Abstract
N-Acetyl-d-glucosamine (GlcNAc) is a major component of bacterial cell wall murein and the lipopolysaccharide of the outer membrane. During growth, over 60% of the murein of the side wall is degraded, and the major products, GlcNAc-anhydro-N-acetylmuramyl peptides, are efficiently imported into the cytoplasm and cleaved to release GlcNAc, anhydro-N-acetylmuramic acid, murein tripeptide (l-Ala-d-Glu-meso-diaminopimelic acid), and d-alanine. Like murein tripeptide, GlcNAc is readily recycled, and this process was thought to involve phosphorylation, since GlcNAc-6-phosphate (GlcNAc-6-P) is efficiently used to synthesize murein or lipopolysaccharide or can be metabolized by glycolysis. Since the gene for GlcNAc kinase had not been identified, in this work we purified GlcNAc kinase (NagK) from Escherichia coli cell extracts and identified the gene by determining the N-terminal sequence of the purified kinase. A nagK deletion mutant lacked phosphorylated GlcNAc in its cytoplasm, and the cell extract of the mutant did not phosphorylate GlcNAc, indicating that NagK is the only GlcNAc kinase expressed in E. coli. Unexpectedly, GlcNAc did not accumulate in a nagK nagEBACD mutant, though both GlcNAc and GlcNAc-6-P accumulate in the nagEBACD mutant, suggesting the existence of an alternative pathway (presumably repressed by GlcNAc-6-P) that reutilizes GlcNAc without the involvement of NagK.
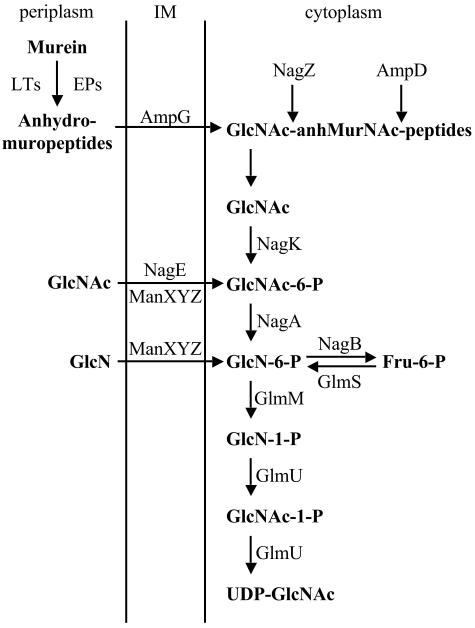
In 1966, Asensio and Ruiz-Amil reported the properties of GlcNAc kinase activity in Escherichia coli and purified the enzyme about 150-fold (1). No further work on the enzyme has been reported, and the nagK gene has not been identified heretofore. The metabolism of GlcNAc and d-glucosamine (GlcN) has been studied for decades. The divergently transcribed nagE and nagBACD operons are involved in the uptake and degradation of the amino sugars. When E. coli grows on GlcNAc-containing medium, GlcNAc is transported by a phosphotransferase system, either NagE or ManXYZ, to generate GlcNAc-6-phosphate (GlcNAc-6-P) (26). GlcNAc-6-P is converted by NagA deacetylase to GlcN-6-P (38). GlcN-6-P enters either the glycolysis pathway via conversion to fructose-6-P by the NagB deaminase (38) or the biosynthesis pathways for murein and lipopolysaccharide by conversion to UDP-GlcNAc by GlmM and GlmU (15-17) (Fig. 1). Hence, NagK is not required for the metabolism of GlcNAc derived from the medium. The other proteins encoded by the nag gene cluster are NagC, a repressor controlling the expression of nagE and the nagBACD operon (24), and NagD, a putative hydrolase, whose activity is not essential for the metabolism of the amino sugars.
FIG. 1.
Scheme of murein recycling and metabolism of GlcNAc. UDP-GlcNAc is the main cytoplasmic precursor of murein and lipopolysaccharide. AmpG is a specific permease for muropeptides. NagE and ManXYZ are phosphoenolpyruvate-dependent phosphotransferases. IM, inner membrane; LTs, lytic transglycosylases; EPs, endopeptidases; Fru-6-P, fructose-6-phosphate.
However, significant amounts of nonphosphorylated GlcNAc are released in the cytoplasm via the murein recycling pathway (4, 37) (Fig. 1). Over half of side-wall murein is broken down each generation by lytic transglycosylases and endopeptidases (7, 9, 11). The principal degradation product is the anhydro-muropeptide GlcNAc-β-1,4-anhydro-N-acetylmuramyl(anhMurNAc)-l-alanyl-γ-d-glutamyl-meso-diaminopimelyl-d-alanine. Muropeptides are imported into the cytoplasm via a specific permease, AmpG (5, 11), and thereafter are cleaved into GlcNAc, anhMurNAc, murein tripeptide, and d-alanine by the combined actions of the NagZ protein (β-N-acetylglucosaminidase), the AmpD protein (anhMurNAc-l-Ala amidase), and the LdcA protein (dl-carboxypeptidase) (4, 10, 12, 33, 37). Murein tripeptide reenters the murein synthesis pathway following direct ligation to UDP-MurNAc by Mpl (18). To a lesser extent, murein tripeptide is cleaved by MpaA, YcjJ, and PepD to yield the individual amino acids (30, 36). Interestingly, the roles of all eight of these enzymes, except PepD, are concerned exclusively with the recycling process.
