were given by Miroux and Walker (43).
Overexpression was performed in 2-liter Erlenmeyer flasks containing 500 ml of tryptone-phosphate broth supplemented with ampicillin (200 μg/ml). The medium in each flask was inoculated with 5 ml of the appropriate overnight culture. The flasks were shaken vigorously in a New Brunswick G10 incubator at 37°C. At an optical density at 590 nm (OD590) of 0.6 to 0.8, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1.0 mM, and shaking was continued for 3 to 5 h. The cells were centrifuged for 20 min at 4,000 × g and 4°C, and the resulting pellets were resuspended in 30 ml of lysis buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 1 mM dithiothreitol [DTT], 2 mM phenylmethylsulfonyl fluoride) and frozen at −20°C until they were used. The following vectors were tested in these experiments: pBAD/HisB, which allowed dose-dependent induction and hence modulation of the expression level (23); pET-43a, which fused the mod genes to the 495-amino-acid NusA protein (25); vector pET-32b(+), which fused the mod genes to thioredoxin (39); and pET-22b(+), which harbored the leader sequence pelB responsible for the export of the proteins (reviewed in reference 71).
Isolation of inclusion bodies.
Cells grown as described above were opened by using a French press at 2,000 lb/in2. The inclusion bodies were separated by centrifugation at 3,000 × g for 20 min at 4°C from the cellular debris remaining in the supernatant (29). The sediment was washed by resuspension in 30 ml of IB wash buffer (50 mM Tris-HCl [pH 7.5], 2 mM EDTA, 100 mM NaCl, 1 to 2 M urea, 0.05% [wt/vol] deoxycholate, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 0.5 mg of lysozyme per ml). This step removed a large portion of the contaminating E. coli proteins, lipids, and nucleic acids but did not solubilize the inclusion bodies. After sedimentation by centrifugation at 10,000 × g for 20 min at 4°C, the supernatant was discarded. The inclusion bodies were used immediately or kept frozen at −20°C until they were used.
Renaturation and purification of the proteins.
Inclusion bodies purified as described above were resuspended in IB denaturation buffer (100 mM Tris [pH 12.0], 2 M urea), conditions that unfolded the proteins only partially, allowing effective renaturation (35). To avoid precipitation of the Mod proteins, the concentration of the proteins was adjusted to 1 mg/ml and the sample was transferred to a dialysis bag (type 20 dialysis membrane; Biomol, Hamburg, Germany). Renaturation was performed by overnight dialysis for 16 h against 1 liter of a buffer containing 50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, and 2 M urea at 4°C. After centrifugation at 20,000× g for 30 min at 4°C to remove insoluble material, the native His-tagged protein was purified further by immobilized-metal affinity chromatography on Ni-nitrilotriacetic acid (QIAGEN, Hilden, Germany) or Talon resin columns (Clontech) by following the manufacturer's instructions. Fractions (2 ml) were collected, and peak fractions were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, pooled, dialyzed, and stored at −20°C. Glycerol was added to a final concentration of 10%. The purified ADP-RTs remained stable for about 6 months. At protein concentrations above 1 mg/ml or after extended storage, the proteins tended to precipitate and gradually lost activity.
Assay of mono-ADP-ribosyltransferase activity.
Activities of the purified recombinant ADP-RTs ModA and ModB were assayed in vitro by radioactive labeling of the target proteins by using 32P-labeled NAD+ as a substrate and the procedures described by Rohrer et al. (52), and the changes were introduced as described by Koch et al. (36). The incubation mixtures (final volume, 100 μl) contained 2.5 to 3 μg of the recombinant transferase and 3.7 × 104 Bq of 32P-labeled NAD (specific activity, 2.96 × 1013 Bq/mmol) in transferase buffer (50 mM Tris-acetate [pH 7.5], 10 mM magnesium acetate, 22 mM NH4Cl, 1 mM EDTA, 10 mM 2-mercaptoethanol, 10% glycerol). To assay residual activities of the mutated enzymes, up to 35 μg of a soluble fraction of E. coli C41(DE3) proteins was added to each test mixture. Since intrinsic proteins of the expressing cells were irreversibly labeled with nonlabeled NAD+, this step led to more comparable band patterns for the different mutants.
A number of ADP-ribosyltransferases are known to transfer the ADP-ribosyl residues not only to proteins but also to guanidino compounds. We used the method of Soman et al. (58, 59) to measure the ADP-ribosylation of p-nitrobenzylidine aminoguanidine (NBAG). Under the test conditions proposed, the acceptor substance NBAG had an absorption maximum at 315 nm and the ADP-ribosylated product had a maximum at 370 nm. An increase in 0.1 OD370 unit corresponded to the formation of 0.05 μmol of product in a 1-ml reaction system.
Site-directed mutagenesis of the modA and modB genes.
For site-directed mutagenesis we used a QuikChange mutagenesis kit from Stratagene and followed the manufacturer's instructions. The correct exchange of target amino acids following mutagenesis was monitored by sequencing the plasmid inserts resulting from the mutagenic exchange. In the procedure a supercoiled double-stranded DNA vector with an insert of interest and two synthetic oligonucleotide primers containing the desired mutations were utilized. The corresponding primer sequences are available upon request.
Testing the toxicity of the mutated ModA and ModB proteins.
It was reported previously (63) that cloning of either modA or modB was lethal to host cells if the vector expression system was leaky. We took advantage of this phenomenon to identify relevant mutations (e.g., mutations within the active site should have reduced or abolished enzyme activity, and hence growth of colonies was a convenient selection system).
For a variant test system to compare the wild-type and mutant enzymes we used an enzyme-linked immunosorbent assay (ELISA)-based test. This system took advantage of the inactive ModB mutant R73A, which was used as a substrate for transribosylation. Specific amounts of ModB mutant R73A were bound to 96-well plates. The plates were incubated with ELISA blocking buffer (0.05% Tween 20 and 0.25% bovine serum albumin in phosphate-buffered saline [PBS] [137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4]) for 1.5 h at room temperature. After each well was washed three times with PBS, ADP-ribosylation was carried out by incubating the wells with the different mutant enzymes and biotinylated NAD+ as a substrate in transferase buffer (50 mM Tris [pH 7.5], 10 mM magnesium acetate, 22 mM NH4Cl, 1 mM EDTA, 10 mM 2-mercaptoethanol, 10% glycerol) for 30 min at room temperature. After horseradish peroxidase-streptavidin (1:500 in ELISA blocking buffer) was added and the preparation was incubated for 30 min at room temperature and washed four times with PBS, 50 μl of TACS-Sapphire (R&D Systems, Wiesbaden, Germany) was added as a substrate. The turnover was measured for 30 min with a μQuant photometer (Bio-Tek Instruments Inc.) at OD630. The results were estimated by using the MikroWin 3.0 software (Mikrotek Laborsystem GmbH).
In vitro transcription.
