Abstract
The processes associated with early events in biofilm formation have become a major research focus over the past several years. Events associated with dispersion of cells from late stage biofilms have, however, received little attention. We demonstrate here that dispersal of Pseudomonas aeruginosa PAO1 from biofilms is inducible by a sudden increase in carbon substrate availability. Most efficient at inducing dispersal were sudden increases in availability of succinate > glutamate > glucose that led to ∼80% reductions in surface-associated biofilm biomass. Nutrient-induced biofilm dispersion was associated with increased expression of flagella (fliC) and correspondingly decreased expression of pilus (pilA) genes in dispersed cells. Changes in gene expression associated with dispersion of P. aeruginosa biofilms were studied by using DNA microarray technology. Results corroborated proteomic data that showed gene expression to be markedly different between biofilms and newly dispersed cells. Gene families that were upregulated in dispersed cells included those for flagellar and ribosomal proteins, kinases, and phage PF1. Within the biofilm, genes encoding a number of denitrification pathways and pilus biosynthesis were also upregulated. Interestingly, nutrient-induced dispersion was associated with an increase in the number of Ser/Thr-phosphorylated proteins within the newly dispersed cells, and inhibition of dephosphorylation reduced the extent of nutrient-induced dispersion. This study is the first to demonstrate that dispersal of P. aeruginosa from biofilms can be induced by the addition of simple carbon sources. This study is also the first to demonstrate that dispersal of P. aeruginosa correlates with a specific dispersal phenotype.
Biofilms are complex, organized communities of bacteria that grow in association with surfaces (62). They can be found at almost any solid-liquid interface, including the inner surfaces of pipes in industrial facilities (16), in domestic plumbing systems (34), on rocks in streams (25), and associated with medical implants (36). One of the most medically important biofilm-forming species is Pseudomonas aeruginosa, which is commonly associated with noscomial infections of the urogenital tract and skin (22) and the lungs of patients suffering from cystic fibrosis (3, 53). Biofilm formation by P. aeruginosa progresses through multiple developmental stages, beginning with attachment to a surface, followed by the immigration and division to form microcolonies, and finally maturation involving expression of matrix polymers (50, 55). Bacteria within each biofilm stage display phenotypes and possess properties that are markedly different from those of the same group growing planktonically (50, 55). In particular, they are much less susceptible to antimicrobial treatments and, as a consequence, are often associated with chronic recurring bacterial infections (26). The developmental life cycle of biofilms comes full circle when biofilm cells disperse (50).
Loss of cells from a biofilm is not restricted to the last stage of biofilm development but may occur continuously at low levels over the course of biofilm formation (2, 23, 50, 51). As biofilm accumulation progresses, cells are released from the biofilm into the bulk liquid, mainly in response to environmental changes. Such a loss of cells from a biofilm may be due to cell lysis or the removal of intact, viable cells. Several mechanisms have been described that result in the removal of intact cells. The removal of intact, viable cells may be nonspecific or passive, such as with sloughing of the biofilm under oxygen-mass transfer rate limitations in thick biofilms or under hydrodynamic stress due to shear generated by moving liquid past the biofilm (2, 13, 44). During sloughing, particles ranging from a few single cells to large aggregates and entire cell clusters are removed from the biofilm. Loss of intact cells from the biofilm may also be associated with chemical factors in the environment. This removal may be active or passive. Chen and Stewart (12) demonstrated that addition of chemicals such as antimicrobial agents to a mixed biofilm of P. aeruginosa and Klebsiella pneumoniae resulted in more than 25% of protein removal from the surface (12). Treatments that caused the loss of more than 25% of the biomass included NaCl and CaCl2; chelating agents; surfactants such as sodium dodecyl sulfate (SDS), Tween 20, and Triton X-100; a pH increase; and lysozyme, hypochlorite, monochloramine, and concentrated urea. Some treatments caused significant killing but not much removal, while other treatments caused removal with little killing (12). Since this study focused on the remaining biofilm, the detachment mechanism is unclear.
Active removal of cells from the biofilm has been associated with either the act of cell division (1) or the active escape of single bacterial cells from the biofilm matrix, with single bacteria swimming away from the biofilm (21, 50). The active escape of single bacterial cells from the biofilm matrix is referred to as dispersion (50). Biofilm dispersion has been suggested to involve phenotypic modifications of the dispersing cells. Single cells dispersed from biofilms display phenotypes that differ not only from those in biofilms (50) but also from established planktonic cultures (1). Such phenotypic adaptation towards dispersion enables bacteria to actively escape from the biofilm matrix. Biofilm dispersion occurs in response to environmental changes and was shown to be induced by oxygen depletion in Pseudomonas putida (2), by lack or depletion of nutrients (17, 38, 52), or by a change in the nutrient composition (step change from a minimal to a complex medium) (32).
Attempts have been made to identify the cellular responses that contribute to this phenomenon of active biofilm dispersion. In 2000, Vats and Lee showed that a surface protein-releasing enzyme (SPRE) produced by Streptococcus mutans is actively involved in the degradation of attachment polymers on tooth surfaces releasing bacteria into the bulk liquid (58). Addition of SPRE has been shown to result in a 20% increase in detachment compared to control samples. Recently, Stoodley et al. (56) showed that cells “twitch” prior to detachment, suggesting that motility is important for dispersal. Furthermore, Jackson et al. (31) described the role of an RNA binding protein, CsrA (carbon storage regulator), that acts both as a repressor of biofilm formation and as an activator of biofilm dispersal in Escherichia coli. Importantly, the effects of CsrA were believed to be mediated by regulation of intracellular glycogen biosynthesis and catabolism (4, 31, 48, 49, 64).
Here, we present evidence that biofilm dispersion can be induced by specific changes in the physiochemical environment. In particular, increases in the concentration of particular carbon sources induced dispersion. Analysis of the dispersed and residual biofilm populations by proteomic and genomic techniques demonstrates that specific metabolic pathways are utilized to generate cells with a specific dispersal phenotype.
MATERIALS AND METHODS
Bacterial strains and media.
The P. aeruginosa strains PAO1, PAO 8023 (crc mutant), and reporter strain PAO1-pfliC::lacZ were used in this study. In addition, P. aeruginosa PA14 and P. aeruginosa isogenic strain PA416 (pilA mutant, pilA::lacZ reporter strain) (43) were used. All strains were grown aerobically in minimal medium (2.56 g of Na2HPO4, 2.08 g of KH2PO4, 1.0 g of NH4Cl, 0.04 g of CaCl2 × 2H2O, 0.5 g of MgSO4 × 7H2O, 0.1 mg of CuSO4 × 5H2O, 0.1 mg of ZnSO4 × H2O, 0.1 mg of FeSO4 × 7H2O, and 0.004 mg of MnCl2 × 4H2O per liter, pH 7.2) at room temperature in shake flasks at 250 rpm. Glutamate (130 mg/liter) was used as the sole carbon source unless otherwise indicated. Biofilms were grown as described below at room temperature in minimal medium containing 1.8 mM glutamate as a carbon source unless otherwise indicated.
Induction of biofilm dispersion.
Biofilms were grown as described previously (50). Briefly, the interior surfaces of silicone tubing of a once-through continuous flow reactor system were used to cultivate biofilms at 22°C. Two reactor types were used in parallel, one being composed of size 15 silicone tubing (Masterflex) with an internal volume of 25 ml (tubing length, 1 m; flow rate, 0.5 ml/min), the other being composed of size 13 tubing with an internal volume of 1 ml (tubing length, 1 m; flow rate, 0.2 ml/min). Biofilms were pregrown for 4 days in minimal medium containing 1.8 mM glutamate as a carbon source. After 4 days of biofilm growth, biofilm dispersion was induced by a sudden 10-fold increase of the carbon concentration in the growth medium, either by increasing the glutamate concentration from 1.8 mM to 19.8 mM or through the addition of either 20 mM succinate, citrate, glucose, or 10 mM ammonium chloride (Sigma-Aldrich, St. Louis, Mo.) to the growth medium. In addition, biofilms were pregrown for 4 days in minimal medium containing 2 mM glucose as a carbon source and biofilm dispersion was induced by either increasing the glucose concentration from 2 to 22 mM or through the addition of either 18 mM glutamate, 20 mM succinate or citrate, or 10 mM ammonium chloride (Sigma-Aldrich) to the growth medium. Dispersion was also induced by depletion of the growth medium of nutrients or by a change in pH (by an increase in pH from 7.2 to 10.2 or by a decrease in pH to 4.2) of the medium. Dispersion was indicated by an increase in turbidity at 600 nm in the effluent from the silicone tubing. The total number of bacteria in the effluent was determined by serial plate counts on Luria-Bertani (LB) agar at 37°C.
Generation of a pfliC::lacZ transcriptional reporter strain.
The generation of the pfliC::lacZ transcriptional reporter gene constructs using a mini-CTX vector and the site-specific integration of the construct at the attB site on the chromosome of P. aeruginosa PAO1 was carried out according to the gene fusion construction method as outlined by Becher and Schweizer (5) and Hoang et al. (29). The sequence of the fliC promoter region was amplified with the following oligonucleotides: 5′-GGTGATTGGATCCAAAGGAC (HindIII restriction site is underlined) and 5′-AATCCAAGCTTTTCTCGAACG (BamHI restriction site is underlined). The resulting 361-bp fragment was digested with BamHI and HindIII endonuclease, gel purified, and subsequently cloned into the BamHI and HindIII restriction sites of the mini-CTX vector. Proper chromosomal integration at the attB site was confirmed as described by Becher and Schweizer (5) and Hoang et al. (29).
Microscopy of the reporter strain during dispersion.