Recently, both cytoplasmic GlcNAc and anhMurNAc derived from cell wall recycling were shown to be reused (23). Cytoplasmic GlcNAc must be phosphorylated to enter the known metabolic pathway of GlcNAc-6-P (Fig. 1). Though one possibility for utilization of murein-derived GlcNAc is for GlcNAc to be exported and then taken up by a specific phosphotransferase system, it was reasonable to assume that NagK is responsible for the phosphorylation of murein-derived GlcNAc. In this communication, the nagK (ycfX) gene is identified, and NagK is characterized and shown to be the only GlcNAc kinase expressed in E. coli to phosphorylate GlcNAc in the cytoplasm. On the basis of the observation that GlcNAc does not accumulate in a nagK nagEBACD mutant, we suggest that an unknown pathway must exist to utilize GlcNAc in the absence of NagK and NagA and that the pathway may be regulated by GlcNAc-6-P.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used here are listed in Table 1. The strains were grown aerobically at 37°C in Luria-Bertani (LB) broth (20), except as indicated. Ampicillin (100 μg/ml), kanamycin (25 μg/ml), tetracycline (10 μg/ml), and chloramphenicol (20 μg/ml) were used as required.
TABLE 1.
E. coli K-12 strains and plasmids used in this work
| E. coli strain or plasmid | Genotype and/or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| MG1655 | F−ilvG rfb-50 rph-1 | 34 |
| XL1-Blue | F′::Tn10 proA+proB+relA1 lacIqendA1 Δ(lacZ)M15 recA1 thi gyrA96 (Nalr) hsdR17(rK− mK+) supE44 lac | Stratagene |
| TP71 | F− mutant lysA opp araD139 rpsL150 deoC1 ptsF25 ftbB5301 rbsR relA1 Δ(argF-lac) | 11 |
| TP71B | TP71 nagB::Kan | 23 |
| TP71BK | TP71 nagB::Kan nagK::Cm | This work |
| TP80 | TP71 nagEBACD::Tet | 23 |
| TP80K | TP71 nagEBACD::Tet nagK::Cm | This work |
| IBPC524 | thi-1 argG6 argE3 his-4 xyl-5 mtl-1 tsx-29 rpsL ΔlacX74 nagA::Cm | 24 |
| TP71A | TP71 nagA::Cm | This work |
| TP71AK | TP71 nagA::Cm ΔnagK | This work |
| Plasmids | ||
| pGEM-T | Ampr | Promega |
| pKD3 | Cmr Ampr | 6 |
| pKD46 | Ampr Para-Red recombinase | 6 |
| pCP20 | Ampr Cmrori(Ts) FLP | 6 |
| pNagK2 | pGEM-T carrying nagK under the control of the Plac promoter | This work |
| pNagK11 | pNagK2 Δ(SacI-PstI) | This work |
Purification and identification of NagK from E. coli wild-type cell extract.
All purification steps were performed at 4°C or the temperature described. E. coli strain MG1655 was grown with vigorous shaking in 5 liters of LB medium at 37°C overnight. Cells (wet weight, 21.1 g) were harvested, washed with PEND50 buffer (50 mM potassium phosphate [pH 7.6], 1 mM EDTA, 0.1 mM GlcNAc, and 1 mM dithiothreitol), and suspended in 50 ml of PEND50, to which phenylmethylsulfonyl fluoride was added (final concentration, 1 mM). The sample was sonicated by a sonifier (Branson Ultrasonics Corp., Danbury, Conn.), and then centrifuged (27,000 × g for 30 min) to remove debris and unbroken cells. Streptomycin sulfate was added to the sample (final concentration, 1.4%), and insoluble material was removed by ultracentrifugation (100,000 × g for 60 min). The supernatant was loaded onto a DEAE-Sephacel column (Amersham Pharmacia, Uppsala, Sweden) which had been equilibrated with PEND50. The column was washed with PEND50 and developed with a linear gradient of 0 to 0.3 M NaCl in PEND50. NagK activity was eluted around 230 mM NaCl. All NagK active fractions were combined, concentrated by Ultrafree 4, 10K (Millipore Corp., Bedford, Mass.), and desalted with PEND1 (the same mixture as PEND50, except it has 1 rather than 50 mM potassium phosphate). The active concentrate was loaded onto a hydroxyapatite column (BIO-GEL HT; Bio-Rad, Hercules, Calif.). The column was washed with PEND1 and developed with a linear gradient of PEND1 to PEND50. NagK was eluted at 20 mM potassium phosphate. Active fractions were combined and loaded onto a MonoQ HR 5/5 column (Amersham Pharmacia) and fractionated by fast pressure liquid chromatography at room temperature. The column was washed with PEND50, and proteins were eluted with a linear gradient of 0 to 0.6 M NaCl. The peak of NagK activity was at 220 mM NaCl. NagK fractions were concentrated and loaded onto a Sephacryl S-200 column (Amersham Pharmacia), and the proteins were separated with 0.15 M NaCl in PEND50 by the fast pressure liquid chromatography system. The retention volume of the peak of the active fractions was 58 ml, and the active fractions were combined and concentrated. Protein concentration was measured by the Bradford assay (Bio-Rad) with bovine serum albumin as a standard. Samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using gels stained with Coomassie brilliant blue R250 (CBB) (29). To determine the N-terminal amino acid sequence of a protein, the protein band of interest was transferred to a Sequi-Blot polyvinylidene difluoride membrane (Bio-Rad) after SDS-PAGE, followed by light staining with CBB to facilitate excision of the protein band. The N-terminal amino acid sequence of the protein was determined with an ABI477 protein sequencer (Applied Biosystems, Foster City, Calif.) at the Tufts University Core Facility.
GlcNAc kinase activity.
