Abstract
Assembly of bacterial flagella is developmentally important during both planktonic cell growth and biofilm formation. Flagellar biogenesis is complex, requiring coordinated expression of over 40 genes, and normally commences during the log-to-stationary transition phase. We describe here a novel membrane-localized regulator, MorA, that controls the timing of flagellar development and affects motility, chemotaxis, and biofilm formation in Pseudomonas putida. MorA is conserved among diverse Pseudomonas species, and homologues are present in all Pseudomonas genomes sequenced thus far. In P. putida, the absence of MorA derepresses flagellar development, which leads to constitutive formation of flagella in the mutant cells in all growth phases. In Pseudomonas aeruginosa, the absence of MorA led to a reduction in biofilm formation. However, unlike the motility of P. putida, the motility of the P. aeruginosa mutants was unaffected. Our data illustrate a novel developmentally regulated sensory and signaling pathway for several properties required for virulence and ecological fitness of Pseudomonas species.
Bacterial cells exist either as free-swimming planktonic cells or in surface-attached communities known as biofilms. In planktonic cell cultures, the log-to-stationary transition phase is marked by the onset of flagellar development, which is a highly complex process involving coordinated expression of over 40 genes in a hierarchical manner (3, 8, 11, 16). Recently, the regulatory control of flagellar biogenesis in Pseudomonas aeruginosa was described in detail (7). In contrast to the three-tier regulation in the multiflagellated organism Escherichia coli, a four-tier hierarchy is present in the monoflagellated organism P. aeruginosa. At the top of the hierarchy are the transcriptional regulators, FleQ, and the alternative sigma factor FliA, which regulate at least 11 operons. Planktonic cells undergo multiple developmental changes during their transition from free-swimming organisms to the surface-attached bacterial communities that constitute biofilms. Appropriate levels of flagellin subunits seem to be a key factor since overexpression of flagellin in E. coli results in reduced adhesion (10). In fully developed biofilms, bacteria like Pseudomonas putida may even lack flagella (22). However, in P. aeruginosa, the role of flagella in biofilm formation still remains an open question as a recent study of biofilm development showed that flagella are not involved in the attachment and that initial microcolony formation occurs by clonal growth (9). Despite the fact that there is a great deal of knowledge concerning the flagellar pathway in diverse bacteria, very little is known about the negative regulation of this process.
Here we describe identification of MorA, a membrane-localized negative regulator of the timing of flagellar formation. In the mutilflagellated organism P. putida, planktonic cells of a morA mutant have constitutive expression of flagella. This property enhances motility and chemotaxis, but it interferes with the ability to form a biofilm in morA mutants. As is the case in P. putida, mutation of morA in the human pathogen P. aeruginosa also leads to reduced biofilm formation. However, no significant effects on motility are seen in P. aeruginosa. The MorA sequence shows that it is conserved in and is unique to the Pseudomonas genomes sequenced thus far.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used in this study are described in Table 1. Transposon mutants of P. putida PNL-MK25 (designated WTPp) were generated by using mTn5-gfp (26) as previously described (1) and were screened for increases in colony diameter in stabbed semisolid agar plates (0.4% [wt/vol] Bacto Agar [Difco]). The hypermotile mutants were designated C3H and B12H. P. putida and P. aeruginosa strains were cultured in Luria-Bertani (LB) medium at 30°C with suitable antibiotics (Table 1). Bacterial growth was measured spectrophotometrically by determining the optical density at 600 nm (OD600).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| P. putida strains | ||
| PNL-MK25 | Wild-type P. putida strain; Cmr Rfr | 1 |
| B12H | PNL-MK25 mutant (morAPp::mTn5-gfp); Cmr Rfr Kmr Gmr | This study |
| C3H | PNL-MK25 mutant (morAPp::mTn5-gfp); Cmr Rfr Kmr Gmr | This study |
| morAPp mutant | PNL-MK25 mutant (morAPp::aacCl); Cmr Rfr Gmr | This study |
| P. aeruginosa strains | ||
| PAO1 | Wild-type P. aeruginosa strain | 7 |
| morAPa mutant | PAO1 mutant (morAPa::aacCl); Gmr | This study |
| Plasmids | ||
| pGB1 | Broad-host-range vector; Ampr Tetr | 5 |
| pGB3 | pGB1 vector carrying GFP; Ampr Tetr | 5 |
| pGB1morA | Full-length morAPp gene with its native promoter cloned into pGB1; Ampr Tetr | This study |
| pUCP19 | Broad-host-range vector; Ampr | 23 |
| pUPMR | Full-length morAPa gene cloned into pUCP19; Ampr | This study |
| pGEM-3Zf(+) | Cloning vector; Ampr | Lab collection |
Cm, chloramphenicol (15 μg/ml); Rf, rifampin (20 μg/ml); Km, kanamycin (15 μg/ml); Gm, gentamicin (20 μg/ml) for PNL-MK25 or 100 μg/ml for (PAO1); Amp, ampicillin (100 μg/ml); Tet, tetracycline (25 μg/ml).