Reporter gene activity was observed with flow cells. Log-phase P. aeruginosa cells (approximately 108 CFU/ml) were inoculated as a 3.0-ml bolus through a septum 4 cm upstream from the flow-cell. Cells attached to the inner surface of the glass coverslip were viewed by transmitted light or epi-UV illumination using an Olympus BX60 microscope (Olympus, Melville, N.Y.) and a ×100 magnification A100PL objective lens or a ×50 magnification ULWD MSPlan long working distance Olympus objective lens. All images were captured with a Magnafire cooled three-chip charge-coupled device camera (Optronics, Inc., Galena, Calif.) and stored as separate digital files for subsequent retrieval and analysis. For bright-field images, images were captured with a 30-ms exposure time while epifluorescence images were captured with a 600-ms exposure time. Under these conditions, nonfluoresescence was detected from P. aeruginosa wild-type cells. As the fluorescent substrate for β-galactosidase, methlyumbelliferyl β-d-galactopyranoside dissolved in N,N-dimethylformamide was added at 0.02 g/liter to the influent medium reservoir during biofilm development.
RNA isolation from biofilm and dispersed cells.
Following induction of dispersal by the sudden increase in the glutamate concentration in the growth medium (from 1.8 to 19.8 mM), dispersed cells were collected at the end of the silicone tubing directly into RNA Protect (QIAGEN, Crawley, United Kingdom) for RNA isolation. The biofilm cells that remained after dispersion were harvested from the interior surface. The harvested cells were suspended in RNA Protect. Total RNA was extracted from the suspended biofilm and planktonic populations using RNeasy RNA isolation kits (QIAGEN) following the manufacturer's instructions.
Gene expression analysis.
Affymetrix GeneChip Oligonucleotide arrays were used to analyze the gene expression of the biofilm and dispersed cell populations. Briefly, biotinylated cRNA samples from planktonic and biofilm P. aeruginosa PAO1 were synthesized and hybridized in triplicates to Pae G1a oligonucleotide arrays (Affymetrix, Inc., Santa Clara, Calif.). These arrays contain probe sets for 5,570 open reading frames and 100 intergenic sequences from the PAO1 strain of P. aeruginosa. Background correction, quantile normalization, and gene expression analysis were carried out with RMAEXPRESS (8). Further analyses, which included unpaired Student's t tests, were performed with MAXDVIEW (available from http://bioinf.man.ac.uk/microarray/maxd/). The microarray data were submitted in a MIAME (Minimum Information About a Microarray Experiment)-compliant format to the ArrayExpress database (www.mged.org/Workgroups/MIAME/miame.html), and an accession number was assigned (E-MEXP-87). To increase statistical robustness, the changes in expression of groups of functionally related genes, rather than individual genes, were interrogated to determine which cellular processes underwent change. Two methods and two annotation sources were used in grouping genes. First, the probe set description available from Affymetrix (http://www.affymetrix.com/) was used to search for distribution patterns of specific gene groups within the global distribution profile of all probe sets on the microarrays. Second, we used gene ontology data from the Pseudomonas Genome Project (57) to categorize sets of top-ranking genes into functional groups (Tables 1 and 2).
TABLE 1.
Genes whose expression was significantly increased in P. aeruginosa cells that were dispersed from the biofilm
| Open reading frame no. | Gene product | Gene ontology | Fold changea |
|---|---|---|---|
| PA5117 | Regulatory protein TypA | Adaptation, protection | −2.65 |
| PA3531 | Bacterioferritin | Adaptation, protection | −3.40 |
| PA3525 | Argininosuccinate synthase | Amino acid biosynthesis and metabolism | −2.07 |
| PA4442 | ATP sulfurylase GTP-binding subunit/APS kinase | Amino acid biosynthesis and metabolism | −2.58 |
| PA0223 | Probable dihydrodipicolinate synthetase | Amino acid biosynthesis and metabolism | −2.70 |
| PA3635 | Enolase | Carbon compound catabolism | −2.17 |
| PA3155 | Probable aminotransferase WbpE | Cell wall/lipopolysaccharide/capsule | −2.39 |
| PA1838 | Sulfite reductase | Central intermediary metabolism | −2.10 |
| PA1338 | γ-Glutamyltranspeptidase precursor | Central intermediary metabolism | −3.11 |
| PA1553 | Probable cytochrome c oxidase subunit | Energy metabolism | −2.02 |
| PA4333 | Probable fumarase | Energy metabolism | −2.03 |
| PA3621 | Ferredoxin I | Energy metabolism | −2.25 |
| PA1584 | Succinate dehydrogenase (B subunit) | Energy metabolism | −2.78 |
| PA1609 | β-Ketoacyl-ACP synthase I | Fatty acid and phospholipid metabolism | −2.32 |
| PA3806 | Conserved hypothetical protein | Hypothetical, unclassified, unknown | −2.06 |
| PA3722 | Hypothetical protein | Hypothetical, unclassified, unknown | −2.11 |
| PA1913 | Hypothetical protein | Hypothetical, unclassified, unknown | −2.19 |
| PA4852 | Conserved hypothetical protein | Hypothetical, unclassified, unknown | −2.29 |
| PA2950 | Hypothetical protein | Hypothetical, unclassified, unknown | −2.51 |
| PA2146 | Conserved hypothetical protein | Hypothetical, unclassified, unknown | −3.02 |
| PA0729 | Hypothetical protein | Hypothetical, unclassified, unknown | −3.34 |
| PA4670 | Ribose-phosphate pyrophosphokinase | Nucleotide biosynthesis and metabolism | −2.23 |
| PA2629 | Adenylosuccinate lyase | Nucleotide biosynthesis and metabolism | −2.36 |
| PA0945 | Phosphoribosylaminoimidazole synthetase | Nucleotide biosynthesis and metabolism | −2.57 |
| PA2550 | Probable acyl coenzyme A dehydrogenase | Putative enzymes | −2.77 |
| PA0224 | Probable aldolase | Putative enzymes | −4.37 |
| PA0718 | Hypothetical protein of bacteriophage Pf1 | Related to phage, transposon, or plasmid | −3.61 |
| PA0722 | Hypothetical protein of bacteriophage Pf1 | Related to phage, transposon, or plasmid | −5.23 |
| PA0720 | Helix destabilizing protein of bacteriophage Pf1 | Related to phage, transposon, or plasmid | −5.29 |
| PA0723 | Coat protein B of bacteriophage Pf1 | Related to phage, transposon, or plasmid | −9.20 |
| PA1148 | Exotoxin A precursor | Secreted factors (toxins, enzymes, alginate) | 2.19 |
| PA4853 | DNA-binding protein Fis | Transcription, RNA processing and degradation | −2.35 |
| PA3743 | tRNA (guanine-N1) methyltransferase | Transcription, RNA processing and degradation | −2.38 |
| PA4275 | Transcription antitermination protein NusG | Transcription, RNA processing and degradation | −3.00 |
| PA3656 | 30S ribosomal protein S2 | Translation, posttranslational modification, degradation | −2.08 |
| PA4563 | 30S ribosomal protein S20 | Translation, posttranslational modification, degradation | −2.19 |
| PA2851 | Translation elongation factor P | Translation, posttranslational modification, degradation | −2.20 |
| PA4268 | 30S ribosomal protein S12 | Translation, posttranslational modification, degradation | −2.34 |
| PA0579 | 30S ribosomal protein S21 | Translation, posttranslational modification, degradation | −2.38 |
| PA2619 | Initiation factor | Translation, posttranslational modification, degradation | −2.63 |
| PA4432 | 30S ribosomal protein S9 | Translation, posttranslational modification, degradation | −3.04 |
| PA3647 | Probable outer membrane protein precursor | Transport of small molecules | −2.02 |
| PA1341 | Probable permease of ABC transporter | Transport of small molecules | −2.07 |
| PA0282 | Sulfate transport protein CysT | Transport of small molecules | −2.08 |
| PA1339 | Probable ATP-binding component of ABC transporter | Transport of small molecules | −2.34 |
| PA0280 | Sulfate transport protein CysA | Transport of small molecules | −2.45 |
| PA1340 | Probable permease of ABC transporter | Transport of small molecules | −3.62 |
Genes with a >2.0-fold change in expression are shown (P < 0.05).
TABLE 2.
