Abstract
The σS subunit of RNA polymerase, the product of the rpoS gene, controls the expression of genes responding to starvation and cellular stresses. Using gene array technology, we investigated rpoS-dependent expression at the onset of stationary phase in Escherichia coli grown in rich medium. Forty-one genes were expressed at significantly lower levels in an rpoS mutant derived from the MG1655 strain; for 10 of these, we also confirmed rpoS and stationary-phase dependence by reverse transcription-PCR. Only seven genes (dps, osmE, osmY, sodC, rpsV, wrbA, and yahO) had previously been recognized as rpoS dependent. Several newly identified rpoS-dependent genes are involved in the uptake and metabolism of amino acids, sugars, and iron. Indeed, the rpoS mutant strain shows severely impaired growth on some sugars such as fructose and N-acetylglucosamine. The rpoS gene controls the production of indole, which acts as a signal molecule in stationary-phase cells, via regulation of the tnaA-encoded tryptophanase enzyme. Genes involved in protein biosynthesis, encoding the ribosome-associated protein RpsV (sra) and the initiation factor IF-1 (infA), were also induced in an rpoS-dependent fashion. Using primer extension, we determined the promoter sequences of a selection of rpoS-regulated genes representative of different functional classes. Significant fractions of these promoters carry sequence features specific for EσS recognition of the −10 region, such as cytosines at positions −13 (70%) and −12 (30%) as well as a TG motif located upstream of the −10 region (50%), thus supporting the TGN0-2C(C/T)ATA(C/A)T consensus sequence recently proposed for σS.
Bacterial cells undergo a variety of morphological and physiological changes as they enter stationary phase. Several global regulators, involved in a complex regulatory network, induce the expression of stationary-phase-responsive genes (reviewed in reference 34). Among these, the rpoS-encoded alternative sigma factor σS plays a major role as a regulator of genes involved in stress responses (32). The σS protein accumulates in stationary phase as well as in response to stress conditions and directs transcription from about 100 genes (41, 47, 85). The high conservation (61) and the tight control of RpoS accumulation in response to a wide range of environmental stimuli are consistent with its crucial role in bacterial physiology (36). rpoS-regulated genes encode a variety of proteins with unrelated physiological functions, and mutations in rpoS have pleiotropic effects. Strains carrying nonfunctional rpoS alleles fail to express acid phosphatase (appA) (91), as well as oxidative stress genes such as superoxide dismutase (sodC) and catalase (katE and katG). rpoS-dependent regulation of oxidative stress genes is often mediated by additional regulatory proteins such as FNR, OxyR, or Fur (69). The rpoS gene also regulates the expression of DNA repair enzymes such as the exonuclease xthA (83), the methyl transferase ada (90), and the nonspecific DNA binding protein dps (3). The σS protein is also required for expression of the acid resistance gadA and gadB genes (21), for the response to either osmotic or temperature shifts (32), and in the expression of genes determining cell morphology, such as bolA (51). Finally, it regulates the expression of several virulence factors in pathogenic Escherichia coli strains e.g., csgBA genes encoding curli (6).
Sigma factors associate with core RNA polymerase (RNAP) to determine specific promoter recognition. The σ70 protein, encoded by the rpoD gene, is the main σ factor in Escherichia coli and is responsible for the transcription of most genes. Alternative σ factors compete with σ70 for core RNA polymerase to form the corresponding holoenzymes (e.g., σS-associated RNAP, or EσS) and direct transcription of genes belonging to specific functional classes (41, 58). Unlike other alternative σ factors, σS recognizes promoters with sequences very similar to σ70-dependent promoters, raising the problem of specific promoter recognition in vivo. Indeed, promoter alignment of rpoS-dependent genes suggests a −10 consensus sequence for EσS [CTATA(A/C)T] basically identical to the Eσ70 consensus (TATAAT) (23, 55). This was confirmed by in vitro systematic evolution of ligands by exponential enrichment (SELEX), which also suggested recognition of an identical −35 sequence (TTGACA) (24). These data are consistent with the strong similarity in the DNA binding domains (2.4 and 4.2) between the two σ factors (57) and with the observation that EσS can initiate transcription at many σ70-dependent promoters in vitro (88). On the other hand, Eσ70 can transcribe some σS-dependent genes in the presence of additional factors or in the absence of specific repressors such as H-NS (6, 22, 87; reviewed in reference 56). Thus, at least for a subclass of promoters, promoter selectivity between Eσ70 and EσS in vivo might be determined by factors other than DNA sequence, such as increased intracellular salt concentrations, degree of DNA supercoiling, modulation by additional regulators (37, 41; reviewed in reference 34), or signal molecules such as the ppGpp alarmone (43, 46). However, specific deviations from the common consensus sequence might favor either form of RNAP; for instance, the presence of a C immediately upstream of the −10 element (−13C [9]), flexibility in the location of the TG motif, and the presence of a C as the first nucleotide of the −10 sequence might favor promoter recognition by EσS (47).