The two assay methods described by Asensio and Ruiz-Amil (1) were employed with the following modifications. For method 1 (for crude mixtures), each enzyme fraction was added to a reaction mixture (final volume, 0.1 ml) containing 1.9 mM GlcNAc, 20 mM ATP, 20 mM MgCl2, and 100 mM Tris-HCl (pH 7.8). A control without ATP was used to correct for GlcNAc deacetylase activity (28). Samples were incubated at 37°C for 1 h. To stop the reaction and precipitate GlcNAc-P, 0.4 ml of 5% ZnSO4 was added, followed by 0.4 ml of 0.15 N Ba(OH)2. Phosphorylated GlcNAc was removed by low-speed centrifugation (300 × g for 5 min), and the remaining GlcNAc was measured by the Morgan-Elson assay (27). The supernatant (0.1 ml) was mixed with 0.1 ml of 10% Na2B4O7 and heated in a boiling-water bath for 7 min. A 0.8 ml-portion of Ehrlich's reagent (91 mg of p-dimethylamino benzaldehyde [Fisher Scientific, Fair Lawn, N.J.], 91 μl of HCl, and 9.909 ml of glacial acetic acid) was then added, and color was developed by incubation for 20 min at 37°C followed by 15 min at room temperature. The absorbance of the sample was measured at 585 nm. For method 2 (for purified NagK), the assay mixture (0.5 ml) contained 100 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 1 mM phosphoenol-pyruvate, 4 mM ATP, 0.2 mM NADH, 4 U of lactate dehydrogenase, 4 U of pyruvate kinase from rabbit muscle (Sigma, St. Louis, Mo.), and 0.23 μg of purified NagK protein. The assay mixture was incubated at 37°C. The reaction was started by the addition of the substrate. The formation of NAD+ was monitored at a wavelength of 340 nm. The apparent Km value was calculated by direct fit to the Lineweaver-Burk plot. One unit is defined as the amount of NagK that phosphorylates 1 μmol of GlcNAc per min at 37°C.
Cloning of NagK.
The nagK DNA fragment from the E. coli chromosome was amplified by PCR using 5′-CATTAAAACAAGGAGCGGC and 5′-AGCATAGCAACCTCTGTTG as primers. The amplified DNA was ligated into the pGEM-T vector (Promega, Madison, Wis.), generating plasmid pNagK2, which carried the nagK gene under the control of the Plac promoter. Though the DNA sequence of the nagK gene on pNagK2 was verified, induction of expression from pNagK2 by the addition of 1 mM isopropyl-β-d-thiogalactoside (IPTG) led to overproduction of a protein larger than expected. The larger protein proved to be the product of an in-frame fusion of lacZ to nagK. To make it out of frame, pNagK2 was digested with two restriction endonucleases, SacI and PstI, which cleave between the start codons of lacZ and nagK. The overhangs of the linear DNA after digestion were filled with deoxynucleoside triphosphate by the Klenow fragment, followed by self-ligation, generating plasmid pNagK11.
Overexpression and purification of recombinant NagK.
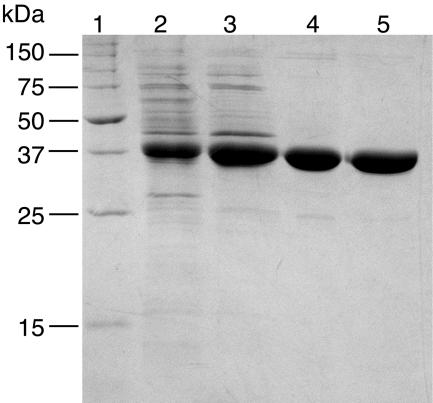
E. coli strain XL1-Blue, carrying pNagK11, was grown at 37°C with vigorous shaking in 1 liter of LB medium containing ampicillin from a 2% inoculum of overnight culture. After the culture grew to a Klett value of 50, IPTG was added to a final concentration of 1 mM, and incubation was continued for 3 h. The cells were harvested by centrifugation, washed with 20 ml of PEND50, suspended in 8 ml of PEND50, broken by sonication, and centrifuged to remove the debris. The supernatant was ultracentrifuged after the addition of streptomycin sulfate (final concentration, 1.4%). SDS-PAGE revealed one overproduced protein present in the cell extract (Fig. 2). The molecular weight of the protein appeared to be the same as that of NagK. The protein was purified to >95% by sequential chromatographic steps with DEAE-Sephacel, hydroxyapatite, and MonoQ as described above for the purification and identification of NagK. Aliquots of purified NagK were frozen in dry ice-ethanol and stored at −80°C. NagK was fully active for months. Thawed samples of NagK retained full activity in 40% glycerol at −20°C for a few months.
FIG. 2.
SDS-PAGE gel (stained with CBB) of samples obtained during purification of recombinant NagK overexpressed by adding the inducer to E. coli strain XL1-Blue carrying pNagK11. Lane 1, molecular mass markers; lane 2, streptomycin sulfate precipitation; lane 3, DEAE-Sephacel; lane 4, hydroxyapatite; lane 5, MonoQ. Lanes 2 to 5 each contained 5 μg of protein.
Deletion of NagK.