Genes whose expression was significantly increased in P. aeruginosa biofilm cells
| Open reading frame no. | Gene product | Gene ontology | Fold changea |
|---|---|---|---|
| PA2386 | l-Ornithine N5-oxygenase | Adaptation, protection | 3.81 |
| PA2401 | Probable non-ribosomal peptide synthetase | Adaptation, protection | 3.74 |
| PA2424 | PvdL, an AMP-binding enzyme, involved in pyoverdine synthesis | Adaptation, protection | 3.70 |
| PA2394 | PvdN, involved in pyoverdine synthesis | Adaptation, protection | 3.45 |
| PA2400 | Probable nonribosomal peptide synthetase | Adaptation, protection | 3.30 |
| PA2425 | PvdG, involved in pyoverdine synthesis | Adaptation, protection | 3.19 |
| PA2392 | PvdP, involved in pyoverdine synthesis | Adaptation, protection | 2.39 |
| PA2399 | Pyoverdine synthetase D | Adaptation, protection | 2.16 |
| PA2395 | PvdO, involved in pyoverdine synthesis | Adaptation, protection | 2.13 |
| PA5427 | Alcohol dehydrogenase | Carbon compound catabolism | 4.44 |
| PA2393 | Probable dipeptidase precursor | Central intermediary metabolism | 3.07 |
| PA4587 | Cytochrome c551 peroxidase precursor | Energy metabolism | 5.95 |
| PA1556 | Probable cytochrome c oxidase subunit | Energy metabolism | 5.58 |
| PA0519 | Nitrite reductase precursor | Energy metabolism | 5.39 |
| PA1555 | Probable cytochrome c | Energy metabolism | 4.69 |
| PA0518 | Cytochrome c-551 precursor | Energy metabolism | 3.44 |
| PA3392 | Nitrous oxide reductase precursor | Energy metabolism | 3.35 |
| PA4470 | Fumarate hydratase | Energy metabolism | 3.30 |
| PA0524 | Nitric oxide reductase subunit B | Energy metabolism | 2.60 |
| PA0511 | Heme d1 biosynthesis protein NirJ | Energy metabolism | 2.07 |
| PA4570 | Hypothetical protein | Unknown | 4.62 |
| PA2381 | Hypothetical protein | Unknown | 4.56 |
| PA4469 | Hypothetical protein | Unknown | 4.35 |
| PA4471 | Hypothetical protein | Unknown | 4.35 |
| PA2412 | Conserved hypothetical protein | Unknown | 3.49 |
| PA5475 | Hypothetical protein | Unknown | 3.42 |
| PA1746 | Hypothetical protein | Unknown | 3.40 |
| PA3572 | Hypothetical protein | Unknown | 3.36 |
| PA2427 | Hypothetical protein | Unknown | 2.99 |
| PA0713 | Hypothetical protein | Unknown | 2.74 |
| PA2033 | Hypothetical protein | Unknown | 2.30 |
| PA2034 | Hypothetical protein | Unknown | 2.24 |
| PA2384 | Hypothetical protein | Unknown | 2.19 |
| PA0802 | Hypothetical protein | Unknown | 2.18 |
| PA2501 | Hypothetical protein | Unknown | 2.03 |
| PA4067 | Outer membrane protein OprG precursor | Membrane proteins | 2.77 |
| PA2413 | Probable class III aminotransferase | Putative enzymes | 4.18 |
| PA2402 | Probable nonribosomal peptide synthetase | Putative enzymes | 4.15 |
| PA2411 | Probable thioesterase | Putative enzymes | 3.46 |
| PA4175 | Pvds-regulated endoprotease, lysyl class | Putative enzymes | 2.34 |
| PA0515 | Probable transcriptional regulator | Transcriptional regulators | 2.39 |
| PA0295 | Probable periplasmic polyamine binding protein | Transport of small molecules | 2.60 |
| PA0198 | Transport protein ExbB | Transport of small molecules | 2.11 |
| PA4143 | Probable toxin transporter | Transport of small molecules | 2.06 |
Genes with a >2.0-fold change in expression are shown (P < 0.05).
Inhibition of biofilm dispersion.
Biofilms were grown as described above. Dispersion was induced by a sudden 10-fold increase of the glutamate concentration in the growth medium as described above. Inhibition of biofilm dispersion was achieved by the addition of two phosphatase inhibitor cocktails (P2850 and P5726; Sigma-Aldrich). Phosphatase inhibitor cocktail I was composed of microcystin LR, cantharidin, and (−)-p-bromotetramisole and has been described as inhibiting the l-isozyme of alkaline phosphatase as well as Ser-Thr phosphatases. Phosphatase inhibitor cocktail II was composed of sodium vanadate, sodium molybdate, sodium tartrate, and imidazole. The cocktail has been described as inhibiting acid and alkaline phosphatases and tyrosine protein phosphatases. Both cocktails were diluted 100-fold as recommended by the supplier (Sigma-Aldrich). The phosphatase inhibitor cocktails were added to the growth medium containing a 10-fold increase in glutamate concentration. The phosphatase inhibitor cocktails were added to the growth medium prior to adding the solution to the biofilms.
2D gel electrophoresis and image analysis.
Total cell extracts were prepared from the dispersed cells and the residual biofilm cells. Crude protein extract was prepared and protein determination was carried out as described previously (50). To determine the phosphoprotein patterns in P. aeruginosa in response to dispersion, Ser/Thr phosphorylated proteins were immunoprecipitated from total cell extracts using polyclonal phospho-(Ser/Thr) antibodies (Cell Signaling Technology, New England BioLabs, Beverly, Mass.). Steady-state biofilm cells were removed from the device and used as controls. First, the polyclonal phospho-(Ser/Thr) antibodies were added to insoluble protein A/G Plus agarose beads (Santa Cruz, Calif.) and incubated on ice for 30 min to allow binding of the antibodies to the agarose beads, thus, immobilizing the antibodies. Unbound antibodies were washed off with Tris-buffered saline (TBS) buffer by centrifugation. A total of 200 μg of cell extract (free of insoluble matter) was added, and the mixture was incubated on a shaker at 4°C overnight. Unbound protein was removed by repeated washing of the insoluble beads in TBS buffer. After centrifugation (1,000 × g, 5 min, 4°C), the pellet was dissolved in urea-based two-dimensional (2D) sample buffer. Immunoprecipitated proteins were separated by 2D polyacrylamide gel electrophoresis (2D/PAGE). 2D/PAGE was conducted according to the principles of O'Farrell (41) as outlined by Görg et al. (27) and as described in detail by Sauer and Camper (51), using a 2D gel system from Genomic Solutions, Inc. (Ann Arbor, Mich.). 2D gels were stained with silver nitrate (7). 2D gels were performed in triplicate for each growth condition to confirm the reproducibility of the protein pattern under planktonic and attached growth conditions. Only differences in protein spots that were reproduced three times are described here. Computational image analysis was carried out by using MELANIE version 3.0 (Genebio, Geneva, Switzerland) as described previously (50).
Protein identification by mass spectrometry.
Protein spots of interest were excised from the gel and digested in situ with trypsin using a ProGest workstation (Genomics Solutions., Inc., Mich.). After digestion for 2 h at 37°C, tryptic peptides were extracted and then desalted with ZipTips (Millipore), and an aliquot of the supernatant was taken for analysis by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry using an Ettan MALDI-TOF Pro (Amersham Pharmacia Biotech, N.J.). Data were obtained according to the following parameters: reflective mode, 20-kV accelerating voltage, 95% grid voltage, and 0.2-ns delay. Trypsin peptides were used as internal standards for every peptide sample to ensure high mass accuracy. Database searches were performed with MASCOT software.
RESULTS
Biofilm dispersion can be induced by nutrients.
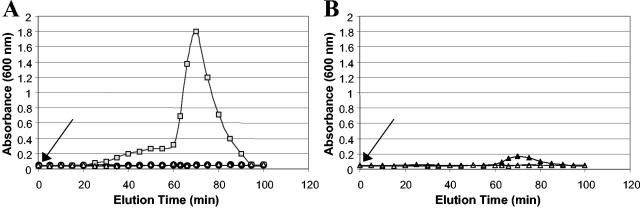
In biofilm dispersion, bacteria actively escape from the biofilm matrix as free planktonic bacteria. While occurring continually at low levels, dispersion is thought to be enhanced as a response to environmental changes such as oxygen limitation, starvation, and changes in pH. To further investigate such shock responses, P. aeruginosa biofilms were allowed to form for 4 days under single-pass flow-through conditions to allow the formation of late-stage biofilms that exceeded the thickness of a monolayer, before biofilm dispersion was induced. Dispersion was induced by changing the growth medium through the addition of 18 mM glutamate, by depletion of the medium of nutrients, or by changes in pH (increase in pH from 7.2 to 10.2 or decrease in pH to 4.2) of the medium. Changes in the turbidity of the eluted medium from the biofilm tube reactor were used as an indicator of dispersal (Fig. 1). Figure 1A shows that while a 3-fold increase in the nutrient concentration was not, however, sufficient to induce dispersion, the level of dispersion was significantly enhanced when glutamate concentrations were increased 10-fold (Fig. 1A). As shown in Fig. 1B, an increase in pH of the growth medium induced some dispersion as compared to the nutrient-induced dispersion, while a decrease in pH of the growth medium to pH 4.2 did not cause biofilm dispersion (data not shown).
FIG. 1.
Induction of biofilm dispersion in response to environmental changes. After 4 days of P. aeruginosa biofilm growth, (A) biofilm dispersion was induced by sudden 3-fold (•) and 10-fold (░⃞) increases of the glutamate concentration in the growth medium. The effluent of untreated biofilms was used as controls (▵). (B) After 4 days of P. aeruginosa biofilm growth, biofilm dispersion was analyzed in response to a change in pH, by an increase in pH change of 3 pH units from pH 7.2 to 10.2 (▴). The effluent of untreated biofilms was used as a control (▵). Dispersion was indicated by an increase of turbidity at 600 nm in the silicone tubing effluent. The arrow indicates addition of nutrients to the growth medium. Biofilms were grown in a biofilm tube reactor, composed of size 15 tubing with an internal volume of 25 ml (tubing length, 1 m; flow rate, 0.5 ml/min), for 4 days in minimal medium containing 1.8 mM glutamate as the sole carbon source. The retention time of the biofilm tube reactor was 50 min.