In this work, we have studied the expression of rpoS-dependent genes at the onset of stationary phase in cell grown in rich medium. We observed rpoS-dependent induction of amino acid and carbohydrate metabolism genes, consistent with the role of RpoS in response to starvation. We show that rpoS controls the production of indole, which plays a role as a signal molecule in stationary phase. Finally, our results show that rpoS also regulates the expression of genes involved in protein synthesis and that of a large number of genes with unknown function, underlining the complexity of the RpoS regulon. We characterized the promoter sequences in a selection of these genes by using primer extension. Our data strongly support the importance of the proposed σS-specific features in the −10 region (−12C, −13C, and TG) as well as the lack of conservation of the −35 promoter element.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and determination of indole production.
The rpoS derivative (EB1.3 [78]) of E. coli strain MG1655 that was used in the present study was obtained by phage P1 transduction from strain MV2792 (93). For RNA extraction, bacterial cultures were grown overnight in Luria broth (LB) medium at 37°C with vigorous aeration, diluted 1:100 in LB medium, and then grown to an optical density at 600 nm (OD600) of 0.2 and rediluted 1:100 in 15 ml of prewarmed LB in 100-ml flasks to avoid carryover of proteins accumulated in late-stationary phase. Samples for RNA isolation, either for gene array and primer extension or for real-time PCR experiments, were collected either at an OD600 of 2.5 to 3, a cell density that in our growth curve corresponds to the onset of stationary phase, or at an OD600 of 0.6 to 0.8 (mid-exponential phase of growth). For growth on limiting sugar concentrations, 1 ml of a bacterial suspension from an overnight culture in LB medium was centrifuged, washed, and resuspended in sterile phosphate-buffered saline. Twenty microliters of bacterial suspensions was used to inoculate 2 ml of M9 medium supplemented with different sugars as the sole carbon source, either at 10 mM (control cultures) or at 0.1 mM (carbon-limiting conditions), and bacteria were grown at 37°C with full aeration for 24 h. Aliquots of the overnight cultures prior to and after centrifugation were plated on L agar plates; plate counts showed very similar bacterial concentrations for strains MG1655 and EB1.3 (ca. 5 × 109 CFU/ml).
For determination of indole production, we followed the method of indole conversion into indigo described in reference 71, with minor modifications. Both strains MG1655 and EB1.3 were transformed with the pStyABB plasmid, which constitutively expresses the styrene monooxygenase styAB genes from Pseudomonas strain S12 (71). In addition to styrene oxidation, the StyAB protein also catalyzes the transformation of indole into indigo, whose blue color can be determined spectrophotometrically at OD620 after lysis of the cells with dimethyl sulfoxide (DMSO) (71). Aliquots (1 ml) of either MG1655/pStyABB or EB1.3/pStyABB cultures grown in LB medium were taken at different times; the bacterial cells were centrifuged and lysed in DMSO, and the presence of indigo was determined spectrophotometrically. In an alternative method, 1-ml supernatants from overnight cultures of either MG1655 or EB1.3 were used to resuspend an equal volume of MG1655/pStyABB culture from the early-exponential phase of growth (OD600, 0.25 to 0.3). The cells were incubated with the spent medium for 30 min at 37°C before DMSO lysis and spectrophotometric determination of indigo production.
RNA isolation, cDNA labeling, and hybridization.