A nagK null mutant (nagK::Cm) was constructed by one-step inactivation of the E. coli chromosomal gene as described before (6, 36), using the primers 5′-CCAGTAAAGGCAGTACATTAAAACAAGGAGCGGCAATGTAGGCTGGAGCTGCTTCG and 5′-AAGCCACCACCAATGACGACCAGGTCAGGGTCAACGATGAATATCCTCCTTAGTTC (the italicized sequences are homologous to the nagK-flanking sequence, and the underlined sequences are homologous to the pKD3 vector sequence flanking the Cmr gene) and pKD3 as the template to synthesize the PCR product used for one-step inactivation. The nagK::Cm mutation was confirmed by PCR with the primers used for cloning NagK, as described in Materials and Methods. nagK::Cm was transduced into strains TP71B and TP80 with T4gt7 phage (20). To construct TP71AK, the Cmr gene in TP71BK was eliminated as described previously (6), i.e., by transforming the strain with the plasmid pCP20, which expresses FLP recombinase, followed by screening for the loss of chloramphenicol resistance. The nagA::Cm mutation from IBPC524 was transduced into the nagB::Kan ΔnagK strain with T4gt7 phage, generating a nagA::Cm ΔnagK strain, TP71AK.
Labeling cells with [6-3H]GlcN and preparation of hot-water extract.
The radioactive labeling and preparation of the cytoplasmic amino sugars derived from cell wall recycling were performed as described previously (23). All strains used in this study carry the nagB mutation, which prevents conversion of GlcN-6-P to fructose-6-P and hence facilitates efficient incorporation of [6-3H]GlcN into murein and lipopolysaccharide (Fig. 1). nagB mutant strains were grown at 37°C in 4 ml of M9 minimal medium containing 0.6% glycerol, 0.1% Casamino Acids, 1 mM MgCl2, 1 μg of thiamine per ml, 20 μg of lysine per ml, and 1 μCi of [6-3H]GlcN (21.6 Ci/mol; NEN Life Science Products, Boston, Mass.) per ml. [6-3H]GlcN is transported by a phosphotransferase system, ManXYZ, to form GlcN-6-P. At mid-log phase, the [6-3H]GlcN-labeled cells were harvested, washed with water, resuspended in water, incubated at 90°C for 5 min, and centrifuged (4,300 × g for 7 min at 4°C). The supernatant (hot-water extract) containing soluble [6-3H]GlcN derivatives was lyophilized and suspended with water.
Analysis of hot-water extracts by HPLC and TLC.
anhMurNAc and UDP-MurNAc-pentapeptide were separated by high-pressure liquid chromatography (HPLC) (23). A mixture of GlcN, GlcN-P, GlcNAc, GlcNAc-P, and UDP-GlcNAc, which did not bind to the HPLC column, was recovered and analyzed by thin-layer chromatography (TLC) as described previously (23). UDP-GlcNAc was defined as that material that remained at the origin in TLC with the basic solvent (80% ethanol, 0.4 N NH4OH) and was resistant to calf intestinal phosphatase. The amount of each compound was normalized with the turbidity of the cell culture.
RESULTS
Identification of NagK.
Confirming the report by Asensio and Ruiz-Amil (1), we detected NagK activity in cell extracts of the E. coli K-12 wild-type strain MG1655. Since NagK was active in the nag gene cluster deletion mutant TP80 (nagEBACD) (Table 2), nagD, the unidentified member of the nag gene cluster, could not be nagK. To identify nagK, the purification of the NagK enzyme from MG1655 cell extract was performed as described in Materials and Methods. NagK was purified over 800-fold by a sequence of four chromatographic steps (Table 3). The final fraction contained two proteins detected by SDS-PAGE gels stained with CBB. The N-terminal sequence of the ∼34-kDa protein band was MYYGFD. A homology search of the E. coli genome with the TagIdent tool (http://us.expasy.org/tools/tagident.html) showed that only the N-terminal sequence of YcfX is identical to the 6-amino-acid sequence. The predicted molecular mass of YcfX is 32.9 kDa. YcfX is coded in a putative lolC-lolD-lolE-ycfX-cobB operon present at 25.4 min on the E. coli chromosome. ycfX appears to be transcribed from a putative σ70-dependent promoter upstream of lolC, and a predicted σ54-dependent promoter exists within the ycfX gene to transcribe cobB. LolCDE is an essential ABC transporter for lipoprotein transport from the inner membrane to the outer membrane (21, 39). YcfX has been shown not to be involved in lipoprotein transport (39). CobB is homologous to the NAD+-dependent protein deacetylase Sir2 (40).
TABLE 2.
NagK activities in nagK::Cm mutants of E. coli
| Strain | Genotype | Sp acta |
|---|---|---|
| TP80 | nagEBACD | 18.3 |
| TP80K | nagEBACD nagK | 0.0 |
| TP71A | nagA | 17.1 |
| TP71AK | nagA nagK | 0.3 |
| TP71B | nagB | 17.3 |
| TP71BK | nagB nagK | 3.4 |
The NagK activity (in units of activity [10−3] per milligram of protein) was measured by method 1 (see Materials and Methods) in cell extracts of cells grown overnight in LB medium at 37°C.
TABLE 3.
Purification of NagK enzyme from E. coli wild-type cell extract
| Purification step | Total protein (mg) | Total activitya (U) | Sp act (U/mg of protein) | Yield (%) | Purification factor |
|---|---|---|---|---|---|
| Cell extract | 2,551 | 101 | 0.04 | 100 | 1 |
| DEAE | 33.9 | 5.9 | 0.17 | 5.9 | 4 |
| HAb | 1.27 | 3.5 | 2.7 | 3.4 | 68 |
| MonoQ | 0.10 | 1.7 | 17.0 | 1.7 | 428 |
| Sephacryl | 0.006 | 0.2 | 33.3 | 0.2 | 842 |
Activities were measured by the Morgan-Elson assay (method 1).
HA, hydroxyapatite.
