Abstract
Brucella abortus cyclic glucan synthase (Cgs) is a 316-kDa (2,831-amino-acid) integral inner membrane protein that is responsible for the synthesis of cyclic β-1,2-glucan by a novel mechanism in which the enzyme itself acts as a protein intermediate. B. abortus Cgs uses UDP-glucose as a sugar donor and has the three enzymatic activities necessary for synthesis of the cyclic polysaccharide (i.e., initiation, elongation, and cyclization). Cyclic glucan is required in B. abortus for effective host interaction and complete expression of virulence. To gain further insight into the structure and mechanism of action of B. abortus Cgs, we studied the membrane topology of the protein using a combination of in silico predictions, a genetic approach involving the construction of fusions between the cgs gene and the genes encoding alkaline phosphatase (phoA) and β-galactosidase (lacZ), and site-directed chemical labeling of lysine residues. We found that B. abortus Cgs is a polytopic membrane protein with the amino and carboxyl termini located in the cytoplasm and with six transmembrane segments, transmembrane segments I (residues 419 to 441), II (residues 452 to 474), III (residues 819 to 841), IV (residues 847 to 869), V (residues 939 to 961), and VI (residues 968 to 990). The six transmembrane segments determine four large cytoplasmic domains and three very small periplasmic regions.
Brucella spp. are gram-negative, facultative, intracellular bacteria that cause a worldwide chronic zoonotic disease known as brucellosis (13). Six species of Brucella with different host specificities and pathogenic characteristics have been described (19, 44). Brucella abortus, the etiological agent of bovine brucellosis, also infects humans, causing undulant fever.
Brucella produces cyclic β-1,2-glucan (43). This compound is a cyclic homopolysaccharide consisting of about 17 to 22 β-1,2-linked glucose units. Recently, isolation of the B. abortus cgs gene, which codes for the cyclic β-1,2-glucan synthase, and characterization of the synthesis of cyclic β-1,2-glucan in this species have been reported (20). B. abortus cgs mutants showed reduced virulence in mice and defective intracellular multiplication, indicating that cyclic glucan is required for an effective host interaction (6). Furthermore, cyclic glucans must be transported into the periplasmic space for activity to occur. Isolation of the B. abortus cyclic glucan transporter gene (cgt) was described recently (38). Cyclic glucan transporter mutations in B. abortus have also affected its interaction with the host. Thus, the presence of cyclic glucan in the periplasmic space in B. abortus is required for complete expression of virulence (38). Because of this, Cgs may be a good target for development of alternative chemotherapy against this pathogen.
B. abortus cyclic glucan synthase (Cgs) is a 316-kDa (2,831-amino-acid) integral inner membrane protein that is responsible for the synthesis of cyclic β-1,2-glucan by a novel mechanism in which the enzyme itself acts as a protein intermediate. Cgs uses UDP-glucose as a sugar donor and has the three enzymatic activities required for synthesis of the cyclic polysaccharide (i.e., initiation, elongation, and cyclization). Initiation consists of the transfer of the first glucose from UDP-glucose to an unidentified amino acid residue of the enzyme. Elongation, catalyzed by a UDP-Glc:β-(1,2) oligosaccharide glucosyltransferase activity, consists of addition of glucose residues to the linear β-(1,2) oligosaccharide linked to the protein. Finally cyclization catalyzes glucan cyclization and release from the protein (7, 20). Consequently, Cgs acts as an inverting processive autoglucosyltransferase.
The C-terminal region of B. abortus Cgs (from amino acid 1,585 to amino acid 2,831) has an overall level of identity of 27% with the Clostridium stercorarium cellobiose and cellodextrin phosphorylases. It is remarkable that this region of the protein is highly conserved, although it is not required for glucan synthesis. It remains to be established whether Cgs has cellobiose and/or cellodextrin phosphorylase enzymatic activity and whether this region is implicated in the in vivo regulation of cyclic glucan synthesis (20).
A key feature of the structure of a membrane protein is its transmembrane topology defined by the disposition of segments of the polypeptide within or to one side or the other of the membrane. In the laboratory we have studied the mechanism of action of the inner membrane protein responsible for the biosynthesis of cyclic glucan. To gain further insight into the structure and mechanism of action of B. abortus Cgs at the molecular level, we studied the membrane topology of the protein. A combination of in silico predictions, a genetic approach consisting of construction of a collection of fusions between Cgs and two sensor enzymes, alkaline phosphatase (AP) and β-galactosidase (βG), and site-directed chemical labeling of lysine residues was used.
It is well known that the signal sequence of alkaline phosphatase can be replaced by export signals derived from other proteins, including those found in integral membrane proteins (32). Alkaline phosphatase-specific activity of fusion proteins closely correlates with the cellular location of the domain in the native protein to which alkaline phosphatase is fused (31). Alkaline phosphatase is enzymatically active only when it is translocated to the periplasm; thus, alkaline phosphatase fusions are active only when they are fused to a periplasmic region of a membrane protein. On the other hand, alkaline phosphatase fused to a cytoplasmic domain yields an inactive cytoplasmically located enzyme (14, 33). In contrast, hybrid proteins containing β-galactosidase exhibit behavior that is reciprocal to that observed with alkaline phosphatase fusions. Fusion of β-galactosidase to cytoplasmic stretches yields hybrid proteins displaying high levels of enzymatic activity (29, 33). Fusions to periplasmic domains are believed not to mediate export of β-galactosidase across the membrane; rather, the enzyme remains embedded within the membrane, preventing proper folding or tetramerization, which results in a lack of enzymatic activity (17). Lee et al. suggested that multiple sites throughout β-galactosidase block export of the enzyme across the membrane (27).