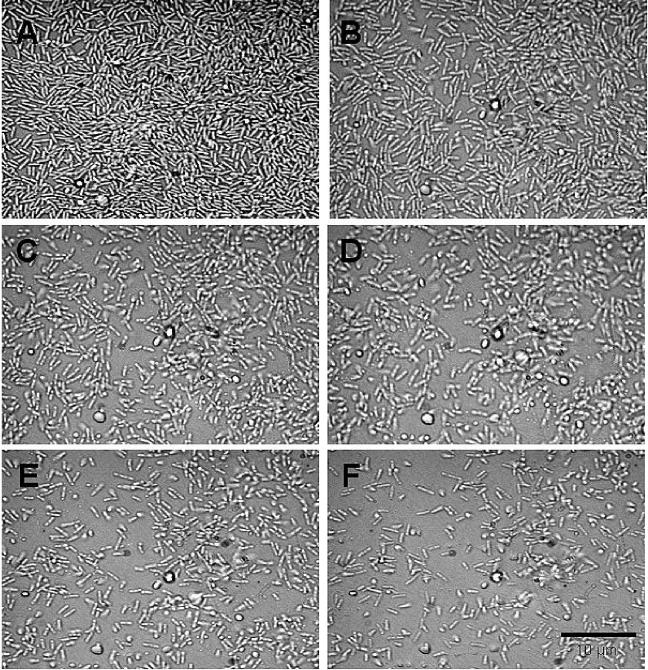
The loss of intact cells from the biofilm associated with chemical factors in the environment can be due to either the act of cell division (1) or the active escape of single bacterial cells from the biofilm matrix (21, 50). We therefore tested whether the increase in turbidity of the eluted medium was associated with cell division or biofilm dispersal. To do so, P. aeruginosa biofilms were grown for 4 days under single-pass flow-through conditions in a flow cells to obtain late-stage biofilms. Within this period of time, biofilms exceeding the thickness of a monolayer were able to develop (Fig. 2A). Dispersion was induced by changing the growth medium through the addition of 18 mM glutamate. Bright-field images were taken of intact biofilms and over the course of nutrient-induced dispersion. Under the conditions, the dispersion event was completed within 45 to 60 min. The microscopic images in Fig. 2 show the same biofilm before (Fig. 2A) and during the nutrient-induced dispersion event (Fig. 2B to F), indicating that the loss of cells from the biofilm was probably due to bacteria actively escaping as single cells from the matrix. No loss of bacteria due to cell division was observed. In addition, we tested the effect of a 10-fold increase in the glutamate concentration on planktonic cells obtained from the effluent of untreated biofilms. Planktonic cells were incubated in the presence of a 10-fold-increased glutamate concentration for a period of 0, 25, 50, 75, 100, and 125 min. At the times indicated, the turbidity (600 nm) and the total number of bacteria (CFU) were determined. No increase in turbidity or cell counts was observed within 100 min (data not shown). However, a 35% increase in cell counts was detected within 125 min (data not shown). Overall, a 10-fold increase in the glutamate concentration resulted in a decrease in the doubling time for P. aeruginosa in suspension from 120 min to 60 min (data not shown). While the findings suggest that the observed increase in turbidity in the effluent after sudden increases in the nutrient concentration (Fig. 1) was due to biofilm dispersion (and not growth), we nevertheless used in the following experiments a biofilm tube reactor, composed of size 13 tubing with an internal volume of 1 ml (flow rate, 0.2 ml/min), to considerably reduce the residence and elution time to 5 min. The reduced residence time ensured completion of dispersion events within 10 min. Thus, the fact that dispersion events were completed within 10 min enabled the differentiation between growth within the planktonic and biofilm population and dispersion.
FIG. 2.
Time course microscopic images demonstrating the influence of a sudden increase in the nutrient concentration on P. aeruginosa PAO1 biofilms. Biofilms were grown in flow cells for 4 days in minimal medium containing 1.8 mM glutamate as the sole carbon source. Biofilm dispersion was induced by a sudden 10-fold increase of the glutamate concentration to 19.8 mM in the growth medium. The microscopic images show the same biofilm before (A) the nutrient-induced dispersion event and 10 min (B), 20 min (C), 30 min (D), 45 min (E), and 60 min (F) following the sudden increase in the nutrient concentration.
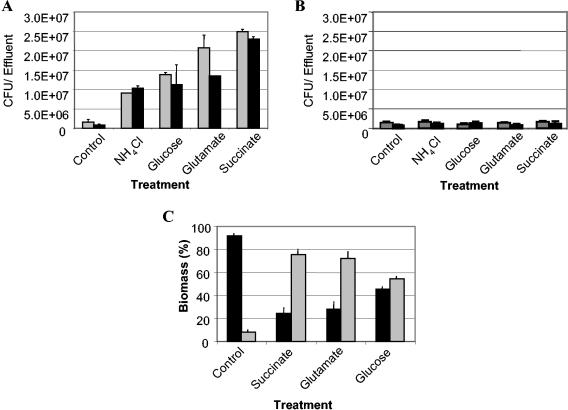
The effectiveness of biofilm dispersion is dependent on the carbon source.
Biofilm dispersion was also induced through the addition of alternate carbon substrates (glutamate, succinate, citrate, or glucose) and ammonium chloride to minimal medium containing either glutamate or glucose as the carbon source (Fig. 3A). While all tested carbon sources were found to induce dispersion under high-nutrient conditions, their effectiveness was variable. Dispersion efficacy was measured by determining the turbidity and the total number of bacteria (CFU) of dispersed and remaining biofilm cells. Untreated biofilms were used as controls. The biofilm mass postdispersion was determined by measuring the turbidity and total number of bacteria (CFU) of untreated biofilms. Treatment with 20 mM succinate was the most effective, followed by citrate (20 mM), glutamate (18 mM), and glucose (20 mM) (Fig. 3A). Since citrate acts as a chelating agent and has been demonstrated to affect overall biofilm structure, this carbon source was not used in subsequent experiments. Succinate-induced biofilm dispersion resulted in an ∼80% reduction in surface-associated biofilm biomass, while treatment with glutamate and glucose resulted in 72 and 54% reductions, respectively (Fig. 3C). Dispersion was also induced by a 10-fold increase in ammonium chloride concentration. This was most effective in growth media that lacked glutamate (carbon and nitrogen source) (Fig. 3A).
FIG. 3.
Influence of various carbon and nitrogen sources on the biofilm dispersion response of Pseudomonas aeruginosa PAO1 (A) and the Pseudomonas aeruginosa crc mutant PAO 8023 (B). Biofilms were grown in a biofilm tube reactor, composed of size 13 tubing with an internal volume of 1 ml (tubing length, 1 m; flow rate, 0.2 ml/min). After 4 days of biofilm growth in minimal medium containing 1.8 mM glutamate (gray bars), biofilm dispersion in P. aeruginosa PAO1 (A) and P. aeruginosa PAO8023 (B) was induced by a sudden increase in the nutrient concentration. The nutrients were added in the following concentrations: 20 mM succinate, 18 mM glutamate, 20 mM glucose, and 20 mM citrate. In addition, biofilm dispersion was induced by a sudden 10-fold increase in the ammonium chloride concentration. The effluent of untreated biofilms was used as controls. The dispersion experiment was repeated with minimal medium containing glucose as the sole carbon source (black bars). Dispersion was indicated by an increase of turbidity at 600 nm in the silicone tubing effluent. The total number of bacteria in the effluent was determined by serial plate counts on LB agar at 37°C. (C) Distribution of biomass of dispersed and remaining cells after nutrient-induced dispersion. Black bars, biomass of remaining biofilm cells; gray bars, biomass of dispersed cells. Biomass was determined by turbidity at 600 nm and by determining the number of viable cells. Untreated biofilms were used as controls.
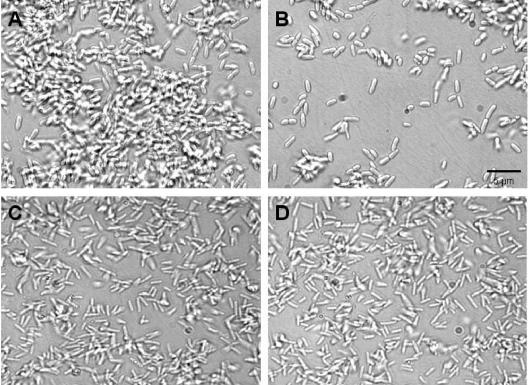
Interestingly, nutrient-induced biofilm dispersion events were observed not only for P. aeruginosa PAO1, but also for P. aeruginosa PA14 (data not shown), P. putida (data not shown), and a pilA mutant of P. aeruginosa (PA416) (Fig. 4A and B). In addition, a crc mutant of P. aeruginosa PAO 8023 with impaired catabolite repression capability (42) was tested for nutrient-induced biofilm dispersion. Recent findings indicated a role of the central carbon flux regulator crc in the formation of biofilms by P. aeruginosa (42). In P. aeruginosa, the Crc protein prevents the utilization of key sugars, such as glucose, when tricarboxylic acid (TCA) cycle intermediates are present. Disruption of crc was shown to dramatically decrease biofilm formation (42). By repeating the experiments with a crc mutant of P. aeruginosa PAO 8023, we showed that there was no dispersion response following changes in nutrient concentration (Fig. 4C and D). Neither the addition of alternate carbon substrates (glutamate, succinate, citrate, or glucose) nor the addition of ammonium chloride induced dispersion (Fig. 3B). The lack of a dispersion response may be related to a biofilm formation defect within the strain (42) resulting in a thin (<5 μm) unstructured biofilm (Fig. 4C and D).
FIG. 4.
Influence of sudden increase in the nutrient concentration on biofilms by two P. aeruginosa mutants, the pilA mutant P. aeruginosa PA416 (A, B) and the crc mutant P. aeruginosa PAO 8023 (C, D). Biofilms were grown in flow cells for 4 days in minimal medium containing 1.8 mM glutamate as the sole carbon source. Biofilm dispersion was induced by a sudden 10-fold increase of the glutamate concentration to 19.8 mM in the growth medium. (A) Intact biofilm of a P. aeruginosa pilA mutant (PA416); (B) remaining biofilm of P. aeruginosa pilA mutant (PA416) after nutrient-induced biofilm dispersion. The microscopic images in panels A and B show the same biofilm before (A) and after (B) nutrient-induced dispersion event. (C) Intact biofilm of a crc mutant of P. aeruginosa (PAO 8023); (D) remaining biofilm of a crc mutant of P. aeruginosa PAO 8023 after nutrient-induced biofilm dispersion.
Nutrient-induced dispersion of biofilm cells correlates with a switch in motility.