To confirm that YcfX is NagK, the ycfX gene was cloned into the expression vector pGEM-T under the control of the Plac promoter. XL1-Blue carrying the plasmid grew slowly even in the absence of inducer, presumably because of an accumulation of a semitoxic level of GlcNAc-6-P (2). Nonetheless, YcfX was overexpressed after induction at mid-log phase. The soluble fraction contained 1,000-fold more NagK activity than was found in wild-type extracts. We conclude that YcfX is NagK. The plasmid containing nagK was named pNagK11.
Characterization of NagK.
To characterize NagK enzymatically, NagK was overproduced in XL1-Blue carrying pNagK11 and purified as described above (Fig. 2). After incubation with purified NagK in the presence of ATP and MgCl2, [6-3H]GlcNAc was converted to a compound which remained at the origin during TLC with a basic solvent. This compound was converted to GlcNAc by treatment with calf intestinal phosphatase, demonstrating the direct phosphorylation of GlcNAc by NagK (data not shown). The activity of the kinase was constant from pH 6.5 to 10. The substrate specificity and kinetics of NagK were determined by method 2, described in Materials and Methods. NagK phosphorylated GlcNAc and glucose. The kinase reaction followed the Michaelis-Menten equation. The calculated Kms for GlcNAc, ATP, and glucose were 342 μM, 896 μM, and 37 mM, respectively, while Vmaxs were 118 μmol of GlcNAc and 24 μmol of glucose per mg of NagK per min. This value for GlcNAc is equivalent to the phosphorylation of 3,900 molecules of GlcNAc by 1 molecule of NagK per min. NagK had very slight activity with GlcN, d-mannose, and N-acetyl-d-mannosamine and no activity with d-galactose, d-mannosamine, or N-acetyl-d-galactosamine (Table 4). While 10 mM GlcNAc-6-P did not inhibit NagK activity when 2 mM GlcNAc was incubated with 14 nM NagK, ADP was a strong inhibitor (Ki was approximately 1.4 mM).
TABLE 4.
Substrate specificity of E. coli NagK
| Substrate | Relative activityb | Kma (mM) | Vmaxa (μM/min/mg) |
|---|---|---|---|
| GlcNAc | 100 | 0.34 | 118 |
| ATP | 0.90 | 108 | |
| Glucose | 21 | 37.2 | 24 |
| Mannose | 0.3 (100) | ND | ND |
| Galactose | <0.1 (50) | ND | ND |
| GlcN | 0.6 (50) | ND | ND |
| d-Mannosamine | <0.1 (20) | ND | ND |
| ManNAcc | 0.4 (50) | ND | ND |
| GalNAcd | <0.1 (1) | ND | ND |
To determine the apparent Km and Vmax, different amounts of substrate were incubated with a 14 nM concentration (0.46 mg/liter) of purified NagK and 4 mM ATP at 37°C and activities measured by method 2 were plotted according to the Lineweaver-Burk diagram. Values of Km and Vmax for ATP were measured with 1 mM GlcNAc. ND, not determined.
The concentrations in parentheses (in millimolar units) were those tested to measure the kinase activities.
ManNAc, N-acetyl-d-mannosamine.
GalNAc, N-acetyl-d-galactosamine.
Deletion of nagK.
A nagK null mutation (nagK::Cm) which retained the promoter of cobB within the C-terminal region of nagK was constructed and transduced into the nagB strain TP71B and the nagEBACD strain TP80, generating TP71BK and TP80K, respectively. The two NagK null mutants grew with about the same doubling times as their parent strains and had normal morphology (data not shown), so under these conditions, NagK is not essential for E. coli growth. TP80 ceased growth in early stationary phase and produced ghost cells (data not shown), presumably due to the toxicity of the accumulated GlcNAc-6-P (2, 38), whereas TP80K, which lacked GlcNAc-6-P (see below), grew normally. As shown in Table 2, TP80K totally lacked NagK activity. TP71AK had a level of activity equal to 2% of that of TP71A, but TP71BK appeared to have a level of activity equal to about 20% of that of the wild type. Since GlcN would register as GlcNAc-P in method 1, which depends on measuring the GlcNAc remaining following precipitation of GlcNAc-P, this apparent activity of TP71BK may be caused by the deacetylation of GlcNAc. We have confirmed that the cell extract of TP71BK, but not that of TP71AK or TP80K, deacetylated [6-3H]GlcNAc by detecting the formation of [3H]GlcN by TLC in the solvent containing n-butanol-acetic acid-water (2:1:1), which separates GlcN from GlcNAc (data not shown). Since NagA is expressed in the cells containing nagB::Km (24), NagA apparently must deacetylate GlcNAc as well as GlcNAc-6-P.
Analysis of [6-3H]GlcN-derived compounds in nagK mutants.
We have observed that in strains lacking nagEBACD, GlcNAc and GlcNAc-6-P derived from murein recycling accumulate (23). If NagK is required for utilization of GlcNAc, then the nagK::Cm mutants should accumulate GlcNAc, lack GlcNAc-6-P, and not recycle GlcNAc. To analyze the cytoplasmic compounds derived from the amino sugars of murein, the wild type and the mutants were labeled with [6-3H]GlcN, and the hot-water extracts of the nagK mutants and their parent strains were analyzed. Table 5 shows the amounts of the various radioactive compounds present in the cytoplasm of the strains examined. As expected, GlcNAc-P was absent or barely detected in the nagK mutants. Though GlcNAc was expected to accumulate in the cytoplasm, neither the nagK nagB mutant nor the nagK nagEBACD mutant accumulated GlcNAc. On the contrary, compared to the nagEBACD mutant, which accumulated GlcNAc and GlcNAc-P, the nagK nagEBACD mutant contained a normal amount of GlcNAc and no GlcNAc-P. Puzzlingly, the amounts of UDP-MurNAc-pentapeptide and UDP-GlcNAc in the nagK nagEBACD mutant were significantly higher than those in the nagEBACD mutant, whereas the amounts in the nagK nagB mutant were less than twofold higher than those in the nagB strain. No other radioactive compound accumulated in the two nagK mutants. These results confirm that cytoplasmic GlcNAc derived from murein recycling is phosphorylated by NagK and also suggest that the GlcNAc is utilized or released by cells lacking NagK.
