Abstract
In vivo cross-linking between native cysteines in the Aer receptor of Escherichia coli showed dimer formation at the membrane anchor and in the putative HAMP domain. Dimers also formed in mutants that did not bind flavin adenine dinucleotide and in truncated peptides without a signaling domain and part of the HAMP domain.
The Escherichia coli chemoreceptors Tsr, Tar, Trg, and Tap share sequence and domain homology. They have a ligand-binding domain in the periplasm and a C-terminal signaling domain that is predominately an extended α-helix, which is folded to form a hairpin structure. The chemoreceptors form dimers (16) and higher-order structures that cluster at the poles of the bacterium (12, 13, 15). The receptors bind ligands in the periplasmic space and signal through the lipid bilayer into the cytosol via a signaling team made up of a trimer of receptor dimers (a squad) and chemotaxis proteins CheA and CheW (1, 9). Aer, the aerotaxis receptor, shares sequence and domain homology with these chemoreceptors in the C-terminal region but displays a unique membrane-binding region and an N-terminal PAS domain (3, 18) (Fig. 1), which binds a flavin adenine dinucleotide (FAD) cofactor (2, 19). In contrast to chemoreceptors, the sensing domain of Aer is in the cytoplasm, and signaling to the C-terminal domain could follow a linear path along the Aer backbone or a more direct transverse path from the PAS domain to the HAMP domain (Fig. 1). One of the unknowns in the structure of the Aer protein is whether the globular PAS domain interferes with the packing of the Aer monomers to form dimers.
FIG. 1.
Cartoon summarizing the putative domains of Aer, the chemotaxis cascade, and the question of dimerization as described in the text. IM, inner membrane; OM, outer membrane; CW, clockwise; CCW, counterclockwise.
By analogy to the three-dimensional structure of the flavin mononucleotide-PAS domain in the Phy3 protein from Adiantum (5), it is likely that the isoalloxazine ring of the FAD molecule binds to the PAS domain of Aer. However, FAD binding to Aer is influenced by regions outside of the PAS domain, since missense mutations in the PAS domain, the F1 region, or the HAMP domain can prevent FAD binding (2, 14, 14a, 19). The role of the membrane-binding region in Aer is unknown. It could have an active role in signal transduction, or a passive role such as maintaining proper registry between the N- and C-terminal domains, stabilizing interactions between Aer monomers, or aligning Aer with components of the electron transport system.
In this study we addressed several related questions. Does the Aer protein form dimers? If so, which regions of Aer are associated in dimers? Is FAD binding required to stabilize dimer formation? Here we show that Aer does form dimers independent of FAD binding and identify regions of Aer that are not required for dimerization.
The Aer protein contains three cysteine residues, two in the membrane anchor (Cys193 and Cys203) and one in the HAMP domain (Cys253). Plasmids pSB83 (pSB20-C193S) and pSB85 (pSB20-C203A) (2) with aer inserts were obtained from J. S. Parkinson (University of Utah) and used to generate paired combinations of amino acid substitutions in Aer (C193S/C203A, C193S/C253A, and C203A/C253A) with the QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, Calif.). The C193S/C203A/C253A triple substitution was created by a second round of mutagenesis with the Aer-C193S/C203A construct as a template. R235C and R235E were similarly constructed by site-directed mutagenesis (14, 14a). Truncated, six-His-tagged constructs of Aer were made by PCR amplification of pAVR2 and subcloned into the EheI-PstI sites of pProEX HTa (6, 14, 19). Each plasmid was sequenced to verify the expected mutation or truncation. FAD binding was determined as reported earlier (19). Aer peptides were expressed at concentrations approximately eightfold above wild-type levels. This was achieved at different isopropyl-β-d-thiogalactopyranoside concentrations for full-length Aer (50 μM) and His6x-Aer2-231 (12.5 μM) and His6x-Aer2-285 (0 μM) constructs. In vivo cross-linking of cysteine residues was initiated with the oxidant copper phenanthroline, a catalyst that cycles between reduced and oxidized forms in the presence of sulfhydryls and oxygen (10). Assays were performed as previously described (8), except that assays were performed at 23°C. Cells were cultured in H1 minimal salts supplemented with 30 mM succinate, 0.1% Casamino Acids, and 100 μg of ampicillin per ml, grown to an optical density at 600 nm of 0.4, and induced with isopropyl-β-d-thiogalactopyranoside for 3 h. Whole cells were incubated with 300 μM Cu(II) (o-phenanthroline)3 for 10 min, and the reaction was quenched with sodium dodecyl sulfate sample buffer containing 2.5 mM N-ethylmaleimide (NEM) but no mercaptoethanol. The concentration of NEM was adequate to prevent artifactual cross-linking during denaturation, since control cells showed no Aer cross-linking when the addition of sample buffer (+NEM) preceded that of copper phenanthroline (data not shown). Aer was identified by a chemiluminescent Western blot assay with rabbit antisera directed against the Aer peptide Aer2-166 as described earlier (19).