The capacity for ADP-ribosylated RNA polymerase to transcribe T4 and E. coli DNA in vitro was tested by using two procedures. For both methods we used T4 genomic DNA as a template to preserve T4-specific modifications and helicity. For the first series of experiments E. coli strain C41(DE3) with plasmid pET-16b(+) (control), p16modA (ModA), or pTKRI (Alt) was grown under induced conditions to an OD590 of 0.65. Cells were harvested and opened with the French press, and the soluble protein fraction was used in the presence of T4 or E. coli DNA. Two micrograms of genomic DNA was incubated with a solution containing 5 μl of 10× RNA polymerase buffer (400 mM Tris-HCl [pH 8.0], 100 mM MgCl2, 500 mM KCl, 5 mM DTT, 0.5 mg of bovine serum albumin per ml), 10 U of RNase inhibitor, 0.5 μl of 25 mM NAD+, and 30 to 110 μg of soluble E. coli protein, and the volume was adjusted to 42 μl with distilled water. The transcription reaction was started with 4 μl of a three-deoxynucleoside triphosphate mixture (5 mM ATP, 5 mM CTP, and 5 mM GTP), 3.75 μl of 1 mM UTP, and 0.25 μl of [α-32P]UTP (0.37 × 106 Bq/μl). After 30 min of incubation at 37°C, the solution was collected on DE81 filters (Whatman) and washed three times with 0.5 M NaH2PO4 buffer (pH 7.0) for 5 min. After drying, the radioactivity incorporated into RNA and retained on the filters was counted in 5 ml of scintillation fluid.
For the second series of experiments the E. coli RNA polymerase was isolated by using the procedure of Burgess and Jendrisak (10). The reaction mixtures (final volume, 0.3 ml) contained 40 mM Tris-HCl (pH 7.9), 10 mM MgCl2, 5 mM 2-mercaptoethanol, 0.1 mM EDTA-Na2, 150 mM KCl, 0.16 mg of bovine serum albumin, 2 μg of T4 wild-type or E. coli DNA, 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, 0.25 mM [α-32P]UTP (10 μCi/μl), and 60 to 170 ng of enzyme. The mixtures were incubated for 10 min at 37°C. Aliquots (50 μl) were applied to DEAE-cellulose filters (Whatman DE81; diameter, 25 mm). The filters were washed and dried as described above, and the radioactivity retained was counted with a scintillation counter by using toluene liquifluor.
RESULTS
Overexpression of ModA and ModB.
Preliminary experiments designed to overexpress the recombinant ModA and ModB proteins revealed two difficulties. First, the gene products were toxic to the cloning host, and second, the recombinant proteins did not remain in solution but accumulated as inclusion bodies. The pET vector system (61, 62) proved to be tight enough to overcome the problem of uninduced expression and enzyme toxicity.
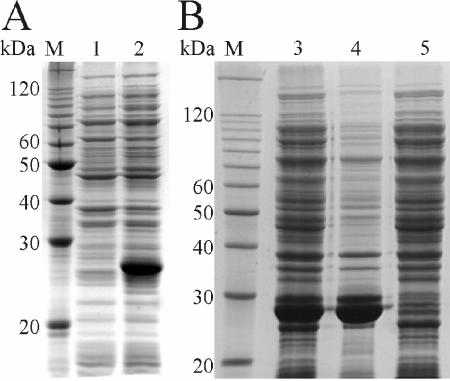
To isolate and work with native enzymes rather than with proteins that have been partially denatured and renatured from inclusion bodies, we tested a number of procedures described previously to circumvent the formation of inclusion bodies (54, 55). We also cloned the modA or modB genes into a number of vectors (Table 1), which allowed fusion of the proteins expressed to solubilization linkers or to the leader sequence pelB or enabled coexpression with the chaperones GroEL and GroES (12, 22) or DnaK (6, 7). Although all proteins were overexpressed, as expected for the vector constructs used, the Mod proteins and their fusions always accumulated as inclusion bodies in the induced cells. The aggregation of the expressed ADP-RTs might have reduced the toxicity since increasing amounts of the ADP-RTs lost the active conformation. This view was supported by the fact that despite the toxicity, about 40% of the cellular protein was ADP-ribosyltransferase within 3 h of induction (Fig. 1).
FIG. 1.
Overexpression of His-ModA (A) and His-ModB (B) in C41(DE3). Overexpression of the proteins was monitored on sodium dodecyl sulfate—13% polyacrylamide gels. (A) Lanes M, molecular weight markers (10-kDa ladder; Gibco-BRL, Gaithersburg, Md.); lane 1, cell extract of p16modA-containing cells prior to induction; lane 2, same as lane 1 but 3 h after induction. (B) Lane 3, cell extract of p16modB-containing cells 3 h after induction; lane 4, insoluble protein fraction of the cell extract shown in lane 3; lane 5, soluble proteins of the cell extract shown in lane 3. For further details see the text.
Purification and renaturation of ModA and ModB.
Because all attempts to prevent protein aggregation failed, the Mod proteins were purified from the corresponding inclusion bodies. The procedure outlined in Materials and Methods at least offered the advantage that the recombinant Mod proteins accumulated in a relatively pure form.
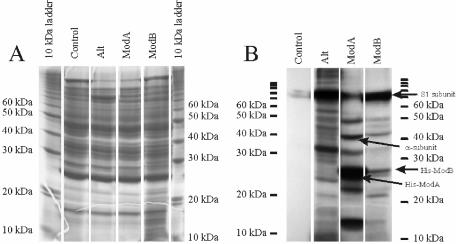
The ADP-ribosylation activities of the proteins were assayed by using the procedure of Rohrer et al. (52) with 32P-labeled NAD+ as a substrate. Figure 2 shows the ribosylation pattern of soluble E. coli proteins ADP-ribosylated by Alt, ModA, or ModB after sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography. The ADP-ribosylation catalyzed by Alt served as a control (36). The patterns of ADP-ribosylation obtained after reactions with Alt, ModA, or ModB were different; only a few bands comigrated and therefore might have represented identical proteins. The apparent molecular masses of the radioactively labeled bands ranged from 16 to 72 kDa for both ModA and ModB. There were 8 to 11 labeled proteins targeted by each ribosyltransferase, and the extent of labeling by ModA was more significant (Fig. 2). The Alt enzyme ADP-ribosylated at least 20 proteins, and it remains to be determined whether all of the proteins carrying a radioactive label intervene in phage replication and, if so, what the consequence of ADP-ribosylation of each protein is. ADP-ribosylation of the RNA polymerase α subunit in the Alt channel appeared to be relatively weak. There may have been two reasons for this: first, the α subunit represented a small portion of all other proteins ADP-ribosylated in the reaction, and second, in contrast to ModA, which labeled both α subunits, Alt modified only one of the two subunits present in the enzyme. Because the three ADP-RTs exhibited different target protein patterns, it seemed reasonable to assume that the enzymes work independently. Self-ADP-ribosylation of ModA and ModB was also observed in the reactions. Below we discuss the differences between these results and those of Skorko and his collaborators (57).
FIG. 2.