TABLE 5.
Amino sugar-containing compounds present in the cytoplasm of the mutants
| Compound | Amt of 6-3H-labeled compound (cpm [103]) in hot-water extracts
|
|||
|---|---|---|---|---|
| nagB mutant | nagB nagK mutant | nagEBACD mutant | nagEBACD nagK mutant | |
| GlcN | 3.8 | 3.1 | <0.1 | 2.3 |
| GlcN-P | 4.4 | 0.7 | <0.1 | 0.5 |
| GlcNAc | 13.4 | 12.2 | 139.4 | 13.6 |
| GlcNAc-P | 24.0 | 0.7 | 129.9 | <0.1 |
| anhMurNAc | <0.1 | <0.1 | 1.4 | 0.9 |
| UDP-MurNAc-pentapeptide | 5.4 | 7.3 | 2.9 | 23.9 |
| UDP-GlcNAc | 16.8 | 27.3 | <0.1 | 42.5 |
To examine whether the lack of accumulation of GlcNAc was due to leakage from the cells, chase experiments were done as described in reference 23. The increase in the amount of radioactivity in the spent medium from the nagEBACD nagK mutant during the chase was similar to that from the nagEBACD cells (data not shown), indicating that GlcNAc was not released to the medium. We can also exclude the possibility that as soon as GlcNAc is leaked out, it is imported by ManXYZ or NagE, because GlcNAc-P, the product of the uptake of GlcNAc, is not detectable in the nagK mutants. Hence, these results strongly suggest that an alternative pathway to utilize GlcNAc in the absence of NagK must exist.
DISCUSSION
During exponential growth, GlcNAc is released in the cytoplasm of E. coli, due to murein recycling. Here, we have identified the kinase which is responsible for phosphorylation of free GlcNAc in the cytoplasm, NagK. The absence of GlcNAc kinase activity and the total or near-total absence of GlcNAc-P from the two nagK mutants proves that NagK is the only GlcNAc kinase expressed in E. coli.
NagK is a 303-amino-acid protein which belongs to the ROK (repressor, open reading frame, kinase) family (35). NagK lacks the N-terminal DNA-binding helix-turn-helix motif which is conserved in the transcription regulators of the ROK family (35), such as NagC from E. coli (25) and XylR from Bacillus subtilis (13). NagK contains the typical ATP-binding site (G4FDIGGT10) of glucokinase near the N terminus (19) and has a very low level of similarity to mammalian GlcNAc kinase (8, 14). NagK is conserved in most gamma-proteobacteria, such as species of the Salmonella, Vibrio, and Haemophilus genera. Since those gram-negative bacteria contain the homologous enzymes required for murein recycling (4), the NagK homologs presumably phosphorylate GlcNAc.
Recombinant NagK was overproduced, purified, and characterized. Of the candidate sugars tested here, NagK phosphorylated only GlcNAc and glucose. Since the Km for glucose was 100-fold higher than that for GlcNAc, NagK should be specific for GlcNAc under physiological conditions. The properties of NagK, such as its substrate specificity, its inhibition by ADP, and its behavior during purification steps, indicate that the protein studied here is identical to the enzyme (EC 2.7.1.59) partially purified and studied 38 years ago by Asensio and Ruiz-Amil (1). Though no other GlcNAc kinase gene has been reported to occur in bacteria, C. Brigham and M. Malamy (personal communication) have recently identified a ROK family kinase, RokA, in Bacteroides fragilis, which phosphorylates both glucose and GlcNAc with approximately the same affinity: the Km for glucose is 0.6 mM, and the Km for GlcNAc is 0.8 mM. Thus, compared to RokA (23% identity to E. coli NagK), NagK has a much more strict substrate specificity.
A strain lacking the nag cluster nagEBACD cannot metabolize GlcNAc because it lacks NagA, the GlcNAc-6-P deacetylase required to form the essential intermediate, GlcN-6-P. As a result, GlcNAc-6-P and GlcNAc accumulate in the cells, and growth into late log phase results in the lysis of many cells, a possible indirect effect of the high level of GlcNAc-6-P in the cells. The accumulation of GlcNAc in the nagEBACD mutant in log phase is not due to the inhibition of NagK by GlcNAc-6-P, because GlcNAc-6-P does not inhibit NagK. Since the activity of NagK is much greater than the rate of the production of GlcNAc by murein recycling, the large pool of GlcNAc suggests that GlcNAc-6-P is being dephosphorylated. There is other indirect evidence for the dephosphorylation of GlcNAc-6-P. The nagEBACD mutant lysed upon addition of GlcNAc to cells growing in the glycerol minimal medium, whereas the nagEBACD nagK mutant was able to grow under these conditions. GlcNAc from the medium is phosphorylated by the mannose phosphotransferase system during uptake. If GlcNAc-6-P is the cause of cell lysis, then the pool of GlcNAc-6-P must remain relatively low in the nagEBACD nagK mutant when GlcNAc is added to the culture, which implies dephosphorylation. In the absence of NagA deacetylase, GlcNAc-6-P is likely to be dephosphorylated to GlcNAc and subsequently utilize the alternative pathway postulated here. On the other hand, in the nagEBACD mutant, GlcNAc is (re)phosphorylated by NagK, resulting in cells with a near-lethal concentration of GlcNAc-6-P. The mechanism of killing by GlcNAc-6-P remains to be determined but is presumably related to the lack of UDP-GlcNAc in the nagEBACD mutant cells (Table 5).