Motility and chemotaxis studies.
To study swimming motility, semisolid LB agar plates (0.4% [wt/vol] Bacto Agar) were inoculated with 2 μl of an overnight bacterial culture. After incubation for 18 h at 30°C, the movement of the bacteria away from the inoculation point was determined relative to the movement of WTPp, as previously described (18). To study chemotaxis, the chemotaxis assay was adapted from the assay described by Shi et al. (24). Overnight cultures were washed and resuspended in half-strength Stanier's mineral medium (25) at an OD600 of 1.0. One milliliter of culture was then mixed with 0.4% (wt/vol) soft agar prepared in half-strength Stanier's medium and poured onto a petri plate with a 1% agarose plug containing the chemoattractant (100 mM aspartic acid) mounted in the center.
DNA manipulations and analyses.
Genomic DNA was isolated as previously described (27). Chromosomal sequences flanking the transposon insertion sites in B12H and C3H were cloned and sequenced as previously described (1). The WTPp morA gene was designated morAPp, and its homologue in P. aeruginosa PAO1 was designated morAPa (GenBank accession number NP_253291). Restriction enzyme-digested DNA were electrophoresed and transferred onto Amersham Hybond-N nylon membranes by using standard protocols (20). A 1.1-kb morA-specific sequence was amplified from a subclone by using vector primers, labeled, and detected by using a DIG High Prime DNA labeling and detection kit (Roche Diagnostics) according to the manufacturer's instructions.
Generation of knockout mutants.
A partial morAPp gene fragment was amplified with primers PmF1 (5′-GTT ATT GCT CGC GTT GCT GTT CTG G-3′) and PmR1 (5′-TTT CCC AGA AGT ATT CGC CCA TCT G-3′), cloned into pGEM-3Zf(+), and then digested with ClaI. The restriction site was then blunted with DNA polymerase I (Klenow fragment), and a gentamicin (aacC1) cassette was then cloned into it. The knockout construct was electroporated (18 kV/cm) into WTPp, and the double-crossover knockout strain (morAPp mutant) was selected with gentamicin (5). A similar approach was used to generate the morAPa mutant with primers PmF2 (5′-TGT ACA TGC TGC TGC GCC WGC-3′) and PmR2 (5′-CAC GTC GAT GGG GAA CTG CTT GAG GTA G-3′) by using the XhoI site for insertion of the gentamicin cassette.
Complementation studies.
The primers used to amplify the full-length morAPp gene together with its native promoter were PpmF (5′-GGC GCC GGG GAA GAA GGA CAG C-3′) and PpmR (5′-GAC AGA TGT GCG GTG CTT TTG CC-3′). PCR amplification was performed by using a mixture of Taq and Vent polymerases, and the product was cloned into pGB1 (5). The resultant construct, pGB1morA, was introduced into both WTPp and the morAPp mutant. The primers used to amplify the full-length morAPa gene were PamFH (5′-CCC AAG CTT GGG CAT CCT AAC TGT CTA C-3′) and PamRX (5′-GCT CTA GAG CCA TCA TGT CGC GGA CA-3′), and the product was cloned into pUCP19 (23). The resultant construct, pUPMR, was introduced into both PAO1 and the morAPa mutant.
Microscopy studies.
For video microscopy, overnight cultures were diluted 1:50 in fresh LB medium before 10 μl was mounted on a hemocytometer and covered with a coverslip. The samples were viewed at a magnification of ×660 with a Nikon Eclipse E-600 microscope, and the movement of the cells was recorded by using a charge-coupled device camera connected to a Hauppauge WinTV-PVR-250 video capture card. For transmission electron microscopy (TEM), cells were harvested at various times, washed with 1% NaCl, and preserved in 2% formaldehyde. One drop of a cell suspension was placed onto a Formvar-coated copper grid (150 mesh). One drop of 2% (wt/vol) phosphotungstic acid (pH 7.0) was then added and left for 2 min. The grids were viewed with a Philips CM10 transmission electron microscope operating at 100 kV.
Biofilm assays.
The biofilm formation assay was adapted from the assay of O'Toole and Kolter (13). The assay was performed in polystyrene round-bottom tubes by using brain heart infusion (Difco) medium for P. aeruginosa PAO1 strains and LB medium for P. putida PNL-MK25; the media were inoculated with 1:100 dilutions from overnight cultures. After inoculation, the cultures were incubated for various times at 30°C with shaking at 100 rpm. At various times, nonadherent cells were removed by rinsing with distilled water. Biofilms were stained with a 0.1% (wt/vol) crystal violet solution for 1 h, and this was followed by rinsing with distilled water. Bound crystal violet was solubilized in 95% ethanol and quantitated by determining the OD595 as a measure of biofilm formation.
Adherent cells in the biofilms formed on polystyrene plastic surfaces were also viewed by transmission light microscopy by using a previously described method (6). Biofilms were formed on polystyrene strips placed in overnight cultures diluted 1:100. At the desired end point a strip was rinsed with LB medium and then stained with 0.1% (wt/vol) crystal violet. Attached bacteria were then examined at a magnification of ×630 by using a Leica DM LB microscope.