Ribosylation patterns of E. coli proteins as catalyzed by T4 ribosyltransferases Alt, ModA, and ModB. (A) Proteins separated on a 13% polyacrylamide gel stained with Coomassie brilliant blue. (B) Autoradiograph of panel A on Fuji X-ray film, exposed for 4 to 9 days. The strong band in the Alt lane at about 70 kDa represents, at least in part, the transribosylated Alt protein. Other Alt-labeled proteins migrating in this area are S1, as well as β and β′, two subunits of RNA polymerase. The positions of the α subunit, His-ModA, and His-ModB are indicated on the right.
Determination of ADP-ribosyltransferase activity of ModA and ModB.
Using the procedure of Soman et al. (58, 59), we measured the transfer onto the artificial substrate NBAG by purified ModA with increasing concentrations of NAD+ between 0 and 10 mM, and we obtained a Vmax of 20 mol of product/mol of ModA/h. The corresponding Km was 258 μM. However, when the NAD+ concentration was kept constant at 10 mM and the concentration of NBAG was allowed to increase from 0 to 0.4 mM, the Vmax was 5.34 mol of product/mol of ModA/h, and the corresponding Km was 219 μM. Studying a presumed ModA transferase fraction isolated from T4-infected cells, Skorko et al. (57) obtained a Km of 14.3 μM for NAD+ under conditions under which excess E. coli RNA polymerase was present. The differences in these data may be due to the different test conditions following the isolation procedures used for ModA preparation in the two laboratories (that is, partially purified enzyme from T4-infected cells versus overexpressed recombinant enzyme that was purified to homogeneity). Also, transribosylation of ModA led to loss of a portion of the enzymatic activity in the NBAG test, which might have been different for the two enzyme preparations.
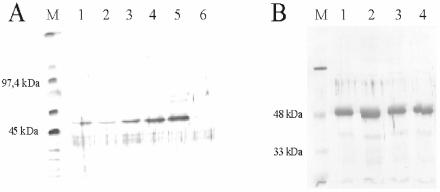
For ModB, the NBAG test was unsatisfactory. The greater tendency of this enzyme for precipitation at elevated protein concentrations may have been a reason for the failure of the ModB enzyme assay. To obtain further evidence that ModB is indeed an ADP-RT, we modified the ADP-ribosylation test, taking advantage of biotinylated NAD+ (bNAD+), as monitored with horseradish peroxidase-linked streptavidin. Figure 3A shows the results of a transribosylation assay with bNAD+ as the substrate. As in a comparable test with radioactive NAD+, the signal intensity increased with increasing amounts of bNAD+.
FIG. 3.
Transribosylation of ModB determined with different amounts of bNAD+. (A) Western blot of Trx-ModB (48 kDa) transribosylated with bNAD+ (Pierce, Rockford, Ill.) as the substrate and separated on a 13% polyacrylamide gel. The biotinylated ADP-ribosyl residue was detected with horseradish peroxidase-coupled streptavidin. Lane M, protein marker; lane 1, 1 pM bNAD+ in the reaction mixture; lane 2, 6.25 pM bNAD+; lane 3, 10 pM bNAD+; lane 4, 50 pM bNAD+; lane 5, 100 pM bNAD+; lane 6, control (no bNAD+). (B) Western blot developed with a mono-ADP-ribosyl antibody. Lane M, molecular weight marker; lane 1, 1 pM bNAD+; lane 2, 5 pM bNAD+; lane 3, 10 pM bNAD+; lane 4, control without external addition of bNAD+. The positive reaction with the antibody in lane 4 indicates that ModB was transribosylated during overexpression with cellular NAD+ as the substrate.
This assay system could be modified further to test the ADP-RT activity of ModA and ModB mutants with an ELISA. To do this, streptavidin-horseradish peroxidase was replaced by mono-ADP-ribosyl antibodies kindly supplied by K. K. McMahon, Texas Tech University, Lubbock. Figure 3B shows that transribosylation of ModB occurred at all concentrations tested, including in the absence of added bNAD+. This result not only confirmed that ModB indeed transferred an ADP-ribosyl compound but also showed that the enzyme was already ADP-ribosylated during overexpression by unlabeled, cellular NAD+, leading to a relatively homogeneous antibody reaction in all channels. Radioactive or biotinyl labeling in the enzyme assays appeared to reach only those sites not yet occupied by unlabeled cellular NAD+. ADP-ribosyltransferase labeling reactions were more efficient with added protein substrates (i.e., RNA polymerase or a soluble extract of E. coli proteins) not previously exposed to the enzymes.
Transcription experiments with altered and modified E. coli RNA polymerase.
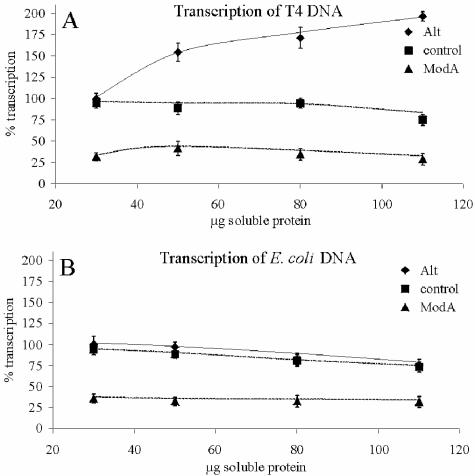
Two sets of in vitro transcription experiments were performed with polymerases modified by the recombinant enzymes Alt and ModA. In the first study, we grew E. coli strain C41(DE3) carrying either plasmid pET-16b(+) (control), p16modA (modA inserted), or pTKRI (alt inserted) under induced conditions. Except for the controls, the growing cells produced the transferase Alt or ModA. Transcription was performed with increasing amounts of the soluble protein fraction. Aliquots were taken, and the amount of RNA synthesized was determined by measuring the incorporation of [α-32P]UTP. As Fig. 4 shows, the Alt-modified RNA polymerase synthesized more RNA on the T4 DNA template than the ModA-modified enzyme synthesized. With E. coli DNA there was no difference between the transcription experiments performed with unmodified polymerase and the transcription experiments performed with Alt-modified polymerase. In both cases, transcription decreased as the amount of protein from the Alt-modified or unmodified extract increased.
FIG. 4.
Transcription of T4 (A) or E. coli DNA (B) with native RNA polymerase or with the Alt- or ModA-modified enzymes. For comparison, the amount of radioactivity incorporated in the presence of 30 μg of soluble protein plus wild-type T4 DNA (5 μg) as a template was defined as 100%. The values are the averages from three independent experiments. The initial steep increase in transcript synthesis with T4 DNA as the template, determined with Alt-modified RNA polymerase, reflects rapid recognition of T4 early promoters and transcription initiation (60). For further details see the text.
