Abstract
In Escherichia coli the response regulator SprE (RssB) facilitates degradation of the sigma factor RpoS by delivering it to the ClpXP protease. This process is regulated: RpoS is degraded in logarithmic phase but becomes stable upon carbon starvation, resulting in its accumulation. Because SprE contains a CheY domain with a conserved phosphorylation site (D58), the prevailing model posits that this control is mediated by phosphorylation. To test this model, we mutated the conserved response regulator phosphorylation site (D58A) of the chromosomal allele of sprE and monitored RpoS levels in response to carbon starvation. Though phosphorylation contributed to the SprE basal activity, we found that RpoS proteolysis was still regulated upon carbon starvation. Furthermore, our results indicate that phosphorylation of wild-type SprE occurs by a mechanism that is independent of acetyl phosphate.
Two-component systems, comprised of a sensor kinase and a response regulator, are commonly used by bacteria to sense and respond to environmental signals. Typically, a phosphotransfer is employed for signal transduction. In a canonical system, such as EnvZ/OmpR in Escherichia coli, the histidine kinase autophosphorylates in response to an environmental signal and then transfers the phosphate to a conserved aspartic acid in the N-terminal domain of the response regulator (11). Once activated by phosphorylation, OmpR and a majority of response regulators modulate transcription of effector genes through the activity of a C-terminal output domain. In most cases examined to date, signal transduction is blocked by mutations that alter either the conserved histidine of the kinase or the conserved aspartic acid of the response regulator (11).
A more unusual E. coli response regulator is SprE (RssB), where the C-terminal effector domain works as a proteolytic facilitator rather than as a transcription factor. SprE binds to the stationary-phase sigma factor RpoS, which is responsible for the transcription of genes involved in protecting the cell from a variety of stresses (10). By forming a quaternary complex with the ClpXP protease, SprE facilitates RpoS degradation (13, 30). Moreover, RpoS proteolysis is regulated: RpoS is rapidly degraded by ClpXP in exponentially growing cells, but degradation stops as cells enter stationary phase (14, 29). This regulation is thought to be mediated by changes in SprE activity since the levels and activity of ClpXP are invariant (24, 30).
Changes in RpoS stability occur when E. coli experiences carbon starvation, a condition that bacteria frequently endure in nature (14, 29), and SprE regulates this stabilization. Strains lacking SprE have elevated levels of RpoS that do not increase further during the transition from logarithmic growth to stationary phase (17, 19). RpoS is also stabilized during osmotic shock, high cell density, and starvation for other nutrients like ammonia and phosphate, although regulated synthesis may also contribute to RpoS levels under these conditions (10).
Although the C-terminal output domain of SprE is unique among response regulators, its N-terminal domain is highly conserved, including the aspartic acid at position 58 that is crucial for signal transduction. Phosphorylation of this residue is a critical feature of response regulator control, and available evidence suggests that SprE is no exception. Biochemical studies have demonstrated that Asp58 is the only site of SprE phosphorylation (3) and that SprE-P is considerably more active than SprE in vitro (13, 30). In vivo experiments with both E. coli and Salmonella enterica serovar Typhimurium have also demonstrated that when the site of SprE phosphorylation is mutated, RpoS levels increase (2, 6, 16). However, several issues complicate these studies. First, multicopy plasmids were used to express the mutant SprE protein in these studies, and thus interpretation of the results is complicated by the antisigma activity of SprE; SprE can bind and sequester RpoS, and this activity is quite pronounced when SprE levels are high (2). Second, Asp58 of SprE was replaced with several different amino acids, Pro, Arg, Asp, and Gln, and each of these amino acid replacements affected RpoS levels differently. All these mutations block SprE phosphorylation, but since they confer different phenotypes they must also affect aspects other than phosphorylation (2, 16). Finally, these in vivo studies did not examine how these mutations affected signal transduction under starvation conditions (1, 2, 6). It has nevertheless been postulated that SprE phosphorylation orchestrates changes in RpoS levels in response to environmental conditions. In particular, it has been suggested that when nutrients are abundant SprE is phosphorylated to facilitate RpoS degradation, while SprE dephosphorylation signals starvation and allows RpoS to accumulate (2, 10, 12, 16, 20).
SprE is an orphan response regulator, and despite many searches, no specific kinase for SprE has been found (6, 23). To better understand the role of phosphorylation in starvation signal transduction, we mutated the conserved aspartic acid (D58) to alanine on the chromosomal sprE gene. If SprE phosphorylation was the starvation signal for controlling RpoS stability, then we would expect RpoS stability and levels in this mutant to remain constantly high under all conditions, including starvation, just as they do in the sprE null strain (19). Instead we found that the RpoS response to carbon starvation in the mutant was remarkably similar to that observed in the wild type. SprE phosphorylation is important for setting the steady-state levels of RpoS, particularly when SprE levels are low during rapid growth, but it does not play an important role in signaling starvation for carbon.