One or more cysteine residues in native Aer cross-linked in vivo when E. coli cells were oxidized with copper phenanthroline, indicating that the residues are in close proximity and that Aer forms an oligomer consisting of at least a dimer (Fig. 2A, lanes 1 and 2). At higher concentrations of sample, a proteolytic fragment of Aer was evident that dimerized with monomeric Aer (Fig. 2A). The fragment apparently occurred in vivo, since it was present when whole cells were disrupted in the presence of a protease inhibitor cocktail (catalog no. 1836153; Roche, Indianapolis, Ind.). To quantitate the percentage of cross-linking, the linear range of the chemiluminescent Western blot assays was determined by serial dilutions (Fig. 2A, lanes 2 to 8). Integrated areas were averaged and plotted. The apparent percentage of cross-linking decreased until the sample was diluted to the linear range. At 10 min, wild-type Aer showed an average of 19% cross-linking (Fig. 2B, lanes 1 and 2; Table 1). To determine which native cysteine(s) was cross-linked, Aer constructs containing a single cysteine residue were tested for the ability to cross-link. We inferred that these constructions maintained a native conformation, since none of the cysteine mutations interfered with aerotaxis signaling, confirming an earlier report by Bibikov et al. (2). There was no evidence of cross-linking between cognate Cys193 residues in the Aer-C203A/C253A protein, suggesting that these residues are not close enough to form a covalent bond after oxidation (Fig. 2B, lanes 3 and 4). However, 5% of Cys203 residues in Aer-C193S/C253A and 14% of Cys253 residues in Aer-C193S/C203A cross-linked within 10 min, indicating a close proximity between these pairs of cysteine residues (Fig. 2B, lanes 7 to 10). This suggests that the membrane anchor (Cys203) and the HAMP domain (Cys253) of two monomers are proximal and are involved in dimerization. Since Cys253 is predicted to be cytosolic, the oxidant must be able to cross the inner membrane and oxidize this sulfhydryl, despite competition from intracellular reduced glutathione.
FIG. 2.
Images of chemiluminescent Western blots showing in vivo dimerization of Aer constructs after a 10-min treatment with the oxidant copper phenanthroline. (A) Serial dilutions of wild-type Aer. Lane 1 is the 0-min control, and lanes 2 to 8 are 10-min time points diluted as follows: lanes 2 and 3, undiluted sample; lanes 4 and 5, twofold dilution; lanes 6 and 7, fourfold dilution; lane 8, eightfold dilution. Mw, molecular weight markers; M, monomeric form of Aer; F, a 37-kDa proteolytic fragment of full-length Aer; M-M, dimerization between monomeric Aer proteins; M-F, dimerization between Aer monomer and proteolytic fragment. (B) Lanes: 1 and 2, wild-type Aer at 0 and 10 min; 3 and 4, control without cysteines (Aer-C193S/C203A/C253A) at 0 and 10 min; 5 and 6, Aer-C193 (Aer-C203A/C253A) at 0 and 10 min; 7 and 8, Aer-C203 (Aer-C193S/C253A) at 0 and 10 min; 9 and 10, Aer-C253 (Aer-C193S/C203A) at 0 and 10 min; 11 and 12, His6x-Aer2-285 at 0 and 10 min; 13 and 14, His6x-Aer2-231 at 0 and 10 min.
TABLE 1.
