Abstract
The virulent Lactobacillus plantarum myophage LP65 was isolated from industrial meat fermentation. Tail contraction led to reorganization of the tail sheath and the baseplate; a tail tube was extruded. In ultrathin section the phage adsorbed via its baseplate to the exterior of the cell, while the tail tube tunneled through the thick bacterial cell wall. Convoluted membrane structures were induced in the infected cell. Progeny phage was detected 100 min postinfection, and lysis occurred after extensive digestion of the cell wall. Sequence analysis revealed a genome of 131,573 bp of nonredundant DNA. Four major genome regions and a large tRNA gene cluster were observed. One module corresponded to DNA replication genes. Helicase/primase and two replication/recombination enzymes represented the only links to T4-like Myoviridae from gram-negative bacteria. Another module corresponded to the structural genes. Sequence relatedness identified links with Listeria phage A511, Staphylococcus phage K, and Bacillus phage SPO1. LP65 structural proteins were identified by two-dimensional proteome analysis and mass spectrometry. The putative tail sheath protein showed a shear-induced change in electrophoretic migration behavior. The genome organization of the structural module in LP65 resembled that of Siphoviridae from the lambda supergroup.
There is hardly any other biological system where horizontal gene transfer had such a rampant influence on genome evolution as in phages (19). It is therefore very difficult to base a natural taxonomy of phages on genomics data. Phage taxonomy is currently based on phage tail and capsid morphology, the chemical type of genetic material, and some biological characteristics (1). Phage biologists are increasingly uneasy about basing phage taxonomy on tail morphology as the primary taxonomical criterion. Different proposals have recently been made as to how taxonomy could be based on the growing amount of phage sequence data. One proposal was to subdivide the phage genomes into their constituent modules, classify the individual modules according to sequence similarity, and describe the phages by a type of code bar where each number refers to a specific allele (gene cluster) for each individual module (25). However, if the number of different alleles for a given module is great and the number of modules composing a phage genome is substantial, the system will quickly become clumsy. Another approach takes a radically different position. It ignores the modular structure of phage genomes and crunches the sequences of phage genomes into a computer program that develops a proteome tree for the phages (49). This approach hinges on the hypothesis that there is, on average, a relatively good association between different modules constituting a phage. Proux et al. finally proposed to base the taxonomy of phages on the comparative genomics of the structural genes, and mainly the head gene cluster (47), which is probably the oldest and, in our experience, the most conserved phage module (8).
All three proposals are based on phylogeny concepts for phage evolution. Since phages (like all viruses) do not belong to the universal tree of life, we cannot take the principles of Darwinian evolution for granted. In addition, our knowledge of the sequence diversity of phage genomes is still very fragmentary. In fact, the first metagenomics analyses of viral genomes in the ocean suggested a substantial diversity of viral genomes in the environment, with the majority of the sequences finding no database matches (6). Based on statistical calculations, it was proposed that phage DNA sequences might represent the largest share of the DNA sequence space in the biosphere (48). However, these estimations contrast with the sequencing of the few marine phage genomes, which revealed links to described phage types at least at the level of genome organization. T4-, T7-, and lambda-like phages were detected among marine phages (10, 43). Sequencing projects painted distinct pictures for phages infecting different bacteria. For example, a bewildering diversity of phage types was observed for mycobacteria (44). In contrast, dairy phages constituted a relatively close-knit group (7). In addition, some phage types showed a wide evolutionary reach. For example, a lambda-like supergroup of Siphoviridae (phages with flexible, noncontractile tails) was defined in gram-negative and gram-positive bacteria and even in Archaea (8, 46). Similarly, T4-like Myoviridae (phages with contractile tails) (37) showed a wide distribution ranging from γ-proteobacteria to cyanobacteria (18, 50). Surprising degrees of sequence conservation were observed for T4-like head genes over this phylogenetic range.
Myoviridae with large genomes were also reported in gram-positive bacteria (21), and the sequence of one representative was reported recently (39). The genome sequence and biological characterization of Lactobacillus plantarum myovirus LP65, presented in the present communication, defines it as a close relative of Staphylococcus phage K, Listeria phage A511, and Bacillus phage SPO1. Over the structural module, the SPO1-like genus of the family Myoviridae shares a closer relationship with Siphoviridae from the lambda supergroup than with other genera of Myoviridae. The tail morphology, used in the current taxonomy system of the International Committee on Taxonomy of Viruses (ICTV) for the classification of tailed phages (order Caudovirales), therefore does not represent their phylogenetic relationships.
MATERIALS AND METHODS
Bacteriophage isolation.
L. plantarum strains were grown aerobically on MRS medium at 30°C. Homogenized meat samples at different fermentation steps were supplemented with MRS and incubated overnight for phage amplification. The incubated samples were centrifuged, sterile filtered a on 0.45-μm-pore-size mini-Sartorius filter (Millian), and reinoculated with 1% (vol/vol) of a defined test strain MRS medium supplemented with CaCl2. Serial passages were performed until a clear lysis was obtained. Phages were isolated by two rounds of plaque purification.
Phage purification.
One liter of MRS medium was inoculated with the specified L. plantarum strain, grown to an optical density at 600 nm of 0.1, and then infected at a multiplicity of infection (MOI) of 3. NaCl was added to the lysate, yielding a final concentration of 0.5 M, and incubated 1 h at 4°C. After centrifugation at 14,500 × g (16 min at 4°C) in a Sorvall RC5B centrifuge, polyethylene glycol 6000 was added to the supernatant to a final concentration of 10% (wt/vol). The lysate was incubated overnight at 4°C with gentle stirring. Polyethylene glycol-precipitated phages were collected by centrifugation at 13,000 × g for 16 min; the resulting pellets were resuspended in 3 ml of phage buffer (20 mM Tris-HCl [pH 7.4], 100 mM NaCl, 10 mM MgSO4) and loaded on a discontinuous CsCl gradient (1.35, 1.53, and 1.65 g of CsCl/ml). The gradients were centrifuged at 4°C in an SW55 rotor at 40,000 rpm for 3 h in a Beckman L8-60 M ultracentrifuge. Purified phages were recovered with a syringe and dialyzed against phage buffer.
Ultrathin sectioning.
Samples (15 ml) of infected cells were collected at 0, 10, 30, 60, 80, 100, 120, and 150 min postinfection (p.i.), centrifuged, and fixed by immersion in a solution of 2.5% (vol/vol) glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4). The liquid samples were encapsulated in agar gel tubes and postfixed with 2% (wt/vol) osmium tetroxide and 0.04% (wt/vol) ruthenium red. After dehydration in a graded ethanol series (70, 80, 90, 96, and 100% [vol/vol] ethanol), the samples were embedded in Spurr resin (Spurr-ethanol, 2:1; 100% ethanol). After polymerization of the resin (70°C, 48 h), ultrathin sections were cut with a Reichert OMU2 ultramicrotome. The sections were stained with aqueous uranyl acetate and lead citrate and were examined by transmission electron microscopy (Philips CM12; 80 kV).
Electron microscopy.
A drop of the phage suspension was applied to a Formvar-carbon-coated copper grid for 5 min, then removed with a pipette and immediately replaced with either 2% (vol/vol) ammonium molybdate at pH 7.0, 2% (vol/vol) phosphotungstic acid, or a solution of 3% (vol/vol) uranyl acetate. After 1 min, the liquid was removed with a filter paper. The grids were examined in a Philips CM12 transmission electron microscope at 80 kV.
DNA techniques.
Purified phages were treated with proteinase K at a final concentration of 1 mg/ml for 2 h at 37°C. DNA was extracted twice with phenol-chloroform and was precipitated with 2 volumes of ethanol. After centrifugation, pellets were washed with 70% (vol/vol) ethanol and resuspended in 50 μl of Tris-EDTA. DNA was digested with restriction enzymes according to the instructions provided by the manufacturer.
For Southern hybridization, restriction enzyme-digested phage DNA was separated on 0.8% (wt/vol) agarose gels. Transfer to a Hybond membrane was done according to the Amersham protocol. Probes were labeled with α-32P by using the random-primed DNA labeling system (Boehringer Mannheim) and were purified with Nuc Trap probe purification columns (Stratagene). Prehybridization and hybridization conditions were carried out in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-5 mM EDTA (pH 8)-0.1% (vol/vol) sodium dodecyl sulfate (SDS)-25% (wt/vol) skim milk overnight at 68°C. The membranes were then rinsed with 2× SSC and washed for 1 h at 65°C with 2× SSC and 0.1% (vol/vol) SDS, then for 1 h in 0.5× SSC and 0.1% SDS, and finally for 15 min in 0.1× SSC and 0.1% SDS. The membranes were dried and then exposed to BIOMAX-MS autoradiography film (Kodak).
For pulsed-field gel electrophoresis, blocks of 1% (wt/vol) agarose containing 106 PFU of the indicated phage were incubated overnight in a lysis buffer (0.05 M EDTA, 10 mM Tris [pH 8.0], 1% [vol/vol] SDS, and 50 μg of proteinase K) and washed three times for 1 h in 10 ml of 10 mM Tris [pH 8.0]-1 mM EDTA. The blocks were then analyzed by electrophoresis in a 1% (wt/vol) agarose low-melting-point gel in 0.5% TBE (45 mM Tris-borate and 1 mM EDTA) for 20 h at 6 V/cm and 14°C, with a pulse time of 1 to 20 s, in a CHEF-DRII apparatus from Bio-Rad. The gel was stained for 30 min with ethidium bromide (0.5 μg/ml).
Protein techniques.
The proteins from untreated phage, phage sonicated for 30 s at a duty cycle of 40%, or phage sheared 10 times through a syringe were separated on an SDS-12% polyacrylamide gel at 10 mA. Protein markers from Bio-Rad were used for molecular mass calibration.