RNA isolation and Northern blot analysis.
Total RNA were extracted by using Trizol reagent (Life Technologies) in accordance with the manufacturer's instructions. For Northern analysis, 8 μg of total RNA was electrophoresed in a 1% phosphate-buffered agarose gel (20) and then transferred onto a positively charged nylon membrane. The nylon membrane was UV cross-linked and stained with methylene blue to ensure equal loading of RNA. The degenerate primers used to amplify a 0.8-kb WTPp fliC gene fragment were FliC-DF (5′-ATG GCT TTA ACA GTA AAC AC-3′) and FliC-DR (5′-CAG AGT CTG CTG CTT GGT C-3′). The primers used to amplify a 0.9-kb PAO1 fliC (accession number NP_249783) gene fragment were FliC-PAO-DF (5′-CGG TTC CAA CGC CTA CGA GAC C-3′) and FliC-PAO-DR (5′-AGT CGG TGT CCT TGA TGC GGC-3′). Labeling of gene fragments, hybridization, and detection with the DIG system (Roche Diagnostics) were performed in accordance with the manufacturer's instructions.
PAGE analysis of total protein.
WTPp and morAPp mutant cell pellets were resuspended in a solution containing 20 mM Tris, 20 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 10 mg of lysozyme per ml and then incubated on ice for 30 min. Triton X-100 was then added to a final concentration of 1% (vol/vol), and the mixture was incubated for 15 min. Prior to electrophoresis, samples were denatured by addition of both sodium dodecyl sulfate (SDS) and β-mercaptoethanol to final concentrations of 1%, followed by heating at 95°C for 10 min. Samples were electrophoresed on an SDS-18% polyacrylamide gel electrophoresis (PAGE) gel and stained with Coomassie blue (20). For protein sequencing, proteins from SDS-PAGE gels were first transferred to a Bio-Rad polyvinylidene difluoride membrane and subjected to automated Edman degradation by using an Applied Biosystems 494 protein sequencer. For quantitative time of flight mass spectrometry, protein bands were excised from the SDS-PAGE gel and subjected to tryptic digestion, and the samples were injected into a Micromass Q-TOF-2 mass spectrometer.
MorA protein localization by Western analysis.
The WTPp and morAPp mutant strains were transformed with pGB3 (5) expressing green fluorescent protein (GFP). Crude membrane preparations were prepared from both WTPp and the morAPp mutant by using a previously described method (12). Anti-MorA polyclonal antibodies against part of MorAPp (amplified with primers MorPASB [5′-CCT CCC GGA TCC CTG ATC GAG CTG TCG TTG C-3′] and MorENDH [5′-CCT CCC AAG CTT TTA GTC GAA CAT GAA CAG CGC-3′]) were raised in rabbits. Rabbit anti-GFP immunoglobulin G was obtained from Molecular Probes. Goat anti-rabbit immunoglobulin G, conjugated with alkaline phosphatase (Promega), was used as the secondary antibody for detection.
Nucleotide sequence accession number.
The nucleotide sequences of the P. putida PNL-MK25 morA gene reported in this paper have been deposited in the GenBank database under accession number AY323811.
RESULTS AND DISCUSSION
Disruption of morAPp enhances swimming motility and chemotaxis.
We used a genetic approach to identify negative regulators of swimming motility. Based on our hypothesis that knockout of such a gene should lead to enhanced motility, WTPp transposon mutants (Table 1) were screened for enhanced swimming motility through 0.4% (wt/vol) soft agar (18). Two transposon insertion mutant strains of P. putida, C3H and B12H (Table 1), which exhibited over 2.5-fold-enhanced motility, were identified (data not shown).
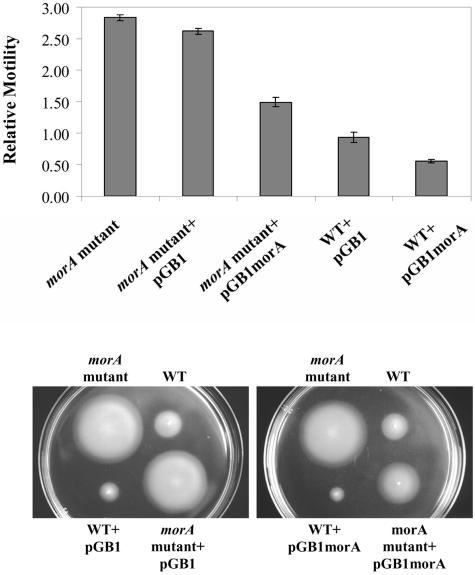
Sequences flanking the transposon insertion sites in B12H and C3H were cloned from genomic DNA (27) and sequenced. In both mutants, the transposon was inserted 27 bp apart in the same gene. Since an apparent loss of gene function led to enhanced motility, we designated the gene the motility regulator (morA) gene. Southern analysis indicated that morAPp was a single-copy gene (data not shown). Hence, targeted knockout of morAPp was performed to verify the mutant phenotype. The morAPp knockout strain (Table 1) showed about a threefold increase in swimming motility through 0.4% soft agar compared to WTPp (Fig. 1), which was similar to the results for C3H and B12H. Chemotaxis assays (24) showed that the morAPp mutant exhibited a more rapid chemotactic response to aspartate than WTPp exhibited (data not shown).