Summary of cross-linking data
| Aer construct | Position(s) where cysteine is present | FAD binding | Signaling | Avg % cross-linkinga |
|---|---|---|---|---|
| Wild type | 193, 203, 253 | + | + | 19 ± 3 |
| C193S/C203A | 253 | + | + | 14 ± 2 |
| C193S/C253A | 203 | + | + | 5 ± 2 |
| C203A/C253A | 193 | + | + | 0 |
| C193S/C203A/C253A | + | + | 0 | |
| His6x-Aer2-285b | 193, 203, 253 | + | − | 27 ± 1 |
| His6x-Aer2-231b | 193, 203 | − | − | 12 ± 2 |
| R235Cc | 193, 203, 235, 253 | − | − | Yesd |
| R235Ec | 193, 203, 253 | − | − | Yesd |
The HAMP domain is critical for signaling and stabilizing FAD binding in Aer (2, 14, 14a). Reciprocally, the FAD cofactor might likewise stabilize the structure of the HAMP domain and in turn the dimeric structure. Such conformational stabilization by cofactors and ligands has been shown for other proteins. For example, the apo form of photoactive yellow protein (the PAS prototype) is more accessible to deuterium exchange and prone to precipitation (7, 11) than the holoprotein, and the unliganded form of the PAS B domain of the aryl hydrocarbon receptor can be displaced from hsp90 by certain aromatic ligands that bind to the cofactor-binding pocket (4). Two Aer proteins containing substitutions in the HAMP domain, R235C and R235E, which abolish FAD binding to the PAS domain (14, 14a), formed dimers in the presence of the oxidant, supporting the hypothesis that dimerization is independent of the FAD cofactor (Table 1). Two His-tagged truncated Aer constructs were tested for dimer formation. One of these constructs included the PAS domain, the HAMP domain, and the proximal signaling region (His6x-Aer2-285). The other construct (His6x-Aer2-231) was missing a segment corresponding to the second amphipathic region (AS-2) of other HAMP domains, and it did not bind FAD (6, 14). Both of these constructs cross-linked in response to copper phenanthroline, exhibiting 12 and 27% cross-linking at 10 min for His6x-Aer2-231 and His6x-Aer2-285, respectively (Fig. 2B, lanes 11 to 14; Table 1). This indicates that dimer formation does not require the C-terminal signaling domain, the full HAMP domain, or FAD binding.
Taken together (Table 1), these data indicate that Aer forms dimers or oligomers typical of other chemoreceptors. Although it is possible that Aer dimers form labile, transient complexes, it is likely that these complexes are stable. Wild-type Aer still formed cross-linked dimers when cells were incubated with copper phenanthroline for 20 min at 4°C (D. Amin and M. S. Johnson, unpublished observation). At temperatures below 18°C, there is a lipid phase transition that limits lateral movement in the membrane of E. coli (17).
At a minimum, the dimer interface includes the membrane anchor and the HAMP domain and is not dependent on the signaling domain or FAD binding (Fig. 3). Cys253, which is predicted to be in the AS-2 subdomain of the HAMP domain, is apparently at the dimer interface, but this region is not required for dimerization, since His6x-Aer2-231 was able to form a dimer. Presumably, the presence of the complete HAMP domain (His6x-Aer2-285) would enhance association of the Aer monomers. Still unknown is the contribution, if any, of the PAS domain and the F1 region to stabilization of the dimeric structure. The F1 region is thus far unique to Aer, but it is found in all Aer homologues and appears to be crucial for Aer function (2; M. Kang and M. S. Johnson, unpublished observation). PAS domains were thought to be strictly dimerization domains before their role in light, oxygen, redox, and voltage sensing was established (20). Clearly, dimerization between the Aer-PAS domains is possible, and such interactions could serve to stabilize the dimer interface.
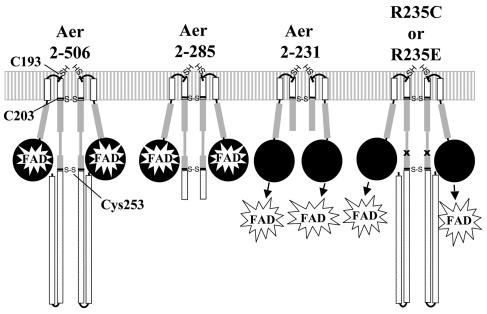
FIG. 3.
Cartoon summarizing cross-linking studies with the constructs described in the text. Relative placement of native cysteines is shown on a cartoon backbone of Aer. Arrows pointing toward FAD represent an inability of these constructs to bind FAD. The letter x shows the relative positions of R235C and R235E replacements. Cross-linking was not tested in an Aer protein containing a single cysteine at residue 235 (Aer-C193S/C203A/C253A/R235C).