Peptide mapping was performed as described previously (12). Protein bands were cut and soaked in 125 mM Tris HCl (pH 6.8)-0.1% (wt/vol) SDS-1 mM EDTA. Each band was transferred in a slot of a one-dimensional (1-D) conventional SDS-12% polyacrylamide gel electrophoresis (PAGE) gel. Ten microliters of Staphylococcus aureus V8 protease (Sigma) at a concentration of 1 μg/μl mixed with 10% (vol/vol) glycerol was added to the slot. Migration was performed at 35 mA for 30 min. The current was switched off for 30 min, followed by a 5-h electrophoresis run. The peptides were visualized with a commercial silver-staining kit.
For 2-D electrophoresis, purified phages (200 μg) were heated at 98°C for 20 min. After 10 min at room temperature, preparations were solubilized in an extraction buffer (1:1) containing 8 M urea, 2 M thiourea, 1% (vol/vol) NP-40, 2% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 20 mM dithioerythrol (DTE), 2% immobilized pH gradient (IPG) buffer, and bromophenol blue for 4 h at room temperature. IPG dry strips (18 cm; Pharmacia) from pH 4 to 7 were rehydrated overnight with the solubilized proteins. Isoelectric focusing was conducted on an IPGphor instrument (Pharmacia Biotech) for 24 h (60,000 V/h). Strips were equilibrated for the second dimension in 5 ml of buffer containing 50 mM Tris-HCl (pH 6.8), 6 M urea, 30%(vol/vol) glycerol, 2% (wt/vol) SDS, and 1% (wt/vol) DTE. After 10 min at room temperature, strips were transferred for another 10 min to the equilibration buffer, where 1% (wt/vol) DTE had been replaced with 2.5% (wt/vol) iodoacetamide and 0.5% (wt/vol) bromophenol blue. The strips were then transferred onto the surface of an SDS-10 to 20% polyacrylamide gel. Protein separation was done in a Protean Plus apparatus (Bio-Rad) at 40 mA/gel at 4°C. Protein were then fixed in 12% (vol/vol) trichloroacetic acid, stained with Coomassie brillant blue G-250, and destained with 7% (vol/vol) acetic acid.
Mass spectrometry (MS).
The protein spots of interest were excised from the gel and washed in aqueous 50% (vol/vol) acetonitrile followed by 0.1 M NH4HCO3. The excised proteins were reduced in 10 mM DTE (45 min at 56°C) and alkylated in 55 mM iodoacetamide (30 min at room temperature). In-gel trypsin digestion was performed in 50 mM NH4HCO3-5 mM CaCl2, and proteins were incubated overnight at 37°C. Resulting tryptic peptides were extracted in 50% (vol/vol) acetonitrile (ACN)-0.1% (vol/vol) trifluoroacetic acid and desalted on a ZipTip C18 column (Millipore).
For nanoelectrospray ion trap (nano-ESI) MS (tandem MS [MS/MS]), tryptic peptides were resuspended in 10 μl of methanol hydroxide-H2O-acetyl hydroxide (50:50:0.1), and 2 μl was loaded on an Au-Pd-coated capillary (Protana). The capillary was positioned by using a binocular on the nano-ES interface of the Q-Tof2 mass spectrometer (Micromass, Elstree, United Kingdom). This MS instrument was equipped with an orthogonal ES source (Z-spray) operating in positive-ion mode. Argon was used as the collision gas for the MS/MS experiments, with the pressure set at 4 mPa. Data acquisition and analysis were performed by using MassLynx 3.5 software (Micromass). [Glu]-fibrinopeptide was used for mass calibration.
Sequencing.
DNA of the purified phage LP65 was isolated, sheared to a size of about 1 kb, shot gun cloned into plasmid pUC18 in Escherichia coli, and sequenced to eightfold coverage at GATC (Konstanz, Germany).
Open reading frames (ORFs) were predicted with the FrameD program (Toulouse Bioinfo INRA) by using ATG, GTG, and TTG as possible start codons and a minimum size of 30 amino acids. Nucleotide and predicted amino acid sequences were compared in the GenBank, EMBL, PIR-Protein, SWISS-PROT, and PROSITE databases. Additional database searches were conducted using BLAST (4) and PSI-BLAST at the National Center for Biotechnology Information and FASTA (27). A search for tRNA genes was conducted using the tRNAscan-SE program (29). The motif search was performed by using Pfam.
Preliminary sequence data for phage SPO1 were obtained from the Pittsburgh Bacteriophage Institute website at http://pbi.bio.pitt.edu. Sequencing of SPO1 was accomplished with support from the National Institutes of Health.
Nucleotide sequence accession number.
The genome from phage LP65 was deposited at GenBank under accession number AY682195.
RESULTS
L. plantarum phage ecology.
L. plantarum is involved in the fermentation of vegetables (sauerkraut, pickles) (30, 54), coffee, meat (38, 51), and silage. In addition, L. plantarum is a commensal in the alimentary tracts of mammals, including humans (24). We screened the Nestlé Culture Collection for L. plantarum phages. One phage was isolated from silage of an Italian farm (LP45), another phage was obtained from maize fermentation yielding chicha, a traditional South American beverage (LP76 isolated in Ecuador), 15 phages were isolated from fermented coffee in Columbia (CC1 to CC15), and 1 phage was isolated from meat (salchichon salami in Spain; phage LP65). Two isolates (LP43 and LP78) were of unspecified origin. Sixteen phage isolates were Siphoviridae (10). Four isolates (LP43, LP65, CC12, CC13) showed the morphology of Myoviridae (Fig. 1).
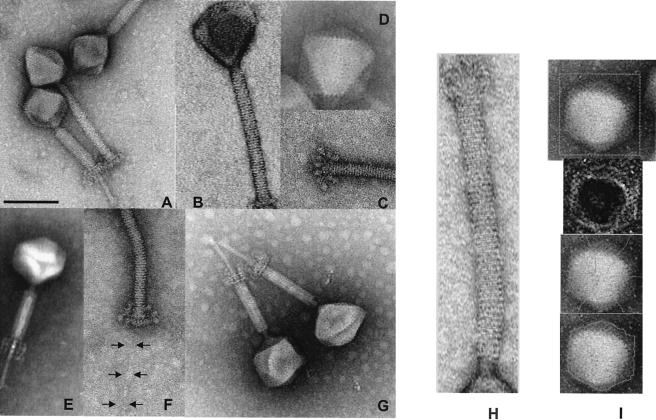
FIG. 1.
Negative-staining electron microscopy of CsCl-purified L. plantarum phage LP65. (A) Comparison of LP65 with contracted and noncontracted tails. (B) Striation of the tail sheath. (C) Baseplate of LP65 with noncontracted tail. (D) Icosahedral faces of the phage head. (E) Visualization of a structure within the interior of the tail. (F) Visualization of the tail fiber extending from the baseplate. (G) Transformation of the baseplate after tail contraction. (H) Image analysis of the tail: determination of the average distance of the tail stacks. (I) Image analysis of the head. From top to bottom: original image, processed gradient image, top of the polygon with medians, segmentation of the polyhedron without geometrical constraints. The negative stain was either ammonium molybdate coupled with bacitracin (C, D, F, G, and I), uranyl acetate (A, B, and H), or phosphotungstic acid (E). Bar, 100 nm. For the other dimensions, see Table 2.
LP65 was chosen for more-detailed investigation. The phage was isolated in 1992 from a Spanish salami factory that encountered problems of salami discoloration after 2 weeks of fermentation. Phage LP65 infected L. plantarum and Carnobacterium sp. strains associated with the fermented meat. The L. plantarum reference myophage fri (21, 51) infected the same L. plantarum strains as LP65, but none of the Carnobacterium sp. strains. Phages LP65, LP45, LP76, and LP78 showed distinct, nonoverlapping host ranges on L. plantarum strains LP65, LP45, LP76, and LP78, respectively (the phage names were actually derived from the code names of the strains on which they were isolated). LP65 also infected the recently sequenced L. plantarum strain WCFS1 (24).
LP65 morphology.
The contraction of the tail defined LP65 as a myovirus (Fig. 1A). Negative-staining electron microscopic pictures were obtained with different salts. Uranyl acetate allowed better resolution of the tail striation (Fig. 1B), while ammonium molybdate permitted better visualization of the baseplate (Fig. 1C) and head structure (Fig. 1D). Phosphotungstic acid stained a structure within the contracted tail (Fig. 1E). The polygonal head is linked via a neck structure to the tail (Fig. 1A, G, and H), which ends with a baseplate structure and a thin tail fiber (Fig. 1F). Upon contraction, the baseplate changes its conformation substantially (compare Fig. 1F and G), it moves up with the shortened tail, and a tail tube extends from the tail sheath (Fig. 1G). The tube showed a faint terminal swelling (Fig. 1A), and phages were frequently observed to adhere by their tube ends, sometimes building large phage bouquets.
The distance between the individual stacks of the tail was determined to be 4.3 ± 0.4 nm (mean + standard deviation) (n = 53) (Fig. 1H). The stacks showed globular structures, possibly representing the individual tail protein monomers. The tail length in the extended conformation was 193 ± 8 nm, and the width was 19.5 ± 1.2 nm when stained with uranyl acetate (n = 68). The corresponding dimensions for molybdate-stained phages were smaller, but not substantially so (e.g., 186 ± 8 nm for the tail length) (n = 30).
Globular structures were visualized at the end of the extended tail. The globules are connected with a stalk to the tail structure in a grape-like fashion (Fig. 1C and F). In contracted phages, a dramatic reorganization of the entire tail structure was observed (Fig. 1A). The length of the tail decreased to 115 ± 5 nm, while the width increased to 25 ± 1 nm (uranyl acetate values) (n = 42). The parallel striation was replaced by an oblique chevron-like pattern (Fig. 1A). A 59- ± 6-nm-long tube with a width of 11 + 1 nm extended from the contracted tail (n = 25). The baseplate also showed a substantial structural change: three parallel interconnected planes perpendicular to the tail could be discerned (Fig. 1G).
