Abstract
The locus of enterocyte effacement (LEE), which includes five major operons (LEE1 through LEE4 and tir), enables enterohemorrhagic Escherichia coli (EHEC) O157:H7 to produce attaching and effacing lesions on host cells. Expression of LEE2, LEE3, and tir is positively regulated by ler, a gene located in LEE1. Transcriptional regulation of the esp operon (LEE4), however, is not well defined. Transposon mutagenesis was used to identify transcriptional regulators of the esp operon by screening for mutants with increased β-galactosidase activity in an EHEC O157:H7 strain harboring an esp::lac transcriptional fusion. All mutants with significant increases in β-galactosidase activity had transposon insertions in hha (hha::Tn). Specific complementation of the hha::Tn mutation with a plasmid-encoded copy of hha reduced β-galactosidase activity to the level expressed in the parental esp::lac strain. Purified Hha, however, bound poorly to the esp promoter, suggesting that Hha might repress the transcription of a positive regulator of esp. Transposon mutagenesis of a Δhha esp::lac strain expressing elevated levels of β-galactosidase resulted in ler mutants with reduced β-galactosidase activity. Purified Hha bound to the ler promoter with a higher affinity, and complementation of a Δhha mutation in a Δhha ler::lac strain repressed β-galactosidase activity to the level expressed in a ler::lac strain. A positive regulatory role of ler in esp expression was demonstrated by specific binding of Ler to the esp promoter, reduced expression of β-galactosidase in Δler esp::lac strains with and without hha, and severalfold-increased transcription of ler and espA in strains lacking hha. These results indicate that hha-mediated repression of ler causes reduced expression of the esp operon.
Enterohemorrhagic Escherichia coli (EHEC) O157:H7, a Shiga toxin-producing E. coli, causes a broad spectrum of diseases, including uncomplicated diarrhea, hemorrhagic colitis, and hemolytic uremic syndrome (21). In addition to Shiga toxins, which act on vascular endothelial cells to produce hemolytic uremic syndrome, EHEC O157:H7 harbors genes mediating its adherence to intestinal epithelial cells by a characteristic attaching-and-effacing mechanism (31). The attaching and effacing lesions are characterized by the loss of microvilli and the formation of pedestals of polymerized actin and other cytoskeletal elements underneath and around the infecting bacterial cells (10). The attaching and effacing phenotype requires concerted action of several genes contained within a pathogenicity island, called the locus of enterocyte effacement (LEE) (32). LEE encodes a type III secretion system, an outer membrane protein called intimin, a translocated intimin receptor called Tir, and secreted proteins EspA, EspD, and EspB. The genes espA, espD, and espB, which encode Esp proteins, are organized into a single operon (2) and are secreted by the type III secretion apparatus. EspA forms a novel filament-like structure which is essential for early bacterial attachment to epithelial cells and translocation of EspB and Tir into host cells (11, 22).
Nucleotide sequence analysis of the LEE region of EHEC O157:H7 strain EDL 933 (37) revealed the presence of 41 open reading frames (ORFs), most of which are organized into five polycistronic operons named LEE1 through LEE4 and tir. The genetic organization of the LEE-encoded ORFs of EHEC O157:H7 is similar to that reported for the LEE of enteropathogenic E. coli (EPEC) O127:H6 (13). The Ler protein, encoded by the gene ler of the LEE1 operon, is essential for transcriptional activation of the LEE2, LEE3, and tir operons, which encode many of the protein factors required for the production of the attaching and effacing phenotype by EHEC and EPEC strains on host cells (12). Ler has also been shown to be a weak regulator of esp operon (LEE4) expression in EPEC (28).
Ler shows some similarities with H-NS (histone-like nucleoid-structuring protein) and StpA (suppressor of td− phenotype A) of Salmonella enterica and E. coli, predominantly with the DNA-binding domain near the carboxy terminus (3, 13). Binding of Ler to the LEE2 regulatory region is required for activation of both the LEE2 and LEE3 operons (43), and Ler counteracts the negative regulation exerted by some H-NS proteins on the expression of LEE2 and LEE3 in EPEC (6). However, the genetic factors required for expression of the esp operon in EHEC O157:H7 are not fully understood.
The objective of this study was to identify transcriptional regulators of esp (LEE4). Our approach was to use transposon mutagenesis of EHEC O157:H7 strain 86-24 Δstx2 Δlac carrying an esp::lac transcriptional fusion to identify mutants with altered β-galactosidase activity compared to the parent strain and then characterize the genes altered by the transposon insertions. With this strategy, hha was identified as an indirect negative regulator of esp. The gene hha is implicated in the negative regulation of α-hemolysin expression in pathogenic E. coli (33). We provide several lines of data that show that Hha represses ler transcription and Ler is a positive regulator of esp expression.
(A preliminary account of this work was presented at the 102nd Annual Meeting of the American Society for Microbiology, Salt Lake City, Utah, 19 to 23 May 2002 [V. K. Sharma and R. W. Morgan, Abstr. 102nd Annu. Meet. Am. Soc. Microbiol., abstr. B269, p.78, 2002].)
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. All E. coli strains were propagated on Luria-Bertani (LB) agar at 37°C. For liquid cultures, well-isolated colonies from LB agar plates were inoculated into LB broth and incubated at 37°C unless stated otherwise in an orbital shaker at 200 rpm. Dulbecco's modified Eagle's medium (DMEM) containing 0.45% glucose was purchased from Invitrogen, Carlsbad, Calif.. Medium were supplemented, when required, with ampicillin at 50 μg/ml or kanamycin at 50 μg/ml.
TABLE 1.
Bacterial strains and plasmidsa
| Strain or plasmid | Relevant genotype and description | Source or reference |
|---|---|---|
| E. coli | ||
| EHEC 86-24 | stx2+ EHEC strain (serotype O157:H7) | 18 |
| EHEC Δstx2 | 86-24 deleted of stx2 | This study |
| EHEC Δstx2 Δlac | 86-24 deleted of stx2 and lac operon | This study |
| EHEC esp::lac | 86-24 Δstx2 Δlac containing chromosomal esp::lac transcriptional fusion | This study |
| EHEC esp::lac/TnKn | Transposon mutant of EHEC esp::lac | This study |
| EHEC esp::lac hha::TnKn | 86-24 esp::lac containing transposon TnKn in hha | This study |
| EHEC esp::lac Δhha | 86-24 esp::lac deleted of hha | This study |
| EHEC Δhha | 86-24 deleted of hha | This study |
| TOP 10 | endA1 recA1 hsdR17(rK− mK−) supE44 φ80dlacZΔM15Δ(lacZYA-argF) | Invitrogen |
| DH10B | endA1 recA1 hsdR17(rK− mK−) supE44 φ80dlacZΔM15Δ(lacZYA-argF) | Gibco-BRL |
| BL21 | F−ompT hsdS(rB− mB−) gal dcm | Amersham Biosciences |
| Plasmids | ||
| pCR2.1 | Cloning vector | Invitrogen |
| pBR322 | Cloning vector | 4 |
| pAM450 | Suicide vector | 27 |
| pUC18 | Cloning vector | 45 |
| pGEX-4T-3 | Protein expression vector | Amersham Biosciences |
| pSM71 | pAM450 derivative used for deleting stx2 in EHEC 86-24 | This study |
| pSM80 | pAM450 derivative used for deleting the lac operon in 86-24 Δstx2 | This study |
| pSM102 | pAM450 derivative used for constructing an esp::lac transcriptional fusion | This study |
| pSM116 | pBR322 containing a cloned copy of hha | This study |
| pSM122 | pAM450 derivative used for deleting hha | This study |
| pSM138 | pGEX-4T-3 carrying an hha ORF for producing GST-Hha fusion protein | This study |
| pSM142 | pCR2.1 containing esp promoter | This study |
| pSM143 | pCR2.1 containing hilA promoter | This study |
| pSM146 | pCR2.1 containing ler promoter | This study |
| pSM163 | pGEX-4T-3 carrying ler ORF for producing GST-Ler fusion protein | This study |
| pSM188 | pAM450 derivative used for deleting ler | This study |
| pSM197R | pCR2.1 containing hha | This study |
| pSM212 | pAM450 derivative used for constructing an esp::lac transcriptional fusion | This study |
Detailed descriptions of the bacterial strains and plasmids listed in this table are provided in Materials and Methods.
Primer design, PCR amplification, and DNA sequencing.