How does E. coli utilize GlcNAc without NagK and NagA?
When nagK was deleted from the nag cluster mutant, not only did GlcNAc-P disappear, but the cells grew normally and the GlcNAc level returned to normal, indicating that an alternate pathway for recycling GlcNAc had been derepressed. Examination of Table 5 reveals that a significant amount of GlcN was detected in both the nagK nagB mutant and the nagK nagEBACD mutant relative to that in the nagEBACD mutant. This suggests that when phosphorylation of GlcNAc is not possible due to the lack of NagK, the first step in utilization of cellular GlcNAc may be deacetylation. In 1956, Roseman detected GlcNAc deacetylase activity in an E. coli cell extract (28). A gene coding GlcNAc deacetylase, however, has not been reported for any organism. To be metabolized further following the deacetylation of GlcNAc, the resulting GlcN must be phosphorylated. A GlcN-specific kinase has been identified in Vibrio cholerae (22), but no homolog exists in the E. coli genome. However, it has been reported that GlcN is phosphorylated by mannose-fructose kinase, although its GlcN kinase activity is only 5% of that of mannose-fructose kinase activity and the Km for GlcN is 5 mM (31). Recently, mannose-fructose kinase has been shown to be encoded by yajF (32). Thus, one possible hypothetical pathway would involve deacetylation of GlcNAc followed by phosphorylation to produce the normal intermediate, GlcN-6-P.
Is the alternative pathway regulated?
Considering the large accumulation of GlcNAc and GlcNAc-P in the nagEBACD mutant (23) and the lack of accumulation of GlcNAc in the nagK nagEBACD mutant, the alternative pathway must be inactive in the nagEBACD mutant and become active only in the nagK nagEBACD mutant lacking GlcNAc-6-P. This observation suggests that the pathway is repressed by NagK or GlcNAc-6-P. NagK belongs to the ROK family but lacks an N-terminal DNA-binding domain, indicating that NagK is unlikely to bind DNA and regulate genes involved in the pathway directly. GlcNAc-6-P is known to be not only an inducer of the nagBACD operon (24) but also the allosteric activator of NagB deaminase (3). Hence, the pathway is likely to be repressed transcriptionally or enzymatically by the high concentration of GlcNAc-6-P present in normal cells and especially in the nagEBACD mutant. Efforts are under way to determine the alternative pathway for utilization of free GlcNAc and its regulation.
Acknowledgments
We are very grateful to Jacqueline Plumbridge for the gift of the nag mutants. We thank Michael Malamy and Debabrata RayChaudhuri for their critical reading of the manuscript.
This work was supported in part by Public Health Service grant GM51610 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Asensio, C., and M. Ruiz-Amil. 1966. N-acetyl-d-glucosamine kinase. II. Escherichia coli. Methods Enzymol. 9:421-425. [Google Scholar]
- 2.Bernheim, N. J., and W. J. Dobrogosz. 1970. Amino sugar sensitivity in Escherichia coli mutants unable to grow on N-acetylglucosamine. J. Bacteriol. 101:384-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calcagno, M., P. J. Campos, G. Mulliert, and J. Suastegui. 1984. Purification, molecular and kinetic properties of glucosamine-6-phosphate isomerase (deaminase) from Escherichia coli. Biochim. Biophys. Acta 787:165-173. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, Q., H. Li, K. Merdek, and J. T. Park. 2000. Molecular characterization of the β-N-acetylglucosaminidase of Escherichia coli and its role in cell wall recycling. J. Bacteriol. 182:4836-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, Q., and J. T. Park. 2002. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J. Bacteriol. 184:6434-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodell, E. W. 1985. Recycling of murein by Escherichia coli. J. Bacteriol. 163:305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinderlich, S., M. Berger, M. Schwarzkopf, K. Effertz, and W. Reutter. 2000. Molecular cloning and characterization of murine and human N-acetylglucosamine kinase. Eur. J. Biochem. 267:3301-3308. [DOI] [PubMed] [Google Scholar]
- 9.Höltje, J.-V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holtje, J. V., U. Kopp, A. Ursinus, and B. Wiedemann. 1994. The negative regulator of β-lactamase induction AmpD is a N-acetyl-anhydromuramyl-l-alanine amidase. FEMS Microbiol. Lett. 122:159-164. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs, C., L. J. Huang, E. Bartowsky, S. Normark, and J. T. Park. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J. 13:4684-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs, C., B. Joris, M. Jamin, K. Klarsov, J. Van Beeumen, D. Mengin-Lecreulx, J. van Heijenoort, J. T. Park, S. Normark, and J. M. Frere. 1995. AmpD, essential for both β-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-l-alanine amidase. Mol. Microbiol. 15:553-559. [DOI] [PubMed] [Google Scholar]
- 13.Kreuzer, P., D. Gärtner, R. Allmansberger, and W. Hillen. 1989. Identification and sequence analysis of the Bacillus subtilis W23 xylR gene and xyl operator. J. Bacteriol. 171:3840-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ligos, J. M., T. L. de Lera, S. Hinderlich, B. Guinea, L. Sanchez, R. Roca, A. Valencia, and A. Bernad. 2002. Functional interaction between the Ser/Thr kinase PKL12 and N-acetylglucosamine kinase, a prominent enzyme implicated in the salvage pathway for GlcNAc recycling. J. Biol. Chem. 277:6333-6343. [DOI] [PubMed] [Google Scholar]