To explore the head dimensions, the original image was transformed into a gradient image (17), which allowed a definition of the top of the polygon and the medians (Fig. 1I). Segmentation of the head was then done without any geometrical constraints or by using a polygonal or a parabolic model for the edges (17). The edge length was determined to be 60.8 ± 3.1 nm (molybdate staining) (n = 45) and did not differ substantially between methods. Triangular and more complicated polygonal surfaces were displayed by many LP65 particles (Fig. 1D). On these surfaces, chain mail-like circular structures could be identified (Fig. 1A, G, and I).
Infection cycle.
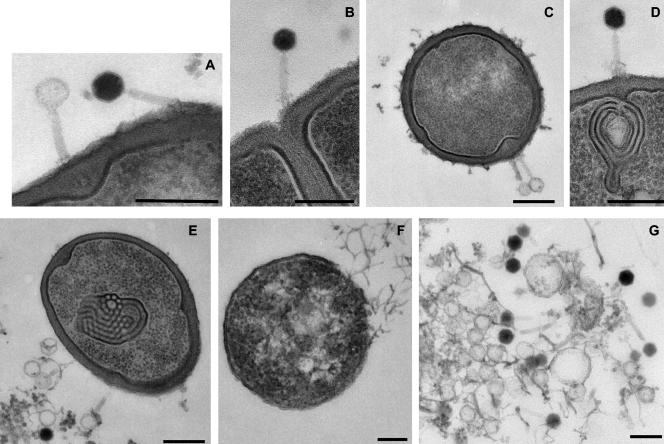
LP65 yields clear plaques of about 1 mm. Cells were infected with an MOI of 3 at an optical density of 0.1 at 600 nm. The first progeny extracellular phage was detected at 100 min p.i. A final titer of 5 × 109 PFU of extracellular phage/ml was reached at 130 min p.i. Phage proteins became visible on the background of cellular proteins at 80 min p.i. (data not shown). Cells were recovered at the different time points and investigated by ultrathin section electron microscopy. In the early infection phases, two types of phage particles were adsorbed to the cell wall (Fig. 2A). One particle type had an electron-dense head interior, suggesting a phage head filled with DNA. A small rim of less electron-dense material probably represents the isometric hexagonal protein shell of the phage capsid. The other adsorbed phage type had an electron-translucent head suggestive of a phage that had already ejected its DNA. The adsorbed phages showed a diffuse broadening of the tail near the cell wall, which probably represents the baseplate of the contracted tail contacting the cell surface (Fig. 2B and D). In several ultrathin sections, an extension of the tail was seen tunneling through the cell wall layer (Fig. 2A and C). We interpret this structure as the tail tube of the contracted tail. The end of this tube comes close to the cell membrane, probably to mediate transport of the DNA into the cytoplasm. The cell wall underlying the phage adsorption site frequently bulged into the cytoplasm (Fig. 2A and C). As the phages showed a tendency to adsorb next to the septum of dividing cells (Fig. 2B), one might wonder whether the bulge represents an early septum formation step. Targeting dividing cells would ensure a metabolically active cell. A frequent sign of early infection was membrane convolutions inside the infected cell (Fig. 2D and E). They resembled the structures observed in L. plantarum exposed to bacteriocins, suggestive of a cellular stress reaction to phage infection (13). Alternatively, the physical penetration of the phage DNA itself might also cause the convolution of the membranes. Mesosomes from L. plantarum showed a distinct ultrastructure (22), and we failed to detect the membrane convolutions in a large series of uninfected L. plantarum cells. At later times of infection, the cell wall had largely disappeared and the cytoplasm showed a central clearing containing structures suggestive of phage heads (Fig. 2F). DNA-filled and empty phage heads were clearly observed in the cytoplasm released from lysed cells (Fig. 2G).
FIG. 2.
Ultrathin section of LP65-infected L. plantarum cells. (A) LP65 with an empty and a full head adsorbed to the cell wall. Note the small protein shell surrounding the DNA-filled head of the right phage particle and the tail tube of the left particle crossing the cell wall. (B) Phage adsorbing near the septum of a cell, showing the baseplate contacting the surface of the cell wall. (C) Adsorbing LP65 particles. Note again the tail tube and the bulging of the cell wall in the cytoplasm. (D) LP65 making contact via the baseplate with the cell wall and the underlying convolution of cell membranes. (E) Cell containing an intracytoplasmic membrane convolution with phage ghosts adsorbed to the cell wall. (F) A cell that has lost most of its cell wall and is about to release its cytoplasmic contents at the top right. Structures suggestive of phage particles are at the central clearing of the cell. (G) Extruded cytoplasm from a lysed cell with filled heads and empty preheads. Bars, 250 nm.
Phage LP65 DNA analysis.
Myophages LP65 and fri, which could be propagated on the same lysogenic host strain, L. plantarum LP65, showed clearly distinct restriction patterns (Fig. 3A), which differed also from the DNA of the siphophage LP651 induced with mitomycin C from the lysogen (Fig. 3B). Superinfection with phage LP65 did not induce this prophage, since screening of several hundred electron microscopic fields of a phage LP65 preparation produced on the lysogenic strain revealed not a single siphophage.
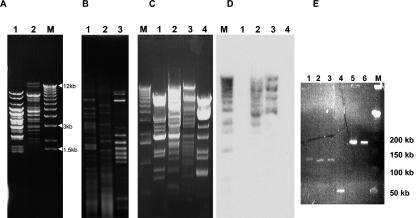
FIG. 3.
DNA analysis of L. plantarum phages. (A) XbaI digestion of DNA from myophage fri (lane 1) and LP65 (lane 2). M: size marker (1-kb lambda DNA ladder; Invitrogen). (B) AccI digests of DNA from phage fri (lane 1), LP65 (lane 2), and the mitomycin C-induced resident prophage LP651 (lane 3) from the propagating L. plantarum strain 65. (C) HindIII digest of DNA from siphophages LP45 (lane 1) and LP76 (lane 4) and myophages fri (lane 2) and LP65 (lane 3). M: size marker (as in A). (D) Southern blot corresponding to the HindIII digest in panel C, probed with radiolabeled phage LP65 DNA. The marker was revealed with labeled lambda DNA. (E) Pulsed-field gel electrophoresis of L. plantarum myophages LP43 (lane 1), LP57 (lane 2), and LP65 (lane 3), beta-4-like coliphage JS122.1 (lane 4), and T4-like coliphages JS148 (lane 5) and RB33 (lane 6). M: size marker (phage lambda concatemers, 50-kb DNA ladder; Promega).
The siphophages LP45 (silage; Italy) and LP76 (beverage; Ecuador) showed restriction patterns distinct from each other and from the myophages LP65 and fri (Fig. 3C). In Southern hybridization, labeled LP65 DNA cross-hybridized with phage fri DNA, but not with DNA from siphophages LP45 and LP76 (Fig. 3D). Pulsed-field gel electrophoresis demonstrated for phages LP65, LP43, and fri an estimated genome size of 130 kb, well below the ca.170-kb genome size of T4-like Myoviridae from E. coli, but far above the 50-kb genome size of beta 4-like siphophage from E. coli (Fig. 3E).
LP65 protein analysis.
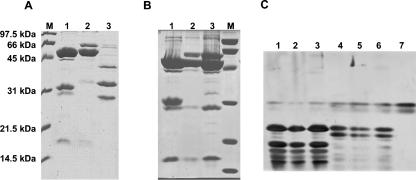
On SDS-PAGE gels, CsCl gradient-purified phage LP65 showed two major proteins of 55 and 33 kDa and minor proteins of about 100, 40, 30, and 17 kDa (Fig. 4A). In contrast, myophage fri showed two major proteins of 65 and 55 kDa but lacked a major 33-kDa protein (Fig. 4A). The latter pattern corresponds to that reported for Myoviridae from gram-positive bacteria (21, 28). Interestingly, mechanical shearing of phage LP65 led to disappearance of the 33-kDa protein and the appearance of a 65-kDa protein (Fig. 4B; compare lane 1 with lanes 2 and 3). Peptide mapping of the V8 protease-digested 33-kDa (Fig. 4C, lane 4) and 65-kDa (Fig. 4C, lanes 5 and 6) proteins showed an identical digestion pattern, suggesting two different conformations of the same protein with grossly distinct migration properties on SDS-PAGE gels induced by shearing and concomitant tail contraction. In 2-D gels, untreated LP65 showed major 65- to 55-kDa proteins that migrated as streaks, but no major 33-kDa protein (Fig. 5). Note that phage LP65 was treated with urea, a chemical known to induce tail contraction in T4-like phages (5), before 2-D electrophoresis, again suggesting a link between the 33- to 65-kDa protein transition and the tail contraction process.
FIG. 4.
1-D protein analysis of L. plantarum phages. (A) SDS-PAGE of untreated CsCl-purified myophages LP65 (lane 1) and fri (lane 2) and siphophage LP45 (lane 3). M: broad-range protein marker (Bio-Rad). (B) Effects of ultrasound treatment (lane 2) and syringe shearing (lane 3) of LP65 on the protein pattern in SDS-PAGE gels compared to that for the untreated preparation (lane 1). (C) V8 peptide maps of the 55-kDa proteins of the three LP65 preparations for which results are shown in panel B, lanes 1 to 3 (lanes 1 to 3), the 33-kDa protein from LP65 for which results are shown in panel B, lane 1 (lane 4), and the 65-kDa proteins from LP65 for which results are shown in panel B, lanes 2 and 3 (lanes 5 and 6). The V8 digestion represents untreated (lanes 1 and 4), ultrasound-treated (lanes 2 and 5), or syringe-sheared (lanes 3 and 6) LP65 particles. V8 protease is shown in lane 7.
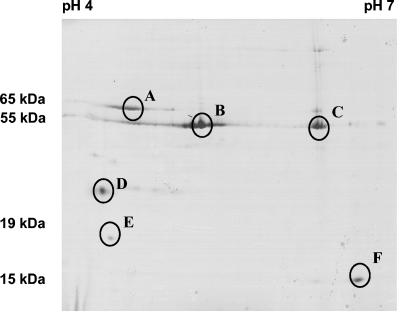
FIG. 5.