MATERIALS AND METHODS
Bacterial strains.
Strains are listed in Table 1. Standard microbiological techniques were used for strain construction (25). The sprED58A allele from HME6mutSsprED58A was introduced by P1 transduction into MC4100 trpC::tet, since trpC is linked to sprE, by selecting on minimal glucose medium and then screening for the loss of the EcoRV site, which is present only in the wild-type sprE allele (see below). The Δ(ackA-pta) allele is linked to zej223::Tn10 and was introduced by P1 transduction into various strains by selection on tetracycline and screening for lack of growth on acetate minimal medium.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | 4 |
| CNP119 | MC4100 λφ(rpoS750′-′lacZ) | 26 |
| CNP153 | CNP119 sprE::tet | This study |
| CNP209 | CNP119 sprED58A | This study |
| CNP211 | CNP119 Δ(ackA-pta) zej223::Tn10 | This study |
| CNP212 | CNP211 sprED58A | This study |
| CNP217 | CNP119 trpC::tet | This study |
| HME6mutS | W3110 Δ(argF-lac)U169 ΔmutS::amp galKTYR145UAG λc1857Δ(cro-bioA) | 5 |
Media and growth conditions.
Luria-Bertani (LB) and minimal glucose M63 media were prepared as described previously (25). For nitrogen starvation, minimal M63 was made without ammonia. For phosphate starvation, morpholinepropanesulfonic acid minimal medium was made without phosphate as described previously (23). Bacteria were grown at 37°C unless otherwise noted.
λ Red-mediated recombination.
Homologous recombination with single-stranded oligonucleotides was carried out as described previously (5). The lagSprED58 oligonucleotide (CAGTTTAAGCCCGTTCATTCGTGGCATCGCGATAGCACATATCATCAGGTCTGGAGTGAAACCTCCCAGC) was used to change the Asp58 to Ala. This mutation also disrupted an EcoRV site (disrupted site is underlined). After the expression of the λ Red genes of HME6mutS was induced, 5 pmol of the lagSprED58A oligonucleotide was used for electroporation. Electroporated cells were diluted and grown on LB agar at 32°C. Individual colonies were screened for mixed recombinants by partial loss of an EcoRV site as determined by EcoRV (New England Biolabs) digestion of PCR products that amplify the entire sprE open reading frame. Mixed recombinant colonies were purified to allow segregation, and then individual colonies were screened for complete loss of the EcoRV site. The nucleotide exchange was confirmed by DNA sequencing by the Princeton University Department of Molecular Biology Synthesis and Sequencing Facility.
Starvation and exit-from-starvation experiments.
For gradual carbon starvation, cultures were grown overnight in M63 medium with 0.4% glucose and then diluted 1:100 in M63 with 0.01% glucose. Starting at an optical density at 600 nm (OD600) of 0.1 and continuing until the OD600 plateaued at 0.5, samples were taken approximately every 30 min and prepared for Western blot analysis as described below. For immediate carbon and nitrogen starvations, cultures were grown overnight in M63 with 0.4% glucose and then diluted 1:100 in fresh medium. When the OD600 reached somewhere between 0.3 and 0.4, logarithmic-phase samples were taken to be processed for Western blot analysis as described below. The remaining cells were pelleted for 7 min in 15-ml conical tubes at 1,800 × g in a Durafuge 200 centrifuge (Precision Scientific), washed once in prewarmed medium lacking the appropriate nutrient, and incubated in prewarmed starvation medium with aeration at 37°C (23). Samples for Western blot analysis were taken at times indicated. For exit-from-starvation experiments, cultures were grown overnight in M63 with 0.01% glucose. After confirmation that the OD600 had plateaued at 0.3, an aliquot was removed and the half-life of RpoS was determined as indicated below. Glucose was then added to a final concentration of 0.4%, and the cells were allowed to resume growth at 37°C while being shaken for 1 min. After 1 min, the half-life of RpoS was determined as described below.
Stability assays.