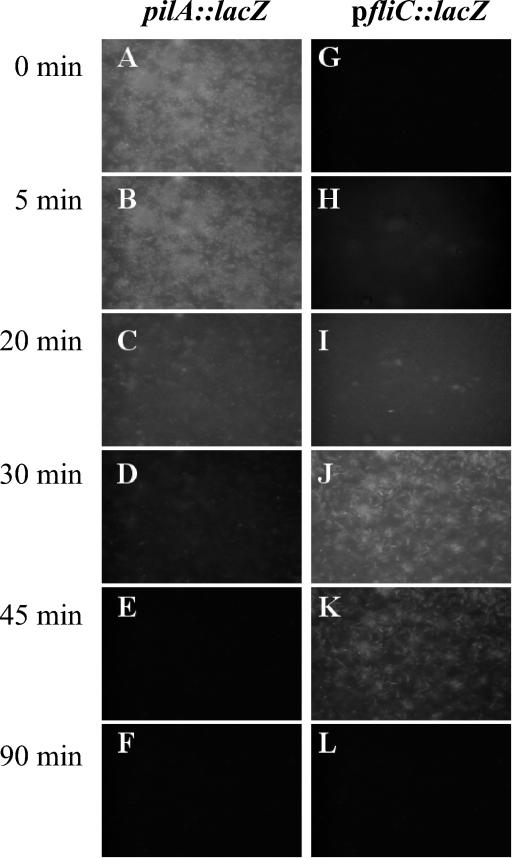
It has been suggested that an important change associated with biofilm formation by P. aeruginosa is the loss of flagella and the production of pili (43, 47). Since in biofilm dispersion, bacteria must actively escape from the biofilm matrix, we tested whether biofilm dispersion involved production of flagella and loss of pili. To accomplish this, we made use of two reporter strains, P. aeruginosa PAO1-pfliC::lacZ and a P. aeruginosa PA14 pilA mutant (P. aeruginosa PA416, pilA::lacZ) to monitor fliC and pilA gene expression during nutrient-induced biofilm dispersion. The pilA gene is required for type IV pilus biogenesis, while fliC is involved in flagellar biogenesis. Biofilms of PA416 and PAO1-pfliC::lacZ were grown for 4 days to obtain late-stage biofilms. Biofilm formation was accompanied by the activation of the pilA gene in PA416. Activation of pilA gene expression was monitored by the onset of lacZ reporter gene activity by fluorescence microscopy (Fig. 5). Within 30 min of induction of biofilm dispersion by increased glutamate, decreased pilA-dependent β-galactosidase activity was observed (Fig. 5) and the biofilm bacteria “twitched” prior to detaching (not shown). Disappearance of fluorescence indicated a decrease in pilA expression and thus a decrease in pilin production in the remaining biofilm population (Fig. 5). Collected dispersed cells were nonfluorescent (data not shown) and also showed decrease pilA-dependent β-galactosidase activity (Fig. 6).
FIG. 5.
Influence of nutrient-induced dispersion in P. aeruginosa biofilms on pilA::lacZ and pfliC::lacZ reporter gene expression over time. Biofilms were grown in flow cells for 4 days in minimal medium containing 1.8 mM glutamate as the sole carbon source. The microscopic images in the left panel show the same biofilm (pilA::lacZ) over the course of biofilm dispersion. The microscopic images in the right panel show the same biofilm (pfliC::lacZ) over the course of biofilm dispersion. Biofilm dispersion was induced by a sudden 10-fold increase of the glutamate concentration to 19.8 mM in the growth medium.
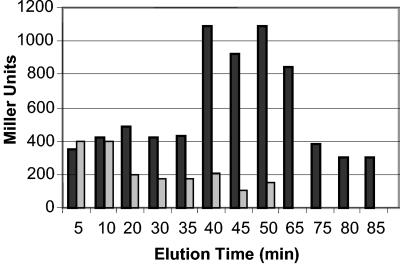
FIG. 6.
Determination of β-galactosidase activity with the Miller assay in dispersed biofilm cells. Biofilms were grown in flow cells for 4 days in minimal medium containing 1.8 mM glutamate as the sole carbon source. Biofilm dispersion was induced by a sudden 10-fold increase of the glutamate concentration to 19.8 mM in the growth medium. Dispersed cells were collected from the effluent from flow cells. Quantitation of β-galactosidase activity in the two reporter strains, P. aeruginosa pilA::lacZ and P. aeruginosa pfliC::lacZ transposon was carried out with the Miller assay (40). Gray bars, β-galactosidase activity of P. aeruginosa pilA::lacZ reporter strain; black bars, β-galactosidase activity of P. aeruginosa pfliC::lacZ reporter strain during nutrient-induced dispersion.
PAO1-pfliC::lacZ reporter strain cells fluoresced in the planktonic phase but lost this ability over the course of biofilm development. This indicated that flagellum production may have ceased upon surface attachment (Fig. 5). Within 30 min of induction of biofilm dispersion, PAO1-pfliC::lacZ biofilm cells became fluorescent and fliC-dependent β-galactosidase activity was detectable once again (Fig. 5). Activation of fliC-dependent β-galactosidase indicated reexpression of the fliC gene. fliC gene expression (or β-galactosidase activity) was detectable for approximately 30 min during the dispersion response, after which the residual biofilm cells returned to a nonfluorescent state (Fig. 5). The disappearance of fluorescence from the shocked biofilm could be attributed to fluorescent PAO1-fliC reporter strain cells leaving the surface, indicating that during biofilm dispersion, bacteria actively escape as single cells from the matrix (Fig. 2). Dispersed cells also showed increased β-galactosidase activity when tested with the Miller assay (Fig. 6).
Depletion of glutamate or a change in pH was insufficient to initiate significant dispersion as well as a notable change in biofilm pilA or fliC fluorescence. This suggested that transcription of genes involved in motility is regulated in response to a nutritional gradient: the greater the gradient the greater the change in transcription.
Differences in gene expression between biofilm and dispersed cells.
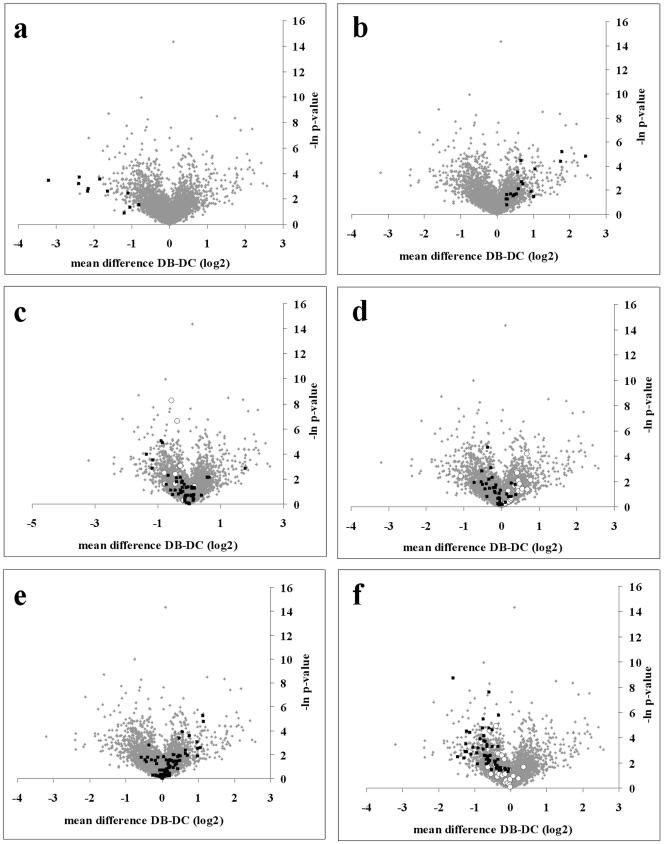
Microarray chips were used in triplicate to compare the patterns of gene expression for biofilm and dispersed cells. Following a direct comparison of the gene expression of dispersed and biofilm cells, the expression of 92 of a total of 5,549 gene sequences present on the Affymetrix Pseudomonas aeruginosa PA01 Genechip was shown to have altered (P value < 0.05). The most highly expressed genes in the dispersed and biofilm population of cells were determined, and those that passed a t test significance cutoff (P < 0.05) are shown in Tables 1 and 2. In the dispersed population, 47 genes had a greater than twofold change in expression (Table 1). The majority of the genes were related to cell growth and metabolism. (For example, levels of expression of five 30S ribosomal protein genes were increased.) A total of eight gene sequences were classified as hypothetical, and their function is currently unknown (Table 1). Additionally, seven gene sequences were classified as “probable” and their likely identities were based upon homology to proteins of known function in other related species. In addition, by filtering for changes in expression of groups of functionally related genes in the Affymetrix probe set description, distribution patterns showing trends in up- or down-regulation were clearly visible (Fig. 7A to F). For dispersed cells, various groups of related genes were shown to be significantly upregulated. These included those associated with the production of ATP synthase and kinases (Fig. 7C) and the expression of flagella (Fig. 7D). The expression of all 11 genes belonging to PF1 phage was also increased (Fig. 7A).
FIG. 7.
t test volcano plots showing the difference in gene expression of gene groups in biofilm and dispersed populations. Plots show (A) phage PF1 genes in dispersed cells; (B) nitric oxide, nitrate, and nitrite metabolism genes (nir, nos, and nor genes) expressed by biofilm cells; (C) ATP synthase (▪) and kinase genes (○); (D) expression of fimbria genes (○) and flagellar genes (▪); (E) expression of secreted factors by biofilm cells; and (F) expression of ribosomal protein genes (▪) and cell division genes (○). Global expression profiles of all probe sets on the microarrays are indicated by gray dots. DC, dispersed biofilm cells (cells that were dispersed from the biofilm); DB, biofilm remaining after nutrient-induced dispersion.