2-D protein analysis of phage LP65. Coomassie-stained 2-D gel electrophoresis of CsCl-purified phage LP65. The first dimension used a linear pH 4 to 7 IPG strip gel (Pharmacia), and the second used a 10 to 20% gradient SDS-PAGE gel. Proteins analyzed by nano-ESI-MS (MS/MS) are identified by circles.
The phage proteins with lower molecular weights yielded relatively clear spots at acidic pHs, while a protein of about 15 kDa focused at a neutral pH (Fig. 5). To clarify the origins of the proteins separated by 2-D analysis, MS/MS analysis was done. Streak A, migrating at 65 kDa, yielded tryptic peptides derived from gp 103, the likely tail sheath protein (see below) of LP65. About half of the predicted 64,672-Da protein was covered by peptide sequences including N- and C-terminal sequences.
Streak B, migrating at 55 kDa, provided peptides covering two-thirds of gp 109, the likely major capsid protein with a predicted molecular size of 56,293 Da (Fig. 5). Spot C also gave peptides corresponding to gp 109; no chemical modifications that could explain the changed focusing behavior of this protein were identified by MS/MS.
Spot D gave peptides from gp 51, lacking database matches and encoded downstream of the DNA replication gene cluster. In contrast, spots E and F provided peptides from gp 91 and gp 102, respectively, representing a possible tail fiber and a tail protein, respectively.
LP65 genome sequence.
The LP65 DNA was sheared and cloned as approximately 1-kb fragments. After an average eightfold sequencing coverage was attained, the phage genome was assembled into a single large contig of 131,573 bp. On this contig 165 open reading frames with more than 30 codons each were identified (Table 1). Several ORFs were probably created by mutations, either by the introduction of a premature stop codon (ORFs 84 and 85) or by the insertion of HNH endonuclease genes, which are frequently intron associated in phages from lactic acid bacteria (16). Endonuclease genes carried on ORFs 114, 126, and 143 were inserted into the likely large-subunit terminase gene, a gene encoding a putative membrane protein, and a possible transcriptional regulator gene, respectively. Fifteen ORFs started with an unusual initiation codon (GTG or TTG).
TABLE 1.
Database matches of the predicted open reading frames of L. plantarum phage LP65
| ORFa | Startb | Stopc | Lengthd (aa) | Hit descriptione | % Identityf,g | E valueg,h |
|---|---|---|---|---|---|---|
| orf1* | 181 | 273 | 31 | |||
| orf2 | 692 | 1006 | 105 | Listeria innocua plasmid pLI100 | 30 | −3 |
| orf3 | 1087 | 1305 | 73 | |||
| orf4* | 2366 | 2563 | −66 | |||
| orf5 | 4071 | 3523 | −183 | |||
| orf6 | 4337 | 4101 | −79 | |||
| orf7 | 4547 | 4356 | −64 | |||
| orf8 | 4913 | 4563 | −117 | |||
| orf9 | 5350 | 4916 | −145 | L. plantarum prophage Lp1 gene 31; DNA replication; phage SPO1 g 153 | 45 | −15 |
| orf10 | 5739 | 5380 | −120 | |||
| orf11 | 6046 | 5873 | −58 | |||
| orf12* | 6297 | 6049 | −83 | |||
| orf13 | 6682 | 6308 | −125 | |||
| orf14 | 7115 | 6753 | −121 | |||
| orf15 | 7725 | 7276 | −150 | L. plantarum prophage Lp1 gene 30: DNA replication | 34 | −5 |
| orf16 | 7866 | 7738 | −43 | |||
| orf17 | 8113 | 7880 | −78 | |||
| orf18 | 8409 | 8131 | −93 | |||
| orf19 | 8730 | 8452 | −93 | |||
| orf20 | 9249 | 8749 | −167 | |||
| orf21 | 9638 | 9351 | −96 | |||
| orf22 | 9864 | 9667 | −66 | |||
| orf23 | 10879 | 9881 | −333 | |||
| orf24 | 11041 | 10883 | −53 | |||
| orf25 | 11262 | 11053 | −70 | |||
| orf26 | 11827 | 11288 | −180 | |||
| orf27 | 12093 | 11830 | −88 | |||
| orf28 | 12620 | 12333 | −96 | L. plantarum phage φg1e; transcription? Gene Rorf95 | 34 | −6 |
| orf29 | 12818 | 12645 | −58 | |||
| orf30 | 13210 | 12821 | −130 | |||
| orf31 | 13501 | 13223 | −93 | |||
| orf32 | 14304 | 13912 | −131 | |||
| orf33 | 14791 | 14375 | −139 | |||
| orf34 | 15553 | 14864 | −230 | |||
| orf35 | 15875 | 15558 | −106 | |||
| orf36 | 16340 | 15894 | −149 | Melanoplus entomopoxvirus, unknown genes in Drosophila, mouse, human | 32 | −13 |
| orf37 | 16786 | 16352 | −145 | L. plantarum prophage Lp2 gene 26; DNA replication? | 49 | −31 |
| orf38 | 17344 | 17063 | −94 | |||
| orf39 | 18061 | 17357 | −235 | |||
| orf40 | 19101 | 18427 | −225 | |||
| orf41 | 19862 | 19098 | −255 | Phage SPO1 g 160 | ||
| orf42* | 21115 | 20102 | −338 | EPS sugar transferase of Streptococcus thermophilus | 29 | −20 |
| orf43 | 21590 | 21204 | −129 | |||
| orf44 | 22125 | 21604 | −174 | |||
| orf45* | 22866 | 22132 | −245 | Phage K g 101 | 32 | −11 |
| orf46* | 23393 | 22863 | −177 | |||
| orf47 | 23830 | 23408 | −141 | orf43; putative tail lysin of Lactococcus bacteriophage 4268 | 44 | NS |
| orf48 | 23996 | 23847 | −50 | |||
| orf49* | 25091 | 24063 | −343 | Prophage pi2 gene 34 of Lactococcus lactis | 29 | −15 |
| orf50* | 25550 | 25224 | −109 | |||
| orf51 | 26106 | 25570 | −179 | Structural protein (2-D,MS) | ||
| orf52 | 26703 | 26185 | −173 | |||
| orf53 | 27971 | 26754 | −406 | gp117 mycophage CJW1: RecA protein (recombinase A); phage K g 93 | 30 | −36 |
| orf54 | 29143 | 27983 | −387 | |||
| orf55* | 29682 | 29203 | −160 | |||
| orf56 | 30412 | 29741 | −224 | Phage SPO1 gene 93 | ||
| orf57 | 30947 | 30465 | −161 | |||
| orf58 | 31568 | 30969 | −200 | |||
| orf59 | 34783 | 31817 | −989 | DNA polymerase phage SPO1 gene 111 and 113, phage K g 86/88/90 | 34 | −87 |
| orf60 | 36081 | 34918 | −388 | Transposase; Enterococcus faecium | 39 | −64 |
| orf61 | 36659 | 36213 | −149 | |||
| orf62 ? | 37300 | 36656 | −215 | Phage SPO1 gene 91 | ||
| orf63* | 38580 | 37301 | −426 | Phage K g 98 | 25 | |
| orf64 | 39544 | 38657 | −296 | |||
| orf65 | 39969 | 39604 | −122 | |||
| orf66* | 40297 | 39956 | −114 | |||
| orf67 | 41056 | 40316 | −247 | Deoxyguanosine kinase of Lactobacillus plantarum | 49 | −54 |
| orf68 | 42139 | 41075 | −355 | DNA primase dnaG phage, phage K g 76 | 29 | −25 |
| orf69 | 42870 | 42154 | −239 | |||
| orf70 | 44859 | 42943 | −639 | gp46 recombination endonuclease phages RB69, RB49, SPO1 43 and 87, K g 74 | 30 | −78 |
| orf71* | 45202 | 44888 | −105 | |||
| orf72 | 45729 | 45205 | −175 | |||
| orf73* | 46254 | 45844 | −137 | Cgl2159 cell division membrane protein of Corynebacterium glutamicum | 31 | 0.072 |
| orf74* | 46545 | 46273 | −91 | Conserved hypothetical protein of Corynebacterium diphtheriae | 35 | 0.067 |
| orf75 | 47613 | 46546 | −356 | gp47 recombination endonuclease of T4-like E. coli. phage RB49: SPO1 g 84, K g 72 | 28 | −24 |
| orf76 | 49119 | 47629 | −497 | gp41 DNA primase-helicase of phage RB69; SPO1 gene 82, K g 71 | 25 | −27 |
| orf77 | 49556 | 49299 | −86 | |||
| tRNA | 49688 | 49760 | tRNA-Gly | |||
| tRNA | 49777 | 49847 | tRNA-Thr | |||
| tRNA | 49855 | 49927 | tRNA-Trp | |||
| orf78 | 50791 | 50462 | −110 | gp37 Streptococcus mitis phage SM1: replication-regulation? | 45 | −12 |
| tRNA | 50751 | 50824 | 24 | tRNA-Pseudo | ||
| orf79 | 51300 | 51467 | 56 | |||
| tRNA | 51456 | 51529 | tRNA-Phe | |||
| tRNA | 51684 | 51757 | tRNA-Leu | |||
| tRNA | 51768 | 51840 | tRNA-Leu | |||
| tRNA | 51953 | 52025 | tRNA-Leu | |||
| tRNA | 52803 | 52874 | tRNA-Asn | |||
| tRNA | 52977 | 53064 | tRNA-Ser | |||
| tRNA | 53205 | 53276 | tRNA-Ile | |||
| tRNA | 53541 | 53613 | tRNA-Arg | |||
| tRNA | 53655 | 53737 | tRNA-Pseudo | |||
| orf80 | 53982 | 53842 | −47 | |||
| tRNA | 53992 | 54067 | tRNA-Arg | |||
| orf81 | 55514 | 54411 | −368 | |||