In this study, we elucidated the membrane topology of B. abortus Cgs. Here we present experimental evidence that Cgs is a polytopic membrane protein with six transmembrane segments (TMSs) and with the amino and carboxyl termini located in the cytoplasm.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. Escherichia coli was grown at 37°C in Luria-Bertani broth (39). Brucella strains were grown at 37°C in brucella broth (Difco Laboratories, Detroit, Mich.). If necessary. media were supplemented with appropriate antibiotics at the following concentrations: ampicillin, 100 μg/ml for E. coli and 50 μg/ml for B. abortus; tetracycline, 20 μg/ml for E. coli; and nalidixic acid, 5 μg/ml for B. abortus.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Phenotype or genotypea | Reference or source |
|---|---|---|
| Strains | ||
| E. coli K-12 strain DH5α-F′IQ | F′φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK+ mK+) phoA supE44 λ−thi-1 gyrA96 relA1/F′ proAB+lacIqZΔM15 zzf::Tn5(Kmr) | 46 |
| E. coli S17.1 (λpir) | λ lysogenic S17-1 derivative producing π protein for replication of plasmids carrying oriR6K (Nals) | K. N. Timmis |
| B. abortus 2308 | Virulent, field isolated; wild type, Nalr Eryr | 40 |
| Plasmids | ||
| pGemT-easy | Cloning vector for PCR products | Promega |
| pBBR1MCS-4 | Broad-host-range cloning vector (Ampr) | 23 |
| pBBK | pBBR1MCS-4 with the KpnI site removed (Ampr) | This study |
| pBA19 | 19-kb fragment of genomic DNA of B. abortus S19 containing cgs of B. abortus in pVK102 (Tcr) | 20 |
| pBA20 | 11.3-kb XhoI-BamHI fragment of pBA19 containing cgs of B. abortus in pBBK (Ampr) | This study |
| pBA21 | Derivative of pBA20 with a 467-bp AgeI-SacI fragment of α-peptide-coding region of β-galactosidase of pBBR1MCS-4 deleted (Ampr) | This study |
| pSWFII | phoA sandwich fusion construction vector (Ampr) | 16 |
| pGTZ | 3.0-kb ′lacZ fragment in pGemT-easy (Ampr) | This study |
| pBA | Derivative of pBA20 containing 1.4-kb ′phoA fragment (Ampr)b | This study |
| pBZ | Derivative of pBA21 containing 3.0-kb ′lacZ fragment (Ampr)c | This study |
All the protocols with live Brucella strains were performed in a biosafety level 3 laboratory facility.
Plasmid construction.
Plasmid pBBR1MCS-4 (23) was digested with KpnI, treated with T4 DNA polymerase (New England Biolabs, Beverly, Mass.), and religated, yielding pBBK. An 11.3-kb XhoI-BamHI fragment from pBA19 (20), containing the cgs gene and its promoter, was ligated into pBBK digested with XhoI and BamHI; the resulting plasmid was designated pBA20.
The phoA gene (encoding alkaline phosphatase) lacking its transcription and translation initiation sequences, the region encoding its signal sequence, and five additional amino acids (designated ′phoA) from pSWFII (16) was excised on an XbaI fragment and ligated into the same site of pBA20; the resulting plasmid was designated pBA (Fig. 1A).
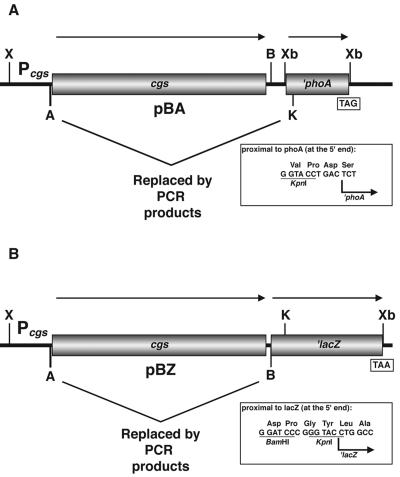
FIG. 1.
Structure of vectors used in the generation of cgs-′phoA and cgs-′lacZ translational gene fusions. (A) pBA contains both the cgs gene with its own promoter and the ′phoA gene from pSWFII (16). (B) pBZ contains both the cgs gene with its own promoter and the ′lacZ gene amplified by PCR with pAB2001 (1) as the template. The AvrII-KpnI and AvrII-BamHI fragments containing the complete cgs gene from pBA and pBZ, respectively, were replaced by PCR products containing different lengths of the 5′ end of cgs gene. The direction of the arrows indicates the direction of gene transcription. TAG and TAA are the translation stop codons of ′phoA and ′lacZ, respectively. The linker regions between cgs and ′phoA and between cgs and ′lacZ are shown in the insets. Pcgs, cgs promoter. Restriction sites: X, XhoI; B, BamHI; K, KpnI; A, AvrII; Xb, XbaI.
In order to avoid annealing of primer LACZ (primer information is available upon request) to the α-peptide coding region for β-galactosidase of pBBR1MCS-4, a 467-bp AgeI-SacI fragment from pBA20 was deleted. The resulting plasmid was designated pBA21. The lacZ gene (encoding β-galactosidase) lacking the start points of transcription and translation and six additional amino acids (designated ′lacZ) was amplified by PCR by using Pfu DNA polymerase (Promega, Madison, Wis.), primers NTlacZ and CTlacZ (see the supplemental material), which introduced BamHI and XbaI sites into the flanking regions, and pAB2001 (1) as the template. The PCR product was cloned into pGemT-easy (Promega), yielding pGTZ. The ′lacZ gene was excised on a BamHI-XbaI fragment from pGTZ and ligated into the same sites of pBA21; the resulting plasmid was designated pBZ (Fig. 1B).
Construction of cgs-′phoA and cgs-′lacZ translational fusions.
Two series of fusion plasmids were constructed, in which various lengths of the 5′ end of the cgs gene amplified by PCR were fused to the phoA and lacZ genes by molecular replacement in pBA and pBZ, respectively (Fig. 1). The 5′-end primer used in each PCR was NCGS containing the AvrII restriction site (see the supplemental material). The individual 3′-end primers used to generate fusions at positions 39, 156, 448, 597, 880, 926, 967, 1001, 1106, 1815, 2095, and 2565 contained the KpnI restriction site (cgs-′phoA fusions) or the BamHI restriction site (cgs-′lacZ fusions) (see the supplemental material). The PCR products were cloned into pGemT-easy, and the resulting plasmids were digested with AvrII-KpnI and with AvrII-BamHI and cloned into pBA and pBZ, respectively, digested with the same enzymes. In this way the AvrII-KpnI and AvrII-BamHI fragments containing the complete cgs gene from pBA and pBZ, respectively, were replaced by the 5′-end fragments of cgs amplified by PCR (Fig. 1). The resulting plasmids were designated pBA and pBZ with a number indicating the position of the Cgs endpoint amino acid. In a similar way, the fusion proteins were designated with a number indicating the Cgs endpoint amino acid and a capital letter (A for AP fusions and Z for βG fusions). Constructs were sequenced to verify the correct frame of the fusions with primers PHOA (annealing to phoA) and LACZ (annealing to lacZ) (see the supplemental material). In the constructs in which the correct frame was verified, the amplified DNA was fully sequenced to confirm PCR fidelity.