In a complementary set of experiments, cells carrying the plasmids were grown under noninducing and inducing conditions. About 5 g of centrifuged cells of one of the strains was extracted, and both the unmodified and the Alt- and ModA-modified polymerases were isolated (10). The transcription activities of the purified enzymes were tested as described above with either T4 or E. coli DNA as the template. The results of these experiments are presented in Table 2. Similar to the results shown in Fig. 4, the Alt-modified E. coli RNA polymerase incorporated about twice as much radioactivity into RNA with T4 DNA as the unmodified enzyme incorporated. This observation confirmed previous experiments which provided evidence that the Alt-catalyzed ADP-ribosylation of RNA polymerase enhances transcription from T4 early promoters (60, 73). On the other hand, the ModA-modified polymerase incorporated only 72% of the radioactivity that was found in the test with the unmodified enzyme and T4 DNA, underlining the negative influence of the ModA-catalyzed modification on the transcription from T4 early DNA. With E. coli DNA, all three enzymes seemed to be less active; however, the Alt-modified enzyme remained the most active of the three polymerases tested.
TABLE 2.
Radioactivity incorporated in in vitro transcription experimentsa
| Template | Radioactivity incorporated (Bq)
|
|
|---|---|---|
| T4 DNA | E. coli DNA | |
| Control (no RNA polymerase) | 94 | 34 |
| Wild-type BL21 polymerase | 9,155 | 2,744 |
| Alt-modified polymerase | 17,029 | 3,419 |
| ModA-modified polymerase | 6,576 | 2,619 |
For details see Materials and Methods.
Mutagenesis of ModA and ModB.
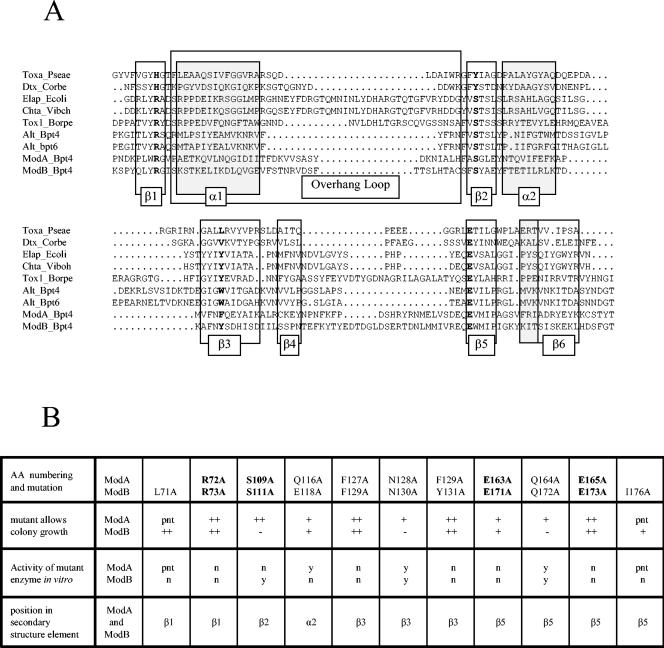
An alignment of the amino acid sequences (2) of ADP-RTs ModA (200 amino acids) and ModB (207 amino acids) of bacteriophage T4 revealed 25% identical and 47% homologous amino acids. This likely reflected an evolutionary relationship between the two phage enzymes. A comparison of the primary structures with all other ADP-RTs in the GenBank database revealed no significant sequence similarities. However, secondary structure alignments (4) provided evidence of structural homologies characteristic for the enzymes of the ribosyltransferase family. The emerging crystal structures provide increasing evidence that not only secondary structures but also tertiary structures are conserved among members of the mono-ADP-RT family (24, 64), and the active site motif R-S-EXE (1, 11, 13, 16, 65, 66, 69) is typical of arginine-specific ribosyltransferases (Fig. 5A shows the orientation).
FIG. 5.
Secondary structure alignment of ADP-ribosyltransferases of eukaryotic and prokaryotic origin, including the enzymes encoded by bacteriophage T4 (73). (A) Active site motif R-S-EXE (boldface) is composed of amino acid residues residing in or around sheets β1, β2, and β5. These secondary structure elements are arranged and stabilized close to each other, together with the nicotinamide portion of NAD+ and the arginine side chain of the acceptor protein (24, 64) (PDB accession codes 1QS2 and 1GIQ). Amino acids in the overhang loop, reduced to simple turns in some enzymes, in part might be involved in interactions with the target protein (5). Secondary structure elements making up the complex catalytic domain and the NAD+ binding pocket are enclosed in boxes. The following enzymes are shown: Toxa_Pseae, exotoxin A of P. aeruginosa; Dtx_Corbe, diphtheria toxin of C. diphtheriae; Elap_Ecoli, heat-labile enterotoxin A of E. coli; Chta_Vibch and Chta_Viboh, cholera toxin of V. cholerae; Tox1_Borpe, pertussis toxin of B. pertussis; Alt_Bpt4, Alt protein of phage T4 (amino acid 471); Alt_Bpt6, Alt protein of phage T6 (amino acid 473); ModA_Bpt4, ModA protein of phage T4 (amino acid 65); and ModB_Bpt4, ModB protein of phage T4 (amino acid 67) (for the T4 enzymes the number of the first amino acid in the alignment is indicated in parentheses). The enzyme codes used in this figure (e.g., Toxa_Pseae or ModB_Bpt4) are the accession codes for the sequences aligned (ENTREZ protein data library). (B) Estimated colony growth of E. coli C41(DE3) transformed with plasmids carrying the mutated genes and grown in the presence of ampicillin and IPTG (enzyme expression induced). Colony growth is indicated as follows: ++, regular colony size; +, smaller colonies. A minus sign indicates that the enzyme remains toxic despite the mutation introduced. The activity of a mutant in the in vitro ribosylation test is indicated as follows: y, yes; n, no. pnt indicates that the position was not tested in ModA; β1, β2, β3, β5, and α2 indicate the secondary structure elements that the corresponding position is associated with. For further details see the text. AA, amino acid.
With the aim of testing the sites of putative structural homology and in few cases their close proximity, we performed site-directed mutagenesis experiments with amino acids conserved among the T4 mod gene products and the other ADP-RTs. Mutants defective in NAD+ binding or in the transferase reaction allowed bacterial growth. The colony sizes under induced and noninduced growth conditions reflected the residual toxicity and, hence, were related to the relative remaining activities of the enzymes. Control experiments with the original pET-16b(+) plasmid in C41(DE3) revealed no difference in colony size under induced and noninduced conditions.
To assay the putative active sites of the Mod enzymes, we replaced nine of the amino acids in ModA with alanine. The results of these experiments are presented in Fig. 5B. The exchanges that reduced the toxicity to colony growth were R72A, S109A, and E165A, as well as F127A and F129A, both of which were located in the β3 region. Other mutations (e.g., Q116A, N128A, E163A, and Q164A) reduced the toxicity of the overexpressed ModA to a minor extent, and only small colonies were observed in the test.