| orf82 | 56461 | 56916 | 152 | |||
| orf83 | 57045 | 57815 | 257 | lp_0559 nucleoside transporter? L. plantarum | 42 | −54 |
| orf84 | 57808 | 58242 | 145 | NAD-responsive transcriptional regulator nadR of Lactococcus lactis | 57 | −41 |
| orf85 ? | 58243 | 58915 | 224 | NAD-responsive transcriptional regulator nadR of Lactococcus lactis | 57 | −71 |
| orf86 | 58918 | 59451 | 178 | ADP-ribose pyrophosphatase of L. plantarum:phage SPO1 gene 156 | 34 | −16 |
| orf87 | 59515 | 59763 | 83 | |||
| orf88* | 61212 | 59818 | −465 | Lysin of L. plantarum phage φgle | 40 | −85 |
| orf89* | 61616 | 61233 | −128 | Holin, lp_2399 prophage Lp2 of L. plantarum | 47 | −11 |
| orf90 | 65934 | 61711 | −1408 | Phage SPO1 gene 69: vrlC protein of Dichelobacter nodosus, phage K g 65 | 32 | −49 |
| orf91* | 66461 | 65946 | −172 | Phage SPO1 gene 68, phage K g 64 | 31 | −12 |
| orf92 | 68511 | 66841 | −557 | Phage SPO1 gene 39:yomE of Bacillus phage SPBc2 | 25 | −3 |
| orf93 | 68758 | 68516 | −81 | |||
| orf94 | 70998 | 68815 | −728 | Phage SPO1 gene 67:ygfG of Lactococcus lactis, phage K g 63 | 41 | −27 |
| orf95 | 72445 | 71045 | −467 | Phage SPO1 gene 66; tail protein gp137 of Mycobacteriophage Bxz1: K g 62 | 33 | −46 |
| orf96 | 72901 | 72524 | −126 | BH0963 of Bacillus halodurans | 28 | −5 |
| orf97 | 79569 | 73231 | −2113 | Tail fiber, hemagglutinin of Streptococcus epidermidis:SPO1 g 63, K g 58 | 24 | −27 |
| orf98 | 83463 | 79642 | −1274 | Lactobacillus bacteriophage adh: tail protein, phage K g 55 | 31 | −15 |
| orf99 | 83975 | 83529 | −149 | Tail fiber of Staphylococcus aureus phage φ12, phage K g 54 | 28 | −5 |
| orf100 | 84672 | 84055 | −206 | ORF 9 of Listeria of phage A511 | 25 | −3 |
| orf101 | 85050 | 84748 | −101 | |||
| orf102 | 85628 | 85161 | −156 | ORF 8 of Listeria phage A511: Staphylococcus phage Twort: SPO1 g 58, K g50 | 56 | −43 |
| orf103 | 87467 | 85644 | −608 | Major tail sheath protein of Listeria A511: phage Twort, SPO1 g 57, K g 49 | 35 | −86 |
| orf104 | 87689 | 87495 | −65 | |||
| orf105 | 88584 | 87682 | −301 | ORF 6 of Listeria phage A511 | 26 | −12 |
| orf106 | 89265 | 88585 | −227 | ORF 5 of Listeria phage A511; phage K g 47 | 30 | −10 |
| orf107 | 90145 | 89252 | −298 | ORF 4 of Listeria phage A511; phage K g 46 | 29 | −20 |
| orf108 | 91078 | 90155 | −308 | ORF 3 of Listeria phage A511:Bacillusphage SPO1 gene 53, K g 45 | 36 | −39 |
| orf109 | 92725 | 91181 | −515 | Phage K g 44 | 41 | −85 |
| orf110 | 93774 | 92815 | −320 | Phage SPO1 gene 15 | ||
| orf111 | 94680 | 93844 | −279 | ORF 1 of Listeria phage A511: of phage SPO1 gene 49, phage K g 42 | 43 | −41 |
| orf112 | 96345 | 94738 | −536 | Phage of SPO1 gene 48: gp34 Streptomyces phage phi-C31, phage K g 41 | 37 | −67 |
| orf113 | 97895 | 96429 | −489 | terL large subunit terminase of Xylella prophage: SPO1 g 34, K g35 | 42 | −103 |
| orf114 | 98933 | 97971 | −321 | Group I intron endonuclease ? Lactococcus lactis phage Tuc2009, phage Twort | 29 | −23 |
| orf115 | 99487 | 99212 | −92 | Phage SPO1 gene 34, phage K gene 35 | 51 | −23 |
| orf116 | 99614 | 99471 | −48 | |||
| orf117 | 100085 | 99627 | −153 | Phage K gene 34 | 24 | |
| orf118 | 100362 | 100069 | −98 | |||
| orf119 | 100832 | 100437 | −132 | Coiled-coil protein of Pyrobaculum | 27 | −3 |
| orf120 | 101021 | 101569 | 183 | Non-heme iron-binding ferritin of Lactococcus lactis | 35 | −4 |
| orf121 | 101790 | 102371 | 194 | lp_0302 extracellular protein of L. plantarum (lysin phage TP901-1) | 33 | −23 |
| orf122 | 102577 | 103074 | 166 | lp_3342 low-temperature requirement C protein of L. plantarum | 48 | −37 |
| orf123 | 103542 | 105341 | 600 | uvsW helicase-ATPase of phage RB69; SPO1 gene 80, K gene 69 | 24 | −22 |
| orf124 | 105407 | 106342 | 312 | |||
| orf125 | 106431 | 107210 | 260 | lp_0259 integral membrane protein of L. plantarum | 68 | −90 |
| orf126 | 107290 | 108501 | 404 | HNH endonuclease of Lactobacillus phage A2;phage Twort and SPO1 gene 112 | 37 | −10 |
| orf127 | 108590 | 108814 | 75 | Phage SPO1 gene 15, leucine zipper motif | 30 | −4 |
| orf128 | 108912 | 109628 | 239 | DNA modification protein, phage Mu MOM-like | 30 | −7 |
| orf129 | 109792 | 110085 | 98 | Phage SPO1 gene 64 | ||
| orf130 | 110072 | 110497 | 142 | Phage SPO1 gene 64 | ||
| orf131 | 110497 | 111066 | 190 | Phage K gene 59 | 31 | −12 |
| orf132 | 111066 | 111776 | 237 | Phage K gene 61 | 34 | −10 |
| orf133 | 111833 | 112198 | 122 | Single-strand binding protein of Staphylococcus phage PVL | 34 | −3 |
| orf134 | 112228 | 115416 | 1063 | Phage SPO1 gene 62: cell wall hydrolase of Clostridium perfringens, K g56 | 30 | −11 |
| orf135 | 115494 | 115751 | −86 | Gp52 protein of Yersinia pestis phage φA1122 | 59 | −4 |
| orf136 | 115754 | 116257 | 168 | |||
| orf137 | 116328 | 117917 | 530 | SPO1 gene 119, phage K gene 18 | 25 | |
| orf138 | 118359 | 119039 | 227 | |||
| orf139 | 119059 | 119469 | 137 | |||
| orf140 | 119536 | 119949 | 138 | |||
| orf141 | 120023 | 120568 | 182 | |||
| orf142 | 120630 | 120848 | 73 | |||
| orf143 | 121037 | 122101 | 355 | HNH homing endonuclease of L. casei phage A2: SPO1 gene 112, K g89 | 36 | −20 |
| orf144 | 122131 | 122424 | 98 | arpR-like transcriptional regulators of Streptococcus thermophilus phages, K g17 | 42 | −16 |
| orf145 | 122579 | 123052 | 158 | |||
| orf146 | 123162 | 123260 | 33 | |||
| orf147 | 123241 | 123504 | 88 | |||
| orf148 | 123523 | 123804 | 94 | |||
| orf149 | 123817 | 124203 | 129 | |||
| orf150 | 124320 | 124688 | 123 | |||
| orf151 | 124739 | 125176 | 146 | lp_0654 prophage Lp1 protein 31 of L. plantarum | 31 | −8 |
| orf152 | 125207 | 125491 | 95 | |||
| orf153 | 125546 | 125713 | 56 | |||
| orf154 | 125716 | 126159 | 148 | |||
| orf155 | 126169 | 126615 | 149 | |||
| orf156 | 126629 | 127087 | 153 | |||
| orf157 | 127098 | 127538 | 147 | gp51 of Listeria phage A118 | ||
| orf158 | 127552 | 127881 | 110 | |||
| orf159 | 128531 | 129070 | 180 | |||
| orf160 | 129138 | 129407 | 90 | |||
| orf161 | 129656 | 129790 | 45 | |||
| orf162 | 129856 | 130220 | 121 | |||
| orf163 | 130221 | 130384 | 54 | |||
| orf164 | 130484 | 130987 | 168 | |||
| orf165 | 130980 | 131387 | 136 |
An asterisk indicates an unusual start codon (GTG or TTG); a question mark suggests that an ORF was created by a mutational process.
Base pair number for the start position of the deduced gene.
Base pair number for the stop position of the deduced gene.
A minus sign indicates a leftward orientation on the map shown in Fig. 6.
Matches with B. subtilis phage SPO1 are boldfaced, matches with Listeria phage A511 are underlined and italicized; matches with Staphylococcus phage K are italicized and boldfaced. g, gene number.
Percent amino acid identity.
Boldfaced values indicate that phage K matches were the highest.
Logarithm of the E value for the match. NS, not significant.
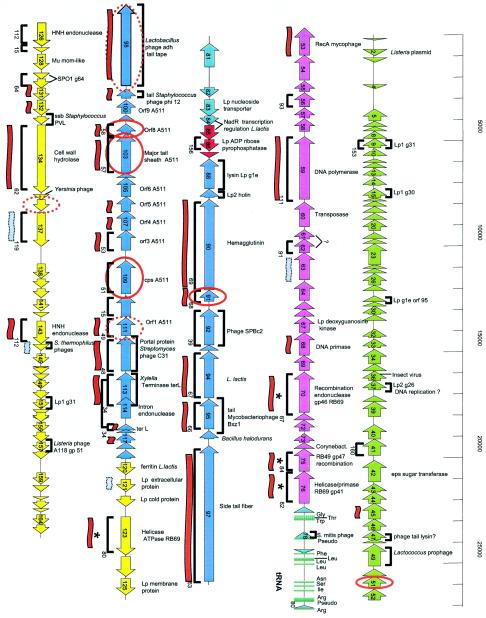
Three major genome regions could be differentiated upon analysis of the distribution of the coding sequences on the two DNA strands (Fig. 6). The first large cluster is represented by the leftward-oriented ORFs 5 to 76. ORF 5 is followed by 3.5 kb of gene-poor DNA, and ORF 76 is preceded by a large cluster of 14 tRNA genes and a small group of 6 rightward-oriented genes. The second large cluster is constituted by ORFs 88 to 119, which are again all leftward oriented. The third cluster consists of the rightward-oriented ORFs 120 to 165.