FIG. 1.
morAPp mutant exhibits enhanced swimming motility. The swimming motility of the wild type (WT) and the morAPp mutant in semisolid agar (0.4% [wt/vol] agar) was examined. The morAPp gene driven by its native promoter was expressed in the wild-type and morAPp mutant strains in trans on a broad-host-range plasmid (pGB1morA). The wild-type strain was P. putida parental strain PNL-MK25. The results are based on three independent experiments, each with five replicates.
Complementation of the morAPp mutant strain with the morAPp gene driven by its native promoter (pGB1morA) (Table 1) resulted in partial restoration of the wild-type motility phenotype; i.e., the motility was reduced, but it was not back to wild-type levels (Fig. 1). MorAPp expression in the morAPp mutant resulted in a 50% decrease in motility, while MorAPp expression in WTPp led to an almost 40% decrease (Fig. 1). These results suggested that there was strict control of morAPp dosage within the cells; a slight perturbation in gene copy number resulted in measurable differences in motility.
MorAPp restricts the timing of flagellar development in planktonic cells.
The growth rates of the mutant and wild-type cells were found to be comparable (data not shown). Video microscopy was performed to examine the movement of individual cells from planktonic cultures at the early log phase (OD600, 0.3), the mid-log phase (OD600, 1.0), and the log-to-stationary transition phase (OD600, 1.7). The majority of the morAPp mutant cells were highly motile in all three growth phases, whereas most of the WTPp cells were nonmotile (see supplemental material at http://plantmiclab.science.nus.edu.sg).
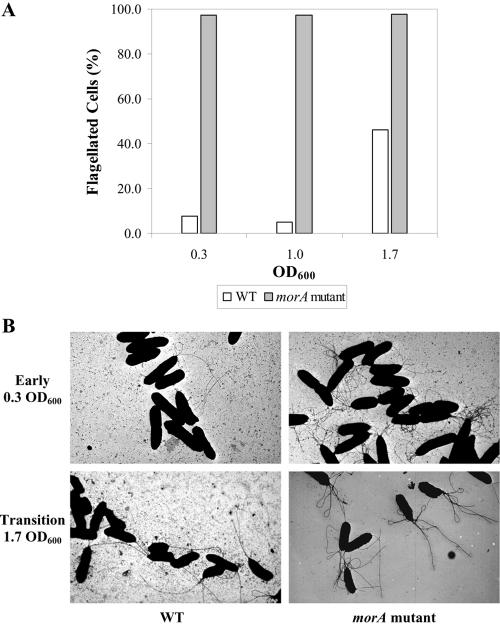
TEM ultrastructural studies were then performed to ascertain whether the changes in bacterial movement were due to changes in flagellar structure or to the rotation frequency of the flagellar apparatus. Based on counts of 400 cells each, almost 98% of the morAPp mutant cells possessed well-developed flagella in all three phases of growth (Fig. 2A). In contrast, less than 10% of WTPp cells were flagellated at the early log and mid-log phases, and flagellation increased only during the log-to-stationary transition phase to about 50% (Fig. 2). The localization of the flagella remained polar for both strains, but the mutant cells were also hyperflagellated compared to the WTPp cells (Fig. 2B). The increase in the number of flagella per cell was observed throughout the growth phases. The TEM studies also showed that both the length of the flagella and the size of the morAPp mutant cells were similar to those of WTPp. Together with the similar growth rates of the strains, these findings suggested that the cell division rates in the two strains were similar. Hence, in morAPp mutants a developmental restriction on the timing of flagellar formation was removed, which resulted in the presence of flagella throughout the growth stages with no effect on cell division or cell size. In comparison, E. coli mutants with mutations in the flagellar master activator FlhD do not form flagella, but cell division continues, giving rise to an extended exponential phase (17). Hence, there is a different pathway for MorAPp, in which its loss allows flagellar production to start from the early growth phase while normal cell division is maintained.
FIG. 2.
morAPp in P. putida PNL-MK25 enhances swimming motility by regulating the timing of flagellar development. (A) Proportion of flagellated cells expressed as a percentage of the total number of wild-type (WT) and morAPp mutant cells counted by TEM. Counts were based on an average of 400 cells. The wild-type strain was P. putida parental strain PNL-MK25. (B) TEM of wild-type and morAPp mutant cells at the early log and log-to-stationary transition growth stages. Magnification, ×7,200.
MorA affects biofilm formation in both P. putida and P. aeruginosa.