The Western blot sample “t = 0 min” was taken before the addition of chloramphenicol (1). Then, chloramphenicol was added to a final concentration of 1.15 mg/ml and subsequent samples were taken at the times indicated, while the cultures were incubated at 37°C. The 1-ml samples were added directly to 50 μl of cold trichloroacetic acid and chilled on ice for over 3 h. Next, the samples were pelleted at 4°C, washed with 500 μl of cold acetone, and then resuspended in sodium dodecyl sulfate loading buffer (25) in a volume equal to an OD600/10 and subjected to electrophoresis and Western blot analysis.
Western blot analysis.
Except for stability assay samples, 1-ml samples were prepared by pelleting whole cells and immediately resuspending them in sodium dodecyl sulfate loading buffer (25) in a volume in milliliters equal to an OD600/10. After resuspension in loading buffer, all samples from the half-life and other experiments were treated in the same fashion. After boiling for 10 min, equal volumes were loaded onto 12% polyacrylamide gels. After electrophoresis, proteins were transferred onto nitrocellulose membranes and probed with either a 1:6,000 dilution of anti-RpoS antibody (our laboratory stock) or a 1:4,000 dilution of SprE antibody (our laboratory stock). For secondary antibody, donkey anti-rabbit immunoglobulin G-horseradish peroxidase conjugate (Amersham Pharmacia Biotech) was used at a 1:6,000 dilution. The bands were detected with the ECL antibody detection kit (Amersham Pharmacia Biotech) and XAR film (Kodak). The intensity of the bands was analyzed with gel image analysis software (Kodak 1D Image Analysis Software or ImageJ Software).
RESULTS
Changes in SprE phosphorylation do not signal carbon starvation.
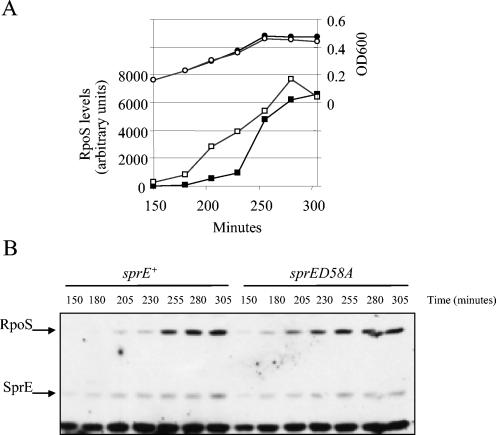
RpoS is thought to accumulate upon carbon starvation because SprE becomes inactive (10, 12, 14, 19). To better understand the role of phosphorylation in vivo, we mutated the Asp58 codon to alanine in the chromosomal allele of sprE, using the λ Red recombination method described by Costantino and Court (5). The conserved aspartic acid residue is the phosphorylation site found in response regulators, and in vitro work has shown that when it is mutated SprE is no longer phosphorylated (3). The mutant strain grew at the same rate as the wild-type isogenic parent in all media used (Fig. 1A compares the growth of the two strains in minimal glucose medium). Western blot analysis (Fig. 1B) also revealed that the D58A mutation does not affect SprE stability. Indeed, SprED58A levels increase concurrently with RpoS levels, as expected if the SprE/RpoS regulatory feedback loop is intact (9, 20, 22).
FIG. 1.
Phosphorylation state of SprE does not regulate accumulation of RpoS in response to gradual carbon starvation. (A) Growth in glucose starvation medium is unaffected in the strain carrying sprED58A (open circles) when compared to the wild type (closed circles). Quantification of RpoS levels from the Western blot shown in panel B is shown. Solid squares, wild-type strain; open squares, mutant strain. (B) Western blot analysis of RpoS and SprE levels during gradual carbon starvation. Samples from a strain carrying either wild-type sprE or the sprED58A allele were prepared during growth in a limiting amount of glucose as described in Materials and Methods. sprE+ refers to strain CNP119, and sprED58A refers to CNP209 (Table 1).
In order to monitor RpoS levels following carbon starvation in strains expressing either wild-type sprE (sprE+) or mutant sprE (sprED58A), cultures from both strains were grown in a limiting amount of glucose (0.01%) so that cell growth would stop early in logarithmic phase (OD600, ∼0.5). This is well below the optical density at which RpoS would accumulate if glucose was not limiting, demonstrating that the increase in RpoS levels in these cultures is not due to density-dependent effects but is rather the result of the depletion of the carbon source. As expected, RpoS levels in the sprE+ background increased as the optical density plateaued (Fig. 1B). Surprisingly, RpoS levels followed the same pattern in the sprED58A background despite modestly increased basal levels in exponential cells (see below). This response to starvation is not what would have been expected if changes in SprE phosphorylation signal starvation. That model would have predicted elevated and unchanging RpoS levels similar to those seen in the signal-blind sprE null mutant (19).