Fusion plasmids were introduced into B. abortus strain 2308 by biparental mating by using E. coli S17.1(λpir) as the donor strain. Transconjugants were selected in plates containing ampicillin (50 μg/ml), nalidixic acid (5 μg/ml), and 40 μg of 5-bromo-3-indolyl phosphate p-toluidine salt per ml or 40 μg of 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal) per ml.
Alkaline phosphatase and β-galactosidase activities.
B. abortus 2308 strains containing derivatives of pBA and pBZ harboring cgs-′phoA and cgs-′lacZ gene fusions, respectively, were grown in brucella broth with the appropriate antibiotic at 37°C to the mid-exponential phase. Alkaline phosphatase and β-galactosidase activities were assayed in permeabilized cells as described by Manoil (29) and Miller (35), respectively. Each assay was performed in duplicate in at least three separate experiments.
Western blotting.
B. abortus 2308 strains were grown under the conditions described above for the enzyme assays. Cells were collected, the protein concentration was determined by the method of Lowry et al. (28), and 60 μg of total proteins was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (26), followed by electrophoretic transfer to a nitrocellulose membrane (Inmobilon-NC; Millipore). Immunoblotting was performed as previously described (8) with a monoclonal antibody against alkaline phosphatase (Chemicom International, Inc.) or with a monoclonal antibody against β-galactosidase (Promega). Bound antibody was visualized by using horseradish peroxidase-conjugated goat anti-mouse immunoglobulin (DAKO Corp.) and the Supersignal West Pico chemiluminescent substrate detection reagents (Pierce Chemical Co.) according to the manufacturer's instructions. The relative rate of synthesis of the fusion proteins was determined by using the 1D Image Analysis software (Kodak Digital Science).
Cell surface biotinylation.
Cell surface proteins were labeled with the membrane-impermeant biotinylation reagent sulfosuccinimidyl 2-(biotinamido)ethyl-1,3-dithiopropionate (NHS-SS-biotin) (Pierce Chemical Co.), as described previously (11, 15). B abortus strain 2308 harboring pBA20 was grown until the A600 was 1.0. One-milliliter samples of culture bacteria were harvested by centrifugation, washed twice with phosphate-buffered saline (PBS) (pH 8.0), and suspended in 200 μl of PBS (pH 8.0). After this, 10 μl of 1% NHS-SS-biotin was added, and the mixture was incubated at 4°C for 2 or 6 min. After two washes in PBS (pH 8.0)-20 mM glycine, samples were subjected to SDS-PAGE. Proteins were transferred to nitrocellulose membranes and incubated with horseradish peroxidase-conjugated streptavidin (Sigma) or a specific polyclonal antiserum against the Cgs protein (see below). In the latter case, bound antibody was visualized as described above for Western blotting.
Cells from a 1 ml-aliquot from the same culture were suspended in PBS (pH 8.0) containing 5 mM MgCl2 and 1 mM phenylmethanesulfonyl fluoride and disrupted by sonication. Total protein biotinylation was performed with disrupted cells for 6 min at 4°C. After incubation, trichloroacetic acid was added to a final concentration of 10% to precipitate the proteins. The trichloroacetic acid precipitates were analyzed by SDS-PAGE and Western blotting, as described above.
Antibody production.
A recombinant histidine-tagged Cgs protein fragment (from amino acid 540 to amino acid 801) was used to prepare antibody in mice by using a standard scheme of immunization.
Topology prediction.
Hydrophobicity profiles were generated by the method of Kyte and Doolittle (25) by using a sliding window of 21 residues. Topology predictions were carried out by using the TopPred II (12), TMHMM (24), SOSUI (36), and HMMTOP (41, 42) programs. All of these programs are accessible on the World Wide Web (http://bo.expasy.org).
Sequencing.
DNA sequencing was carried out by the dideoxy method with an automated model 373 DNA sequencer (Perkin-Elmer Applied Biosystems Division, Foster City, Calif.) used according to the manufacturer's instructions.
RESULTS
Topology prediction of cyclic glucan synthase.
Hydropathy analysis with the algorithm of Kyte and Doolittle (25) predicted that Cgs possesses at least three extremely hydrophobic regions that could constitute TMSs. The regions of the protein that correspond to the TMSs predicted with the different programs (see below) are indicated in Fig. 2A.
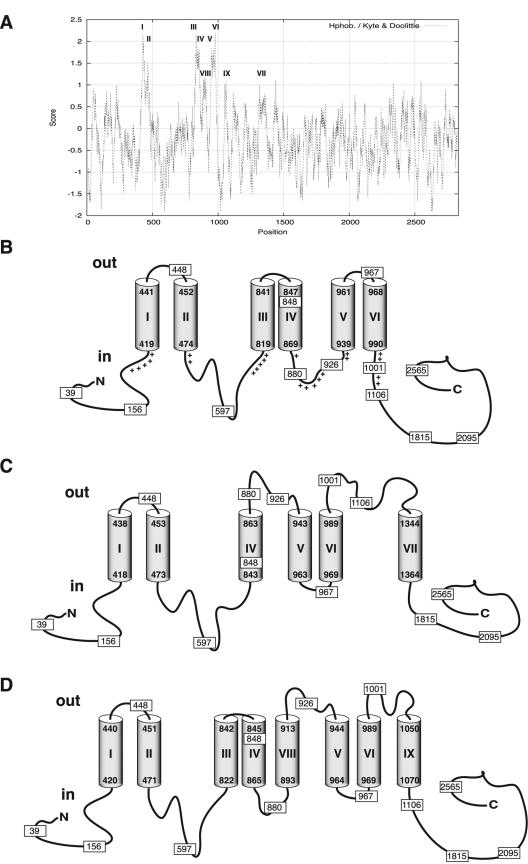
FIG. 2.