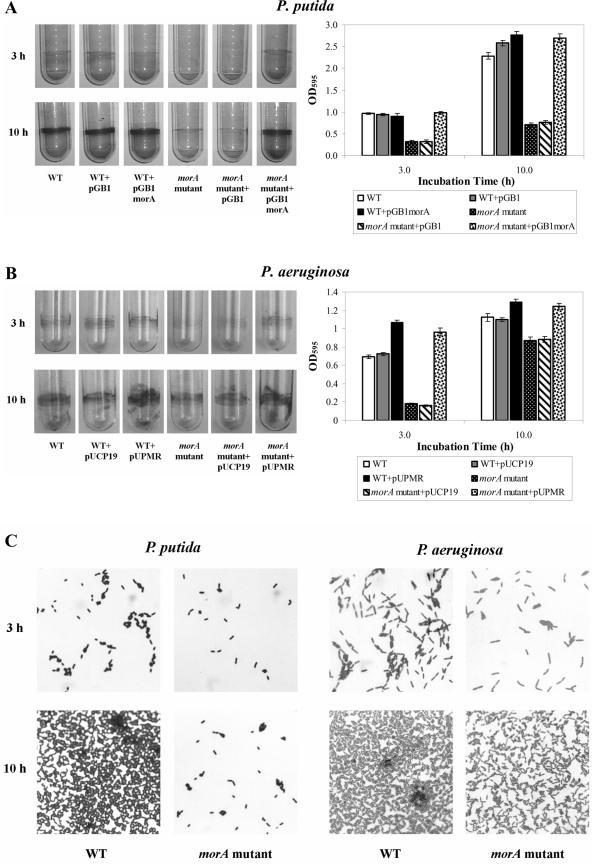
Previous studies have indicated that biofilm formation requires appropriate flagellar biogenesis (15, 21). Therefore, we investigated whether constitutive production of flagella had an effect on biofilm formation. At 3 h, the sizes of the biofilms formed by the noncomplemented mutants were significantly less than the sizes of the biofilms formed by WTPp, as determined by crystal violet staining (Fig. 3A). Adherent cells in the biofilms formed on the polystyrene plastic surfaces were also viewed by transmitted light microscopy. Fewer morAPp mutant cells were found to adhere to the surfaces (Fig. 3C).
FIG. 3.
morA affects biofilm formation in P. putida PNL-MK25 and P. aeruginosa PAO1. (A) Plasmids pGB1 and pGB1morA were introduced into both the wild-type strain (WT) and the morA mutant (morAPp::aacCI mutant strain). Biofilms that formed at 3 and 10 h after inoculation in polystyrene tubes were stained with 0.1% crystal violet. The wild-type strain was P. putida parental strain PNL-MK25. (B) Plasmids pUCP19 and pUPMR were introduced into both the wild-type strain and the morA mutant (morAPa::aacCI mutant strain), and biofilms that formed at 3 and 10 h after inoculation were detected as described above. The wild-type strain (WT) was P. aeruginosa parental strain PAO1. (C) Adherent cells in the biofilms formed on polystyrene surfaces at 3 and 10 h after inoculation were stained with crystal violet and examined by transmitted light microscopy (magnification, ×630). WT, wild type.
At 10 h, the biofilms formed by the morAPp mutant were visible, but they were still smaller than those formed by WTPp (Fig. 3A). The number of mutant cells adhering to the polystyrene surface after 10 h had increased compared to the 3-h biofilm, but it was still considerably less than the number of WTPp cells (Fig. 3A). Complementation of the morAPp mutants with pGB1morA completely restored the biofilm formation phenotype (Fig. 3A). These results suggested that the precocious production of flagella in morAPp mutants was correlated with a reduction in biofilm formation. It has been reported previously that overexpression of flagellin results in decreased adhesion of E. coli to sand (10). We therefore hypothesized that flagella might be necessary in specific quantities and only at defined stages of bacterial adhesion and biofilm formation.
Since motility plays a crucial role in the virulence of several pathogens, we investigated the function of a MorA homolog in the human pathogen P. aeruginosa PAO1. In contrast to disruption of P. putida morAPp, disruption of the morAPa gene in the monoflagellated PAO1 strain had no effect on swimming motility. Additionally, there was no change in the flagellum number or length in the mutant (data not shown). However, the loss of MorAPa led to impairment of biofilm formation in a time-dependent manner similar to that in P. putida; after 3 h, the biofilm formed by the mutant was about 70% smaller than that formed by wild-type PAO1 (Fig. 3B). The differences in biofilm formation gradually narrowed as the bacteria grew, but even at 10 h the morAPa mutant still formed about 20% less biofilm than the wild-type strain PAO1 formed. These results were corroborated by light microscopy of biofilms formed on polystyrene surfaces (Fig. 3C). Complementation of the morAPa mutant with pUPMR (Table 1) restored the phenotype completely. Vector controls did not differ significantly from the corresponding non-vector-containing strains. A significant increase in biofilm formation was observed after 3 h in wild-type cells harboring pUPMR, which we believe was due to the increased expression of morA in the wild-type cells. Based on these results, we concluded that although morAPa is not essential for biofilm formation during prolonged periods, it does play a significant role in the early establishment stages.
morA restricts fliC expression in P. putida and P. aeruginosa.