The primers for PCR amplification of the nucleotide sequences of EHEC O157:H7 strain 86-24 are listed in Table 2. These primer sequences were selected from the published sequence of EHEC O157:H7 EDL 933 (38). Primers were synthesized by Integrated DNA Technologies (Coralville, Iowa). PCR amplifications were performed in 50 μl containing 5 μl of DNA (0.2 μg) and 0.3 μM each of the forward and reverse primers. The AmpliTaq Gold (PE Biosystems, Foster City, Calif.) or Failsafe PCR kit (Epicenter Technologies, Madison, Wis.) was used to amplify DNA fragments of <2.0 and >2.0 kb, respectively, according to the instructions provided by the manufacturers of these kits. PCR-amplified products were purified either with the Qiagen PCR purification kit or by first resolving PCR samples on agarose gels followed by extraction of DNA from agarose slices with a QIAquick gel extraction kit (Qiagen, Valencia, Calif.).
TABLE 2.
Primers used for PCR and sequencinga
| Primer | Nucleotide sequence (5′-3′) | Position (bp)/gene/accession no./reference |
|---|---|---|
| VS174 | CATCTAGAGGAAGCAGTAGACGAAATCG | 19931-19950/L0097/AF125520 |
| VS175 | CATCTAGAGGTCTTCTTCCGGTTTGTTC | 24224-24205/L0105/AF125520 |
| VS177 | ATCCCGGGTGAGGCATAACCTGATTCGTG | 22700-22720/region between stx2 and L0105/AE125520 |
| VS178 | ATCCCGGGTTCATATACAGGTGTTCCTTTTGG | 21466-21443/region between argO and stx2/AE125520 |
| VS244 | ACGTCTAGATGAGCAAGATGCTGAATCACTG | 2688-2667/Z0443/AE005214 |
| VS245 | CGAGTCGACCATTCACCACCCTGAATTGAC | 1174-1194/lacI-Z0442 intergenic region AE005214 |
| VS246 | CGAGTCGACAGAGATTACAAAGTTGAATCGTCAG | 6257-6231/lacA/AE005213 |
| VS247 | ACGTCTAGAACTGTTACAGATGATCCCACTGC | 4685-4706/cynS/AE005213 |
| VS256 | CAGTCTAGACAGGTAAGGTTTATCTG | 15241-15221/escD/AE071034 |
| VS257 | CAGGTCGACTATATACCTTTGATAATTTTC | 13742-13765/sepL-espA intergenic/region AE071034 |
| VS264 | CAGGTCGACGCAACATCCGTTGTTAATGTTGAG | 13726-13704espA/AE071034 |
| VS265 | CAGTCTAGATCCCGGCAGAACGAATACCGTTC | 12256-12278/espD/AE071034 |
| VS266 | CAGGTCGACAATTTCACACAGGATACAGCTATG | 11304-11287lacZ/AE005213 |
| VS268 | CAAATTGCCACTCACCATTCAG | 11100-11121/lacZ/AE005213 |
| VS269 | TTGAATTATGGCCCACACCAG | 8451-8431/lacZ/AE005213 |
| VS270 | TGTATTGTAACAGTGGCCCGAAG | 7854-7876/lacY/AE005213 |
| VS275 | CCGGTTATTTACCAAGGGATATTGC | 13159-13183/espA/AE071034 |
| VS276 | AAATGGCGCCTCTTTACTTGACTG | 14266-14242/sepL/AE071034 |
| VS280 | CAGGTCGACCCTGATAAGCGAAGCGTATCAGGC | 6191-6214/cynX-lacA intergenic region/AE005213 |
| VS303 | CATCTAGAGATTCCGTTCTCCGTTATGC | 3045-3026/acrB/AE005213/AE005225 |
| VS304 | CAGTCGACTCGGACATACTTCTACCCATG | 1630-1650/ybaJ-hha intergenic region/AE005225 |
| VS305 | CAGTCGACACTACTGAACAACATAAAGGTG | 1771-1792/ybaJ/AE005225 |
| VS306 | CAGTCGACTCGCTTTCGGAGCTATAACCG | 1409-1388/ylaD-hha intergenic region/AE005225 |
| VS307 | CATCTAGAGCTGGCAGGAGATAAGGAGGT | 1-20/downstream of ylaC/AE005225 |
| VS308 | ACGGATCCATGGTATTAATATGCAGGATC | 1769-1746/hha/AE005225 |
| VS309 | CAGGATCCCCGGTTATAGCTCCGAAAG | 1389-1409/hha/AE005225 |
| VS317 | ACGTCGACCCGGTTATAGCTCCGAAAG | 1389-1409/hha/AE005225 |
| VS318 | ACGAATTCAATGTCCGAAAAACCTTTAACG | 1637-1619/hha/AE005225 |
| VS319 | CCGTTGAAGTGAAAGACGGTC | 1453-1474/gapA/AE005401 |
| VS320 | AACCACTTTCTTCGCACCAGC | 1638-1618/gapA/AE005401 |
| VS323 | TGTGGATGCCAAAATTGCTG | 13493-13474/espA/AE071034 |
| VS324 | CGCCTTCACTGTTTGCAGATC | 13322-13342/espA/AE071034 |
| VS327 | CATCTAGAACAGGCTCTAAAACGGTTAAC | 41697-41677/LEE/AE071034 |
| VS328 | CAGTCGACGCTTTAATATTTTAAGCTATTAGCG | 40200-40222/LEE/AE071034 |
| VS330 | CATCTAGACTTATCCTCTGGTATGATATCTTC | 38596-38619/LEE/AE071034 |
| VS331 | CAGGATCCATGCGGAGATTATTTATTATG | 40199-40173/lerORF/AE071034 |
| VS332 | CAGTCGACATGTTAAATATTTTTCAGCGG | 39806-39827/lerORF/AE071034 |
| VS339 | AAAGACATGATATCGGCACAGC | 2018-1918/ybaJ/AE005225 |
| VS340 | TGAGCATATTGCGACCTTCG | 1882-1864/ybaJ/AE005225 |
| VS341 | CGGAATTCGTCCAGATGACA | 8153-8168/hilApromoter/AE008831.1/14 |
| VS342 | TGGGGTGTAAATGCTGCTT | 8627-8609/hilApromoter/AE008831.1/14 |
| VS353 | GCTTAACCACCTCATCCTTCG | 13639-13659/espApromoter/AE071034 |
| VS354 | GACTGGTTGCTGGAGAGAGTCC | 13982-13963/espApromoter/AE071034 |
| VS355 | GAATGTATGGACTTGTTGTATGTG | 40139-40162/lerpromoter/AE071034 |
| VS356 | TTTGTTTATGCAATGAGATCTATC | 40526-40505/lerpromoter/AE071034 |
| VS376 | AGTCCATCATCAGGCACATTAG | 39985-40006/lerORF/AE071034 |
| VS371 | CAGGATCCATGAAGAAGATGTTTATGGCGG | 22433-22454/stx2B/AF125520 |
| VS372 | CAGTCGACTCAGTCATTATTAAACTGCACTTCAG | 22702-22676/stx2B/AF125520 |
| VS377 | CATACAACAAGTCCCATACATTC | 40160-41139lerORF/AE071034 |
| VS398 | GAGTCGACCATGAAATAATTAAATGATAACG | 30809-39787/LEE/AE071034 |
| VS415 | GGAGTCGACGAATAAAAACCAAGCTGAACC | 40534-40554/LEE/AE071034 |
| VS426 | GCAGTTCCTTTGCTTCATTG | 39642-39662/LEE/AE071034 |
| VS427 | GCCTTCCTGTAACTCGAATTAAG | 40593-40571/LEE/AE071034 |
The position of the primer sequence represents the location in the published sequence deposited under the indicated accession number at NCBI. Underlined sequences GGATCC, GAATTC, GTCGAC, CCCGGG, and TCTAGA represent restriction sites for BamHI, EcoRI, SalI, SmaI, and XbaI, respectively.
DNA sequencing was performed with a Thermo Sequenase dye terminator cycle sequencing kit (Amersham Pharmacia Biotech Inc., Piscataway, N.J.) and an ABI 377 DNA sequencer (PE Applied Biosystems, Foster City, Calif.).
Deletion of stx2 and lac operons.