Hydropathy analysis and membrane topology prediction for cyclic glucan synthase. Hydrophobicity profiles were generated by the method of Kyte and Doolittle (25) by using a sliding window of 21 residues (A). Alternative topology models were generated by using the programs SOSUI (36) and HMMTOP (41, 42) (B); TopPred II with the GES hydropathy scale (window size, 21 residues) (12) (C); and TopPred II with the KD hydropathy scale (window size, 21 residues) (12) and TMHMM (24) (D). I to IX correspond to the predicted TMSs of Cgs. Cylinders represent membrane-spanning segments. The positions of Cgs-AP and Cgs-βG fusions are indicated by rectangles, and the numbers indicate the numbers of Cgs amino acids fused to the reporter enzyme. The plus signs in panel B indicate positively charged residues in the vicinity of the TMSs.
Topology prediction analyses of Cgs were performed by using the programs SOSUI (36), HMMTOP (41, 42), and TopPred II with the Goldman-Engelman-Steitz (GES) or Kyte-Doolittle (KD) hydropathy scale (window size, 21 residues) (12) and TMHMM (24).
The SOSUI and HMMTOP programs gave similar predictions, and there were only slightly differences concerning the limits of the transmembrane segments (36, 41, 42). Both programs predicted six TMSs (TMSs I to VI) (Fig. 2B). The TopPred II program used with the GES scale also predicted six TMSs with high scores; however, the distribution was different (12). An additional TMS (TMS VII) between residues 1344 and 1364 was predicted instead of TMS III (Fig. 2B and C). Finally, an analysis was performed by using the programs TopPred II with the KD scale and TMHMM (12, 24). Both of these programs predicted, with high scores, eight TMSs (TMSs I, II, III, IV, V, VI, VIII, and IX). As shown in Fig. 2D, two additional TMSs, one between residues 893 and 913 (TMS VIII) and one between residues 1050 and 1070 (TMS IX), were also predicted; however, these programs did not predict TMS VII of Fig. 2C.
The predictions based on this analysis were used to generate the experimental strategy in which the enzymes alkaline phosphatase and β-galactosidase were used as topological reporters.
Determination of the topology of Cgs by means of alkaline phosphatase and β-galactosidase fusions.
A collection of fusions to AP and βG were obtained as described in Materials and Methods. The combined use of the two reporter enzymes had the advantage of providing positive enzymatic activity signals for both periplasmic and cytoplasmic domains. Based on the predicted topological models, AP and βG fusions were constructed at the same amino acid in all putative periplasmic and cytoplasmic regions, as well as in the amino- and carboxyl-terminal domains (Fig. 2). Generation of both AP and βG fusions at exactly the same amino acid permitted evaluation of the subcellular location of a particular amino acid based on a comparison of the two enzymatic activities. In order to avoid possible altered insertion of the protein into the membrane due to a different membrane composition, the activities and expression of the fusion proteins were evaluated in the original background, B. abortus 2308, and not in E. coli.
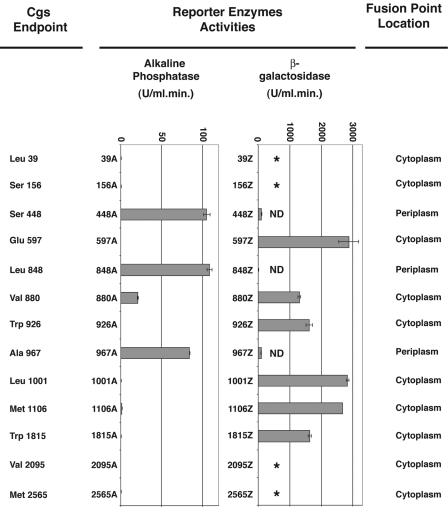
Nine fusions to AP and βG were constructed at positions 448, 597, 848, 880, 926, 967, 1001, 1106, and 1815 (Fig. 2). At positions 39, 156, 2095, and 2565 only fusions to AP were constructed. AP and βG activities were measured in permeabilized cells as described in Materials and Methods; the results are shown in Fig. 3.
FIG. 3.
Activities of Cgs-AP and Cgs-βG fusion proteins. Alkaline phosphatase and β-galactosidase enzyme assays were performed with permeabilized cells of B. abortus strain 2308 carrying the fusion plasmids. The bar graph data are the means and standard errors (in units of enzyme activity) for two separate enzyme activity determinations for a single set of permeabilized cells. Similar results were obtained in three different experiments. Activities were corrected by subtracting the background activity of B. abortus 2308 harboring plasmid pBA or pBZ and were normalized by using the rate of synthesis of the fusion proteins, as described in Materials and Methods. The activities of Cgs-AP fusion proteins having very low activities are as follows: 39A, 0.668 ± 0.007 U; 156A, 0.653 ± 0.04 U; 597A, 0.212 ± 0.072 U; 926A, 0.231 ± 0.042 U; 1001A, 0.776 ± 0.081U; 1106A, 1.5 ± 0.218 U; 1815A, 0.573 ± 0.098 U; 2095A, 0.137 ± 0.045 U; and 2565A, 0.302 ± 0.05 U. The activities of Cgs-βG fusion proteins having low activities are as follows: 448Z, 109.2 ± 3.8 U; 848Z, 20.2 ± 0.1 U; and 967Z, 95.2 ± 2.3 U. An asterisk indicates that a plasmid expressing the fusion was not constructed. ND indicates that the fusion protein was not detected by immunoblotting.