Carbon starvation was also achieved by directly resuspending exponentially growing cells in medium lacking glucose. This procedure arrests growth immediately so we can compare RpoS levels at the same optical density in both exponentially growing and starved cells. RpoS levels were determined by Western blotting before and 1 h after starvation. In both the wild-type and mutant strains, RpoS levels increased substantially upon starvation (Fig. 2, compare log and starved t = 0). In summary, in the presence of either SprE+ or SprED58A, RpoS levels responded in the same manner to carbon starvation.
FIG. 2.
RpoS stability increases in response to carbon starvation independently of SprE phosphorylation. The half-life of RpoS was measured by Western blot analysis of samples taken before (time zero) and after the addition of chloramphenicol. Samples were taken from cells growing exponentially and from cells that had been immediately starved for carbon for 1 h. In both wild-type sprE (A) and sprED58A (B) backgrounds, RpoS is unstable during log phase and then its levels increase and it becomes stabilized upon glucose starvation. Likewise, RpoS-LacZ stability is regulated in a fashion similar to that of RpoS in the sprED58A strain (C). Above each lane, time (in minutes) after the addition of chloramphenicol is shown.
It has previously been reported that RpoS becomes dramatically stabilized upon carbon starvation (14, 29). We therefore tested whether this process was affected in the sprED58A mutant. We monitored the stability of RpoS during logarithmic phase and 1 h after an immediate carbon starvation in both wild-type and mutant strains. During logarithmic growth, RpoS was rapidly degraded in both wild-type and sprED58A backgrounds (Fig. 2, log samples). As shown previously, RpoS was stabilized upon carbon starvation in a sprE wild-type strain (Fig. 2A). Likewise, RpoS was stabilized upon starvation in the sprED58A strain (Fig. 2B). It is known that the RpoS750-LacZ fusion protein is degraded in a SprE-dependent manner just like RpoS in wild-type strains and that this degradation ceases upon carbon starvation (19). Figure 2C shows that the same is true in sprED58A strains. Thus, unphosphorylated SprE still promotes RpoS degradation during logarithmic growth, and changes in SprE phosphorylation do not regulate RpoS or RpoS750-LacZ stability upon the onset of carbon starvation.
Changes in SprE phosphorylation are not required for rapid RpoS degradation upon exit from carbon starvation.
SprE is required to destabilize RpoS upon exit from stationary phase since RpoS levels remain elevated upon exit in a sprE null strain (data not shown). Although the sprED58A mutation does not prevent RpoS stabilization upon entry into stationary phase, it might affect RpoS destabilization upon exit. We added glucose to cells that had been starved overnight (12 h), at an OD600 of 0.3, and then measured the half-life of RpoS. Previous experiments have shown that RpoS is unstable 10 min after the addition of glucose (20), but we were interested in whether RpoS stability would change on a shorter time scale. One minute after the addition of glucose, we collected the initial sample for the stability assay. As discussed above, Western blot analysis of both the wild-type and mutant strains showed that RpoS is stable when the cells are starved and no glucose is added (Fig. 2 and 3). In the wild-type background, RpoS degradation is detectable within 3 min after exposure to glucose (Fig. 3A). Similarly, in the sprED58A background, RpoS is rapidly degraded during the exit from starvation (Fig. 3B). Thus, phosphorylation of SprE is not required for rapid RpoS proteolysis upon exit from stationary phase.
FIG. 3.
RpoS is rapidly degraded upon exit from carbon starvation independently of SprE phosphorylation. The half-life of RpoS was measured by Western blotting of samples taken 1 min after addition of glucose. Samples were taken from sprE+ (A) and sprED58A (B) strains. Samples were taken from cultures that had been starved for glucose and cultures that had been replenished with glucose for 1 min. Above each lane, time (in minutes) after the addition of either glucose or chloramphenicol is shown. (C) Quantification of RpoS levels from the Western blots (A and B), with closed squares representing the wild-type stain and open squares representing the mutant strain.
Changes in SprE phosphorylation are not required for RpoS accumulation during phosphate limitation.
It has also been suggested that RpoS degradation is regulated upon phosphate limitation (23). Phosphate limitation can be achieved by resuspending logarithmically growing cells in media lacking inorganic phosphate. Growth does not stop under these conditions but continues at a severely reduced rate as cells deplete intracellular phosphate pools. Nevertheless, the cells sense phosphate limitation, as verified by the dramatic increase in RpoS levels and the activation of the PhoB/PhoR two-component regulatory system (23, 28).