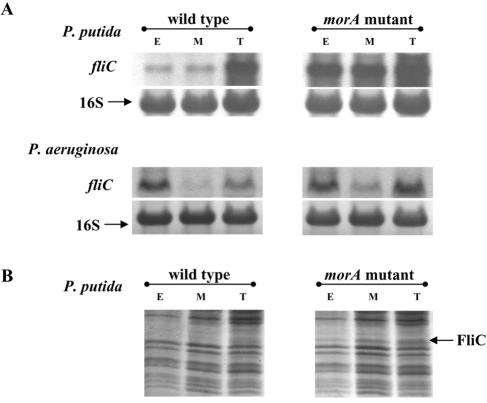
In order to ascertain if the presence of the flagellar apparatus in morAPp mutants was due to increased expression of flagellar genes, studies were carried out to quantitate flagellin (fliC) RNA and protein. Northern analysis (20) showed that the fliC transcript levels in WTPp were very low at the early log and mid-log phases and increased substantially only at the log-to-stationary transition phase (Fig. 4A). In contrast, fliC was constitutively expressed throughout the growth phases in the morAPp mutant strain (Fig. 4A). SDS-PAGE analysis of extracts of morAPp mutant cells showed that flagellin, as confirmed by Edman sequencing and quantitative time-of-flight mass spectrometry (data not shown), was present at all three growth phases (Fig. 4B). In contrast, flagellin was barely detectable only at the log-to-stationary transition phase in WTPp cells. These results suggested that disruption of morAPp resulted in derepression of flagellin expression and, consequently, flagella were constitutively produced. We therefore propose that MorAPp is a key component of the regulatory system that normally restricts the timing of expression of the flagellar biosynthesis pathway to late phases of growth in P. putida.
FIG. 4.
Expression of flagellin (fliC) is deregulated in the morA mutant strains. (A) Northern analysis of P. putida and P. aeruginosa wild-type and morA mutant cells harvested at the early log phase (OD600, 0.3) (lanes E), the mid-log phase (OD600, 1.0) (lanes M), and the log-to-stationary transition phase (OD600, 1.7) (lanes T) with digoxigenin-labeled P. putida and P. aeruginosa fliC probes, respectively. (B) SDS-PAGE analysis of P. putida wild-type and morAPp mutant total protein extracted from cells harvested at the early log, mid-log, and log-to-stationary transition phases.
We also examined flagellin expression levels in PAO1 and the morAPa mutant. In the early log stage, there was no apparent difference in fliC expression in the two strains; slight differences were observed only in the mid-log and log-to-stationary transition stages (Fig. 4A). The difference in the levels of the fliC transcript in PAO1 wild-type and morAPa mutant strains was not significant, unlike the case in P. putida. Also, the hyperflagellation of the morA mutant seen in P. putida was not observed in P. aeruginosa. This suggests that MorA involvement in the flagellum pathway is different in the two species.
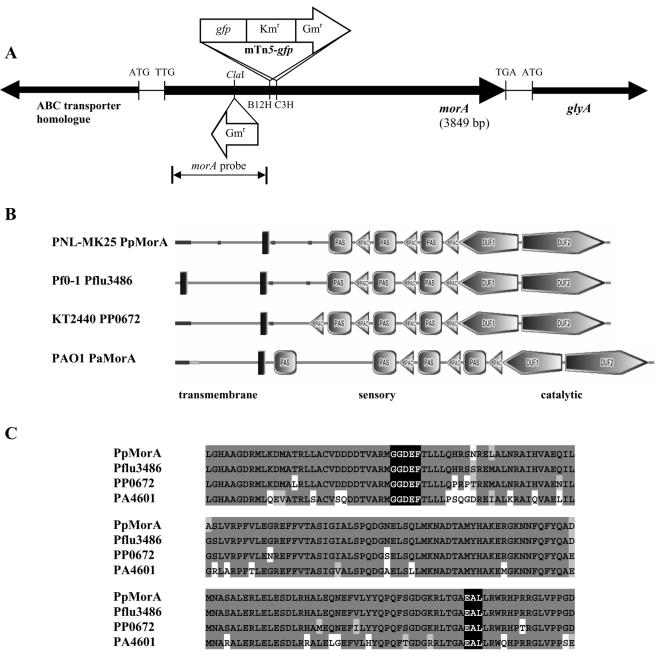
morAPp locus and its predicted protein.
A 6,096-bp region corresponding to the morAPp region was sequenced. morAPp is flanked by a gene encoding a probable ABC transporter (yjjK) and a serine hydroxymethyltransferase gene (glyA) (Fig. 5A). The morAPp gene is 3,849 bp long and is predicted to encode a 1,282-amino-acid polypeptide with a molecular mass of 145 kDa.
FIG. 5.