Microarray analysis of the biofilm population that remained after dispersion showed that 46 genes exhibited a greater than twofold increase in expression (Table 2). The functions of 15 of the 46-upregulated genes were unknown, and these were classified as hypothetical and possessing unknown function (Table 2). Furthermore, 15 genes were classified as “probable” and most-likely identities and functions were assigned. With respect to function, 65% of the genes that were significantly upregulated in the biofilm were ambiguous. Examination of the data from the remaining biofilm cells showed that cells within the biofilm also had increased expression of groups of functionally related genes. For example, genes that are involved in nitric oxide, nitrite, and nitrate metabolism (nir, nos, and nar) were induced significantly (P = 0.05) compared to dispersed cells (Fig. 7B). Additionally, genes that contain the description “secreted factors” (which include alginate biosynthesis genes) were predominantly upregulated in biofilm cells (Fig. 7E). As a functionally similar group, fimbriae were predominantly upregulated in biofilm cells and not in dispersed cells (Fig. 7D). Also of interest were genes that were involved in pyoverdine synthesis and six genes that are known to be involved in the synthesis of the virulence factor pyoverdine were upregulated (Table 2) (20, 39).
Differences in protein phosphorylation patterns between biofilm and dispersed cells.
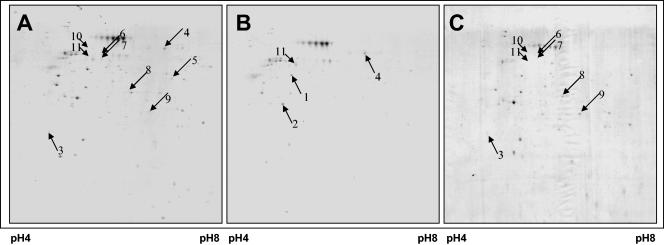
Protein phosphorylation is a ubiquitous process in prokaryotic cells. It alters the function of enzymes, ion channels, and other proteins in response to extracellular stimuli that include temperature, pH, and oxygen, as well as nitrogen and carbon sources. In order to study changes in protein phosphorylation during the forced dispersion of biofilms, a combination of approaches including functional proteomics and immunoprecipitation to enrich for Ser/Thr-phosphorylated proteins were used. Biofilms were grown for 4 days using a biofilm tube reactor after which dispersion was induced by sudden increases in the glutamate concentration. Steady-state biofilm cells were removed from the device and used as controls. 2D images representing immunoprecipitated Ser/Thr-phosphorylated proteins are shown in Fig. 8. Greater than 200 proteins were detected, per 2D image, including those proteins that originated from the polyclonal antibodies deployed. Interestingly, while the patterns of protein phosphorylation were similar between intact biofilms (before dispersion) and the dispersed cells (Fig. 8A and C)—overall, comparisons of the 2D images revealed differential phosphorylation of 15 proteins (7% change in the overall phosphorylation patterns)—dramatic changes were detected in the phosphorylation patterns of the residual biofilm after the dispersion events were complete. Under the conditions, the dispersion event was completed within 10 min. Comparisons of the 2D images revealed a more than threefold decrease in protein spot volume for more than 10 proteins in the residual biofilm populations compared to the phosphorylation patterns of dispersed cells. In addition, comparisons of the 2D images revealed a loss of more than 25 proteins in the residual biofilm population. Overall, these findings indicate that dephosphorylation of proteins had followed dispersion (Fig. 8). Four proteins, found to be less abundant in the residual biofilm population, were identified by MALDI-TOF mass spectrometry (Table 3). Among the identified proteins were elongation factor Tu, a GTP-binding protein that delivers aminoacyl-tRNA to the A site of the ribosome during protein synthesis (19), and ATP synthase α subunit (Table 3; Fig. 8). The latter protein belongs to the family of ATP synthases with ATP synthase α and β subunit family signature that includes the nucleotide-binding site for ATP and ADP. Interestingly, ATP synthase has recently been shown to be phosphorylated (33). In addition, two proteins that were more abundant in the remaining biofilm population compared to dispersed cells were identified by MALDI-TOF mass spectrometry as a probable helicase (PA0799) and a probable transcriptional regulator with AraC family signature (PA2047) (Table 3; Fig. 8). AraC belongs to the AraC/XylS family of prokaryotic positive transcriptional regulators (24). Members of the family are about 300 amino acids long and have three main regulatory functions in common: carbon metabolism, stress response, and pathogenesis (10). In addition, five proteins were identified that were absent in the phosphorylation patterns of remaining biofilms. Among the identified proteins were nitrate reductase β subunit NarH, involved in nitrate metabolism, phosphomannomutase AlgC, and the thiamine biosynthesis protein ThiC; the Tat (twin arginine translocation) pathway signal domain protein and the type III secretion protein, PscC, involved in protein translocation and secretion were also identified (Table 3). The bacterial twin-arginine translocation (Tat) pathway has been recently described for Bacillus subtilis PhoD, a phosphodiesterase containing a twin-arginine signal peptide, as being dependent on the pH gradient across the cytosolic membrane (45). The type III secretion protein PscC shares a bacterial type II and III secretion system protein domain and homologies to type IV pilus biogenesis proteins in P. aeruginosa, P. putida, and Vibrio cholerae.
FIG. 8.
Influence of nutrient-induced dispersion on P. aeruginosa protein phosphorylation patterns. Shown are 2D images of phosphorylated Ser/Thr proteins obtained from P. aeruginosa biofilms before (A) and after (B) the dispersion event. (C) 2D image showing phosphorylated Ser/Thr proteins from dispersed P. aeruginosa cells collected from the effluent. Arrows indicate proteins that were identified by MALDI-TOF mass spectrometry (see Table 3).
TABLE 3.
Identified proteins from 2D gels showing phosphorylated Ser/Thr-proteins obtained from P. aeruginosaa
| Spot no. | Protein identification | pI | Mol mass (kDa) | Change in spot volb | Ser/Thr phosphorylation sitec | ATP/GTP-binding sitec |
|---|---|---|---|---|---|---|
| 1 | ATP synthase, α chain, PA5556 | 5.3 | 55.38 | 3.0 | + | + |
| 2 | Elongation factor Tu, PA4265 | 5.2 | 43.35 | 4.4 | + | + |
| 3 | Biotin carboxylase, PA5436 | 5.6 | 32.37 | 3.2 | + | + |
| 4 | Probable transcriptional regulator, AraC-type DNA-binding domain-containing proteins, PA2047 | 9.3 | 36.6 | 0.3 | + | |
| 5 | Nitrate reductase β-subunit, PA3874 | 5.9 | 36.7 | NAd | + | |
| 6 | Thiamin biosynthesis protein ThiC, PA4973 | 5.9 | 69.6 | NA | + | |
| 7 | Tat (twin arginine translocation) pathway signal domain protein, PA3910 | 5.3 | 59.9 | NA | + | |
| 8 | Phosphomannomutase AlgC, PA5322 | 5.2 | 50.3 | NA | + | |
| 9 | Hypothetical protein, PA3513 | 6.9 | 35.1 | 3.3 | + | |
| 10 | Type III secretion protein PscC, PA1716 | 5.7 | 66.16 | NA | + | |
| 11 | Probable helicase, PA0799 | 8.5 | 47.2 | 0.3 | + | + |
Proteins were identified by peptide mass fingerprinting using tryptic digest and MALDI-TOF mass spectrometry.
Ratio between spot volume in dispersed biofilm cells versus that in remaining biofilm cells.
+, presence of a GTP/ATP binding site or potential Ser/Thr phosphorylation site.
NA, protein spots absent from the remaining biofilm cells.
The identified proteins were analyzed with respect to potential serine and threonine phosphorylation sites using the PhosphoBase v. 2.0 database (http://www.cbs.dtu.dk/databases/PhosphoBase/). All identified proteins harbored potential Ser/Thr phosphorylation sites (Table 3). Furthermore, the amino acids serine and/or threonine within these sites were conserved among members of the respective protein families (data not shown). In addition, several proteins harbored ATP- or GTP-binding sites (Table 3).
Inhibition of biofilm dispersion by phosphatase inhibitors.
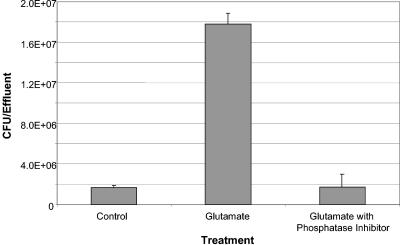
Since glutamate-induced biofilm dispersion resulted in dephosphorylation of Ser/Thr-phosphorylated proteins within the residual biofilm, we then investigated the effects of inhibiting dephosphorylation upon dispersion. Biofilms were allowed to form, as described earlier, and dispersed, as before, by increasing the glutamate concentration but in the presence and absence of various phosphatase inhibitors. Untreated biofilms and biofilms exposed to mixtures of the various phosphatase inhibitors alone served as controls. No dispersion events were detectable for those biofilms that were treated with high glutamate concentrations in the presence of phosphatase inhibitors (Fig. 9), indicating that posttranslational protein modifications such as protein dephosphorylation play an important role in the dispersion response. The results were independent of the type of biofilm tube reactor used.
FIG. 9.
Influence of phosphatase inhibitors on nutrient-induced dispersion in Pseudomonas aeruginosa biofilms. Biofilms were grown in a biofilm tube reactor, composed of size 13 tubing with an internal volume of 1 ml (tubing length, 1 m; flow rate, 0.2 ml/min) for 4 days in minimal medium containing 1.8 mM glutamate as the sole carbon source. Biofilms were then treated with a sudden 10-fold increase of the glutamate concentration or a sudden increase in the glutamate concentration in the presence of phosphatase inhibitors. Untreated biofilms or biofilms treated with phosphatase inhibitors alone were used as controls. Dispersion was indicated by an increase of turbidity at 600 nm in the silicone tubing effluent. The total number of bacteria in the effluent was determined by serial plate counts on LB agar at 37°C.