To evaluate the role of SprE phosphorylation during phosphate limitation, we measured the RpoS half-life in phosphate-deprived cells and compared it to that under carbon-starved conditions in both the wild type and mutant backgrounds. Exponentially growing cells in morpholinepropanesulfonic acid medium were deprived of phosphate by resuspending them in fresh medium lacking this anion, and the RpoS half-life was monitored after the addition of chloramphenicol by Western blotting as described above. Consistent with previous reports (14, 19, 29), our data show that RpoS was stabilized completely after carbon starvation in the wild-type strain. The mutant strain followed the same pattern (Fig. 4, carbon-starved lanes for SprE+ and SprED58A). After 1 h of phosphate limitation, RpoS levels in the sprE+ background were high (Fig. 4, t = 0 phosphate-limited samples). But after protein synthesis was shut off, RpoS degradation continued in the phosphate-limited cultures, though at a slower rate than that seen in logarithmic phase (Fig. 4, phosphate-limited SprE+ lanes). In the sprED58A background, RpoS levels also increased upon phosphate limitation. In this mutant background, RpoS was also degraded, albeit at an even slower rate than in the wild-type sprE background (Fig. 4, phosphate-limited SprED58A samples). In summary, in both the wild-type and mutant sprE backgrounds, RpoS became partially stabilized upon phosphate limitation. This is in contrast to the absolute stabilization that occurs during carbon starvation, suggesting that there are other mechanisms at work to increase RpoS levels during phosphate limitation besides regulation of proteolysis. Our lab has previously reported a PhoB/PhoR-dependent increase in rpoS translation (23).
FIG. 4.
RpoS is partially stabilized during phosphate deprivation in both the sprE+ and sprED58A backgrounds. The half-life of RpoS was measured by Western blot analysis of samples taken before and after addition of chloramphenicol. Samples were taken from cultures that had been immediately starved for 1 h for carbon or phosphate. RpoS behaved similarly in both the sprE+ (A and closed symbols in panel C) and the sprED58A (B and open symbols in panel C) strain backgrounds but was not completely stabilized after phosphate deprivation, as it was after carbon starvation. Above each lane, time (in minutes) after the addition of chloramphenicol is shown. Quantification of the RpoS levels is shown in panel C, with circles representing carbon starvation and squares representing phosphate starvation.
We assessed the role of SprE phosphorylation in the regulation of RpoS stability following ammonia starvation. There was no difference between the RpoS responses in the wild-type and mutant strains (data not shown).
Phosphorylation stimulates SprE activity in vivo.
Although the RpoS starvation response persisted with the mutated SprE, the basal levels of RpoS were higher in the mutated strain than in the wild-type strain in logarithmically growing cells, as seen in Fig. 1. To further investigate this, we compared RpoS levels during logarithmic growth in M63 glucose medium in sprE+, sprED58, and sprE::tet backgrounds. Western blot analysis showed that RpoS levels were higher in the sprED58A background compared to the wild-type background, yet they were not as high as in the sprE null background (Fig. 5A). The same results were seen in cells grown in LB medium (data not shown).
FIG. 5.
In logarithmic phase, phosphorylation of SprE contributes to its basal activity. (A) Western blot of samples taken during logarithmic-phase growth in minimal glucose medium. RpoS levels with SprED58A are at an intermediate level between the wild type and the sprE null strain. (B) Western blot of half-life samples taken during log-phase growth in M63 minimal glucose medium. Above each lane, time (in minutes) after the addition of chloramphenicol is shown. (C) Quantification of the RpoS levels shown in panel B, with the wild-type strain represented by closed squares, the sprED58A strain represented by open diamonds, and the sprE null strain represented by closed triangles. The RpoS half-life was calculated to be 4.7-fold longer with SprED58A than with the wild type, yet there is substantial SprE activity remaining compared to that in the sprE null strain.
To quantitate the effect of phosphorylation on SprE activity, we measured the half-life of RpoS during exponential growth in minimal glucose medium in cells carrying the sprE+, sprED58A, and sprE::tet alleles. Figures 5B and 5C show that RpoS was rapidly degraded, with a half-life of approximately 30 s in the wild-type background. As expected based on previous studies (17, 19), in the sprE null background, the half-life of RpoS was >200 min. In contrast, in the sprED58A background, the RpoS half-life was around 2.5 min, about fivefold longer than in the wild type. Thus, while unphosphorylated SprE effectively targets RpoS for degradation, phosphorylation does increase SprE activity in vivo.
Another phosphate donor besides acetyl phosphate contributes to SprE phosphorylation.