The enzymatic activities of the fusion proteins led to localization of the fusion sites, as summarized in Fig. 3. The lack of AP activity of fusions 880A, 926A, 1001A, and 1106A and the high βG activities of fusions 880Z, 926Z, 1001Z, and 1106Z support the conclusion that the regions of the protein containing residues 880, 926, 1001, and 1106 are located on the cytoplasmic side of the membrane. These results also revealed that the regions of the protein between residues 893 and 913 (TMS VIII) and between residues 1050 and 1070 (TMS IX) are located in the cytoplasm and do not span the membrane, as suggested by the predicted topology in Fig. 2D. Moreover, the high AP activity detected for fusion 967A supports the hypothesis that residue 967 is located in the periplasmic space, which is different from what is proposed in Fig. 2C and D. The lack of AP activity and the high βG activities of fusions at residues 1106 and 1815 do not support the existence of TMS VII (residues 1344 to 1364) as predicted in Fig. 2C. Furthermore, the high AP activity of the fusion at residue 848 and the reporter activities of the fusions to AP and βG at residue 597 support the existence of TMS III (residues 819 to 841), which is predicted in Fig. 2B and D but not in Fig. 2C.
Taken together, all these results indicate that the membrane topology of Cgs agrees with the predicted model shown in Fig. 2B and differs from the models shown in Fig. 2C and D.
Analysis of fusion protein expression.
In order to determine the level of expression of fusion proteins, whole-cell extracts of B. abortus 2308 containing cgs-′phoA and cgs-′lacZ gene fusions were obtained and subjected to immunoblot analysis with monoclonal antibodies against alkaline phosphatase and β-galactosidase as described in Materials and Methods.
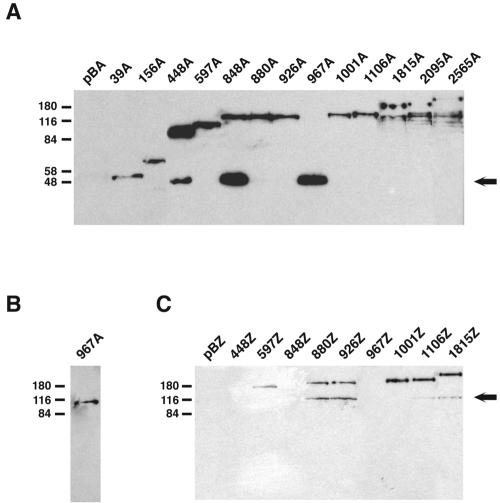
In all cases, the fusion proteins had the expected molecular weight. Fusion proteins 448A, 848A, and 967A appeared to be degraded, producing a protein with an electrophoretic mobility similar to that expected for alkaline phosphatase (∼47 kDa) (Fig. 4A). Similar degradation products have been observed previously in other phoA-based topology studies (2, 9, 31). They are thought to be the result of the action of a periplasmic protease that releases a properly folded AP moiety from the fusion protein. Therefore, the appearance of these degradation products may be a useful indicator of AP export. Accordingly, degradation products that were the size of AP were observed in all samples with high levels of enzyme activity. Such degradation products were absent in samples with low levels of activity. In fusion protein 967A only the degradation product that was the size of AP was observed, indicating that in this case the cleavage was total and not partial, as occurs in fusion proteins 448A and 848A (Fig. 4A). To confirm this result, a whole-cell extract of B. abortus 2308 harboring pBA967 was subjected to immunoblot analysis with a specific polyclonal antiserum against Cgs. A protein with the expected molecular mass (∼108 kDa) corresponding to a Cgs fragment consisting of 967 amino acids was detected (Fig. 4B). Fusion proteins 1815A, 2095A, and 2565A were also partially degraded, but a degradation product that was the size of AP was absent in these fusion proteins (Fig. 4A).
FIG. 4.
Western blot analysis of fusion proteins. Whole-cell extracts of B. abortus 2308 containing derivatives of pBA and pBZ harboring cgs-′phoA and cgs-′lacZ gene fusions, respectively, were subjected to SDS—10% PAGE. Proteins were transferred to nitrocellulose and developed with a monoclonal antibody against AP (A), a specific polyclonal antiserum against Cgs (B), or a monoclonal antibody against β-galactosidase (C). pBA and pBZ are B. abortus 2308 strains carrying control plasmids pBA and pBZ, respectively. Lanes are labeled with the designations of the fusion proteins (Fig. 3). The positions of molecular mass standards (in kilodaltons) are indicated on the left. The arrows on the right indicate degradation products whose sizes are similar to the sizes of AP (∼47 kDa) and β-galactosidase (∼116 kDa).
Fusion proteins 880Z, 926Z, 1106Z, and 1815Z had polypeptides with an electrophoretic mobility similar to that expected for native β-galactosidase (∼116 kDa) (Fig. 4C). These degradation products may imply that cytoplasmic β-galactosidase is liberated by proteolysis of membrane-associated fusion proteins. Fusion proteins 448Z, 848Z, and 967Z did not produce a protein readily detectable by immunoblotting (Fig. 4C); therefore, they were not considered in the analysis. As judged by AP fusion results, residues 448, 848, and 967 are located in the periplasm. Fusions of β-galactosidase to periplasmic domains are believed not to mediate export of β-galactosidase across the membrane; rather, the enzyme remains embedded within the membrane, which prevents proper folding. This could be the reason for the instability of these fusion proteins.
Lysine biotinylation.
To validate the model proposed in Fig. 2B and discard the other predicted models shown in Fig. 2C and D, we used an independent approach based on site-directed chemical labeling of key residues. Selective amino acid labeling with biotin is an alternative approach for identifying external loops. Whether this strategy can be used depends on the location of key residues (generally Lys or Cys). Cgs contains 88 Lys residues, most of which (59 residues) are located in the C-terminal region of the protein between amino acids 990 and 2831. Fifteen Lys residues are located in the N-terminal region from residue 1 to residue 420. According to the model shown in Fig. 2B, no Lys residues are located in the regions of Cgs facing the periplasmic space. However, according to the models shown in Fig. 2C and D, 11 and 2 Lys residues, respectively, are located in the periplasm and accessible to external reagents.