FIG. 6.
Genome sequence of phage LP65. ORF (arrows) and tRNA (small turquoise bars) prediction in the 131,573-bp sequence of the single contig obtained for LP65 DNA. Gene clusters defined by overall genome organization and database matches are marked with different colors (green, left arm; violet, DNA replication module; red, lysis cassette; blue, structural module; yellow, unknown). Brackets above and below the ORFs indicate matches of the deduced protein with predicted proteins from phages of gram-positive bacteria (except SPO1 and A511) andB. subtilis myophage SPO1, respectively. The matching SPO1 genes are indicated with their gene numbers. Red and grey flags below the ORF indicate significant and weak matches with Staphylococcus phage K, respectively. Genes sharing amino acid sequence identity with T4-like myophages are marked with an asterisk. Gene products identified by MS/MS are circled in red. Dotted circles represent identifications with low scores (Sequest scores of <20).
The observed database matches allowed a first tentative subdivision of the LP65 genome into modules (Table 1). ORFs 53 to 76 contained a number of genes with links to DNA replication functions (RecA, DNA polymerase, DNA primase, DNA helicase, and several recombination endonucleases). The recombination enzymes shared significant sequence identity with gp 41, gp 46, and gp 47 from the T4-like E. coli phages RB69 and RB49 (53). These genes were together with the ectopically placed helicase-ATPase encoded by ORF 123, the only links to T4-like Myoviridae from gram-negative bacteria.
ORFs 5 to 52 seem to represent a distinct module unrelated to DNA replication. The most frequent links are with prophages from L. plantarum, suggesting species-specific functions. Another large cluster, ORFs 88 to 119, represents the likely structural genes from phage LP65, with links to Listeria phage A511 (28), Staphylococcus phage K (39), and Bacillus phage SPO1 (Fig. 6). These links allowed a tentative identification of the major tail sheath (ORF 103) and the major capsid protein (ORF 109). A further large gene cluster is represented by ORFs 120 to 165, the only large rightward-oriented genome region (Fig. 6). The database matches do not allow speculations on its function. Relatively prominent were links with genes from L. plantarum (n = 4) and other low-GC-content gram-positive bacteria (n = 5). Six genes, including two encoding HNH endonucleases, shared sequence relatedness with phage SPO1.
Comparative genomics.
Thirty-two LP65 ORFs shared significant sequence identity with Bacillus subtilis phage SPO1 (Table 1; Fig. 6) retrieved from the website of the Pittsburgh Bacteriophage Institute (http://pbi.bio.pitt.edu/). However, the sequence relatedness was observed only at the protein level and was not higher than that shared with Listeria phage A511, which ranged from 25 to 56% amino acid identity (Fig. 7). Thirty LP65 ORFs shared significant protein sequence identity with Staphylococcus phage K. There was substantial overlap among all three phages: 20 ORFs showed sequence relatedness among phages LP65, K, and SPO1. This similarity was especially pronounced over the structural genes. The putative terminase, portal, minor and major head, tail sheath, and a number of structural genes further downstream were well conserved. Variability was introduced by gene replacements (e.g., LP65 ORF 110) and gene insertion/deletions (e.g., LP65 ORF 101).
FIG. 7.
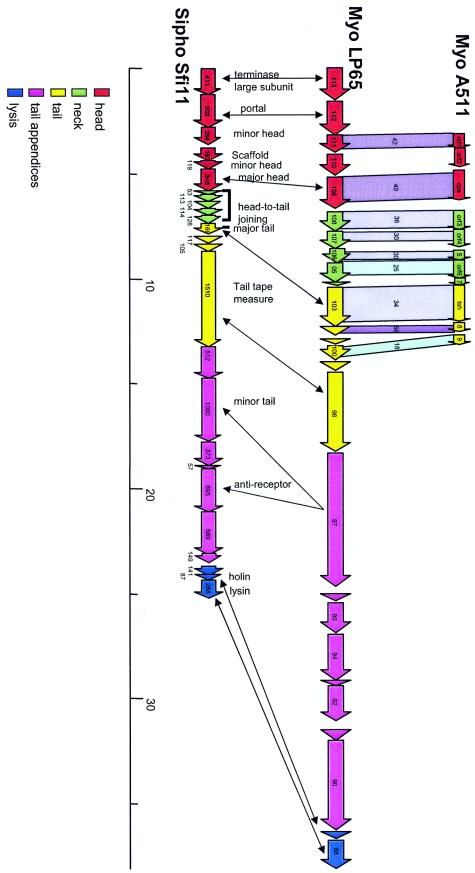
Comparative genomics suggests an Sfi11 Siphovirus-like organization of the putative structural gene cluster from LP65. The deduced structural gene map from LP65 (center) is aligned with the corresponding region from Listeria myophage A511 (top). Genes sharing protein sequence identity are linked by different intensities of shading (numbers give the percent amino acid identity). Likewise, the LP65 gene map is also aligned with the structural gene map from S. thermophilus phage Sfi11, the type phage of a widely distributed class of lambda-like Siphoviridae in low-GC-content gram-positive bacteria. The modular structure of the Sfi11 structural gene cluster is indicated by the color code as analyzed previously (31), and selected genes are indicated with their deduced functions. Double-headed arrows link genes possibly sharing the same functions according to database matches obtained for LP65 ORFs. The colinearity of the gene maps from the phages suggests the modular structure for LP65 and A511 indicated by the color code.
The SPO1-like genus of the family Myoviridae, an ICTV-recognized genus in this phage family, showed no relatedness in genome organization to T4-like Myoviridae; the only links were the sharing of four sequence-related DNA replication genes (Fig. 6). In contrast, fascinating relationships with siphophages from the lambda supergroup (8) were detected (Fig. 7). As in phages from the lambda superfamily, the large-subunit terminase gene in LP65 precedes a putative portal protein-encoding gene. Both LP65 proteins still shared significant protein sequence identity with siphophages, which further substantiates the evolutionary links between these phage groups (Fig. 6). Between the portal and the major capsid gene, LP65 showed two genes, which in the lambda superfamily would correspond to a minor head protein (ORF 111) and a scaffold protein (ORF 110). Actually, in the lambda supergroup, two subtypes were identified: the Sfi21 type, with the portal-protease-head gene constellation, and the Sfi11 type, with the portal-minor head-scaffold-major head gene constellation (8). LP65 appears more related to the latter type, in agreement with the observation that no proteolytic processing of the major head protein was observed by mass spectrometry. The LP65 major head gene (ORF 109) is separated from the major tail protein (ORF 103) by four genes, which in the lambda superfamily represent candidate head-to-tail joining genes. These genes are longer than the corresponding lambda genes, which could reflect the morphologically more prominent neck structure in SPO1-like phages than in lambda-like phages. In phages from the lambda supergroup, the following gene order was observed: major tail gene-two tail genes-a large tail tape measure gene-a further large gene encoding the phage anti-receptor (8, 14). A strikingly similar genome organization is found in LP65. Four genes, one with sequence links to a Staphylococcus siphophage φ12 (20) tail protein, separate the major tail gene from a long gene sharing protein sequence similarity with the tail tape measure gene from the Lactobacillus siphophage adh (3). The next long gene shared links with side tail fiber proteins from E. coli siphophages over two protein domains. Further downstream, seven additional proteins were observed; two were distantly related to potential tail genes from siphophages. This number is greater than that normally found in lambda-like Siphoviridae. This larger number of putative LP65 tail genes should not be surprising in view of the complex tail structure. As in Siphoviridae from low-GC-content gram-positive bacteria, the lysis cassette in LP65 directly followed the structural genes.
Phage LP65 and K comparison.
Phages LP65 and K showed the same overall genome organization, i.e., a central location of the DNA replication and structural gene modules, flanked by long arms of genes at both genome ends transcribed in opposite directions. The structural gene cluster was the most conserved genome region among phages LP65, K, and SPO1. The gene order of the structural module was largely colinear except for an apparent insertion (ORFs 36 to 40 in K) and a rearrangement of possible tail genes (ORFs 56, 59, and 61 from K), which had homologues in a side arm of the LP65 genome (ORFs 131 to 134 in LP65). The two side arms consisted of about 40 ORFs in each of the phages compared and contained mainly genes without database matches, indicating phage strain- or host species-restricted gene functions. The DNA replication module was the second conserved region of phages LP65 and K. In LP65 a helicase gene was found in an apparently ectopic position (ORF 123). Both phages differ markedly with respect to the position of the lysis cassette and the tRNA genes. Intron-associated HNH endonuclease genes were found in both phages, but they were located in different ORFs. We suspect that introns are much more widely distributed in phages than initially anticipated. In fact, HNH endonuclease genes were also associated with introns in phages infecting evolutionarily related bacterial hosts, such as Streptococcus thermophilus. An ecological survey of S. thermophilus phages revealed introns in all phages containing a defined homing sequence in the phage lysin gene (16). While all group I introns of bacteria have been localized to the anticodon position of tRNA genes, phage introns have been detected in a number of distinct genes, demonstrating flexibility in the intron homing process between phages. Closely related phages differ substantially in the distribution of endonuclease genes (e.g., Lactococcus lactis phages bIL170/sk1 [15]; E. coli phages T4/JS98 [11]), suggesting that these elements probably do not play an important functional role but represent selfish DNA hitchhiking phage DNA.