Acknowledgments
We thank J. S. Parkinson for plasmids, K. Watts for standardizing the Western blots, and J. Mello for technical assistance.
This work was supported by grants from the National Institute of General Medical Sciences (GM29481) to B.L.T. and from Loma Linda University to M.S.J.
REFERENCES
- 1.Ames, P., C. A. Studdert, R. H. Reiser, and J. S. Parkinson. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:7060-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibikov, S. I., L. A. Barnes, Y. Gitin, and J. S. Parkinson. 2000. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5830-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibikov, S. I., R. Biran, K. E. Rudd, and J. S. Parkinson. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coumailleau, P., L. Poellinger, J. A. Gustafsson, and M. L. Whitelaw. 1995. Definition of a minimal domain of the dioxin receptor that is associated with Hsp90 and maintains wild type ligand binding affinity and specificity. J. Biol. Chem. 270:25291-25300. [DOI] [PubMed] [Google Scholar]
- 5.Crosson, S., and K. Moffat. 2001. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. USA 98:2995-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrmann, S., Q. Ma, M. S. Johnson, A. V. Repik, and B. L. Taylor. 2004. PAS domain of the Aer redox sensor requires C-terminal residues for native-fold formation and FAD binding. J. Bacteriol. 186:6782-6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoff, W. D., A. Xie, I. H. Van Stokkum, X. J. Tang, J. Gural, A. R. Kroon, and K. J. Hellingwerf. 1999. Global conformational changes upon receptor stimulation in photoactive yellow protein. Biochemistry 38:1009-1017. [DOI] [PubMed] [Google Scholar]
- 8.Hughson, A. G., and G. L. Hazelbauer. 1996. Detecting the conformational change of transmembrane signaling in a bacterial chemoreceptor by measuring effects on disulfide cross-linking in vivo. Proc. Natl. Acad. Sci. USA 93:11546-11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim, S. H., W. Wang, and K. K. Kim. 2002. Dynamic and clustering model of bacterial chemotaxis receptors: structural basis for signaling and high sensitivity. Proc. Natl. Acad. Sci. USA 99:11611-11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobashi, K. 1968. Catalytic oxidation of sulfhydryl groups by o-phenanthroline copper complex. Biochim. Biophys. Acta 158:239-245. [DOI] [PubMed] [Google Scholar]
- 11.Kroon, A. R., W. D. Hoff, H. P. Fennema, J. Gijzen, G. J. Koomen, J. W. Verhoeven, W. Crielaard, and K. J. Hellingwerf. 1996. Spectral tuning, fluorescence, and photoactivity in hybrids of photoactive yellow protein, reconstituted with native or modified chromophores. J. Biol. Chem. 271:31949-31956. [DOI] [PubMed] [Google Scholar]
- 12.Lybarger, S. R., and J. R. Maddock. 2000. Differences in the polar clustering of the high- and low-abundance chemoreceptors of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:8057-8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lybarger, S. R., and J. R. Maddock. 2001. Polarity in action: asymmetric protein localization in bacteria. J. Bacteriol. 183:3261-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma, Q. 2001. HAMP domain and signaling mechanism of the Aer protein. Ph.D. dissertation. Loma Linda University, Loma Linda, Calif.
- 14a.Ma, Q., M. S. Johnson, and B. L. Taylor. Genetic analysis of the HAMP domain of the Aer aerotaxis sensor localizes FAD-binding determinants to the AS-2 helix. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 15.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717-1723. [DOI] [PubMed] [Google Scholar]
- 16.Milligan, D. L., and D. E. Koshland, Jr. 1988. Site-directed cross-linking. Establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J. Biol. Chem. 263:6268-6275. [PubMed] [Google Scholar]
- 17.Overath, P., M. Brenner, T. Gulik-Krzywicki, E. Shechter, and L. Letellier. 1975. Lipid phase transitions in cytoplasmic and outer membranes of Escherichia coli. Biochim. Biophys. Acta 389:358-369. [DOI] [PubMed] [Google Scholar]
- 18.Rebbapragada, A., M. S. Johnson, G. P. Harding, A. J. Zuccarelli, H. M. Fletcher, I. B. Zhulin, and B. L. Taylor. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. USA 94:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Repik, A., A. Rebbapragada, M. S. Johnson, J. O. Haznedar, I. B. Zhulin, and B. L. Taylor. 2000. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 36:806-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]