Acetyl phosphate has been shown to be an effective phosphodonor for response regulators in vitro, and biochemical studies have shown that it can phosphorylate SprE and stimulate activity (13, 27, 30). However, the role of acetyl phosphate in controlling SprE activity in vivo is not well understood. Bouche et al. originally showed that the RpoS half-life increased by threefold in a Δ(ackA-pta) strain, which is deficient in acetyl phosphate production, but signal transduction in response to stress remained intact (3). On the other hand, in S. enterica serovar Typhimurium, RpoS levels were unaffected by a Δ(ackA-pta) mutation (6).
Since the sprED58A mutation provides a tool for studying SprE phosphorylation in vivo, we decided to revisit the role of acetyl phosphate in RpoS regulation. We compared RpoS levels in the wild type, Δ(ackA-pta), sprED58A, and double mutant Δ(ackA-pta) sprED58A strains (Fig. 6). If acetyl phosphate were the sole donor for SprE phosphorylation, then RpoS levels in the absence of acetyl phosphate would be the same as in the sprED58A strain. In minimal medium, RpoS levels were only slightly increased in the sprE+ Δ(ackA-pta) strain. They were much higher in the sprED58A Δ(ackA-pta) background, indicating that acetyl phosphate is not the sole phosphodonor for SprE. Under these conditions, the effect of the Δ(ackA-pta) mutation on RpoS levels was so small it was difficult to determine if this mutation was additive in the double mutant Δ(ackA-pta) sprED58A and also whether acetyl phosphate affects only RpoS stability or some other aspect of RpoS regulation. Thus, although acetyl phosphate is clearly not the only phosphodonor for SprE, we cannot rule out the possibility that it contributes modestly to SprE phosphorylation in minimal glucose medium.
FIG. 6.
The sprED58A mutation is additive with Δ(ackA-pta). Western blot samples were taken from exponentially growing cells in LB and M63 minimal glucose media. The single mutant sprED58A and the double mutant sprED58A Δ(ackA-pta) had higher RpoS levels than the Δ(ackA-pta) single mutant in minimal medium, indicating that acetyl phosphate is not the sole phosphodonor for SprE. In LB medium, the additivity seen in the double mutant compared to the single mutants suggests that the lack of acetyl phosphate affects another mechanism of RpoS regulation, most likely synthesis.
In LB medium, however, RpoS levels in the double mutant were clearly much higher than in the single mutants, Δ(ackA-pta) or sprED58A. This suggests that in LB medium, acetyl phosphate affects RpoS levels in some way other than SprE phosphorylation. Considering the slow growth of the Δ(ackA-pta) strain in LB medium (more pronounced than the slight growth defect in minimal medium), we think it likely that growth-dependent regulation of RpoS synthesis plays a role in these conditions (6).
DISCUSSION
RpoS degradation is facilitated by SprE and regulated by nutrient availability. Since SprE contains a conserved CheY response regulator domain, it has been thought that SprE activity was modulated by phosphorylation (10, 12). Indeed, the activity of SprE is stimulated by phosphorylation in vitro (13, 27, 30). In contrast, we present evidence here that two-component phosphotransfer reactions do not play a significant role in the SprE-mediated control of RpoS stability in response to starvation. When the conserved phosphorylation site is mutated (sprED58A) in the chromosomal gene, RpoS stability is still regulated according to nutrient availability regardless of which nutrient (carbon, phosphate, or nitrogen) is depleted or removed. Moreover, exit from stationary phase, a SprE-dependent process that dramatically reduces cellular RpoS levels in minutes, is largely unaffected as well.
The in vivo activity of SprED58A was surprisingly high; most response regulators show no or little activity without phosphorylation (13, 21, 27, 30). Nonetheless SprE phosphorylation is significant. Although it is not required for signal transduction, it is necessary for maximal SprE activity. The sprED58A mutation increased the half-life of RpoS fivefold in exponentially growing cells. Phosphorylation, therefore, may be important in keeping RpoS levels very low during fast growth.
Our results provide a simple explanation for why genetic screens to identify the SprE phosphodonor have not been successful. We suspect that mutations which eliminate the phosphodonor might not confer a strong phenotype, just as sprED58A confers only a modest defect. While mutations that prevent the production of acetyl phosphate [Δ(ackA-pta)] can increase cellular RpoS levels, our results indicate that the bulk of this increase occurs by a mechanism(s) that is independent of phosphorylation. Thus, the identity of the SprE phosphodonor remains elusive. Identification of this factor(s) will require more sophisticated genetic or biochemical approaches.