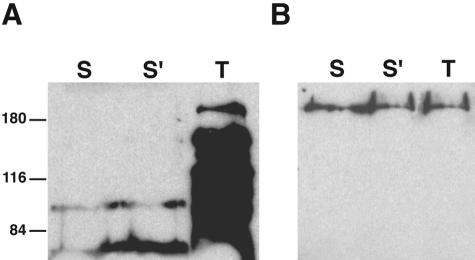
We tested the extracellular accessibility of lysine residues to the membrane-impermeant biotinylation reagent NHS-SS-biotin. This reagent is particularly useful because of its high accessibility to small external loops, and it has been employed recently to characterize the external loop topology in the serotonin transporter (11) and in the acyl-lipid desaturase from Bacillus subtilis (15). Cells of B. abortus 2308 harboring pBA20 were treated with this reagent and analyzed by gel electrophoresis and Western blotting by using horseradish peroxidase-conjugated streptavidin (Fig. 5A), and the same membrane was stripped and reprobed with a specific polyclonal antiserum against the Cgs protein (Fig. 5B). The same procedure was performed with lysed cells. Biotinylated Cgs was identified in lysed cells but not in intact cells (Fig. 5A). As expected, the protein was detected in both cases with the specific anti-Cgs antiserum in 2308 harboring pBA20 (Fig. 5B). As a control, the same experiment was carried out with a cgs mutant (data not shown). In this case, the protein was not detected in lysed or intact cells. These experiments confirmed the proposed model shown in Fig. 2B and eliminated the alternative models shown in Fig. 2C and D.
FIG. 5.
Lysine biotinylation of Cgs. Cells of B. abortus 2308 harboring pBA20 were incubated with the membrane-impermeant biotinylation reagent NHS-SS-biotin (Pierce Chemical Co.), as described in Materials and Methods. Cell surface proteins were biotinylated for 2 min (lane S) or 6 min (lane S′). Total cell proteins were biotinylated for 6 min after cell disruption by sonication (lane T). Biotinylated proteins were subjected to SDS—10% PAGE and developed by Western blotting by using horseradish peroxidase-conjugated streptavidin (A). The same membrane was stripped and reprobed with a specific polyclonal antiserum against the Cgs protein (B). The positions of molecular mass standards (in kilodaltons) are indicated on the left.
DISCUSSION
Understanding the mechanism of action of B. abortus cyclic β-1,2-glucan synthase at the molecular level requires knowledge of the membrane arrangement of the protein. The membrane topology was studied by using a combination of in silico predictions, a genetic approach involving construction of fusions between the cgs gene and the genes encoding alkaline phosphatase (phoA) and β-galactosidase (lacZ), and site-directed chemical labeling of lysine residues. These techniques have been used in the past to elucidate the topology of a number of membrane proteins (10, 15, 18, 29-31, 33, 34).
By using computer analyses, three alternative models of the orientation of Cgs in the membrane were predicted (Fig. 2). Based on these models, alkaline phosphatase and β-galactosidase fusions were constructed in every putative periplasmic and cytoplasmic domain and in the N-terminal and C-terminal regions of the protein. The activities and expression of the fusion proteins were analyzed (Fig. 3 and 4). All the results are in good agreement with the predicted model shown in Fig. 2B and do not support the predicted models shown in Fig. 2C and D. It is worth noting that the data obtained from the in silico analysis and the generation of gene fusions in the desired positions allowed us to reduce the number of fusions required to experimentally determine the membrane topology of this huge protein. Finally, we employed an independent approach based on site-directed chemical labeling of lysine residues (Fig. 5) to validate the proposed model.
All the results together support the topology model of Cgs proposed in Fig. 2B. This model shows that Cgs is a polytopic membrane protein with the amino and carboxyl termini located in the cytoplasm and with six transmembrane segments, TMS I (residues 419 to 441), TMS II (residues 452 to 474), TSM III (residues 819 to 841), TSM IV (residues 847 to 869), TSM V (residues 939 to 961), and TSM VI (residues 968 to 990).
Transmembrane stretches of membrane proteins usually can be identified as long sequences composed mostly of hydrophobic amino acids. However, features of the amino acid residues other than their hydrophobicity may contribute to the topology of proteins in the membrane. It has been shown that polytopic integral membrane proteins from both prokaryotic and eukaryotic organisms generally have a net positive charge in the cytoplasmic polar regions of the protein, which connect transmembrane segments, and a net negative charge in the extracytoplasmic polar domain. This is known as the positive-inside rule and is generally applied to loops smaller than 65 residues (45). It has also been shown that the number of positively charged amino acid residues, rather than the total content of charged residues or the net charge, of a particular region controls its location relative to the membrane (37). All the periplasmic loops of Cgs possess either a net negative charge or no net charge at all, with no more than one positively charged amino acid residue (Table 2). The cytoplasmic region between TMSs IV and V (residues 870 to 938) possesses a net positive charge (Fig. 2B and Table 2). In the cytoplasmic loops longer than 65 residues, the number of positively charged residues and the net charge in the vicinity of the TMSs were calculated (Fig. 2B and Table 2). In some of these regions, although the net charge is not positive, the number of positively charged amino acid residues is high, a fact that probably contributes to the orientation of the corresponding TMSs. Accordingly, the distribution of charged amino acid residues in the proposed topological model of Cgs is consistent with the positive-inside rule.
TABLE 2.
Distribution of charged residues in the proposed topological model of Cgs
| Cgs region | No. of residues | Locationa | qb | q′c |
|---|---|---|---|---|
| 1-418 | 418 | Cyt | ≫ | 5/0 (q = 5) |
| 442-451 | 10 | Per | 1/1 (q = 0) | |
| 475-818 | 344 | Cyt | ≫ | 2/2 (q = 0) 5/1 (q = 4) |
| 842-846 | 5 | Per | 0/0 (q = 0) | |
| 870-938 | 69 | Cyt | 8/6 (q = +2) | |
| 962-967 | 6 | Per | 0/1 (q = −1) | |
| 991-2831 | 841 | Cyt | ≫ | 4/4 (q = 0) |
Cyt, cytoplasmic; Per, periplasmic.
≫, domains contain much more than 65 residues. q is the net charge calculated as follows: K + R/D + E (positively charged residues/negatively charged residues).
q′ is the net charge calculated as follows: K + R/D + E (positively charged residues/negatively charged residues), before or after the TMSs, taking up to 20 amino acid residues into account.