All DNA manipulations were performed by standard methods (40). To create deletions of target genes in the chromosome of EHEC O157:H7 strain 86-24 hha+ lac+, a 1.5-kb sequence located upstream (termed US for upstream sequence) and a 1.5-kb sequence located downstream (termed DS for downstream sequence) of the genes to be deleted were isolated by PCR. The primer sets (VS174-XbaI and VS178-SmaI and VS175-XbaI and VS177-SmaI to amplify US and DS fragments, respectively, for the stx2 deletion; VS244-XbaI and VS245-SalI and VS246-SalI and VS247-XbaI to amplify US and DS fragments, respectively, for the lac deletion) used in PCR amplification incorporated restriction sites for XbaI at the 5′ and SmaI (for stx2 deletion) or SalI (for lac deletion) at the 3′ end of fragments containing the US, and SmaI (for stx2 deletion) or SalI (for lac deletion) at the 3′ and XbaI at the 5′ end of fragments carrying the DS. The 3′ end of the US fragment was joined to the 5′ end of the DS fragment through SmaI (for stx2 fragments) or SalI (for lac fragments) to generate a 3-kb fragment containing the US and DS fragments (US-DS).
The 3-kb US-DS fragment, containing restriction sites for XbaI at its 5′ and 3′ ends, was cloned at the XbaI site of temperature-sensitive, ampicillin-resistance-encoding plasmid pAM450 (27) to generate plasmids pSM71 (for stx2 deletion) and pSM80 (for lac deletion). Plasmid pSM71 was electroporated into EHEC O157:H7 strain 86-24 with a Gene Pulser (Bio-Rad, Richmond, Calif.) at settings of 200 Ω, 25 μF, and 2.5 kV, and an isolate containing pSM71 was cultured under conditions (27) that facilitated exchange of the cloned 3-kb fragment of pSM71 with sequences flanking the stx2 operon to create a deletion of stx2. The chromosomal deletion for stx2 was confirmed by PCR with the primer set VS174 and VS175. Plasmid pSM80 was electroporated into EHEC O157:H7 strain 86-24 Δstx2 to delete the lac operon by the procedure described above. The lac operon deletion in the chromosome was confirmed by PCR with primers VS244 and VS247. In addition, isolates confirmed to have lost the lac operon were tested for β-galactosidase activity.
Determination of β-galactosidase activity.
An overnight culture was diluted 1:100 in LB broth or DMEM and incubated at 37°C. Samples were taken at different time intervals to measure the optical density at 600 nm (OD600) and β-galactosidase activity (30).
Construction of esp::lac and ler::lac transcriptional fusions.
To introduce esp::lac and ler::lac transcriptional fusions into the chromosome of strain 86-24 Δstx2 Δlac, we used the same procedure that we employed for deleting the stx2 and lac operons from the chromosome. Briefly, a 1.5-kb sequence located upstream (US) and a 1.5-kb sequence located downstream (DS) of the start codons for the espA and ler ORFs were isolated by PCR. The primer sets (VS256-XbaI and VS257-SalI and VS264-SalI and VS265-XbaI, for amplifying the US and DS fragments, respectively, for constructing the espA::lac fusion and VS327-XbaI and VS328-SalI and VS330-XbaI and VS398-SalI to amplify the US and DS fragments, respectively, for constructing the ler::lac fusion) used in PCR amplification incorporated restriction sites for XbaI at the 5′ and SalI at the 3′ end of fragments containing the US and SalI at the 3′ and XbaI at the 5′ end of fragments carrying the DS.
The 3′ end of the US fragment was joined to the 5′ end of the DS fragment through SalI to generate a 3-kb US-DS fragment. A 5.1-kb SalI fragment containing the lacZ, lacY, and lacA ORFs (lac cassette) was isolated from strain 86-24 Δstx2 lac+ by PCR with primers VS266-SalI and VS280-SalI and cloned at the SalI site (present at the junction of the US and DS) of the 3-kb US-DS fragment to generate an 8.1-kb US-lac-DS fragment, flanked at its 5′ and 3′ ends by XbaI and containing the lacZ ORF immediately downstream of the espA or ler promoter. The 8.1-kb fragment containing an esp::lac or ler::lac transcriptional fusion was cloned at the XbaI site of pAM450 to produce plasmids pSM102 and pSM212, respectively. Plasmids pSM102 and pSM212 were introduced into strain 86-24 Δstx2 Δlac by electroporation, and isolates containing pSM102 or pSM212 were cultured under conditions (described above) to facilitate integration and excision events for generating espA::lac and ler::lac transcriptional fusions by replacement of chromosomal sequences in the esp and ler regions, respectively, by the sequence present on the 8.1-kb fragment of pSM102 and pSM212, respectively. The presence of a chromosomal esp::lac or ler::lac transcriptional fusion was confirmed by PCR with primers VS276 and VS270 and VS269 and VS275 to amplify the 3.6-kb (escD-lacZ lacY region) and 2.6-kb (lacY lacA-esp region) fragments from the chromosomally generated esp::lac fusion or with primers VS270 and VS427 and VS269 and VS426 to amplify the 3.8-kb (5′ of the ler ORF-lacZ lacY region) and 2.3-kb (lacY lacA-3′ of the ler ORF region) fragments from the chromosomally generated ler::lac fusion, respectively. Isolates carrying the esp::lac or ler::lac fusion were tested for their β-galactosidase activities.
Transposon mutagenesis.
Strain 86-24 Δstx2 Δlac containing a chromosomal esp::lac or ler::lac fusion was transformed with a transposon encoding kanamycin resistance (TnKn; EZ::Tn<KAN-2>Tnp; Epicenter Technologies, Madison, Wis.). Transformed cells were plated on LB agar containing kanamycin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). After overnight incubation at 37°C, plates were visually screened for colonies appearing either lighter or darker blue than the majority of isolates on the plate. Differently colored colonies were selected for quantitative measurement of β-galactosidase activity.
Determination of transposon insertion sites.
Genomic DNAs of transposon mutants were digested with EcoRI (the transposon lacks EcoRI sites) and cloned at the EcoRI site of pUC18 (45). The recombinant plasmids were subjected to DNA sequencing with primers complementary to the 5′ (KAN-2 FP-1) and 3′ (KAN-2 RP-1) ends of the transposon (EZ::Tn<KAN-2>Tnp). The deduced DNA sequences were compared to nucleotide sequences in the database at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov).
Construction of in-frame hha and ler deletions.
A 1.3-kb sequence located upstream (US) and a 1.5-kb sequence located downstream (DS) of hha and a 1.17-kb sequence located upstream (US) and a 1.2-kb sequence located downstream (DS) of ler were isolated by PCR. The primer sets (VS303-XbaI and VS305-SalI and VS306-SalI and VS307-XbaI for amplifying the US and DS fragments, respectively, of hha, and VS327-XbaI and VS415-SalI and VS398-SalI and VS330-XbaI, for amplifying the US and DS fragments, respectively, of ler) used in PCR amplification incorporated restriction sites for XbaI at the 5′ and SalI at the 3′ end of fragments containing the US and SalI at the 3′ and XbaI at the 5′ end of fragments carrying the DS. The 3′ end of fragment US was joined to the 5′ end of fragment DS through SalI to generate 2.8-kb (for the hha deletion) and 2.37-kb (for the ler deletion) US-DS fragments, which were cloned at the XbaI site of pAM450 to generate pSM122 and pSM188, respectively. Plasmids pSM122 and pSM188 were introduced into strain 86-24 Δstx2 Δlac or 86-24 carrying the esp::lac or ler::lac fusion, and an isolate containing pSM122 or pSM188 was cultured under the conditions described above to generate a hha or ler deletion. The presence of the hha or ler deletion was confirmed by PCR with primers VS309 and VS340 or VS426 and VS427, respectively.
Cloning of hha.
The hha gene (380 bp) was isolated by PCR from strain 86-24 with primers VS308 and VS309, and the amplified product was digested with BamHI and cloned at the BamHI site of pBR322 to generate pSM116. A 630-bp fragment containing hha was also isolated from 86-24 with primers VS309 and VS339 and cloned in pCR2.1-TOPO to construct pSM197R. Plasmids pSM116 and pSM197R were used in complementation of hha::TnKn and Δhha mutants of 86-24.
Purification of Hha and Ler.
A 263-bp Hha-encoding fragment was PCR amplified from strain 86-24 with primers VS317 and VS318, and the amplified fragment was digested with SalI and EcoRI and cloned in expression vector pGEX-4T-3 (Amersham Pharmacia Biotech Inc., Piscataway, N.J.) to generate an in-frame translational fusion of hha with the 3′ end of the sequence encoding glutathione S-transferase (GST). The plasmid containing the GST-Hha fusion (pSM138) was introduced into E. coli BL21 (Amersham Pharmacia Biotech Inc., Piscataway, N.J.). The GST-Hha fusion protein from E. coli BL21 containing pSM138 was purified according to the instructions of the supplier of the glutathione-Sepharose matrix used for purifying GST-target protein fusions (Amersham Pharmacia Biotech Inc., Piscataway, N.J.) and by the procedure described previously (41). The Hha moiety was cleaved from GST-Hha fusion protein by thrombin treatment. Hha was stored at −20°C in phosphate-buffered saline containing 50% glycerol.