If phosphorylation of SprE is not the signal controlling RpoS proteolysis, then how is the process regulated? The four factors necessary and sufficient for RpoS degradation are RpoS, SprE, ClpX, and ClpP. RpoS activity is clearly not required since RpoS-LacZ hybrid proteins are degraded in a regulated manner in the presence (Fig. 2C) or absence (our unpublished data) of functional RpoS. During carbon starvation ClpXP actively degrades proteins, and the levels of ClpX and ClpP are comparable to their respective levels in logarithmic phase (7, 24). Although it is possible that ClpX or ClpP activity becomes limiting during carbon starvation this should to some extent be countered by the increase in SprE levels that occurs as cells enter stationary phase (9, 20, 22). Thus, the activity of SprE could be regulated through some as-yet-unknown mechanism. SprE could be covalently modified, interact with another protein, or interact directly with a small molecule. Indeed, there are several examples in Streptomyces of highly conserved response regulators whose activity and regulation are not disrupted by mutations that alter their phosphorylation site. However, the regulatory mechanisms have yet to be elucidated (8, 15, 18).
A SprE-dependent, phosphorylation-independent mechanism for altering RpoS stability has been demonstrated. High levels of RpoS titrate active SprE, and this in turn stabilizes RpoS (1, 20, 23). This is most clearly observed when RpoS synthesis is increased by using an inducible promoter (20). Note, however, that while synthesis of RpoS during phosphate limitation is high enough to bring RpoS levels up to those seen during carbon starvation (Fig. 4), this increase is not sufficient to completely titrate SprE. SprE-mediated degradation of RpoS still occurs under these conditions, albeit at slower rates, even when phosphorylation is blocked by the sprED58A mutation. Clearly both SprE phosphorylation and titration can affect RpoS stability, yet together they do not account for the stabilization that occurs during carbon starvation. Some other regulatory mechanism must operate during carbon starvation which completely arrests RpoS degradation.
It is striking how quickly RpoS degradation commences in starved cells upon glucose addition. Within minutes, RpoS levels decrease to levels comparable to those in logarithmic-phase cells. The speed of this process argues for a simple switch, such as a change in the concentration of some metabolite, as opposed to a more complex process that would involve macromolecular synthesis. We suggest that exit from stationary phase might be a better condition than starvation (entry into stationary phase) to study the regulation of RpoS degradation. Exit offers several experimental advantages: the experiments are simple to control, they do not disrupt the growth, and they allow the timing of the signal to be precisely monitored.
Acknowledgments
We are indebted to Don Court and Nina Costantino for kindly teaching us how to use recombineering with single-stranded nucleotides and for providing strain HME6mutS. We are also grateful to Mark Mandel for sharing unpublished results and Johan Paulsson and the other members of the Silhavy lab for critical reading of the manuscript. We thank Susan DiRenzo for her assistance in preparation of this manuscript.
T.J.S was supported by an NIGMS Award (GM65216).
REFERENCES
- 1.Audia, J. P., and J. W. Foster. 2003. Acid shock accumulation of sigma S in Salmonella enterica involves increased translation, not regulated degradation. J. Mol. Microbiol. Biotechnol. 5:17-28. [DOI] [PubMed] [Google Scholar]
- 2.Becker, G., E. Klauck, and R. Hengge-Aronis. 2000. The response regulator RssB, a recognition factor for sigmaS proteolysis in Escherichia coli, can act like an anti-sigmaS factor. Mol. Microbiol. 35:657-666. [DOI] [PubMed] [Google Scholar]
- 3.Bouche, S., E. Klauck, D. Fischer, M. Lucassen, K. Jung, and R. Hengge-Aronis. 1998. Regulation of RssB-dependent proteolysis in Escherichia coli: a role for acetyl phosphate in a response regulator-controlled process. Mol. Microbiol. 27:787-795. [DOI] [PubMed] [Google Scholar]