Fusion proteins 880A and 926A have fusion joints at the beginning and end, respectively, of the cytoplasmic region between TMSs IV and V (residues 870 to 938) of the protein. They exhibit 21.1 and 0.231 U of AP activity, respectively, whereas AP fusions to periplasmic domains exhibit up to 109 U of activity (Fig. 3). This pattern, in which fusions to the beginning of a cytoplasmic domain exhibit considerably higher AP activity than fusions to the end of the same domain, was also seen with fusions to cytoplasmic domains of the E. coli MalF protein (4, 5). The number of positively charged residues and the net charge of the loop between TMSs IV and V (residues 870 to 938) are 8 and 2, respectively. For the region between residues 870 and 926 these values are 6 and 1, respectively. In contrast, for the region between residues 870 and 880 the number of positively charged residues and the net charge are 1 and 0, respectively (Fig. 2B). As shown by Boyd and Beckwith (4), export of carboxyl-terminally fused alkaline phosphatase is increased when the positive charge of the cytoplasmic domain is reduced with a concomitant increase in AP activity, as observed with fusion protein 880A. Conversely, carboxyl-terminally fused alkaline phosphatase is almost completely retained in the cytoplasm with a reduction in AP activity when the positive charge is increased, as observed with fusion protein 926A.
In the proposed topological model of Cgs (Fig. 2B) all the TMSs are located in the first one-third of the protein (residues 1 to 990), a region of the protein known to be required for the synthesis of cyclic β-1,2-glucan (see above) (20). The six TMSs determine four large cytoplasmic domains and three very small periplasmic regions. According to this topology, the four cytoplasmic domains of the protein are anchored to the membrane by the six TMSs and three small extracytoplasmic domains that keep the TMSs together. This topological structure may contribute to ensuring the correct folding and enzymatic activity of the cytoplasmic domains of the protein. It can also be speculated that the localization of the enzyme in the vicinity of the membrane is important for the transport of its product (cyclic glucan) into the periplasmic space by the action of the cyclic glucan transporter Cgt (which is also an integral inner membrane protein) (38). Experimental evidence has suggested that in Agrobacterium tumefaciens the cyclic glucan synthase (ChvB) interacts with the cyclic glucan transporter (ChvA) (21). Sequence analysis of the amino terminus of Cgs (residues 1 to 400) with the 3D-pssm program (Imperial College of Science, Technology & Medicine [http://www.sbg.bio.ic.ac.uk) (22) revealed a predicted fold similar to the fold of the sigma 70 subunit fragment of E. coli RNA polymerase, the peroxisomal targeting signal-1 receptor, and the tetratricopeptide repeat motif. All these protein folds are involved in protein-protein interactions. The tetratricopeptide repeat is a protein-protein interaction module found in multiple copies in a number of functionally different proteins that facilitates specific interactions with a partner protein(s) (3). Further work is required to establish whether Cgs and Cgt interact with each other in the membrane.
Two of the three enzymatic activities of Cgs (initiation and elongation) use UDP-glucose as a sugar donor. Consequently, at least these two reactions must take place on the cytoplasmic side of the membrane, where UDP-glucose is available. Cyclization, a transglucosylation reaction, may occur on the periplasmic side of the membrane. However, the fact that only small loops of the protein are on the periplasmic side of the membrane and the fact that cgt mutants accumulate cyclic glucan in the cytoplasm (38) support the hypothesis that all Cgs enzymatic activities face the cytoplasmic side of the membrane.
The next step towards understanding the mechanism of action of Cgs at the molecular level is trying to dissociate the three different enzymatic activities of this interesting multi-enzyme. It is thought that the active site(s) of the enzyme must be located in the regions of the protein facing the cytoplasm. This study provides the basis for identification and characterization of the functional domains, as well as identification of the active site(s).
Acknowledgments
This work was supported by grant PICT 2000 no. 01-09194 from the Agencia Nacional de Promoción Científica y Tecnológica, SECyT, Buenos Aires, Argentina, and by grant PIP 2346 from the Consejo Nacional de Investigaciones Científicas y Técnicas, CONICET, Buenos Aires, Argentina. A.E.C. and M.S.R. are fellows of CONICET. N.I.D.I. and R.A.U. are members of the research carrier of CONICET.
We thank M. Ehrmann and J. Beckwith for the kind gift of vector pSWFII. We acknowledge J. J. Cazzulo, University of General San Martín, for critical reading of the manuscript and useful suggestions and Pablo Briones, University of General San Martín, for technical assistance.