DISCUSSION
Continuous dispersal of cells from biofilm communities and their association with climactic collapse involve complex processes which are only beginning to be understood (30, 31, 54). It has been suggested that the transition from a planktonic (free-swimming) mode to growth as a biofilm occurs as a response to the availability of nutrients (9, 15, 38, 46). While nutrient-depleted environments enhance biofilm formation, it has been demonstrated that the availability of nutrients in high concentrations represses the formation of biofilms (15, 18, 31, 38). This finding has been supported by recent studies of B. subtilis, in which biofilm formation was inhibited by high glucose concentrations, probably through the catabolite control protein CcpA (54). Furthermore, differential expression of 40 genes responsive to glucose concentration has been observed in biofilms when grown in the presence of an elevated glucose concentration (54). Similar conclusions were drawn for S. mutans (60) and E. coli (31) when grown under stagnant biofilm growth conditions. Overall, these studies indicate that the formation of biofilms is a complex process of microbial development in response to a decreased availability of nutrients. Based on our findings, however, it appears that biofilm accretion might be regulated indirectly by biofilm dispersion through the availability of nutrients. Thus, the availability of nutrients plays a major role in biofilm dispersion by regulating the formation of new biofilms and limiting them to periods when nutrients are in excess. This is consistent with earlier findings by James et al. (32). The authors demonstrated that biofilm dispersion is induced by a change in the nutrient composition, by a step change from a minimal to a complex medium (32). The finding of massive dispersion events under favorable growth conditions is consistent with current models of biofilm development and the biofilm survival strategy whereby bacteria only benefit nutritionally from the biofilm mode of growth at low nutrient concentrations or under unfavorable growth conditions but abandon this mode of growth when conditions in the bulk liquid become favorable (9, 15, 38, 46).
Regulation of the central carbon flux and catabolism appears to play a major role in biofilm formation (31, 42). Recent findings indicate a major role of the regulation of the central carbon flux and catabolism in formation of biofilms in P. aeruginosa. In P. aeruginosa, the catabolite repression control (Crc) protein has been shown to be essential for biofilm formation and prevents the utilization of key sugars, such as glucose, when TCA cycle intermediates are present (42). Disruption of crc was shown to dramatically decrease biofilm formation, indicating an essential role of Crc for biofilm formation (42). The authors concluded that nutritional cues are integrated by Crc as part of a signal transduction pathway that regulates biofilm development, possibly by controlling the transcription of genes required for type IV pilus biogenesis (42). Our findings indicate that P. aeruginosa biofilm dispersion can be induced by various nutrients, including key sugars and TCA cycle intermediates. Biofilm dispersion was found to be independent of the presence of both glucose and TCA cycle intermediates (see Fig. 3) and independent of the expression of pilin structural genes since the pilA mutant P. aeruginosa (PA416) underwent dispersion in response to high-nutrient conditions (Fig. 4). DNA microarray analysis did not support notions of differential gene expression of the crc gene during the dispersion response.
DNA microarray analysis of the dispersed and residual biofilm populations did however demonstrate a specific dispersal phenotype. Direct comparison of the gene expression of dispersed and biofilm cells revealed the altered expression of 92 of a total of 5,549 gene sequences. Furthermore, DNA microarray analysis demonstrated that dispersion involves changes in expression of whole gene families, such as those involved in pilus and flagellum biosynthesis. The analysis clearly demonstrated that phenotypic adaptation towards dispersion in response to environmental changes correlated with a switch in motility with an increased expression of flagellum biosynthesis genes and a decreased expression of pilus biosynthesis genes. In addition, using transcriptional reporter strains combined with fluorescent microscopy, we demonstrated that dispersion correlated with an increased fliC transcription and decreased pilA transcription. Cells remaining within the biofilm, however, did not upregulate transcription of the fliC gene but instead transcribe pilA. Thus, while biofilm formation is associated with the loss of flagella and the production of pili (43, 47), biofilm dispersion is clearly associated with the loss of pili and the production of flagella. Since biofilm dispersion is independent of the presence of type IV pili (P. aeruginosa PA416 does disperse; see Fig. 4), we suggest, as described by Dow et al. (21), that flagellum-driven motility is a major release mechanism for the active escape of bacteria from the biofilm, thus enabling bacteria to actively escape from the biofilm matrix. Furthermore, our finding of flagellum-driven motility as a major release mechanism are consistent with the differential phosphorylation of ATP synthase α subunit during the dispersion response (Fig. 8). ATP synthase is associated with flagella and flagellar rotation by providing proton motive force via the generation of ATP. This finding is consistent with the increased expression of genes involved in energy metabolism in dispersed cells (Table 1). Flagellum-driven motility as a major release mechanism may also indicate a role of chemotaxis in the dispersion response by sensing chemical gradients or changes in the availability to nutrients in the environment.
Several recent publications have implied that biofilm encounter low levels of oxygen (6, 50, 63). Under such unfavorable conditions, P. aeruginosa is able to use nitrate for both assimilation and anaerobic respiration (35) and a variety of gene families (nir, nos, nor, and cytochrome families) are used. Our finding of increased expression of many nir, nos, nor, and cytochrome genes by cells within the biofilm suggests that P. aeruginosa biofilms are exposed to anaerobic conditions (Tables 1 and 2). The finding is consistent with recent reports by Carterson et al. (11). The identification of nitrate reductase β subunit, NarH, and the hypothetical protein probably involved in nitrate transport from immunoprecipitated Ser/Thr-phosphorylated protein patterns of intact and dispersed biofilm cells is consistent with our finding (Table 3; Fig. 8). Interestingly, nitric oxide reductase is required to modulate and prevent accumulation of toxic nitric oxide, a by-product of anaerobic respiration, and its expression has been noted in P. aeruginosa biofilms in the lung of cystic fibrosis patients (28, 65). As such, in natural situations, the expression of nitrate reductase and certain cytochromes seems to be a biofilm-specific response (14). The dispersal phenotype clearly correlated with a decreased expression of genes associated with the metabolism of nitrogenous materials indicating a reversion of the biofilm phenotype to the planktonic mode of growth. Reversion to the planktonic mode of growth was also apparent by the increased expression of genes involved in the production of phage PF1 from PF1 prophage within the P. aeruginosa genome (dispersed cells). Phage PF1 genes have previously been shown to be upregulated in biofilm cells (61) and dispersing cells (59). Webb et al. (59) proposed that prophage-mediated cell death is an important mechanism for cellular differentiation inside biofilms that facilitates dispersal of a subpopulation of surviving cells. This is consistent with previous findings by Sauer et al. (50) demonstrating the expression of phage proteins in planktonic and dispersed cells, but not in mature, intact biofilm cells. The actual role of PF1 phage in dispersal is still not clear, although it is likely that the increase in metabolism that is associated with dispersion may well initiate upregulation of the prophage genes (59). The finding of a specific dispersal phenotype was also suggested by the proteomic analysis of protein phosphorylation patterns of residual biofilm and dispersed cells. More importantly, we demonstrated a role of posttranslational modification via protein phosphorylation in the dispersion response by demonstrating that inhibition of dephosphorylation prevents the dispersion event. This finding may indicate a role of posttranslational modification in the transduction of nutritional signals that trigger dispersion of P. aeruginosa biofilms.
While some of the genes and gene families have been characterized, many of the genes expressed in the biofilm have yet to be characterized. Their function is unknown, and this is probably a consequence of most studies being conducted on planktonic populations (14). Such a finding is not surprising but adds further support for the need to study biofilm populations, especially as >99% of all bacteria preferentially exist in biofilms (15, 26).
In conclusion, the work presented here demonstrates that biofilm bacteria respond to their outside environment by phenotypic adaptation, probably through a variety of complex regulatory networks that are involved in nutrient-driven dispersal events. Dispersal events lead inexorably to a massive disaggregation of the biofilm matrix and a significant change in structure of P. aeruginosa biofilms. Furthermore, the work here reinforces the concept that bacteria within a biofilm possess a “biofilm-specific phenotype” (37, 50) that is different from that of dispersed biofilm cells and that the phenotypic biofilm characteristics are in part due to posttranslational protein modifications rather than differential gene and protein expression. While we are aware that the use of nutrients for biofilm dispersal is not suitable for commercial applications, the information gained on differential gene expression and posttranslational modifications has shed light on the mechanisms that lead to biofilm dispersion. An understanding of signal transduction in biofilm dispersion by screening for differential gene expression and phosphorylated proteins associated with dispersion may provide new targets for the treatment of biofilm infections and might ultimately lead to novel approaches to biofilm control.
Acknowledgments
This work was supported by grants from the National Institutes of Health (HL073835-01 and A1055521-01) and the National Science Foundation (0311307).
We thank Andrew Hayes and Leanne Wardleworth for Microarray technical support. We thank George A. O'Toole for providing the strains P. aeruginosa PA416 and PAO 0823 and Herbert P. Schweizer for sharing bacterial strains and plasmids, technical advice, and protocols for the generation of the pfliC::lacZ reporter strain in P. aeruginosa.