For Ler purification, a 380-bp fragment was PCR amplified from the strain 86-24 with primers VS331 and VS332, digested with BamHI and SalI, and cloned in pGEX-4T-3 to generate a translational fusion (pSM163) between GST and Ler. The GST-Ler fusion was purified from E. coli BL21 containing pSM163 by the procedure described above. The Hha and GST-Ler protein preparations were analyzed on a sodium dodecyl sulfate-12% polyacrylamide gel to determine their molecular weights and purity. The conditions for polyacrylamide gel electrophoresis, staining, and destaining of gels were as described in the manual for the Mini-Protean II electrophoresis cell (Bio-Rad, Hercules, Calif.). Hha and GST-Ler protein concentrations were estimated with the Bio-Rad protein assay kit (Bio-Rad, Hercules, Calif.).
Electrophoretic mobility shift assays.
The 350-bp esp promoter fragment was isolated with primers VS353 and VS354. Because Hha proteins from E. coli and S. enterica serovar Typhimurium are almost identical in their amino acid sequences, it has been speculated that Hha produced by E. coli should bind to the 450-bp hilA promoter of S. enterica serovar Typhimurium (14). A 450-bp sequence upstream of hilA was also isolated by PCR with primers VS341 and VS342 (14) and used as a positive control in an electrophoretic mobility shift assay. The 388-bp fragment containing the ler promoter was isolated by PCR with primers VS355 and VS356. The fragments containing the espA, hilA, and ler promoters were cloned in the pCR2.1 Topo TA cloning vector to generate plasmids pSM142, pSM143, and pSM146, respectively. The cloned fragments were isolated from recombinant plasmids with EcoRI, resolved on a 4% agarose gel (BMA, Rockland, Maine), and recovered from gel slices. End labeling of the recessed 3′ termini of promoter fragments with [α-32P]dATP, purification of labeled fragments from unincorporated deoxynucleotides, and determination of the binding of labeled fragments to Hha and GST-Ler in mobility shift assays were performed according to previously described procedures (8, 41). A 280-bp stx2B ORF-encoding sequence was also isolated from 86-24 by PCR with primers VS371 and VS372 for cloning in pCR-XL-Topo TA cloning vector to generate pSM156. The 280-bp fragment recovered from pSM156 after EcoRI treatment was used as a nonspecific competitor DNA in the electrophoretic mobility shift assay.
RT-PCR analysis.
EHEC O157:H7 strains 86-24 Δstx2 Δlac and 86-24 Δstx2 Δlac Δhha were cultured in LB broth at 37°C with shaking to an OD600 of 0.8. Bacterial cells were collected by centrifugation at 6,000 × g and processed for RNA isolation with the RNAeasy kit (Qiagen, Valencia, Calif.) according to the directions of the manufacturer. The isolated RNA was treated with 40 U of RNase-free DNase (Stratagene, La Jolla, Calif.) at 37°C, followed by heating of the sample to 95°C for 15 min to inactivate the DNase. The RNA was analyzed on an agarose gel to verify its integrity by observing the presence of distinct 23S and 16S rRNA bands, and the concentration of RNA was determined spectrophotometrically. A single-step reverse transcription (RT)-PCR kit (PE Biosystems, Foster City, Calif.) was used according to the directions of the manufacturer for detecting gene-specific transcripts. Primer sets VS319 and VS320, VS323 and VS324, and VS376 and VS377 facilitated detection of transcripts specific to gapA, espA, and ler, respectively. The samples were analyzed on a 4% Nusieve agarose gel containing ethidium bromide (Cambrex Corporation, East Rutherford, N.J.), and the gel was visualized with the Alpha Innotech Image documentation system (Alpha Innotech Corporation, San Leandro, Calif.).
RESULTS
Construction of an EHEC O157-H7 Δstx2 Δlac mutant carrying an esp::lac fusion.
To identify transcriptional regulators of the esp operon by transposon mutagenesis, EHEC O157:H7 strain 86-24 was deleted of stx2 (to reduce accidental exposure to Stx2) and lac operons (to remove endogenous β-galactosidase activity). The isolates deleted of stx2 produced a 3-kb fragment, and the parent 86-24 stx2+ produced a 4.2-kb fragment (data not shown). Deletion of the lac operon was confirmed by the lack of amplification of lacZ lacY and lacY lacA fragments by PCR with primers VS244 and VS268 and VS269 and VS280, respectively (data not shown). Strain 86-24 Δstx2 deleted of the lac operon produced white colonies on LB agar containing X-Gal and no detectable β-galactosidase activity in an o-nitrophenyl-β-d-galactopyranoside (ONPG)-based assay. Strain 86-24 containing the lac operon, on the other hand, produced blue colonies on X-Gal agar and 327 U of β-galactosidase activity in an ONPG-based assay.
Strain 86-24 Δstx2 Δlac was then used for constructing an esp::lac transcriptional fusion by allelic replacement of the esp gene on the chromosome with the esp::lac fusion present on pSM102. The generation of this fusion in the chromosome was confirmed by PCR with primers VS276 and VS270 and VS269 and VS275. These primers produced amplicons of 3.6 kb (escD-lacZ lacY region) and 2.6 kb (lacY lacA-esp region) from the chromosomally generated esp::lac fusion (data not shown). A graphic representation of the esp::lac fusion and its location relative to some of the other LEE-encoded genes are shown in Fig. 1A.
FIG. 1.
(A) Map of the chromosomal esp::lac transcriptional fusion. The arrow indicates the direction of transcription from the esp promoter (solid rectangle) into the lacZ ORF of the lac cassette. (B) β-Galactosidase activity of EHEC O157:H7 strain 86-24 esp::lac and a transposon (TnKn) mutant of 86-24 esp::lac with and without complementation with plasmid pSM116. Line graphs represent the OD600 values of esp::lac (□), esp::lac/TnKn (Δ), and esp::lac/pSM116 (×). β-Galactosidase activities are shown for the esp::lac (open bars), esp::lac/TnKn (hatched bars), and esp::lac/pSM116 (solid bars) strains. (C) Determination of β-galactosidase activity of the 86-24 Δhha esp::lac (carrying a TnKn insertion in an unknown gene), 86-24 Δhha esp::lac, and 86-24 esp::lac strains. Line graphs represent the OD600 for the esp::lac (□), esp::lac Δhha (▵), and esp::lac Δhha carrying TnKn in an unknown gene (♦) strains. β-Galactosidase activities are shown for the esp::lac (open bars), Δhha esp::lac (hatched bars), and Δhha esp::lac TnKn::unknown gene (solid bars) strains. Bacterial strains were grown in LB broth at 37°C with shaking, and samples were taken at different times to measure the OD600 and β-galactosidase activity with an ONPG-based assay. Error bars indicate standard deviations based on three measurements.
Transposon mutagenesis to identify transcriptional regulators of esp.
Strain 86-24 Δstx2 Δlac carrying an esp::lac fusion was subjected to transposon mutagenesis to identify the gene(s) affecting transcription of the esp operon. A total of five kanamycin-resistant colonies that appeared darker blue on LB agar containing kanamycin and X-Gal from a total of more than 3,000 colonies also produced elevated levels of β-galactosidase activity in ONPG-based assays. The β-galactosidase activity of one representative TnKn mutant selected for further analysis is shown in Fig. 1B. β-Galactosidase production was elevated in the mutant strain by 17-fold (655 U h−1) compared to the parent 86-24 hha+ esp::lac strain (39 U h−1) after 4.5 h of growth. These results indicated that the transposon insertion abolished the function of a gene product that was involved in negative regulation of the esp::lac fusion.
Nucleotide sequence analysis of the DNA at the transposon insertion sites in TnKn mutants of 86-24 Δstx2 Δlac esp::lac, expressing β-galactosidase at elevated levels, resulted in identification of an ORF with 100% homology to hha in four and yaaJ in one of the five independent mutant isolates. The gene hha, implicated in the negative regulation of α-hemolysin in pathogenic E. coli (33), encodes a 72-amino-acid (8.6-kDa) protein and is a member of the Hha/YmoA/RmoA family of proteins, which modulate virulence gene expression (Hha and YmoA) (9, 33) and conjugative transfer of broad-host-range plasmid R100 (RmoA) (35).