REFERENCES
- 1.Becker, A., M. Schmidt, W. Jager, and A. Puhler. 1995. New gentamicin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene 162:37-39. [DOI] [PubMed] [Google Scholar]
- 2.Blank, T. E., and M. S. Donnenberg. 2001. Novel topology of BfpE, a cytoplasmic membrane protein required for type IV fimbrial biogenesis in enteropathogenic Escherichia coli. J. Bacteriol. 183:4435-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blatch, G. L., and M. Lassle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932-939. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, D., and J. Beckwith. 1989. Positively charged amino acid residues can act as topogenic determinants in membrane proteins. Proc. Natl. Acad. Sci. USA 86:9446-9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, D., C. Manoil, and J. Beckwith. 1987. Determinants of membrane protein topology. Proc. Natl. Acad. Sci. USA 84:8525-8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briones, G., N. Inon de Iannino, M. Roset, A. Vigliocco, P. S. Paulo, and R. A. Ugalde. 2001. Brucella abortus cyclic beta-1,2-glucan mutants have reduced virulence in mice and are defective in intracellular replication in HeLa cells. Infect. Immun. 69:4528-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briones, G., N. Inon de Iannino, M. Steinberg, and R. A. Ugalde. 1997. Periplasmic cyclic 1,2-beta-glucan in Brucella spp. is not osmoregulated. Microbiology 143:1115-1124. [DOI] [PubMed] [Google Scholar]
- 8.Burnett, W. N. 1981. “Western Blotting:” electrophoretic transfer of proteins from sodium dodecyl sulphate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Ann. Biochem. 112:195-203. [DOI] [PubMed] [Google Scholar]
- 9.Calamia, J., and C. Manoil. 1990. Lac permease of Escherichia coli: topology and sequence elements promoting membrane insertion. Proc. Natl. Acad. Sci. USA 87:4937-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caldwell, A. M., and R. L. Smith. 2003. Membrane topology of the ZntB efflux system of Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:374-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, J. G., S. Liu-Chen, and G. Rudnick. 1998. Determination of external loop topology in the serotonin transporter by site-directed chemical labeling. J. Biol. Chem. 273:12675-12681. [DOI] [PubMed] [Google Scholar]
- 12.Claros, M. G., and G. von Heijne. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10:685-686.7704669 [Google Scholar]
- 13.Corbel, J. M. 1997. Brucellosis: an overview. Emerg. Infect. Dis. 2:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derman, A. I., and J. Beckwith. 1991. Escherichia coli alkaline phosphatase fails to acquire disulfide bonds when retained in the cytoplasm. J. Bacteriol. 173:7719-7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diaz, A. R., M. C. Mansilla, A. J. Vila, and D. de Mendoza. 2002. Membrane topology of the acyl-lipid desaturase from Bacillus subtilis. J. Biol. Chem. 277:48099-48106. [DOI] [PubMed] [Google Scholar]
- 16.Ehrmann, M., D. Boyd, and J. Beckwith. 1990. Genetic analysis of membrane protein topology by a sandwich gene fusion approach. Proc. Natl. Acad. Sci. USA 87:7574-7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Froshauer, S., G. N. Green, D. Boyd, K. McGovern, and J. Beckwith. 1988. Genetic analysis of the membrane insertion and topology of MalF, a cytoplasmic membrane protein of Escherichia coli. J. Mol. Biol. 200:501-511. [DOI] [PubMed] [Google Scholar]
- 18.Gandlur, S. M., L. Wei, J. Levine, J. Russell, and P. Kaur. 2004. Membrane topology of the DrrB protein of the doxorubicin transporter of Streptomyces peucetius. J. Biol. Chem. 279:27799-27806. [DOI] [PubMed] [Google Scholar]
- 19.Hoyer, B. H., and N. B. McCullough. 1968. Homologies of deoxyribonucleic acids from Brucella ovis, canine abortion organisms, and other Brucella species. J. Bacteriol. 96:1783-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iñón de Iannino, N., G. Briones, M. Tolmasky, and R. A. Ugalde. 1998. Molecular cloning and characterization of cgs, the Brucella abortus cyclic beta(1-2) glucan synthetase gene: genetic complementation of Rhizobium meliloti ndvB and Agrobacterium tumefaciens chvB mutants. J. Bacteriol. 180:4392-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iñón de Iannino, N., and R. A. Ugalde. 1989. Biochemical characterization of avirulent Agrobacterium tumefaciens chvA mutants: synthesis and excretion of beta-(1-2)glucan. J. Bacteriol. 171:2842-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelley, L. A., R. M. MacCallum, and M. J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:499-520. [DOI] [PubMed] [Google Scholar]
- 23.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 24.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 25.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Lee, C., P. Li, H. Inouye, E. R. Brickman, and J. Beckwith. 1989. Genetic studies on the inability of beta-galactosidase to be translocated across the Escherichia coli cytoplasmic membrane. J. Bacteriol. 171:4609-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 29.Manoil, C. 1991. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 34:61-75. [DOI] [PubMed] [Google Scholar]
- 30.Manoil, C. 1990. Analysis of protein localization by use of gene fusions with complementary properties. J. Bacteriol. 172:1035-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manoil, C., and J. Beckwith. 1986. A genetic approach to analyzing membrane protein topology. Science 233:1403-1408. [DOI] [PubMed] [Google Scholar]
- 32.Manoil, C., and J. Beckwith. 1985. TnphoA: a transposon probe for protein export signals. Proc. Natl. Acad. Sci. USA 82:8129-8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manoil, C., J. J. Mekalanos, and J. Beckwith. 1990. Alkaline phosphatase fusions: sensors of subcellular location. J. Bacteriol. 172:515-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mazur, A., J. E. Krol, M. Marczak, and A. Skorupska. 2003. Membrane topology of PssT, the transmembrane protein component of the type I exopolysaccharide transport system in Rhizobium leguminosarum bv. trifolii strain TA1. J. Bacteriol. 185:2503-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Mitaku, S., and T. Hirokawa. 1999. Physicochemical factors for discriminating between soluble and membrane proteins: hydrophobicity of helical segments and protein length. Protein Eng. 12:953-957. [DOI] [PubMed] [Google Scholar]
- 37.Nilsson, I., and G. von Heijne. 1990. Fine-tuning the topology of a polytopic membrane protein: role of positively and negatively charged amino acids. Cell 62:1135-1141. [DOI] [PubMed] [Google Scholar]
- 38.Roset, M. S., A. E. Ciocchini, R. A. Ugalde, and N. Iñón de Iannino. 2004. Molecular cloning and characterization of cgt, the Brucella abortus cyclic beta-1,2-glucan transporter gene, and its role in virulence. Infect. Immun. 72:2263-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Sangari, F. J., J. M. Garcia-Lobo, and J. Aguero. 1994. The Brucella abortus vaccine strain B19 carries a deletion in the erythritol catabolic genes. FEMS. Microbiol. Lett. 121:337-342. [DOI] [PubMed] [Google Scholar]
- 41.Tusnady, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 42.Tusnady, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 43.Ugalde, R. A. 1999. Intracellular lifestyle of Brucella spp. Common genes with other animal pathogens, plant pathogens, and endosymbionts. Microbes Infect. 1:1211-1219. [DOI] [PubMed] [Google Scholar]
- 44.Verger, J. M., F. Grimont, P. A. D. Grimont, and M. Crayon. 1985. Brucella, a monospecific genus as shown by deoxyribonucleic acid hybridization. Int. J. Syst. Bacteriol. 35:292-295. [Google Scholar]
- 45.von Heijne, G. 1986. The distribution of positively charge residues in bacterial inner membrane proteins correlates with the transmembrane topology. EMBO J. 5:3021-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]