REFERENCES
- 1.Allison, D. G., D. J. Evans, M. R. W. Brown, and P. Gilbert. 1990. Possible involvement of the division cycle in dispersal of Escherichia coli from biofilms. J. Bacteriol. 172:1667-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Applegate, D. H., and J. D. Bryers. 1991. Effects on carbon and oxygen limitations and calcium concentrations on biofilm removal processes. Biotechnol. Bioeng. 37:17-25. [DOI] [PubMed] [Google Scholar]
- 3.Bagge, N., M. Schuster, M. Hentzer, O. Ciofu, M. Givskov, E. P. Greenberg, and N. Hoiby. 2003. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and β-lactamase and alginate production. Antimicrob. Agents Chemother. 48:1175-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker, C. S., I. Morozov, K. Suzuki, T. Romeo, and P. Babitzke. 2002. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol. Microbiol. 44:1599-1610. [DOI] [PubMed] [Google Scholar]
- 5.Becher, A., and H. P. Schweizer. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. BioTechniques 29:948-952. [DOI] [PubMed] [Google Scholar]
- 6.Beenken, K. E., P. M. Dunman, F. McAleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins, and M. S. Smeltzer. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186:4665-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blum, H., H. Beier, and H. J. Gross. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93-99. [Google Scholar]
- 8.Bolstad, B. M., R. A. Irizarry, M. Astrand, and T. P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185-193. [DOI] [PubMed] [Google Scholar]
- 9.Bowden, G. H., and Y. H. Li. 1997. Nutritional influences on biofilm development. Adv. Dent. Res. 11:81-99. [DOI] [PubMed] [Google Scholar]
- 10.Bustos, S. A., and R. F. Schleif. 1993. Functional domains of the AraC protein. Proc. Natl. Acad. Sci. USA 90:5638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carterson, A. J., K. Sauer, M. R. Parsek, D. J. Hassett, J. R. Schurr, A. Frisk, and M. J. Schurr. 2004. Pseudomonas aeruginosa requires the anaerobic transcriptional regulators anr and dnr for biofilm formation. Abstr. 104th Gen. Meet. Am. Soc. Microbiol., abstr. J.-033, p. 354. American Society for Microbiology, Washington, D.C.
- 12.Chen, X., and P. S. Stewart. 2000. Biofilm removal caused by chemical treatments. Water Res. 34:4229-4233. [Google Scholar]
- 13.Choi, Y. C., and E. Morgenroth. 2003. Monitoring biofilm detachment under dynamic changes in shear stress using laser-based particle size analysis and mass fractionation. Water Sci. Technol. 47:69-76. [PubMed] [Google Scholar]
- 14.Costerton, J. W. 2002. Anaerobic biofilm infections in cystic fibrosis. Mol. Cell 10:699-700. [DOI] [PubMed] [Google Scholar]
- 15.Costerton, J. W., Z. Lewandowski, D. Caldwell, D. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 16.Costerton, J. W., Z. Lewandowski, D. DeBeer, D. Caldwell, D. Korber, and G. James. 1994. Biofilms, the customized microniche. J. Bacteriol. 176:2137-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalaquis, P. J., D. E. Caldwell, J. R. Lawrence, and A. R. McCurdy. 1989. Detachment of Pseudomonas fluorescens from biofilms on glass surfaces in response to nutrient stress. Microb. Ecol. 18:199-210. [DOI] [PubMed] [Google Scholar]
- 18.Danhorn, T., M. Hentzer, M. Givskov, M. R. Parsek, and C. Fuqua. 2004. Phosphorus limitation enhances biofilm formation of the plant pathogen Agrobacterium tumefaciens through the PhoR-PhoB regulatory system. J. Bacteriol. 186:4492-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daviter, T., H. J. Wieden, and M. V. Rodnina. 2003. Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J. Mol. Biol. 332:689-699. [DOI] [PubMed] [Google Scholar]
- 20.Déziel, E., Y. Comeau, and R. Villemur. 2001. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183:1195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dow, J. M., L. Crossman, K. Findlay, Y. Q. He, J. X. Feng, and J. L. Tang. 2003. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. USA 100:10995-11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drenkard, E. 2003. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 5:1213-1219. [DOI] [PubMed] [Google Scholar]
- 23.Fux, C. A., S. Wilson, and P. Stoodley. 2004. Detachment characteristics and oxacillin resistance of Staphyloccocus aureus biofilm emboli in an in vitro catheter infection model. J. Bacteriol. 186:4486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallegos, M.-T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geesey, G. G., W. T. Richardson, H. G. Yeomans, R. T. Irvin, and J. W. Costerton. 1977. Microscopic examination of natural sessile bacterial populations from an alpine stream. Can. J. Microbiol. 23:1733-1736. [DOI] [PubMed] [Google Scholar]
- 26.Gilbert, P., T. Maira-Litran, A. J. McBain, A. H. Rickard, and F. W. Whyte. 2002. The physiology and collective recalcitrance of microbial biofilm communities. Adv. Microb. Physiol. 46:202-256. [PubMed] [Google Scholar]
- 27.Görg, A., C. Obermaier, G. Boguth, A. Harder, B. Scheibe, R. Wildgruber, and W. Weiss. 2000. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 6:1037-1053. [DOI] [PubMed] [Google Scholar]
- 28.Hassett, D. J., J. Cuppoletti, B. Trapnell, S. V. Lymar, J. J. Rowe, S. S. Yoon, G. M. Hilliard, K. Parvatiyar, M. C. Kamani, D. J. Wozniak, S. H. Hwang, T. R. McDermott, and U. A. Ochsner. 2002. Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv. Drug Deliv. Rev. 54:1425-1443. [DOI] [PubMed] [Google Scholar]
- 29.Hoang, T. T., A. J. Kutchma, A. Becher, and H. P. Schweizer. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression. Plasmid 43:59-72. [DOI] [PubMed] [Google Scholar]
- 30.Jackson, D. W., J. W. Simecka, and T. Romeo. 2003. Catabolite repression of Escherichia coli biofilm formation. J. Bacteriol. 184:3406-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.James, G. A., D. R. Korber, D. E. Caldwell, and J. W. Costerton. 1995. Digital image analysis of growth and starvation responses of a surface-colonizing Acinetobacter sp. J. Bacteriol. 177:907-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanekatsu, M., H. Saito, K. Motohashi, and T. Hisabori. 1998. The beta subunit of chloroplast ATP synthase (CF0CF1-ATPase) is phosphorylated by casein kinase II. Biochem. Mol. Biol. Int. 46:99-105. [DOI] [PubMed] [Google Scholar]
- 34.Kerr, C. J., K. S. Osborn, A. H. Rickard, G. D. Robson, and P. S. Handley. 2003. Biofilms in water distribution systems, p. 757-776. In M. Duncan and N. J. Horan (ed.), Water and wastewater engineering. Academic Press, London, United Kingdom.
- 35.Kerschen, E. J., V. R. Irani, D. J. Hassett, and J. J. Rowe. 2001. snr-1 gene is required for nitrate reduction in Pseudomonas aeruginosa PAO1. J. Bacteriol. 183:2125-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khoury, A. E., K. Lam, B. Ellis, and J. W. Costerton 1992. Prevention and control of bacterial infections associated with medical devices. ASAIO J. 38:M174-M178. [DOI] [PubMed] [Google Scholar]
- 37.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 38.Marshall, J. C. 1988. Adhesion and growth of bacteria at surfaces in oligotrophic habitats. Can. J. Microbiol. 34:503-506. [Google Scholar]
- 39.Meyer, J.-M., A. Neely, A. Stintzi, C. Georges, and I. A. Holder. 1996. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 64:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 41.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 42.O'Toole, G. A., K. A. Gibbs, P. W. Hager, P. V. Phibbs, Jr., and R. Kolter. 2000. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 182:425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 44.Picioreanu, C., M. C. van Loosdrecht, and J. J. Heijnen. 2001.Two-dimensional model of biofilm detachment caused by internal stress from liquid flow. Biotechnol. Bioeng. 72:205-218. [PubMed] [Google Scholar]
- 45.Pop, O., U. Martin, C. Abel, and J. P. Muller. 2002. The twin-arginine signal peptide of PhoD and the TatAd/Cd proteins of Bacillus subtilis form an autonomous Tat translocation system. J. Biol. Chem. 277:3268-3273. [DOI] [PubMed] [Google Scholar]
- 46.Pratt, L. A., and R. Kolter. 1999. Genetic analyses of bacterial biofilm formation. Curr. Opin. Microbiol. 2:598-603. [DOI] [PubMed] [Google Scholar]
- 47.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romeo, T., M. Gong, M. Y. Liu, and A.-M. Brun-Zinkernagel. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 175:4744-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sabnis, N., H. Yang, and T. Romeo. 1995. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J. Biol. Chem. 270:29096-29104. [DOI] [PubMed] [Google Scholar]
- 50.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sauer, K., and A. K. Camper. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183:6579-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sawyer, L. K., and S. W. Hermanowicz. 1998. Detachment of biofilm bacteria due to variations in nutrient supply. Water Sci. Technol. 37:211-214. [Google Scholar]
- 53.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 54.Stanley, N. R., R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2003. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J. Bacteriol. 185:1951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 56.Stoodley, P., L. Hall-Stoodley, J. D. Boyle, H. M. Lappin-Scott, and J. W. Costerton. 2001. Growth and detachment of cell clusters from mature mixed-species biofilms. Appl. Environ. Microbiol. 67:5608-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stover, C. K., X.-Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 58.Vats, N., and S. F. Lee. 2000. Active detachment of Streptococcus mutans cells adhered to epon-hydroxylapatite surfaces coated with salivary proteins in vitro. Arch. Oral Biol. 45:305-314. [DOI] [PubMed] [Google Scholar]
- 59.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 62.Wimpenny, J. W. T. 2000. An overview of biofilms as functional communities, p. 1-24. In D. G. Allison, P. Gilbert, H. M. Lappin-Scott, and M. Wilson (ed.). Community structure and co-operation in biofilms. Cambridge University Press, Cambridge, United Kingdom.
- 63.Xu, K. D., P. S. Stewart, F. Xia, C.-T. Huang, and G. A. McFeters. 1989. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64:4035-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang, H., M. Y. Liu, and T. Romeo. 1996. Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J. Bacteriol. 178:1012-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoon, S. S., R. F. Hennigan, G. M. Hilliard, U. A. Ochsner, K. Parvatiyar, M. C. Kamani, H. L. Allen, T. R. DeKievit, P. R. Gardner, U. Schwab, J. J. Rowe, B. H. Iglewski, T. R. McDermott, R. P. Mason, D. J. Wozniak, R. E. Hancock, M. R. Parsek, T. L. Noah, R. C. Boucher, and D. J. Hassett. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3:593-603. [DOI] [PubMed] [Google Scholar]