To confirm the effect of hha on esp expression, the isolate 86-24 Δstx2 Δlac esp::lac hha::TnKn was transformed with pSM116 (pBR322 containing a cloned copy of hha). Transformants had reduced β-galactosidase activity, similar to that produced by the parent 86-24 hha+ esp::lac, confirming that Hha acted as a negative regulator of esp expression (Fig. 1B).
Effect of growth medium and osmolarity on esp transcription.
Strain 86-24 Δstx2 Δlac esp::lac was deleted of hha by the allelic replacement method (described above) with pSM122. The hha deletion was confirmed by the lack of hha amplification in PCR with primers VS309 and VS340 (data not shown). Since expression of LEE-encoded genes has been shown to be enhanced by growth of EHEC O157:H7 in minimal medium of high osmolarity (2), we cultured 86-24 Δstx2 Δlac esp::lac with and without hha in LB broth and minimal medium (DMEM) of low or high osmolarity to determine if hha-mediated repression of the esp operon is more pronounced in LB broth than in DMEM.
Samples from these cultures were evaluated for the production of β-galactosidase at various time intervals. After 5 h of growth, strain 86-24 Δhha esp::lac produced severalfold-higher β-galactosidase activity in LB (74-fold higher; Fig. 2A) or DMEM (281-fold higher; Fig. 2B) containing 100 mM NaCl compared to the parent 86-24 esp::lac carrying a functional copy of hha. Although the β-galactosidase activity of 86-24 Δhha esp::lac was higher in LB (13-fold higher) and DMEM (38-fold higher) containing 400 mM NaCl than the parent 86-24 esp::lac, the higher salt concentration appeared to have a negative effect on growth and the expression of β-galactosidase activity. It is apparent from these results that growth of the 86-24 esp::lac strain lacking hha in DMEM of low or high osmolarity resulted in higher levels of β-galactosidase than were produced in LB broth of low or high osmolarity. DMEM containing a high salt concentration also resulted in 3.6-fold-higher β-galactosidase activity of the parent 86-24 esp::lac than DMEM containing low salt concentrations.
FIG. 2.
Effect of growth medium and NaCl on the expression of β-galactosidase activity. EHEC O157:H7 strains 86-24 esp::lac and 86-24 Δhha esp::lac were cultured in LB broth (A) or DMEM (B) in the presence of 100 or 400 mM NaCl. Samples from cultures grown at 37°C (with shaking at 200 rpm) were taken at the indicated times for measuring the OD600 and β-galactosidase activity with an ONPG-based assay. The OD600 is represented as line graphs for the esp::lac fusion in 100 mM (▵) or 400 mM (▴) NaCl and the Δhha esp::lac strain in 100 mM (□) or 400 mM (▪) NaCl). β-Galactosidase activities are shown as solid (100 mM NaCl) or open (400 mM NaCl) bars for esp::lac and small checkerboard bars (100 mM NaCl) or large checkerboard bars (400 mM NaCl) for the Δhha esp::lac fusion strain. Error bars indicate standard deviations based on three measurements.
Binding of hha to the esp promoter.
Amino acid sequence analysis of Hha from 86-24 revealed no classical helix-turn-helix motif (5), although a 10-amino-acid domain (SAADHRLAEL) with no known function is present at the C terminus (14). Hha of 86-24 exhibited 100, 99, 82, and 50% homology to Hha of E. coli, S. enterica serovar Typhimurium (14), YmoA of Yersinia enterocolitica (9), and RmoA of E. coli (35), respectively. Purified Hha was incubated with a 350-bp end-labeled fragment containing the previously mapped esp promoter region (2), and the reaction mixture was analyzed on a nondenaturing polyacrylamide gel. As shown in Fig. 3, Hha bound poorly to the esp promoter fragment, as only a small fraction of the promoter was bound at the highest concentration of Hha (3.5 × 10−10 mol). On the other hand, as a control, Hha bound to the hilA promoter fragment with a higher affinity, and more than half of the promoter fragment was bound to Hha at the highest concentration of the protein tested (3.5 × 10−10 mol).
FIG. 3.
(A) Detection of Hha binding to DNA fragments with gel shift assays. 32P-labeled DNA fragments (7,500 cpm) containing the espA, ler, and hilA promoter regions were incubated with 0 mol (lane 0), 5.8 × 10−11 mol (lane 1), 1.16 × 10−10 mol (lane 2), 1.74 × 10−10 mol (lane 3), and 3.5 × 10−10 mol (lane 4) of Hha. Samples containing promoter DNA and purified protein were incubated at 30°C for 30 min and resolved on a nondenaturing polyacrylamide gel, and DNA bands were visualized by autoradiography. Vertical lines on the right indicate the positions of free and bound DNA, and arrows on the left indicate the positions of two complexes formed by the binding of Hha to the ler promoter.
Identification of the gene directly affected by hha.
The results described above suggested that Hha represses esp transcription indirectly, that is, that Hha may repress a transcriptional activator of esp. To identify genes having a positive effect on esp transcription in the absence of Hha, a hha deletion was constructed in strain 86-24 Δstx2 Δlac esp::lac. The resulting strain, 86-24 Δhha esp::lac, had elevated β-galactosidase activity (Fig. 1C), similar to 86-24 esp::lac hha::TnKn. Strain 86-24 Δstx2 Δlac Δhha esp::lac was subjected to transposon mutagenesis, which resulted in the identification of seven kanamycin-resistant colonies that were light blue in color and one kanamycin-resistant colony appearing white out of about 4,000 colonies that were dark blue on kanamycin-X-Gal-LB agar plates.
Analysis of the DNA at the transposon insertion sites in three of seven light blue TnKn isolates resulted in identification of an ORF with 100% homology to the ler gene of EHEC O157:H7 (13). The white colony isolate had the transposon inserted in lacY, and of the remaining four light blue isolates, two isolates had transposon insertions in truA, one in tyrU, and one in fruR. Since ler is known to be a positive regulator of several LEE- and non-LEE-encoded genes, we selected one of the three Δhha esp::lac ler::TnKn isolates to determine if hha directly affects ler transcription. The expression of β-galactosidase activity in the Δhha esp::lac ler::TnKn isolate was similar to that in the 86-24 Δstx2 Δlac esp::lac strain containing a wild-type copy of hha (Fig. 1C).
Downregulation of ler expression by hha.
An isolate of strain 86-24 Δstx2 Δlac containing a ler::lac transcriptional fusion was constructed with the allelic replacement plasmid pSM212. The presence of a chromosomal ler::lac fusion was confirmed by PCR with primers VS270 and VS427 and VS269 and VS426, which resulted in the amplification of a 3.8-kb (5′ ler ORF-lacZ lacY region) and a 2.3-kb (lacY lacA-3′ ler ORF region) fragment, respectively, from the chromosomally generated ler::lac fusion (data not shown). Strain 86-24 Δstx2 Δlac ler::lac was then deleted of hha by the allelic replacement method (described above) with plasmid pSM122. The hha deletion was confirmed by the lack of hha amplification by PCR with primers VS309 and VS340 (data not shown). As shown in Table 3, 86-24 Δhha ler::lac produced 22% more β-galactosidase than the parent 86-24 ler::lac. The introduction of pSM197R, a pCR2.1 derivative carrying a cloned copy of hha, into 86-24 Δhha ler::lac reduced its β-galactosidase activity to a level 23% lower than that of the parent 86-24 hha+ ler::lac strain.
TABLE 3.
Downregulation of ler expression by hhaa
| Strain 86-24 genotype | Plasmidb | β-Galactosidase activity/A600 | % changec |
|---|---|---|---|
| ler::lac | 1,389.6 ± 36.3 | 0 | |
| Δhha ler::lac | 1,700 ± 46.0 | +22 | |
| Δhha ler::lac | pSM197R | 1,080 ± 39.1 | −23 |
| Δhha ler::lac | pCR2.1 | 1,646 ± 89.1 | +18 |
The effect of hha on the expression of ler was monitored by determination of the β-galactosidase activity of a chromosomal ler::lac fusion in strain 86-24 and its derivatives containing or deleted of hha. Strains were cultured in LB broth, and samples for monitoring β-galactosidase activity and A600 were taken after 270 min of growth.
pSM197R; harboring a cloned copy of hha in pCR2.1, was used for complementing the lack of hha function in Δhha ler::lac strains.
Increases (+) and decreases (−) in the level of β-galactosidase activity compared to that produced by the parent ler::lac fusion strain are shown. The assay variability was within 5% of the mean of three independent measurements.
Interaction of Hha with the ler promoter region.