DISCUSSION
At the level of morphology, L. plantarum phage LP65 resembled Bacillus SPO1 phages (40-42), but the phages differed in tail length (Table 2). In contrast to SPO1, a tail fiber was observed in LP65. A further contact between phage LP65 and the cell surface is apparently mediated by the baseplate in its contracted conformation. SPO1 (40) and LP65 demonstrated a marked reorganization of the baseplate with tail contraction, but the initial and subsequent baseplate forms clearly differ from that of T4-like Myoviridae in gram-negative bacteria (23). After tail contraction, a tube extends from the tail. Thin-section electron microscopy demonstrated that the LP65 tail tube tunnels through the cell wall. The tube then probably contacts the cell membrane, but not much is known about the process of DNA injection by phage infecting lactic acid bacteria. Prophages from dairy bacteria encode membrane proteins that interfere with the siphophage DNA injection process (34). Gene inactivation studies identified bacterial membrane proteins that apparently served as sites for DNA injection by siphophages (32). Biochemical and genetic experiments combined with thin-section electron microscopy might provide further insights into these early steps of phage infection. Notably, phage LP65 can infect lysogenic L. plantarum cells, including the sequenced WCFS1 strain, which contains three prophages (24). LP65 appears obligately lytic and does not have a genetic switch region. It would thus not be suppressed by superinfection immunity exerted by a CI-like repressor. However, LP65 apparently also defies the superinfection exclusion systems of the resident siphovirus prophages (52).
TABLE 2.
Comparison of morphometric measurements for B. subtilis phage SPO1 and L. plantarum phage LP65
| Bacteriophage | Lengtha (nm) ± SD of:
|
||
|---|---|---|---|
| Extended tail | Contracted tail | Inner tube | |
| SPO1 | 140 ± 2.1 | 63.4 ± 3.7 | 142 ± 3.8 |
| LP65 | 192.9 ± 8.1 | 115.3 ± 5.3 | 58.9 ± 6.3 |
The presence of several phage genomes in the same L. plantarum host cell opens possibilities for gene exchange between the superinfecting virulent myophage and the resident temperate siphophages. However, Southern hybridization and genome sequence analysis did not reveal recent gene exchanges between the phages investigated, the criterion for recent exchange being high-level DNA sequence identity between L. plantarum myo- and siphophage genes. One reason for this lack of gene exchange could be that the superinfection of the lysogen with LP65 apparently did not lead to activation of the resident prophages. Previously, it had been established that the WCFS1 prophages are not mitomycin C inducible, suggesting inactivating mutations in the prophages (52). However, several LP65 genes shared protein sequence relatedness with temperate phages from L. plantarum or the bacterial host. This observation suggests that lateral gene transfer between myo- and siphophages and the L. plantarum host occurred in a more distant past.
SPO1-like myophages from gram-positive bacteria present a new genome organization that differs clearly from that of the other established genera of Myoviridae and recently sequenced unclassified Myoviridae. Apparently, a contractile tail structure can be built by phages that do not share overall genome or structural gene organization (e.g., T4-, P2-, Mu-, and SPO1-like myophages). This observation raises the possibility that the contractile tails in Myoviridae have evolved several times and are the product of convergent evolution. Notably, the comparative-genomics analysis of the structural gene cluster from myovirus LP65 suggested a more recent shared ancestor with lambda-like Siphoviridae than with Myoviridae outside of the SPO1 group. This raises the possibility that a basic lambda-like structural gene cluster can also acquire and adapt supplementary genes encoding a contractible tail sheath and baseplate proteins. The increasing morphological complexity is reflected by the larger number of putative tail genes in LP65 than in the structural gene cluster in Siphoviridae from the lambda supergroup. Apparently, not all structural genes in LP65 and phage K are concentrated in a single structural gene cluster, as suggested by the location of at least one major structural gene directly downstream of the DNA replication module. Such a situation is already known from T4-like phages, where the baseplate genes come in two separate clusters (baseplate hub and baseplate wedge genes) and the tail fiber genes are encoded elsewhere in the genome (36, 37). Only 8 to 10 structural proteins were identified in phages LP65 and K. On the basis of the genome analysis, one might expect at least 22 structural proteins in LP65. Even more structural proteins were identified in SPO1 by labeling experiments (41). Detailed proteome analysis for SPO1-like phage types is thus clearly warranted.
Genome size also sets the SPO1-like genus of the family Myoviridae apart from the other genera of Myoviridae (33). Mu- and P2-like Myoviridae have substantially smaller genomes of 37 to 34 kb. T4-like phages have larger genomes (164 to 169 kb in E. coli). The T4-like vibriophage KVP40 shows an even larger genome of 245 kb (36), as does the unclassified Pseudomonas myophage KZ (280 kb) (35). In contrast, phage SPO1 with 133,399 bp (http://pbi.bio.pitt.edu/), phage LP65 with 131,573 bp, and phage K with 127,395 bp (37) have substantially smaller genomes. SPO1 has a terminal redundancy of 12.6 kb (45), which is present as a direct repeat, yielding a genome of about 145 kb. The pulsed-field gel electrophoretic data from LP65 make such a large terminal redundancy unlikely. In fact, the nonredundant DNA size from LP65 determined by sequencing is close to the estimated overall genome size. However, we do not have direct experimental evidence that the terminal DNA segments determined by sequencing correspond to the actual ends of the LP65 genome. These ends can in principle be obtained by direct sequencing using the phage DNA as a template and primers placed at the ends of the assembled sequence. In fact, the genome size of SPO1-like phages is close to that of the large Bacillus siphophage SPBc2 (134 kb) (26). However, no sequence relatedness links phages LP65 and K with siphophage SPBc2, while many LP65 genes showed matches with lambda-like siphophages from low-GC-content gram-positive bacteria demonstrating much smaller genomes than SPO1-like myophages.
Comparative genomics in the SPO1 phage group allowed some tentative inferences. As in the case of Siphoviridae from low-GC-content gram-positive bacteria (8), Myoviridae from the same group of bacteria showed graded relatedness. Myophages LP65 and fri (21, 51) infecting the same bacterial species (L. plantarum) shared extensive DNA sequence identity, as demonstrated by cross-hybridization in Southern blot analysis. Lactobacillus phage LP65 shared extensive protein sequence identity, mainly over the structural genes, with Bacillus phage SPO1 and Staphylococcus phage K, but not DNA sequence identity. Finally, over the structural genes, LP65 shared a related genome map with Siphoviridae from the lambda superfamily (31), but only weak protein sequence identity.
As in the case of other phage groups (e.g., the Sfi21-like Siphoviridae, the T4-like Myoviridae), the structural genes represent the most conserved genome cluster in SPO1-like Myoviridae, followed by the DNA replication genes. This central core of the genome shows some flexibility: gene replacements, insertions, and deletions are apparently permitted, as well as gene relocations into other genome regions. In contrast, large nonconserved gene clusters are found at the right and left arms of the linear SPO1-like phage genomes. In fact, the LP65 gene clusters downstream of the DNA replication module nearly lacked links to phage K and SPO1, while a number of links were observed with the bacterial host and its prophages. This region might thus code for genus- or even species-specific gene functions. The LP65 gene cluster upstream of the structural genes showed more matches with entries from the database than the other LP65 genome arm, but no function could be proposed for this segment. The corresponding genome arm in phage K largely lacked database matches (39).
The analysis of the SPO1-like phage genomes extends the evolutionary reach of the lambda-like structural gene module and provides a fascinating bridge between Siphoviridae and Myoviridae. Such links had already been suggested by chimeric prophages combining lambda-like siphovirus head genes with P2-like myovirus tail genes (2). These two morphologically differentiated groups of phages are thus apparently in genetic contact. In contrast, phages classified into different genera of Myoviridae do not share much genomic similarity, and only a few genes, restricted to DNA replication functions, were exchanged between SPO1- and T4-like Myoviridae. These observations question the usefulness of phage tail morphology as the primary criterion in phage taxonomy. Myoviridae are certainly not a natural group of phages, but a group artificially united by a phage tail contraction process that most likely evolved independently several times. Should we therefore abandon the current ICTV-backed phage taxonomy? The answer is probably no. At the moment we cannot recommend replacing the morphology-based taxonomy with a genomics-based taxonomy. First, a taxonomic system does not necessarily have to reflect the phylogenetic relationships of its constituents. Second, we do not yet know whether a genomics-based taxonomy will lead us to a practical classification scheme. In fact, we do not yet have a precise idea about the genetic complexity of the global phage population (9, 19, 48). The independent sequencing of Myoviridae with large genomes in low-GC-content gram-positive bacteria identified recurrent principles of genome organization in these phages. From a taxonomic viewpoint, this result is most interesting: the genetic diversity of phages at least in this restricted group of bacterial hosts is apparently not so great that it defies taxonomical approaches. The link of the structural genes of SPO1-like phages with those from the lambda supergroup suggests that we might finally address only a limited number of basic types of phage genome organization in many bacterial groups.
Acknowledgments
We thank Anne Bruttin, Barbara Marchesini, and Walter Gaier for help in the early phase of the work, Carlos Canchaya for help in bioinformatic analysis, Corinne Vachier (ENS Cachan, Cachan, France) for help in image quantification, and Chris Blake for critical reading of the manuscript. Preliminary sequence data for phage SPO1 were obtained from the Pittsburgh Bacteriophage Institute website at http://pbi.bio.pitt.edu.
Sequencing of SPO1 was accomplished with support from NIH.