- 4.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 5.Costantino, N., and D. L. Court. 2003. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc. Natl. Acad. Sci. USA 100:15748-15753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunning, C., and T. Elliott. 1999. RpoS synthesis is growth rate regulated in Salmonella typhimurium, but its turnover is not dependent on acetyl phosphate synthesis or PTS function. J. Bacteriol. 181:4853-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damerau, K., and A. C. St. John. 1993. Role of Clp protease subunits in degradation of carbon starvation proteins in Escherichia coli. J. Bacteriol. 175:53-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furuya, K., and C. R. Hutchinson. 1996. The DnrN protein of Streptomyces peucetius, a pseudo-response regulator, is a DNA-binding protein involved in the regulation of daunorubicin biosynthesis. J. Bacteriol. 178:6310-6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson, K. E., and T. J. Silhavy. 2000. SprE levels are growth phase regulated in a sigma(S)-dependent manner at the level of translation. J. Bacteriol. 182:4117-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoch, J. A., and K. I. Varughese. 2001. Keeping signals straight in phosphorelay signal transduction. J. Bacteriol. 183:4941-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenal, U., and R. Hengge-Aronis. 2003. Regulation by proteolysis in bacterial cells. Curr. Opin. Microbiol. 6:163-172. [DOI] [PubMed] [Google Scholar]
- 13.Klauck, E., M. Lingnau, and R. Hengge-Aronis. 2001. Role of the response regulator RssB in sigma recognition and initiation of sigma proteolysis in Escherichia coli. Mol. Microbiol. 40:1381-1390. [DOI] [PubMed] [Google Scholar]
- 14.Lange, R., and R. Hengge-Aronis. 1994. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 8:1600-1612. [DOI] [PubMed] [Google Scholar]
- 15.Molle, V., and M. J. Buttner. 2000. Different alleles of the response regulator gene bldM arrest Streptomyces coelicolor development at distinct stages. Mol. Microbiol. 36:1265-1278. [DOI] [PubMed] [Google Scholar]
- 16.Moreno, M., J. P. Audia, S. M. Bearson, C. Webb, and J. W. Foster. 2000. Regulation of sigma S degradation in Salmonella enterica var typhimurium: in vivo interactions between sigma S, the response regulator MviA(RssB) and ClpX. J. Mol. Microbiol. Biotechnol. 2:245-254. [PubMed] [Google Scholar]
- 17.Muffler, A., D. Fischer, S. Altuvia, G. Storz, and R. Hengge-Aronis. 1996. The response regulator RssB controls stability of the sigma(S) subunit of RNA polymerase in Escherichia coli. EMBO J. 15:1333-1339. [PMC free article] [PubMed] [Google Scholar]
- 18.Otten, S. L., X. Liu, J. Ferguson, and C. R. Hutchinson. 1995. Cloning and characterization of the Streptomyces peucetius dnrQS genes encoding a daunosamine biosynthesis enzyme and a glycosyl transferase involved in daunorubicin biosynthesis. J. Bacteriol. 177:6688-6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pratt, L. A., and T. J. Silhavy. 1996. The response regulator SprE controls the stability of RpoS. Proc. Natl. Acad. Sci. USA 93:2488-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruteanu, M., and R. Hengge-Aronis. 2002. The cellular level of the recognition factor RssB is rate-limiting for sigmaS proteolysis: implications for RssB regulation and signal transduction in sigmaS turnover in Escherichia coli. Mol. Microbiol. 45:1701-1713. [DOI] [PubMed] [Google Scholar]
- 21.Quon, K. C., G. T. Marczynski, and L. Shapiro. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83-93. [DOI] [PubMed] [Google Scholar]
- 22.Ruiz, N., C. N. Peterson, and T. J. Silhavy. 2001. RpoS-dependent transcriptional control of sprE: regulatory feedback loop. J. Bacteriol. 183:5974-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruiz, N., and T. J. Silhavy. 2003. Constitutive activation of the Escherichia coli Pho regulon upregulates rpoS translation in an Hfq-dependent fashion. J. Bacteriol. 185:5984-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schweder, T., K. H. Lee, O. Lomovskaya, and A. Matin. 1996. Regulation of Escherichia coli starvation sigma factor (sigma S) by ClpXP protease. J. Bacteriol. 178:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 26.Sledjeski, D. D., A. Gupta, and S. Gottesman. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15:3993-4000. [PMC free article] [PubMed] [Google Scholar]
- 27.Studemann, A., M. Noirclerc-Savoye, E. Klauck, G. Becker, D. Schneider, and R. Hengge. 2003. Sequential recognition of two distinct sites in sigma(S) by the proteolytic targeting factor RssB and ClpX. EMBO J. 22:4111-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wanner, B. L. 1996. Phosphorous assimilation and control of the phosphate regulon, p. 1357-1381. In F.C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 29.Zgurskaya, H. I., M. Keyhan, and A. Matin. 1997. The sigma S level in starving Escherichia coli cells increases solely as a result of its increased stability, despite decreased synthesis. Mol. Microbiol. 24:643-651. [DOI] [PubMed] [Google Scholar]
- 30.Zhou, Y., S. Gottesman, J. R. Hoskins, M. R. Maurizi, and S. Wickner. 2001. The RssB response regulator directly targets sigma(S) for degradation by ClpXP. Genes Dev. 15:627-637. [DOI] [PMC free article] [PubMed] [Google Scholar]