A 388-bp fragment containing the ler promoter region was incubated with various amounts of purified Hha, and the reaction mixtures were analyzed by nondenaturing polyacrylamide gel electrophoresis. Hha bound to the ler promoter fragment with a higher affinity than to the hilA promoter fragment at equivalent concentrations of Hha (Fig. 3). Moreover, binding of Hha to the ler promoter fragment produced two types of complexes (indicated by the arrows); a fast-migrating complex appeared at lower Hha concentrations (5.8 × 10−11 to 3.5 × 10−10 mol of Hha), and a slow-migrating complex at higher Hha concentrations (1.74 × 10−10 to 3.5 × 10−10 mol of Hha).
Effect of hha on transcription of the esp and ler genes.
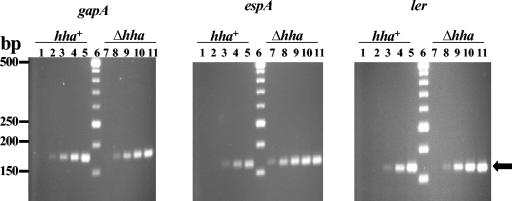
Purified RNAs from strains 86-24 hha+ and 86-24 Δhha were analyzed by RT-PCR to determine the relative amounts of espA- and ler-specific transcripts. A housekeeping gene, gapA, served as a positive control, as its expression was not affected by the presence or absence of hha. As shown in Fig. 4, ler- and esp-specific amplified products were detected at ≥10-fold and ≥100-fold-higher levels, respectively, in a Δhha background than in the hha+ strain. The amount of gapA-specific amplification products was almost identical in both strains.
FIG. 4.
Determination of transcriptional levels of esp and ler with RT-PCR. Total RNA purified from 86-24 Δstx2 Δlac and 86-24 Δstx2 Δlac Δhha, which were grown in LB broth at 37°C (with shaking at 200 rpm), was used in RT-PCR assays containing primer sets for specific amplification and detection of gapA-, esp-, and ler-specific transcripts. gapA, a housekeeping gene, was used as a control. Amplified DNA was resolved on 4% agarose gels containing ethidium bromide, and DNA bands were visualized with the Alpha Innotech Image documentation system. Lanes 1 to 5, RT-PCR conducted in the presence of 7.5 × 10−4, 7.5 × 10−3, 7.5 × 10−2, 7.5 × 10−1, and 7.5 μg, respectively, of total RNA from 86-24 Δstx2 Δlac; lane: 6, DNA size markers (sizes listed on the left); lanes 7 to 11, RT-PCR conducted in the presence of 7.5 × 10−4, 7.5 × 10−3, 7.5 × 10−2, 7.5 × 10−1, and 7.5 μg, respectively, of total RNA from 86-24 Δstx2 Δlac Δhha. The arrow on the right points to the position of amplified products specific for gapA, espA, and ler.
Positive regulatory role of ler in esp gene expression.
The preceding results suggested that hha is a negative regulator of ler and that removal of hha led to elevated esp transcription, presumably by the positive action of Ler. To test this hypothesis, the expression of β-galactosidase from an esp::lac strain was tested in the presence and absence of hha and the presence and absence of ler. The ler deletion was introduced into strains 86-24 Δstx2 Δlac esp::lac and 86-24 Δstx2 Δlac Δhha esp::lac with pSM188 and the allelic replacement method described above. The loss of ler was confirmed by PCR with the primer set VS426 and VS427. The strain deleted of ler produced a 230-bp amplicon, and the strain harboring ler produced a 940-bp amplicon (data not shown). As shown in Table 4, 86-24 Δhha ler+ esp::lac produced 3818% higher β-galactosidase activity than the parent 86-24 hha+ ler+ esp::lac. In contrast, 86-24 Δhha Δler esp::lac showed only a 200% increase in β-galactosidase activity compared to the 86-24 hha+ ler+ esp::lac control strain. These results suggested a positive requirement for ler in the expression of esp genes. The 86-24 Δler Δhha esp::lac strain produced a 200% higher level of β-galactosidase activity than 86-24 hha+ Δler+ esp::lac, suggesting that hha may also have a negative effect, albeit a milder one, on expression of the esp operon.
TABLE 4.
Dependence of esp expression on lera
| Strain 86-24 genotype | β-Galactosidase activity/A600 | % changeb |
|---|---|---|
| hha+ ler+ esp::lac | 29.7 ± 3.6 | 0 |
| Δhha ler+ esp::lac | 1,134 ± 48.4 | +3,818 |
| Δhha Δler esp::lac | 59.4 ± 6.3 | +200 |
| Δler hha+ esp::lac | 29.2 ± 4.2 | −1.69 |
Effect of ler on the expression of esp operon was monitored by the determination of β-galactosidase activity of a chromosomal esp::lac fusion in 86-24 and its derivatives deleted either of ler or hha or both ler and hha. Strains were cultured in LB broth and samples for monitoring β-galactosidase activity and A600 were taken after 270 min of growth.
See Table 3, footnote c. The assay variability was within 4 to 15% of the mean of three independent measurements. High variability was normally seen in samples expressing very low β-galactosidase activity.
Interaction of Ler with the esp promoter region.
A 350-bp end-labeled fragment (5 ng) containing the esp promoter was incubated with 1.5 × 10−11 mol of GST-Ler fusion protein. At this protein concentration (Fig. 5), more than 50% of the labeled fragments were bound (bound target) with the GST-Ler fusion protein, resulting in a single slow-migrating complex (lane 3) compared to the band (free target) that resulted when the labeled fragment was incubated without the Ler protein (lane 1) or with the GST protein only (lane 2). When the labeled promoter fragment (5 ng) was mixed with increasing amounts (10 to 500 ng) of the unlabeled esp promoter fragment (specific competitor) prior to addition of the GST-Ler protein, an increasing amount of labeled DNA migrated as free DNA (lanes 4 to 8), indicating that the GST-Ler protein-esp promoter interactions were specific. On the other hand, addition of 100 to 500 ng of an unlabeled nonspecific DNA (lanes 9 and 10) to the reactions containing the labeled esp promoter fragment and the GST-Ler fusion did not alter the specific interactions between the esp promoter and Ler.
FIG. 5.
Gel shift assay demonstrating specificity of binding of purified Ler to the esp promoter. 32P-labeled esp promoter DNA (5 ng containing 7,500 cpm) was mixed either with unlabeled esp promoter DNA (specific competitor) or an unrelated DNA fragment (nonspecific competitor) and 1.5 × 10−11 mol of purified GST-Ler fusion protein. Lane 1, labeled esp promoter incubated with 2.25 × 10−10 mol of GST protein; lane 2, labeled esp promoter incubated with GST-Ler fusion protein; lanes 3 to 8, labeled esp promoter incubated with GST-Ler and 0, 5, 25, 50, 100, and 500 ng, respectively, of unlabeled esp promoter DNA; lanes 9 and 10, labeled esp promoter incubated with GST-Ler and 100 or 500 ng, respectively, of unlabeled nonspecific competitor DNA. Samples containing promoter DNA and purified protein were incubated at 30°C for 30 min and resolved on a nondenaturing polyacrylamide gel, and DNA bands were visualized by autoradiography. Vertical lines on the right indicate the positions of free and bound DNA.
DISCUSSION
The esp operon of EHEC O157:H7 encodes secreted proteins EspA, EspD, and EspB, and it is analogous to LEE4 of EPEC O126:H6 (26). The Esp proteins are essential for the formation of attaching and effacing lesions by EHEC and EPEC on intestinal epithelial cells (20). The promoter element driving the expression of the esp operon in EHEC is virtually identical to the promoter governing the expression of LEE4 in EPEC, and transcription activities at these promoter regions are dependent on environmental stimuli (2). The transcriptional regulation of several LEE- and non-LEE-encoded operons in EPEC and EHEC is under the positive regulation of Ler, a protein with similarity to the H-NS family of DNA binding proteins (12). Moreover, recent studies have also shown that Ler is absolutely necessary for full transcription of the esp (LEE4) operon in both EPEC and EHEC (12), but the role of ler in transcriptional regulation of the esp operon is not completely understood. Similarly, the requirement of additional, albeit unknown, factors in transcriptional regulation of the esp operon has not been completely ruled out.