Conservation of MorA in Pseudomonas species. (A) Organization of the P. putida PNL-MK25 morA locus. The solid arrows show the directions of the genes. Mutant strains B12H and C3H carried single mTn5-gfp transposon insertions at positions 1453 and 1480 of morA, respectively. The targeted morAPp knockout mutant was generated by inserting a gentamicin cassette at the ClaI site located at position 1121. morAPp is flanked by a probable ABC transporter and a serine hydroxymethyltransferase gene, glyA. The 1.1-kb region used to probe PNL-MK25 genomic DNA is indicated. (B) Domain architecture of MorA family members in Pseudomonas species. The three conserved regions of the predicted MorA proteins are (i) a transmembrane domain(s) (vertical bars), (ii) sensory PAS and PAC domains, and (iii) catalytic GGDEF (DUF1) and EAL (DUF2) domains. The domains were predicted by using the Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de). (C) GGDEF and EAL domains are highly conserved. Alignment of the GGDEF and EAL domains of MorA, Pflu3486, PP0672, and PA4601 was performed by using ClustalW (http://www.ebi.ac.uk/clustalw). The GGDEF and EAL domains are highlighted with black, identical amino acids are highlighted with dark gray, and similar amino acids are highlighted with light gray.
Searches of DNA sequence databases with BLAST (2) showed that morA homologues are present in all Pseudomonas genomes sequenced thus far and that they share a high degree of sequence similarity. Since the genome of P. aeruginosa is available, we compared the morAPa locus with the three flagellar gene regions (7) and found that morAPa is not located within these regions.
The primary structures of predicted MorA proteins from various Pseudomonas species are well conserved, and the sequence similarity values range between 58 and 93%. Members of the Pseudomonas MorA family are (i) present as single copies in the genome, (ii) likely to be membrane localized due to the transmembrane domains, (iii) possess a central sensory domain consisting of PAS-PAC motifs, and (iv) have C-terminal GGDEF and EAL domains (Fig. 5B and C). The N-terminal transmembrane region is more variable (30 to 80% similarity) than the PAS-PAC motifs and the GGDEF and EAL domains (over 80% similarity). PAS-PAC motifs have proposed functions as sensors for light, redox potential, or oxygen concentration (29). The GGDEF domain shows weak similarity to eukaryotic adenylyl cyclases (14), and the GGDEF and EAL motifs have been shown to be involved in cyclic diguanylic acid (c-di-GMP) turnover in Acetobacter xylinum (4, 19). However, using a previously described method (28), we were unable to detect c-di-GMP in crude extracts of WTPp and morAPp mutant strains (data not shown).
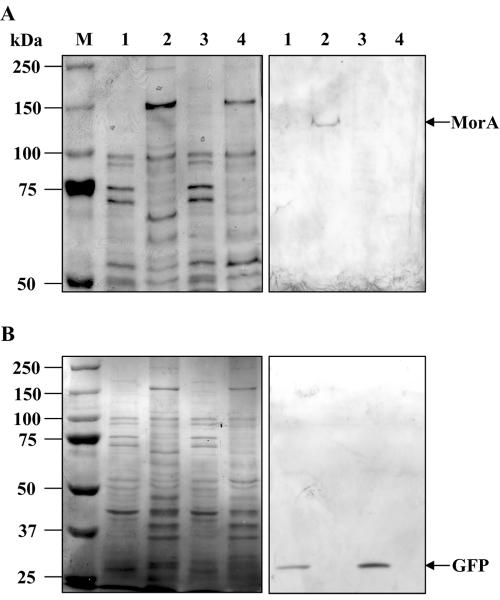
Localization of MorA in P. putida was examined by Western analysis. An ∼140-kDa band was detected in the crude membrane fraction and not in the cytoplasmic fraction of WTPp (Fig. 6A). Additionally, this band was absent in both fractions of the morAPp mutant. To ensure that there was no contamination of the membrane fraction with the cytoplasmic fraction, a cytoplasmic localization control, GFP, was expressed from low-copy-number plasmid pGB3 (5) in both WTPp and the morAPp mutant. Western analysis showed that the GFP protein was detected only in the cytoplasmic fractions of both strains (Fig. 6B). Hence, the results indicate that MorAPp is membrane localized.
FIG. 6.
Membrane localization of P. putida MorA. Coomassie blue staining (left panel) and Western blot analysis (right panel) were performed with duplicate gels. Membranes were probed with anti-MorA antibody (A) or anti-GFP antibody (B) in cytoplasmic and membrane fractions of the P. putida wild type and morAPp mutant. Both strains expressed GFP from plasmid pGB3. Lane M, Bio-Rad Precision Plus protein standards; lane 1, P. putida wild-type cytoplasmic fraction; lane 2, P. putida wild-type membrane fraction; lane 3, P. putida morAPp mutant cytoplasmic fraction; lane 4, P. putida morAPp mutant membrane fraction. The predicted molecular mass of MorA is 145 kDa. The wild-type strain was P. putida parental strain PNL-MK25.
In conclusion, here we describe MorA, a novel membrane-localized negative regulator of motility in the soil microbe P. putida that normally restricts flagellar synthesis and assembly to the late growth stages of the bacterial cells. Disruption of this regulator leads to constitutive maximal expression of flagella during all phases of growth. An indirect consequence of this appears to be impairment of biofilm formation but enhancement of chemotaxis ability. MorA is highly conserved in several Pseudomonas species, but its role in flagellar development and biofilm formation appears to vary between species. Hence, experiments with both microarrays and complementation constructs spanning different regions of the MorA protein are being designed to gain insight into the regulatory mechanism.