In ModB, 11 amino acids were replaced by alanine. Mutations L71A and R73A in region β1, mutations F129A and Y131A in region β3, and the E173A mutation in region β5 allowed full colony growth and therefore largely abolished enzyme activity, while E118A, E171A, and I176A reduced the toxicity of the enzyme and still allowed small colonies to grow (Fig. 5B). Mutation L71A was situated in β1 and contributed to the hydrophobic pocket supporting nicotinamide binding, and mutation I176A was situated close to the EXE motif (EQE in both ModA and ModB). The two corresponding positions in ModA were not assayed. Mutants with the S111A and N130A mutations, as well as the Q172A mutation, in ModB exhibited toxicity close to that of the wild-type enzyme, suggesting that these amino acids alone do not decisively influence NAD+ binding and the transfer reaction. With the exception of S111A and the amino acids additionally tested with ModB, the results of the mutation experiments were similar to those obtained with ModA. In contrast to the nonpolar environment of Ser109 in ModA, Ser111 in ModB is followed by Tyr112, which is essentially able to form a similar hydrogen bond, which may additionally contribute to the stabilization of the intermediate oxocarbenium. Thus, the single mutation S111A did not reduce the activity of ModB to a point where toxicity was completely lost; the consensus motif of the amino acids involved in the stabilization of the intermediate state is STS (SGL in ModA and SYA in ModB), which allows ambiguities in hydrogen bonding, possibly depending on the enzyme and/or acceptor proteins.
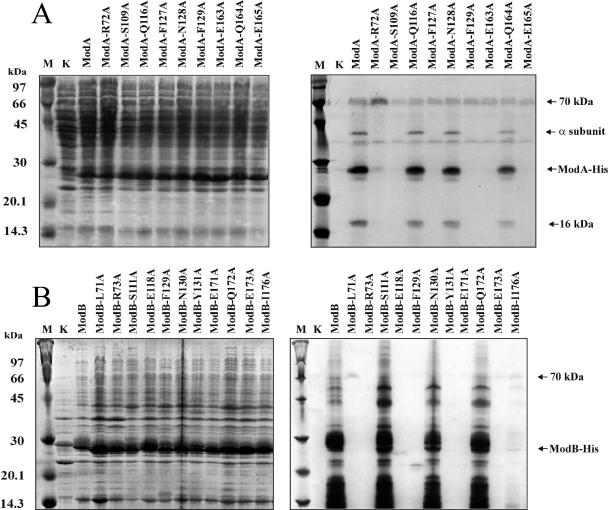
Mutants identified by the toxicity tests were also assayed for the ability to ADP-ribosylate. To do this, the overexpressed wild-type enzymes and the mutant enzymes were partially pu-rified from the corresponding cell lysates and subjected to ribosylation tests as described above. Mutations that did not result in an appreciable reduction in the enzyme toxicity essentially showed ribosylation products similar to those of the wild-type controls for ModA and ModB. Mutant proteins that had largely lost toxicity had also lost the ability to perform transribosylation and the ability to ADP-ribosylate their target proteins (Fig. 6). We concluded that the amino acids which were mutated in this study by site-directed mutagenesis and which showed reduced toxicity are important for the catalytic activity of the two Mod enzymes, and consequently, the alignments of the secondary structures indeed identified the active sites of both ADP-ribosyltransferases. Moreover, the transfer reaction catalyzed by the T4 enzymes must follow the Sn1-type reaction scheme proposed for mono-ADP-ribosyltransferases, which is entirely different from reaction scheme of ligases (40).
FIG. 6.
ADP-ribosyltransferase activity tests performed with ModA (A) and ModB (B) wild-type and mutant enzymes: 13% polyacrylamide gels stained with Coomassie brilliant blue (left gels) and autoradiographs of the stained gels (right gels). To monitor ADP-ribosylation activity, crude cell extracts of the different ModA- or ModB-producing cells were supplemented with soluble E. coli proteins, and in vitro labeling was performed as described in Materials and Methods. Lane M, Rainbow 14C-methylated protein molecular weight markers (Amersham Bioscience); lane K, controls consisting of an E. coli C41(DE3) cell lysate supplemented with radioactively labeled NAD+ but not with cell extracts of ModA- or ModB-producing cells. In the other lanes enzyme-producing cell extracts were added as indicated at the top.
DISCUSSION
In this paper we describe overexpression and isolation of the ADP-ribosyltransferases ModA and ModB of bacteriophage T4. Although both proteins were purified by partial denaturation from inclusion bodies, the renatured enzymes were active and modified defined sets of host proteins. Similar to the T4 Alt enzyme (36) and to other ribosyltransferases (47), both Mod proteins also perform a transribosylation reaction. As shown above, ADP-ribosyltransfer is catalyzed during enzyme synthesis in overexpressing cells with cellular NAD+ as a substrate. Since ADP-ribosylation is not reversible (36, 57), two precautions are necessary to monitor the reaction. First, radioactivity with the highest specific activity must be used to detect residual positions on cellular proteins that were modified with unlabeled cellular NAD+ during enzyme overexpression. Second, soluble cell extracts or purified proteins not previously exposed to the enzyme need to be added to the reaction mixtures. Skorko et al. (57), using uniformly labeled [14C]NAD+, added 40 μg of external RNA polymerase core enzyme to their reaction mixtures. Consequently, only two radioactive bands, the α subunit and a smaller protein most probably representing the transribosylated ModA protein, appeared on the autoradiographs. In contrast, our procedure added up to 35 μg of a soluble extract of all E. coli proteins (the exact amount depended on the number of protein bands to be resolved per gel lane) and labeled with 32P-labeled NAD+ three- to fourfold the specific activity used by Skorko and his collaborators (57). Therefore, the seemingly different results were likely due to the different proteins added to the reaction mixtures and were not caused by major differences between the enzymes or the reactions catalyzed.
In a previous study transcription of T4 early promoters by unribosylated and ADP-ribosylated (Alt-modified) E. coli RNA polymerase was assayed (60). T4 promoters resided on a 5-kb promoter probe vector. When integrated into a plasmid vector, not only does the T4 promoter lose phage-specific hydroxymethylation and glucosylation, but on small supercoiled plasmid DNA it also has a helicity that is quite different from that of the linear 169-kb genomic DNA with its high A-T skew. Since glucosylation might contribute to hydrogen bonding and hence to the structure of T4 DNA (for a review see reference 42), both factors are putatively important for correct T4 promoter recognition and viral transcription regulation. T4 early and middle-mode promoters harbor several glucosylated hydroxymethyl cytosine residues at defined positions (cf. T4 promoter logos in reference 42).
Aware of the limitations of different transcription test systems that also lack most of the T4-specific transcription factor and auxiliary protein environment, we complemented the data obtained previously by performing in vitro transcription experiments with Alt- and ModA-modified RNA polymerase and by using native T4 DNA as a template. The results show that under both experimental conditions, the Alt-induced modification of RNA polymerase increased transcription from T4 DNA by about 50%. In contrast, the ModA-induced modification reduced transcription from T4 DNA, as well as transcription from E. coli DNA. These results support previous findings (60, 73) that ADP-ribosylation catalyzed by Alt enhances transcription from T4 early promoters, possibly by outcompeting transcription initiated at host promoters. The ModA-catalyzed modification of host RNA polymerase prevents transcription from host promoters that carry an UP element (18, 53) and from T4 early promoters (21). However, since residual transcription was observed on all templates after ModA modification of RNA polymerase and since the initially low transcription of E. coli DNA was hardly reduced upon modification (Table 2), we concluded that the ModA-induced modification is not sufficient to prevent host transcription. This result also supports previous reports that the cessation of host transcription is mediated mainly by the T4 Alc protein (17). In contrast, the ModA-directed ribosylation reaction may condition E. coli RNA polymerase for the interaction with T4-encoded auxiliary factors, such as AsiA and MotA for middle-mode transcription, as well as gp55, gp33, gp45, and gp44/62 for late transcription, redirecting RNA polymerase to transcribe from T4 middle (27, 48) and late (26, 74, 75, 76) promoters.