In order to identify transcriptional factors affecting transcription of the esp operon, strain 86-24 Δstx2 Δlac carrying an esp::lac transcriptional fusion was subjected to transposon mutagenesis. A transposon mutant that produced 17-fold more β-galactosidase than the parent esp::lac fusion strain was identified. The maximum expression of β-galactosidase was observed as bacterial cells entered the stationary phase. Nucleotide sequence analysis of DNA flanking the transposon insertion site resulted in the identification of hha. The hha gene encodes an 8.5-kDa protein (Hha) that is implicated in the regulation of α-hemolysin in pathogenic E. coli (33) and S. enterica serovar Typhimurium (14). A plasmid-cloned copy of hha reduced the highly increased expression of the esp::lac fusion in the hha::TnKn mutant to the level expressed in the parent 86-24 esp::lac strain, confirming that hha acted as a negative regulator of esp gene expression.
We also observed that 86-24 esp::lac Δhha produced severalfold-higher β-galactosidase activity than the parent esp::lac strain in LB broth or DMEM-HEPES of low or high osmolarity. With a highly sensitive assay for measuring β-galactosidase activity, Beltrametti et al. (2) observed only a fourfold increase in the level of β-galactosidase activity of an esp::lac fusion strain of EHEC O157:H7 in DMEM-HEPES or in high-osmolarity M9 minimal medium. In our studies, we noticed that 86-24 esp::lac Δhha produced 294-fold more β-galactosidase activity than the parent esp::lac strain after 5 h of growth in DMEM-HEPES (100 mM NaCl), indicating that the presence of hha downregulated the expression of the esp genes in the esp::lac strain. Although the growth rates of the 86-24 esp::lac and esp::lac Δhha strains were reduced in DMEM-HEPES of high osmolarity (400 mM NaCl), the merely fourfold increase in the β-galactosidase activity of the parent strain compared to the 138-fold increase in the hha mutant strain after 5 h of growth in high-osmolarity DMEM-HEPES reiterates the negative role of hha in esp gene expression.
Hha homologs are involved in the regulation of virulence genes in Y. enterocolitica (YmoA) (9) and conjugative plasmid transfer in E. coli (RmoA) (35) and constitute an Hha-YmoA-RmoA family of proteins. These proteins are environment-dependent modulators of gene expression, and mutations in the genes encoding these proteins produce pleiotropic effects. Since environment dependency and pleiotropism are also characteristics of H-NS, a well-characterized nucleoid-associated protein (1), Hha and related proteins are also classified as nucleoid-associated proteins. Experimental evidence supporting the nucleoid association of Hha is implicit in findings that hha mutants display alterations in reporter plasmid topology (7) and overexpression of Hha increases the frequency of transposition of insertion elements in E. coli (7, 29).
Recent studies have demonstrated that Hha acts as a negative modulator of gene expression either by binding directly to a specific sequence located in the promoter region of a target gene (hilA regulation in S. enterica serovar Typhimurium) (14) or by oligomerizing with H-NS protein before binding to a specific regulatory sequence in the target gene (hemolysin expression in E. coli) (25, 34). However, the Hha protein purified from strain 86-24 bound poorly to the fragment containing the esp promoter in a gel shift assay but showed greater affinity for the fragment containing the hilA promoter region of S. enterica serovar Typhimurium. The poor binding of Hha to the esp promoter observed in gel shift assays and only a 200% increase in the expression of β-galactosidase of 86-24 Δhha Δler esp::lac compared to 86-24 Δler hha+ esp::lac might be indicative of poor binding of Hha to the esp promoter under in vivo conditions. However, the lack of efficient binding of Hha to the esp promoter could not be attributed to the promoter sequence that we selected for use in the electrophoretic mobility shift assay because this promoter region was identified based on primer extension data that clearly defined the transcriptional start site immediately upstream of espA and on the ability of this promoter to direct the expression of lacZ fusions, providing strong evidence that this promoter directed transcription of the polycistronic espADB mRNA (2).
A second promoter, located upstream of sepL, has also been demonstrated to direct the synthesis of a polycistronic transcript extending from sepL through espD that may be processed into a sepL monocistronic mRNA and a larger espADB transcript (23, 28, 36). However, the relative contribution of transcripts from the sepL promoter versus the espA promoter in the synthesis of EspA, EspD, and EspB is unclear. Roe et al. (39) also reported that expression of the esp::lac translation fusion was detected only when this fusion was transcribed from the sepL promoter, but that study involved partial comparison of nonisogenic isolates of E. coli O157:H7, and it is unclear if the promoter regions of these isolates are identical to those studied here or as reported by Beltrametti et al. and Kresse et al. (2, 23). It is reasonable to assume that both promoters can direct transcription of espADB but that these promoters may have different strengths and may be under different forms of regulation. The apparent direct interaction between Ler and the espA promoter that we have demonstrated in our studies provides additional evidence that this promoter likely has an important role in esp expression.
One possible explanation for the lack of Hha binding to the esp promoter with a higher affinity could be an absolute need by Hha to form nucleoprotein complexes with H-NS or H-NS-like proteins in order for it to bind and completely repress transcription from the esp promoter. Data from transcriptional studies involving esp::lac transcriptional fusions and identification of putative H-NS-binding sites in the esp promoter region of EHEC O157:H7 EDL933 have implicated H-NS in the negative regulation of esp operon (2).
An alternative explanation is that Hha represses the expression of another gene that serves as a positive regulator of esp transcription. We identified transposon mutants of 86-24 Δhha esp::lac that no longer expressed high levels of β-galactosidase activity. In fact, the β-galactosidase activity of these mutants was similar to the level expressed by 86-24 hha+ ler+ esp::lac. These mutants carried transposon insertions in ler, suggesting that Hha might repress ler, and repression of ler by Hha results in reduced expression of esp. A negative effect of Hha on the transcription of ler was demonstrated by the following observations. (i) Increased production of β-galactosidase activity in 86-24 Δhha ler::lac. This increase in the β-galactosidase activity of Δhha ler::lac (1.2-fold higher than that of the ler::lac strain), however, was much lower than that observed in the Δhha esp::lac strain (17-fold higher than the esp::lac strain). A small increase in β-galactosidase activity of the Δhha ler::lac strain could be attributed to the fact that the parent ler::lac fusion strain produced a very high basal level of β-galactosidase activity. A higher basal-level expression of β-galactosidase activity from the ler::lac fusion strain has also been observed in other studies (44).
(ii) The presence of pSM197R (pCR2.1 carrying a cloned copy of hha) in 86-24 Δhha ler::lac reduced β-galactosidase activity to a level lower (presumably due to the presence of multiple copies of the plasmid-cloned hha) than that expressed by the parent 86-24 ler::lac strain. (iii) RT-PCR assays showed a ≥10-fold increase in the level of ler-specific transcripts in 86-24 carrying an in-frame hha deletion. (iv) Purified Hha bound to the ler promoter with the highest affinity. Hha not only bound to the ler promoter but also generated a binding pattern that was strikingly different from the one observed with the hilA promoter of S. enterica serovar Typhimurium (14). Hha formed a single complex with the hilA promoter, while two discrete complexes of different sizes were observed for the ler promoter at different concentrations of Hha.
Although we have not presented direct evidence, the pattern of Hha binding to the ler promoter suggests a possible cooperative mode of binding, which presumably requires multimerization of Hha. The cooperative mode of DNA binding is exhibited by regulatory proteins that contain an N-terminal oligomerization domain to recognize and bind to specific regulatory sequences for modulating the transcriptional activity of downstream genes (19, 24). N-terminal domains possessing structural features facilitating adoption of coiled-coil formation for protein oligomerization have been identified in H-NS and H-NS-like proteins (3) and Hha (23, 32).
A positive regulatory role of ler in expression of the esp genes was corroborated by the findings that strain 86-24 Δhha esp::lac deleted of ler could no longer produce higher levels of β-galactosidase activity and purified Ler exhibited specific binding to the esp promoter. Previous reports have shown that H-NS plays a negative role in the transcription of esp genes and that putative H-NS binding sites exist in the promoter region of the esp operon (2). Bustamante et al. (6) demonstrated that Ler acts as an antirepressor of H-NS and presumably activates the expression of the divergently transcribed LEE2 and LEE3 operons by competing with H-NS for binding to the promoter element located upstream of LEE2. The activation of esp expression observed in EHEC 86-24 Δhha, which also showed increased expression of Ler, could also result from the antirepressor properties of Ler that facilitate preferential binding of Ler over H-NS to the esp promoter region.
In summary, the results presented in this study clearly demonstrates that Ler acts as a positive regulator of esp expression and the level of esp expression is regulated by the degree of repression exerted by Hha on ler transcription. Several regulatory factors that affect the expression of ler in both EPEC and EHEC have been identified. In EPEC, expression of ler is under the positive regulation of the Per regulon (28), and the quorum-sensing E. coli regulator A (QseA) activates the transcription of ler in both EPEC and EHEC (42). Transcription of ler in EPEC is also dependent on integration host factor (15), Fis (16), and BipA (17). Since quorum sensing appears to be the major pathway that activates the expression of ler in EHEC O157:H7, a better understanding of the factors that modulate the expression of hha and facilitate enhanced expression of ler would increase our understanding of virulence gene expression in EHEC O157:H7.