- 15.Mengin-Lecreulx, D., and J. van Heijenoort. 1996. Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli. J. Biol. Chem. 271:32-39. [DOI] [PubMed] [Google Scholar]
- 16.Mengin-Lecreulx, D., and J. van Heijenoort. 1994. Copurification of glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase activities of Escherichia coli: characterization of the glmU gene product as a bifunctional enzyme catalyzing two subsequent steps in the pathway for UDP-N-acetylglucosamine synthesis. J. Bacteriol. 176:5788-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mengin-Lecreulx, D., and J. van Heijenoort. 1993. Identification of the glmU gene encoding N-acetylglucosamine-1-phosphate uridyltransferase in Escherichia coli. J. Bacteriol. 175:6150-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mengin-Lecreulx, D., J. van Heijenoort, and J. T. Park. 1996. Identification of the mpl gene encoding UDP-N-acetylmuramate:l-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J. Bacteriol. 178:5347-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer, D., C. Schneider-Fresenius, R. Horlacher, R. Peist, and W. Boos. 1997. Molecular characterization of glucokinase from Escherichia coli K-12. J. Bacteriol. 179:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria, p. 268-274. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Narita, S., K. Tanaka, S. Matsuyama, and H. Tokuda. 2002. Disruption of lolCDE, encoding an ATP-binding cassette transporter, is lethal for Escherichia coli and prevents release of lipoproteins from the inner membrane. J. Bacteriol. 184:1417-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park, J. K., L. X. Wang, and S. Roseman. 2002. Isolation of a glucosamine-specific kinase, a unique enzyme of Vibrio cholerae. J. Biol. Chem. 277:15573-15578. [DOI] [PubMed] [Google Scholar]
- 23.Park, J. T. 2001. Identification of a dedicated recycling pathway for anhydro-N-acetylmuramic acid and N-acetylglucosamine derived from Escherichia coli cell wall murein. J. Bacteriol. 183:3842-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plumbridge, J. A. 1991. Repression and induction of the nag regulon of Escherichia coli K-12: the roles of nagC and nagA in maintenance of the uninduced state. Mol. Microbiol. 5:2053-2062. [DOI] [PubMed] [Google Scholar]
- 25.Plumbridge, J. A. 1989. Sequence of the nagBACD operon in Escherichia coli K12 and pattern of transcription within the nag regulon. Mol. Microbiol. 3:505-515. [DOI] [PubMed] [Google Scholar]
- 26.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1996. Phosphoenolpyruvate:carbohydrate phosphotransferase systems, p. 1149-1174. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 27.Reissig, J. L., J. L. Strominger, and L. F. Leloir. 1955. A modified colorimetric method for the estimation of N-acetylamino sugars. J. Biol. Chem. 217:959-966. [PubMed] [Google Scholar]
- 28.Roseman, S. 1957. Glucosamine metabolism. I. N-acetylglucosamine deacetylase. J. Biol. Chem. 226:115-124. [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schmidt, D. M. Z., B. K. Hubbard, and J. A. Gerlt. 2001. Evolution of enzymatic activities in the enolase superfamily: functional assignment of unknown proteins in Bacillus subtilis and Escherichia coli as l-Ala-d/l-Glu epimerases. Biochemistry 40:15707-15715. [DOI] [PubMed] [Google Scholar]
- 31.Sebastian, J., and C. Asensio. 1972. Purification and properties of the mannokinase from Escherichia coli. Arch. Biochem. Biophys. 151:227-233. [DOI] [PubMed] [Google Scholar]
- 32.Sproul, A. A., L. T. Lambourne, D. J. Jean-Jacques, and H. L. Kornberg. 2001. Genetic control of manno(fructo)kinase activity in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:15257-15259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Templin, M. F., A. Ursinus, and J. V. Holtje. 1999. A defect in cell wall recycling triggers autolysis during the stationary growth phase of Escherichia coli. EMBO J. 18:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian, G., and W. K. Maas. 1994. Mutational analysis of the arginine repressor of Escherichia coli. Mol. Microbiol. 13:599-608. [DOI] [PubMed] [Google Scholar]
- 35.Titgemeyer, F., J. Reizer, A. Reizer, and M. H. Saier, Jr. 1994. Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology (Reading) 140:2349-2354. [DOI] [PubMed] [Google Scholar]
- 36.Uehara, T., and J. T. Park. 2003. Identification of MpaA, an amidase in Escherichia coli that hydrolyzes the γ-d-glutamyl-meso-diaminopimelate bond in murein peptides. J. Bacteriol. 185:679-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Votsch, W., and M. F. Templin. 2000. Characterization of a β-N-acetylglucosaminidase of Escherichia coli and elucidation of its role in muropeptide recycling and β-lactamase induction. J. Biol. Chem. 275:39032-39038. [DOI] [PubMed] [Google Scholar]
- 38.White, R. J. 1968. Control of amino sugar metabolism in Escherichia coli and isolation of mutants unable to degrade amino sugars. Biochem. J. 106:847-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yakushi, T., K. Masuda, S. Narita, S. Matsuyama, and H. Tokuda. 2000. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2:212-218. [DOI] [PubMed] [Google Scholar]
- 40.Zhao, K., X. Chai, and R. Marmorstein. 2004. Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Escherichia coli. J. Mol. Biol. 337:731-741. [DOI] [PubMed] [Google Scholar]