REFERENCES
- 1.Ackermann, H. W. 1998. Tailed bacteriophages: the order Caudovirales. Adv. Virus Res. 51:135-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, G. E., D. Angeles, N. Tran-Dinh, and N. K. Verma. 2002. Complete genomic sequence of SfV, a serotype-converting temperate bacteriophage of Shigella flexneri. J. Bacteriol. 184:1974-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altermann, E., J. R. Klein, and B. Henrich. 1999. Primary structure and features of the genome of the Lactobacillus gasseri temperate bacteriophage φadh. Gene 236:333-346. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arisaka, F., J. Engel, and H. Klump. 1981. Contraction and dissociation of the bacteriophage T4 tail sheath induced by heat and urea. Prog. Clin. Biol. Res. 64:365-379. [PubMed] [Google Scholar]
- 6.Breitbart, M., P. Salamon, B. Andresen, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rohwer. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. USA 99:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brüssow, H. 2001. Phages of dairy bacteria. Annu. Rev. Microbiol. 55:283-303. [DOI] [PubMed] [Google Scholar]
- 8.Brüssow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-222. [DOI] [PubMed] [Google Scholar]
- 9.Brüssow, H., and R. W. Hendrix. 2002. Phage genomics: small is beautiful. Cell 108:13-16. [DOI] [PubMed] [Google Scholar]
- 10.Chibani-Chennoufi, S., A. Bruttin, M. L. Dillmann, and H. Brüssow. 2004. Phage-host interaction: an ecological perspective. J. Bacteriol. 186:3677-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chibani-Chennoufi, S., C. Canchaya, A. Bruttin, and H. Brüssow. Comparative genomics of the T4-like Escherichia coli phage J598: implications for the evolution of T4 phages. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 12.Cleveland, D. W., S. G. Fischer, M. W. Kirschner, and U. K. Laemmli. 1977. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J. Biol. Chem. 252:1102-1106. [PubMed] [Google Scholar]
- 13.Cuozzo, S. A., P. Castellano, F. J. Sesma, G. M. Vignolo, and R. R. Raya. 2003. Differential roles of the two-component peptides of lactocin 705 in antimicrobial activity. Curr. Microbiol. 46:180-183. [DOI] [PubMed] [Google Scholar]
- 14.Desiere, F., S. Lucchini, and H. Brüssow. 1999. Comparative sequence analysis of the DNA packaging, head, and tail morphogenesis modules in the temperate cos-site Streptococcus thermophilus bacteriophage Sfi21. Virology 260:244-253. [DOI] [PubMed] [Google Scholar]
- 15.Desiere, F., C. Mahanivong, A. J. Hillier, P. S. Chandry, B. E. Davidson, and H. Brussow. 2001. Comparative genomics of lactococcal phages: insight from the complete genome sequence of Lactococcus lactis phage BK5-T. Virology 283:240-252. [DOI] [PubMed] [Google Scholar]
- 16.Foley, S., A. Bruttin, and H. Brüssow. 2000. Widespread distribution of a group I intron and its three deletion derivatives in the lysin gene of Streptococcus thermophilus bacteriophages. J. Virol. 74:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fournier, D. 2002. Analyse de bacteriophages. Ecole d'ingenieurs de Genève, Geneva, Switzerland.
- 18.Hambly, E., F. Tetart, C. Desplats, W. H. Wilson, H. M. Krisch, and N. H. Mann. 2001. A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc. Natl. Acad. Sci. USA 98:11411-11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendrix, R. W. 2003. Bacteriophage genomics. Curr. Opin. Microbiol. 6:506-511. [DOI] [PubMed] [Google Scholar]
- 20.Iandolo, J. J., V. Worrell, K. H. Groicher, Y. Qian, R. Tian, S. Kenton, A. Dorman, H. Ji, S. Lin, P. Loh, S. Qi, H. Zhu, and B. A. Roe. 2002. Comparative analysis of the genomes of the temperate bacteriophages φ11, φ12 and φ13 of Staphylococcus aureus 8325. Gene 289:109-118. [DOI] [PubMed] [Google Scholar]
- 21.Jarvis, A. W., L. J. Collins, and H. W. Ackermann. 1993. A study of five bacteriophages of the Myoviridae family which replicate on different gram-positive bacteria. Arch. Virol. 133:75-84. [DOI] [PubMed] [Google Scholar]
- 22.Kakefuda, T., J. T. Holden, and N. M. Utech. 1967. Ultrastructure of the membrane system in Lactobacillus plantarum. J. Bacteriol. 93:472-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karam, J. D. 1994. Molecular biology of bacteriophage T4. ASM Press, Washington, D.C.
- 24.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence, J. G., G. F. Hatfull, and R. W. Hendrix. 2002. Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J. Bacteriol. 184:4891-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazarevic, V., A. Dusterhoft, B. Soldo, H. Hilbert, C. Mauel, and D. Karamata. 1999. Nucleotide sequence of the Bacillus subtilis temperate bacteriophage SPβc2. Microbiology 145:1055-1067. [DOI] [PubMed] [Google Scholar]
- 27.Lipman, D. J., and W. R. Pearson. 1985. Rapid and sensitive protein similarity searches. Science 227:1435-1441. [DOI] [PubMed] [Google Scholar]
- 28.Loessner, M. J., and S. Scherer. 1995. Organization and transcriptional analysis of the Listeria phage A511 late gene region comprising the major capsid and tail sheath protein genes cps and tsh. J. Bacteriol. 177:6601-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, Z., F. Breidt, V. Plengvidhya, and H. P. Fleming. 2003. Bacteriophage ecology in commercial sauerkraut fermentations. Appl. Environ. Microbiol. 69:3192-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lucchini, S., F. Desiere, and H. Brussow. 1999. Comparative genomics of Streptococcus thermophilus phage species supports a modular evolution theory. J. Virol. 73:8647-8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucchini, S., J. Sidoti, and H. Brussow. 2000. Broad-range bacteriophage resistance in Streptococcus thermophilus by insertional mutagenesis. Virology 275:267-277. [DOI] [PubMed] [Google Scholar]
- 33.Maniloff, J., and H. W. Ackermann. 1998. Taxonomy of bacterial viruses: establishment of tailed virus genera and the order Caudovirales. Arch. Virol. 143:2051-2063. [DOI] [PubMed] [Google Scholar]
- 34.McGrath, S., G. F. Fitzgerald, and D. van Sinderen. 2002. Identification and characterization of phage-resistance genes in temperate lactococcal bacteriophages. Mol. Microbiol. 43:509-520. [DOI] [PubMed] [Google Scholar]
- 35.Mesyanzhinov, V. V., J. Robben, B. Grymonprez, V. A. Kostyuchenko, M. V. Bourkaltseva, N. N. Sykilinda, V. N. Krylov, and G. Volckaert. 2002. The genome of bacteriophage φKZ of Pseudomonas aeruginosa. J. Mol. Biol. 317:1-19. [DOI] [PubMed] [Google Scholar]
- 36.Miller, E. S., J. F. Heidelberg, J. A. Eisen, W. C. Nelson, A. S. Durkin, A. Ciecko, T. V. Feldblyum, O. White, I. T. Paulsen, W. C. Nierman, J. Lee, B. Szczypinski, and C. M. Fraser. 2003. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J. Bacteriol. 185:5220-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, E. S., E. Kutter, G. Mosig, F. Arisaka, T. Kunisawa, and W. Ruger. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67:86-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montel, M. C. 2000. Fermented foods—fermented meat products, p. 744-753. In R. K. Robinson, C. A. Batt, and P. D. Patel (ed.), Encyclopedia of food microbiology. Academic Press, San Diego, Calif.
- 39.O'Flaherty, S., A. Coffey, R. Edwards, W. Meaney, G. F. Fitzgerald, and R. P. Ross. 2004. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+C content. J. Bacteriol. 186:2862-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker, M. L., and F. A. Eiserling. 1983. Bacteriophage SPO1 structure and morphogenesis. I. Tail structure and length regulation. J. Virol. 46:239-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker, M. L., and F. A. Eiserling. 1983. Bacteriophage SPO1 structure and morphogenesis. III. SPO1 proteins and synthesis. J. Virol. 46:260-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parker, M. L., E. J. Ralston, and F. A. Eiserling. 1983. Bacteriophage SPO1 structure and morphogenesis. II. Head structure and DNA size. J. Virol. 46:250-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paul, J. H., M. B. Sullivan, A. M. Segall, and F. Rohwer. 2002. Marine phage genomics. Comp. Biochem. Physiol. B 133:463-476. [DOI] [PubMed] [Google Scholar]
- 44.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 45.Perkus, M. E., and D. A. Shub. 1985. Mapping the genes in the terminal redundancy of bacteriophage SPO1 with restriction endonucleases. J. Virol. 56:40-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfister, P., A. Wasserfallen, R. Stettler, and T. Leisinger. 1998. Molecular analysis of Methanobacterium phage ψM2. Mol. Microbiol. 30:233-244. [DOI] [PubMed] [Google Scholar]
- 47.Proux, C., D. van Sinderen, J. Suarez, P. Garcia, V. Ladero, G. F. Fitzgerald, F. Desiere, and H. Brussow. 2002. The dilemma of phage taxonomy illustrated by comparative genomics of Sfi21-like Siphoviridae in lactic acid bacteria. J. Bacteriol. 184:6026-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohwer, F. 2003. Global phage diversity. Cell 113:141. [DOI] [PubMed] [Google Scholar]
- 49.Rohwer, F., and R. Edwards. 2002. The Phage Proteomic Tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tetart, F., C. Desplats, M. Kutateladze, C. Monod, H. W. Ackermann, and H. M. Krisch. 2001. Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages. J. Bacteriol. 183:358-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trevors, K. E., R. A. Holley, and A. G. Kempton. 1983. Isolation and characterization of a Lactobacillus plantarum bacteriophage isolated from a meat starter culture. J. Appl. Bacteriol. 54:281-288. [Google Scholar]
- 52.Ventura, M., C. Canchaya, M. Kleerebezem, W. M. de Vos, R. J. Siezen, and H. Brussow. 2003. The prophage sequences of Lactobacillus plantarum strain WCFS1. Virology 316:245-255. [DOI] [PubMed] [Google Scholar]
- 53.Yeh, L. S., T. Hsu, and J. D. Karam. 1998. Divergence of a DNA replication gene cluster in the T4-related bacteriophage RB69. J. Bacteriol. 180:2005-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoon, S. S., R. Barrangou-Poueys, F. Breidt, Jr., T. R. Klaenhammer, and H. P. Fleming. 2002. Isolation and characterization of bacteriophages from fermenting sauerkraut. Appl. Environ. Microbiol. 68:973-976. [DOI] [PMC free article] [PubMed] [Google Scholar]