The ADP-RTs of bacteriophage T4 have different primary structures and catalyze partially overlapping reactions (36, 63). Secondary structure alignment and mutation analyses showed that the phage enzymes are structurally and functionally related to the bacterial exotoxins and to a number of regulatory proteins, which are also members of this enzyme family. Several of the toxins are known to be encoded by prophages (67, 70). Although the Alt, ModA, and ModB genes had been identified as nonessential genes under laboratory conditions (20), the ADP-ribosylation of a large number of host proteins supports the view that these enzymes play an active role in advancing the phage replicative cycle in natural environments. To fully appreciate the role of these enzymes and their contributions to the regulation of the replicative processes, it would be desirable to further identify the target proteins and to study in detail the consequences of the phage-induced modifications. Does the modification of the target proteins change their reactions, or alternatively, are they simply inactivated to, e.g., stop host synthesis? The identification of a complete NAD+ salvage pathway in the T4-like vibriophage KVP40 (41) underlines the importance of NAD+ and NAD-dependent ribosyltransferases for viral development. With the recombinant T4-encoded ADP-ribosyltransferases in hand, there are ways to study in vitro and in more detail the consequences of modification of a number of host proteins, including RNA polymerase (ModA), and of the S1 protein (ModB).
Acknowledgments
We are grateful to Eric Miller, North Carolina State University, for thoughtful discussions about the manuscript and to K. K. McMahon, Texas Tech University, Lubbock, for her gift of mono-ADP-ribosyl antibodies. We also thank U. Aschke for expert technical assistance and V. Salniene (Institute of Biochemistry, Vilnius, Lithuania) for cloning modB into the vectors, supporting the mutation experiments.
This work was funded by Deutsche Forschungsgemeinschaft grant RU 123 22-2 (to W R.) and by grant 0311499 (Cooperation in Biotechnology with Lithuania) from the BMWFT to W.R. and R.N.
REFERENCES
- 1.Aktories, K., M. Jung, J. Bohmer, G. Fritz, J. Vandekerckhove, and I. Just. 1995. Studies on the active-site structure of C3-like exoenzymes: involvement of glutamic acid in catalysis of ADP-ribosylation. Biochimie 77:326-332. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl, ed. 1993. Current protocols in molecular biology. Greene Publishing and Wiley-Interscience, New York, N.Y.
- 4.Bazan, J. F., and F. Koch-Nolte. 1997. Sequence and structural links between distant ADP-ribosyltransferase families. Adv. Exp. Med. Biol. 419:99-107. [DOI] [PubMed] [Google Scholar]
- 5.Bell, C. E., and D. Eisenberg. 1996. Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide. Biochemistry 35:1137-1149. [DOI] [PubMed] [Google Scholar]
- 6.Blum, P., J. Ory, J. Bauernfeind, and J. Krska. 1992. Physiological consequences of DnaK and DnaJ overproduction in Escherichia coli. J. Bacteriol. 174:7436-7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blum, P., M. Velligan, N. Lin, and A. Matin. 1992. DnaK-mediated alterations in human growth hormone protein inclusion bodies. Bio/Technology 10:301-304. [DOI] [PubMed] [Google Scholar]
- 8.Boni, I. V., D. M. Isaeva, M. L. Musychenko, and N. V. Tzareva. 1991. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 19:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bortell, R., T. Kanaitsuka, L. A. Stevens, J. Moss, J. P. Mordes, A. A. Rossini, and D. L. Greiner. 1999. The RT6 Art2 family of ADP-ribosyltransferases in rat and mouse. Mol. Cell. Biochem. 193:61-68. [PubMed] [Google Scholar]
- 10.Burgess, R. R., and J. J. Jendrisak. 1975. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry 14:4634-4638. [DOI] [PubMed] [Google Scholar]
- 11.Burnette, W. N., V. L. Mar, B. W. Platler, J. D. Schlotterbeck, M. D. McGinley, K. S. Stoney, M. F. Rohde, and H. R. Kaslow. 1991. Site-specific mutagenesis of the catalytic subunit of cholera toxin: substituting lysine for arginine 7 causes loss of activity. Infect. Immunol. 59:4266-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cieplak, W., Jr., D. J. Mead, R. J. Messer, and C. C. Grant. 1995. Site-directed mutagenic alteration of potential active-site residues of the A subunit of Escherichia coli heat-labile enterotoxin. Evidence for a catalytic role for glutamic acid 112. J. Biol. Chem. 270:30545-30550. [DOI] [PubMed] [Google Scholar]
- 14.Colland, F., G. Orsini, E. N. Brody, H. Buc, and A. Kolb. 1998. The bacteriophage T4 AsiA protein: a molecular switch for sigma 70-dependent promoters. Mol. Microbiol. 27:819-829. [DOI] [PubMed] [Google Scholar]
- 15.Collier, R. J. 1975. Diphtheria toxin: mode of action and structure. Bacteriol. Rev. 39:54-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domenighini, M., and R. Rappuoli. 1996. Three conserved consensus sequences identify the NAD-binding site of ADP-ribosylating enzymes, expressed by eukaryotes, bacteria and T-even bacteriophages. Mol. Microbiol. 21:667-674. [DOI] [PubMed] [Google Scholar]
- 17.Drivdahl, R. H., and E. M. Kutter. 1990. Inhibition of transcription of cytosine-containing DNA in vitro by the alc gene product of bacteriophage T4. J. Bacteriol. 172:2716-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaal, T., W. Ross, E. E. Blatter, H. Tang, X. Jia, V. V. Krishnan, N. Assa-Munt, R. H. Ebright, and R. L. Gourse. 1996. DNA-binding determinants of the alpha subunit of RNA polymerase: novel DNA-binding domain architecture. Genes Dev. 10:16-26. [DOI] [PubMed] [Google Scholar]
- 19.Goff, C. G. 1974. Chemical structure of a modification of the Escherichia coli ribonucleic acid polymerase alpha polypeptides induced by bacteriophage T4 infection. J. Biol. Chem. 249:6181-6190. [PubMed] [Google Scholar]
- 20.Goff, C. G., and J. Setzer. 1980. ADP ribosylation of Escherichia coli RNA polymerase is nonessential for bacteriophage T4 development. J. Virol. 33:547-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldfarb, A., and Palm, P. 1981. Control of promoter utilization by bacteriophage T4-induced modification of RNA polymerase alpha subunit. Nucleic Acids Res. 9:4863-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goloubinoff, P., A. A. Gatenby, and G. H. Lorimer. 1989. GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature 337:44-47. [DOI] [PubMed] [Google Scholar]