Acknowledgments
This work was supported by grants R-154-000-145-112 and R-154-000-114-112 from the Academic Research Fund, National University of Singapore. W.K.C. is funded by a research scholarship awarded by the National University of Singapore.
We thank Moshe Benziman and Haim Wienhouse, Department of Biological Chemistry, Institute of Life Sciences, Hebrew University of Jerusalem, Jerusalem, Israel, for their generous gift of crude c-di-GMP.
REFERENCES
- 1.Adaikkalam, V., and S. Swarup. 2002. Molecular characterization of an operon, cueAR, encoding a putative P1-type ATPase and a MerR-type regulatory protein involved in copper homeostasis in Pseudomonas putida. Microbiology 148:2857-2867. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amsler, C. D., M. Cho, and P. Matsumura. 1993. Multiple factors underlying the maximum motility of Escherichia coli as cultures enter post-exponential growth. J. Bacteriol. 175:6238-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausmees, N., R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. Lindberg. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 204:163-167. [DOI] [PubMed] [Google Scholar]
- 5.Bloemberg, G. V., G. A. O'Toole, B. J. J. Lugtenberg, and R. Kolter. 1997. Green fluorescent protein as a marker for Pseudomonas spp. Appl. Environ. Microbiol. 63:4543-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bomchil, N., P. Watnick, and R. Kolter. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 185:1384-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50:809-824. [DOI] [PubMed] [Google Scholar]
- 8.Givskov, M., L. Eberl, G. Christioansen, M. J. Benedik, and S. Molin. 1995. Induction of phospholipase and flagellar synthesis in Serratia liquefaciens is controlled by expression of the flagellar master operon flhD. Mol. Microbiol. 15:445-454. [DOI] [PubMed] [Google Scholar]
- 9.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 10.Landini, P., and A. J. B. Zehnder. 2002. The global regulatory hns gene negatively affects adhesion to solid surfaces by anaerobically grown Escherichia coli by modulating expression of flagellar genes and lipopolysaccharide production. J. Bacteriol. 184:1522-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 12.Minghetti, K. C., V. C. Goswitz, N. E. Gabriel, J. J. Hill, C. A. Barassi, C. D. Georgiou, S. I. Chan, and R. B. Gennis. 1992. Modified, large-scale purification of the cytochrome o complex (bo-type oxidase) of Escherichia coli yields a two heme/one copper terminal oxidase with high specific activity. Biochemistry 31:6917-6924. [DOI] [PubMed] [Google Scholar]
- 13.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 14.Pei, J., and N. V. Grishin. 2001. GGDEF domain is homologous to adenylyl cyclase. Proteins 42:210-216. [DOI] [PubMed] [Google Scholar]
- 15.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 16.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39:1595-1609. [DOI] [PubMed] [Google Scholar]
- 17.Prüß, B. M., and P. Matsumura. 1997. Cell cycle regulation of flagellar genes. J. Bacteriol. 179:5602-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robleto, E. A., I. Lopez-Hernandez, M. W. Silby, and S. B. Levy. 2003. Genetic analysis of the AdnA regulon in Pseudomonas fluorescens: nonessential role of flagella in adhesion to sand and biofilm formation. J. Bacteriol. 185:453-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinberger-Ohana, R. Mayer, S. Braun, E. de Vroom, G. A. van der Marel, J. H. van Boom, and M. Benziman. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279-281. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 21.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sauer, K., and A. K. Camper. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183:6579-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-112. [DOI] [PubMed] [Google Scholar]
- 24.Shi, W., Z. Yang, Y. Geng, L. E. Wolinsky, and M. A. Lovett. 1998. Chemotaxis in Borrelia burgdorferi. J. Bacteriol. 180:231-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanier, R. Y., N. J. Palleroni, and M. Doudoroff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159-271. [DOI] [PubMed] [Google Scholar]
- 26.Suarez, A., A. Guttler, M. Stratz, L. H. Staendner, K. N. Timmis, and C. A. Guzman. 1997. Green fluorescent protein-based reporter systems for genetic analysis of bacteria including monocopy applications. Gene 196:69-74. [DOI] [PubMed] [Google Scholar]
- 27.Syn, C. K. C., and S. Swarup. 2000. A scalable protocol for the isolation of large-sized genomic DNA within an hour from several bacteria. Anal. Biochem. 278:86-90. [DOI] [PubMed] [Google Scholar]
- 28.Weinhouse, H., S. Sapir, D. Amikam, Y. Shilo, G. Volman, P. Ohana, and M. Benziman. 1997. c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 416:207-211. [DOI] [PubMed] [Google Scholar]
- 29.Zhulin, I. B., B. L. Taylor, and R. Dixon. 1997. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 22:331-333. [DOI] [PubMed] [Google Scholar]