Acknowledgments
We thank Robert Morgan for technical assistance and Julia Ridpath for DNA sequencing. We are grateful to Shirley Halling, Steven Carlson, and Thad Stanton for critical reading of the manuscript and Allison O' Brien for providing temperature-sensitive plasmid pAM450.
The use of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
REFERENCES
- 1.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 2.Beltrametti, F., A. U. Kresse, and C. A. Guzman. 1999. Transcriptional regulation of the esp genes of enterohemorrhagic Escherichia coli. J. Bacteriol. 181:3409-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertin, P., N. Benhabiles, E. Krin, C. Laurent-Winter, C. Tendeng, E. Turlin, A. Thomas, A. Danchin, and R. Brasseur. 1999. The structural and functional organization of H-NS-like proteins is evolutionarily conserved in gram-negative bacteria. Mol. Microbiol. 31:319-329. [DOI] [PubMed] [Google Scholar]
- 4.Bolivar, F., R. L. Rodriguez, M. C. Betlach, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. I. Ampicillin-resistant derivatives of the plasmid pMB9. Gene 2:75-93. [DOI] [PubMed] [Google Scholar]
- 5.Brennan, R. G., and B. W. Matthews. 1989. The helix-turn-helix DNA binding motif. J. Biol. Chem. 264:1903-1906. [PubMed] [Google Scholar]
- 6.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664-678. [DOI] [PubMed] [Google Scholar]
- 7.Carmona, M., C. Balsalobre, F. Munoa, M. Mourino, Y. Jubete, F. De la Cruz, and A. Juarez. 1993. Escherichia coli hha mutants, DNA supercoiling and expression of the haemolysin genes from the recombinant plasmid pANN202-312. Mol. Microbiol. 9:1011-1018. [DOI] [PubMed] [Google Scholar]
- 8.Chodosh, L. A. 1987. Mobility shift DNA-binding assay using gel electrophoresis, p. 12.2.1-12.2.11. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. D. Seidmann, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Greene Publishing Associates and Wiley Interscience, New York, N.Y.
- 9.de la Cruz, F., M. Carmona, and A. Juarez. 1992. The Hha protein from Escherichia coli is highly homologous to the YmoA protein from Yersinia enterocolitica. Mol. Microbiol. 6:3451-3462. [DOI] [PubMed] [Google Scholar]
- 10.Donnenberg, M. S., J. B. Kaper, and B. B. Finlay. 1997. Interactions between enteropathogenic Escherichia coli and host epithelial cells. Trends Microbiol. 5:109-114. [DOI] [PubMed] [Google Scholar]
- 11.Ebel, F., T. Podzadel, M. Rohde, A. U. Kresse, S. Kramer, C. Deibel, C. A. Guzman, and T. Chakraborty. 1998. Initial binding of Shiga toxin-producing Escherichia coli to host cells and subsequent induction of actin rearrangements depend on filamentous EspA-containing surface appendages. Mol. Microbiol. 30:147-161. [DOI] [PubMed] [Google Scholar]
- 12.Elliott, S. J., V. Sperandio, J. A. Giron, S. Shin, J. L. Mellies, L. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 14.Fahlen, T. F., R. L. Wilson, J. D. Boddicker, and B. D. Jones. 2001. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 183:6620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedberg, D., T. Umanski, Y. Fang, and I. Rosenshine. 1999. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34:941-952. [DOI] [PubMed] [Google Scholar]
- 16.Goldberg, M. D., M. Johnson, J. C. Hinton, and P. H. Williams. 2001. Role of the nucleoid-associated protein Fis in the regulation of virulence properties of enteropathogenic Escherichia coli. Mol. Microbiol. 41:549-559. [DOI] [PubMed] [Google Scholar]
- 17.Grant, A. J., M. Farris, P. Alefounder, P. H. Williams, M. J. Woodward, and C. D. O'Connor. 2003. Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC). Mol. Microbiol. 48:507-521. [DOI] [PubMed] [Google Scholar]
- 18.Griffin, P. M., S. M. Ostroff, R. V. Tauxe, K. D. Greene, J. G. Wells, J. H. Lewis, and P. A. Blake. 1988. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann. Intern. Med. 109:705-712. [DOI] [PubMed] [Google Scholar]
- 19.Harrison, S. C., and A. K. Aggarwal. 1990. DNA recognition by proteins with the helix-turn-helix motif. Annu. Rev. Biochem. 59:933-969. [DOI] [PubMed] [Google Scholar]
- 20.Jarvis, K. G., J. A. Giron, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA. 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karmali, M. A. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knutton, S., J. Adu-Bobie, C. Bain, A. D. Phillips, G. Dougan, and G. Frankel. 1997. Downregulation of intimin expression during attaching and effacing enteropathogenic Escherichia coli adhesion. Infect. Immun. 65:1644-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kresse, A. U., F. Beltrametti, A. Muller, F. Ebel, and C. A. Guzman. 2000. Characterization of SepL of enterohemorrhagic Escherichia coli. J. Bacteriol. 182:6490-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupas, A. 1996. Coiled coils: new structures and new functions. Trends Biochem. Sci. 21:375-382. [PubMed] [Google Scholar]
- 25.Madrid, C., J. M. Nieto, S. Paytubi, M. Falconi, C. O. Gualerzi, and A. Juarez. 2002. Temperature- and H-NS-dependent regulation of a plasmid-encoded virulence operon expressing Escherichia coli hemolysin. J. Bacteriol. 184:5058-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKee, M. L., and A. D. O'Brien. 1996. Truncated hemorrhagic Escherichia coli (EHEC) O157:H7 intimin (EaeA) fusion proteins promote adherence of EHEC strains to Hep-2 cells. Infect. Immun. 64:2225-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 29.Mikulskis, A. V., and G. R. Cornelis. 1994. A new class of proteins regulating gene expression in enterobacteria. Mol. Microbiol. 11:77-86. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 31.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nieto, J. M., M. Carmona, S. Bolland, Y. Jubete, F. de la Cruz, and A. Juarez. 1991. The hha gene modulates haemolysin expression in Escherichia coli. Mol. Microbiol. 5:1285-1293. [DOI] [PubMed] [Google Scholar]
- 34.Nieto, J. M., C. Madrid, E. Miquelay, J. L. Parra, S. Rodriguez, and A. Juarez. 2002. Evidence for direct protein-protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of proteins. J. Bacteriol. 184:629-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nieto, J. M., A. Prenafeta, E. Miquelay, S. Torrades, and A. Juarez. 1998. Sequence, identification and effect on conjugation of the rmoA gene of plasmid R100-1. FEMS Microbiol. Lett. 169:59-66. [DOI] [PubMed] [Google Scholar]
- 36.O'Connell, C. B., E. A. Creasey, S. Knutton, S. Elliott, L. J. Crowther, W. Luo, M. J. Albert, J. B. Kaper, G. Frankel, and M. S. Donnenberg. 2004. SepL, a protein required for enteropathogenic Escherichia coli type III translocation, interacts with secretion component SepD. Mol. Microbiol. 52:1613-1625. [DOI] [PubMed] [Google Scholar]
- 37.Perna, N. T., G. F. Mayhew, G. Posfai, S. Elliott, M. S. Donnenberg, J. B. Kaper, and F. R. Blattner. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perna, N. T., G. Plunkett, 3rd, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 39.Roe, A. J., H. Yull, S. W. Naylor, M. J. Woodward, D. G. Smith, and D. L. Gally. 2003. Heterogeneous surface expression of EspA translocon filaments by Escherichia coli O157:H7 is controlled at the posttranscriptional level. Infect. Immun. 71:5900-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Sharma, V. K., C. J. Hackbarth, T. M. Dickinson, and G. L. Archer. 1998. Interaction of native and mutant MecI repressors with sequences that regulate mecA, the gene encoding penicillin binding protein 2a in methicillin-resistant staphylococci. J. Bacteriol. 180:2160-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sperandio, V., C. C. Li, and J. B. Kaper. 2002. Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect. Immun. 70:3085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sperandio, V., J. L. Mellies, R. M. Delahay, G. Frankel, J. A. Crawford, W. Nguyen, and J. B. Kaper. 2000. Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol. Microbiol. 38:781-793. [DOI] [PubMed] [Google Scholar]
- 44.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]