- 23.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han S., J. A. Craig, C. D. Putnam, N. B. Carozzi, and J. A. Tainer. 1999. Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex. Nat. Struct. Biol. 6:932-936. [DOI] [PubMed] [Google Scholar]
- 25.Harrison, R. G. 2000. Expression of soluble heterologous proteins via fusion with NusA protein. Innovations 11:4-7. [Google Scholar]
- 26.Herendeen, D. R., G. A. Kassavetis, and E. P. Geiduschek. 1992. A transcriptional enhancer whose function imposes a requirement that proteins track along DNA. Science 256:1298-1303. [DOI] [PubMed] [Google Scholar]
- 27.Hinton, D. M., R. March-Amegadzie, J. S. Gerber, and M. Sharma. 1996. Characterization of pre-transcription complexes made at a bacteriophage T4 middle promoter: involvement of the T4 MotA activator and the T4 AsiA protein, a sigma 70 binding protein, in the formation of the open complex. J. Mol. Biol. 256:235-248. [DOI] [PubMed] [Google Scholar]
- 28.Hinton, D. M., and S. Vuthoori. 2000. Efficient inhibition of Escherichia coli RNA polymerase by the bacteriophage T4 AsiA protein requires that AsiA binds first to free sigma70. J. Mol. Biol. 304:731-739. [DOI] [PubMed] [Google Scholar]
- 29.Hlodan, R., S. Craig, and R. H. Pain. 1991. Protein folding and its implications for the production of recombinant proteins. Biotechnol. Genet. Eng. Rev. 9:47-88. [PubMed] [Google Scholar]
- 30.Honjo, T., Y. Nishizuka, and O. Hayaishi. 1968. Diphtheria toxin-dependent adenosine diphosphate ribosylation of aminoacyl transferase II and inhibition of protein synthesis. J. Biol. Chem. 243:3553-3555. [PubMed] [Google Scholar]
- 31.Horvitz, H. R. 1974. Bacteriophage T4 mutants deficient in alteration and modification of the Escherichia coli RNA polymerase. J. Mol. Biol. 90:739-750. [DOI] [PubMed] [Google Scholar]
- 32.Igarashi, K., and A. Ishihama. 1991. Bipartite functional map of the E. coli RNA polymerase alpha subunit: involvement of the C-terminal region in transcription activation by cAMP-CRP. Cell 65:1015-1022. [DOI] [PubMed] [Google Scholar]
- 33.Iglewski, B. H., and D. Kabat. 1975. NAD-dependent inhibition of protein synthesis by Pseudomonas aeruginosa toxin. Proc. Natl. Acad. Sci. USA 72:2284-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katada, T., and M. Ui. 1982. ADP ribosylation of the specific membrane protein of C6 cells by islet-activating protein associated with modification of adenylate cyclase activity. J. Biol. Chem. 257:7210-7216. [PubMed] [Google Scholar]
- 35.Khan, R. H., K. B. Rao, A. N. Eshwari, S. M. Totey, and A. K. Panda. 1998. Solubilization of recombinant ovine growth hormone with retention of native-like secondary structure and its refolding from the inclusion bodies of Escherichia coli. Biotechnol. Prog. 14:722-728. [DOI] [PubMed] [Google Scholar]
- 36.Koch, T., A. Raudonikiene, K. Wilkens, and W. Rüger. 1995. Overexpression, purification, and characterization of the ADP-ribosyltransferase gpAlt of bacteriophage T4: ADP-ribosylation of E. coli RNA polymerase modulates T4 “early” transcription. Gene Expr. 4:253-264. [PMC free article] [PubMed] [Google Scholar]
- 37.Koch, T., and W. Rüger. 1994. The ADP-ribosyltransferases gpAlt of bacteriophages T2, T4, and T6: sequencing of the genes and comparison of their products. Virology 203:294-298. [DOI] [PubMed] [Google Scholar]
- 38.Kolb, A., J. M. Hermoso, J. O. Thomas, and W. Szer. 1977. Nucleic acid helix-unwinding properties of ribosomal protein S1 and the role of S1 in mRNA binding to ribosomes. Proc. Natl. Acad. Sci. USA 74:2379-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LaVallie, E. R., E. A. DiBlasio, S. Kovacic, K. L. Grant, P. F. Schendel, and J. M. McCoy. 1993. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Bio/Technology 11:187-193. [DOI] [PubMed] [Google Scholar]
- 40.Lehman, I. R. 1974. DNA ligase: structure, mechanism, and function. Science 186:790-797. [DOI] [PubMed] [Google Scholar]
- 41.Miller, E. S., J. F. Heidelberg, J. A. Eisen, W. C. Nelson, A. S. Durkin, A. Ciecko, T. V. Feldblyum, O. White, T. Paulsen, W. C. Nierman, J. Lee, B. Szczypinski, and C. M. Frazer. 2003. The complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J. Bacteriol. 185:5220-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller, E. S., E. Kutter, G. Mosig, F. Arisaka, T. Kunisawa, and W. Rüger. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67:86-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 44.Mosig, G., N. E. Colowick, and B. C. Pietz. 1998. Several new bacteriophage T4 genes, mapped by sequencing deletion endpoints between genes 56 (dCTPase) and dda (a DNA-dependent ATPase-helicase) modulate transcription. Gene 223:143-155. [DOI] [PubMed] [Google Scholar]
- 45.Moss, J., J. C. Osborne, P. H. Fishman, Jr., S. Nakaya, and D. C. Robertson. 1981. Escherichia coli heat-labile enterotoxin. Ganglioside specificity and ADP-ribosyltransferase activity. J. Biol. Chem. 256:12861-12865. [PubMed] [Google Scholar]
- 46.Moss, J., and M. Vaughan. 1988. ADP-ribosylation of guanyl nucleotide-binding regulatory proteins by bacterial toxins. Adv. Enzymol. Relat. Areas Mol. Biol. 61:303-379. [DOI] [PubMed] [Google Scholar]
- 47.Okamoto, K., K. Okamoto, A. Miyama, T. Tsuji, T. Honda, and T. Tiwatani. 1988. Effect of substitution of glycine for arginine at position 146 of the A1 subunit on biological activity of Escherichia coli heat-labile enterotoxin. J. Bacteriol. 170:2208-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ouhammouch, M., K. Adelman, S. R. Harvey, G. Orsini, and E. N. Brody. 1995. Bacteriophage T4 MotA and AsiA proteins suffice to direct Escherichia coli RNA polymerase to initiate transcription at T4 middle promoters. Proc. Natl. Acad. Sci. USA 92:1451-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ovchinnikov, Y. A., V. M. Lipkin, N. N. Modyanov, O. Y. Chertov, and Y. V. Smirnov. 1977. Primary structure of alpha-subunit of DNA-dependent RNA polymerase from Escherichia coli. FEBS Lett. 76:108-111. [DOI] [PubMed] [Google Scholar]
- 50.Pallen, M. J., A. C. Lam, N. J. Loman, and A. McBride. 2001. An abundance of bacterial ADP-ribosyltransferases: implications for the origin of exotoxins and their human homologues. Trends Microbiol. 9:302-307. [DOI] [PubMed] [Google Scholar]
- 51.Pope, M. R., S. A. Murrell, and P. W. Ludden. 1985. Covalent modification of the iron protein of nitrogenase from Rhodospirillum rubrum by adenosine diphosphoribosylation of a specific arginine residue. Proc. Natl. Acad. Sci. USA 82:3173-3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rohrer, H., W. Zillig, and R. Mailhammer. 1975. ADP-ribosylation of DNA-dependent RNA polymerase of Escherichia coli by an NAD+: protein ADP-ribosyltransferase from bacteriophage T4. Eur. J. Biochem. 60:227-238. [DOI] [PubMed] [Google Scholar]
- 53.Ross, W., S. E. Aiyar, J. Salomon, and R. L. Gourse. 1998. Escherichia coli promoters with UP elements of different strengths: modular structure of bacterial promoters. J. Bacteriol. 180:5375-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schein, C. H., and M. Noteborn. 1988. Formation of soluble recombinant proteins in Escherichia coli is favoured by lower growth temperature. Bio/Technology 6:291-294. [Google Scholar]
- 55.Schein, C. H. 1991. Optimizing protein folding to the native state in bacteria. Curr. Opin. Biotechnol. 2:746-750. [DOI] [PubMed] [Google Scholar]
- 56.Shall, S. 1995. ADP-ribosylation reactions. Biochimie 77:313-318. [DOI] [PubMed] [Google Scholar]
- 57.Skorko, R., W. Zillig, H. Rohrer, H. Fujiki, and R. Mailhammer. 1977. Purification and properties of the NAD+:protein ADP-ribosyltransferase responsible for the T4-phage-induced modification of the alpha subunit of DNA-dependent RNA polymerase of Escherichia coli. Eur. J. Biochem. 79:55-66. [DOI] [PubMed] [Google Scholar]
- 58.Soman, G., J. F. Miller, and D. J. Graves. 1984. Use of guanylhydrazones as substrates for guanidine-specific mono-ADP-ribosyltransferases. Methods Enzymol. 106:403-410. [DOI] [PubMed] [Google Scholar]
- 59.Soman, G., K. B. Tomer, and D. J. Graves. 1983. Assay of mono ADP-ribosyltransferase activity by using guanylhydrazones. Anal. Biochem. 134:101-110. [DOI] [PubMed] [Google Scholar]
- 60.Sommer, N., V. Salniene, E. Gineikiene, R. Nivinskas, and W. Rüger. 2000. T4 early promoter strength probed in vivo with unribosylated and ADP-ribosylated Escherichia coli RNA polymerase: a mutation analysis. Microbiology 146:2643-2653. [DOI] [PubMed] [Google Scholar]
- 61.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 62.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 63.Tiemann, B., R. Depping, and W. Rüger. 1999. Overexpression, purification, and partial characterization of ADP-ribosyltransferases modA and modB of bacteriophage T4. Gene Expr. 8:187-196. [PMC free article] [PubMed] [Google Scholar]
- 64.Tsuge, H., M. Nagahama, H. Nishimura, J. Hisatsune, Y. Sakaguchi, Y. Itogawa, N. Katunuma, and J. Sakurai. 2003. Crystal structure and site-directed mutagenesis of enzymatic components from Clostridium perfringens iota-toxin. J. Mol. Biol. 325:471-483. [DOI] [PubMed] [Google Scholar]
- 65.Tsuji, T., T. Inoue, A. Miyama, and M. Noda. 1991. Glutamic acid-112 of the A subunit of heat-labile enterotoxin from enterotoxigenic Escherichia coli is important for ADP-ribosyltransferase activity. FEBS Lett. 291:319-321. [DOI] [PubMed] [Google Scholar]
- 66.Tweten, R. K., J. T. Barbieri, and R. J. Collier. 1985. Diphtheria toxin. Effect of substituting aspartic acid for glutamic acid 148 on ADP-ribosyltransferase activity. J. Biol. Chem. 260:10392-10394. [PubMed] [Google Scholar]
- 67.Uchida, T., D. M. Gill, and A. M. J. Pappenheimer. 1971. Mutation in the structural gene for diphtheria toxin carried by temperate phage. Nat. New Biol. 233:8-11. [DOI] [PubMed] [Google Scholar]
- 68.Urbauer, J. L., M. F. Simeonov, R. J. Urbauer, K. Adelman, J. M. Gilmore, and E. N. Brody. 2002. Solution structure and stability of the anti-sigma factor AsiA: implications for novel functions. Proc. Natl. Acad. Sci. USA 99:1831-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Damme, J., M. Jung, F. Hofmann, I. Just, J. Vandekerckhove, and K. Aktories. 1996. Analysis of the catalytic site of the actin ADP-ribosylating Clostridium perfringens iota toxin. FEBS Lett. 380:291-295. [DOI] [PubMed] [Google Scholar]
- 70.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 71.Wickner, W., A. J. Driessen, and F. U. Hartl. 1991. The enzymology of protein translocation across the Escherichia coli plasma membrane. Annu. Rev. Biochem. 60:101-124. [DOI] [PubMed] [Google Scholar]
- 72.Wilkens, K., and W. Rüger. 1996. Characterization of bacteriophage T4 early promoters in vivo with a new promoter probe vector. Plasmid 35:108-120. [DOI] [PubMed] [Google Scholar]
- 73.Wilkens, K., B. Tiemann, F. Bazan, and W. Rüger. 1997. ADP-ribosylation and early transcription regulation by bacteriophage T4. Adv. Exp. Med. Biol. 419:71-82. [DOI] [PubMed] [Google Scholar]
- 74.Williams, K. P., G. A. Kassavetis, and E. P. Geiduschek. 1987. Interactions of the bacteriophage T4 gene 55 product with Escherichia coli RNA polymerase. Competition with Escherichia coli sigma 70 and release from late T4 transcription complexes following initiation. J. Biol. Chem. 262:12365-12371. [PubMed] [Google Scholar]
- 75.Williams, K. P., R. Muller, W. Rüger, and E. P. Geiduschek. 1989. Overproduced bacteriophage T4 gene 33 protein binds RNA polymerase. J. Bacteriol. 171:3579-3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wong, K., and E. P. Geiduschek. 1998. Activator-sigma interaction: a hydrophobic segment mediates the interaction of a sigma family promoter recognition protein with a sliding clamp transcription activator. J. Mol. Biol. 284:195-203. [DOI] [PubMed] [Google Scholar]
- 77.Zhang, Y., E. L. Pohlmann, C. M. Halbleib, P. W. Ludden, and G. P. Roberts. 2001. Effect of PII and its homolog GlnK on reversible ADP-ribosylation of dinitrogenase reductase by heterologous expression of the Rhodospirillum rubrum dinitrogenase reductase ADP-ribosyl transferase-dinitrogenase reductase-activating glycohydrolase regulatory system in Klebsiella pneumoniae. J. Bacteriol. 183:1610-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]