Cells were harvested by centrifugation, and RNA was isolated with the RNeasy mini kit (QIAGEN). Total-RNA purification was performed by on-column DNase I digestion according to the manufacturer's instructions. RNA samples were quantified by using a spectrophotometer (260 nm), checked by gel electrophoresis, and stored at −80°C until further use. For microarray hybridization, 25 μg of RNA samples was used as a template for reverse transcriptase to produce cDNA labeled with either Cy3- or Cy5-dCTP (Cyscribe first-strand cDNA kit; Amersham Biosciences). Four different array hybridizations were performed using RNA samples extracted from two independent cultures under analogous conditions. To correct for possible differences in Cy3 and Cy5 dye incorporation, the cDNAs were labeled with Cy3 dye in two hybridizations and with Cy5 dye in the other two (dye swapping). For each of the four experiments, the Cy3- and Cy5-labeled cDNAs, products of the reverse transcriptase reactions, were pooled, purified by using QIAGEN Qiaquick spin columns, and concentrated with a Microcon-30 concentrator (Millipore) prior to addition of the hybridization buffer. For our investigation we used the E. coli K-12 V2 array (MWG), which contains 4,286 genes (http://www.mwg-biotech.com). Hybridizations were run overnight at 42°C in the buffer supplied, and subsequent washing steps with SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) were performed at room temperature (22°C).
Procedure for microarray data analysis.
Sixteen-bit tagged-image format file (TIFF) images produced by the scan of the microarray slides were analyzed with Affymetrix 428 Scanalyse and Jaguar software (version 2; Affymetrix), and data were managed by using Microsoft Excel. No ribosomal DNA spots are available on the MWG array slides for normalization. Thus, normalization was computed by using Jaguar software (average method): the mean values of an array are corrected by a factor which equalizes the global intensity of the signals resulting from each channel. An average background was determined for each individual hybridization experiment from the background values given by Jaguar for each spot with a specific dye. The normalized intensities of the spots were sorted according to the signals obtained for the wild-type MG1655 control samples (Cy3- or Cy5-labeled cDNA), and values lower than 1.3 times the average background were eliminated. This subtraction was performed in order to eliminate spots with nonsignificant expression levels in the wild-type strain, which were therefore unsuitable for addressing down-regulation due to the rpoS mutation. Several known rpoS-dependent genes (e.g., bolA, katE) fell into this category and were not further considered in our analysis, despite a high wild-type/rpoS ratio. Data from the four array experiments were flagged, pooled, and sorted to identify reproducible spots. Genes corresponding to spots with a wild type/rpoS signal ratio (R) of >2.5 in at least three out of four hybridizations, which never showed an R of <1 and whose final average ratio was ≥2.5, were considered down-regulated in the rpoS strain and are listed in Table 2.
TABLE 2.
Genes induced at the onset of stationary phase with σs -dependent expression as identified by microarray analyses
| Gene | Gene product | Functional classa | Mean WT valueb | Ratio (WT/rpoS) SDc | Regulationd |
|---|---|---|---|---|---|
| Starvation and stress response genes | |||||
| dpsS | DNA protection protein, bacterial ferritin | SSR | 1,597 | 2.5 ± 0.4 | IHFS, OxyRS, rssBTC, MMC, PQ, NaSal, Lrp, BG, DHCP, OS |
| osmES | Stress-inducible outer membrane lipoprotein | SSR | 1,728 | 9.6 ± 2.1 | FisS, NaSal, Lrp, OS |
| osmYS | Hyperosmotically inducible periplasmic protein | SSR | 424 | 4.4 ± 1.3 | CRPS, LrpS, IHFS, H-NSS, arcBTC, MMC, Lrp, Rcs, BG, DHCP |
| psiF | Induced by phosphate starvation | 656 | 4.8 ± 0.4 | ompR/envZTC, CD | |
| sodCS | Superoxide dismulase precursor (Cu-Zn) | SSR | 459 | 2.7 ± 0.5 | uvrYTC, arcBTC |
| Protein synthesis | |||||
| rpsVS | 30S ribosomal subunit protein S22 | PS | 3,466 | 5.2 ± 2.8 | FISS, IHFS, ppGppS |
| infA | Protein chain initiation factor IF-1 | PS | 1,011 | 2.7 ± 0.8 | |
| Amino acid transport and metabolism | |||||
| artP | Arginine transport, ATP-binding component | AATM | 714 | 2.6 ± 0.7 | PQ, NaSal, Lrp |
| artI | Arginine transport, periplasmic binding protein | AATM | 346 | 3.8 ± 1.9 | PQ, Lrp |
| ansP | L-Asparagine permease | AATM | 572 | 2.5 ± 0.4 | DHCP |
| ilvD | Dihydroxyacid dehydratase | AATM | 364 | 4.8 ± 2.1 | ompR/envZTC |
| tnaA | Tryptophanase | AATM | 7,808 | 13.7 ± 4.3 | phoQTC, ompR/envZTC, CRP, uvrYTC, arcATC, arcBTC, rcsBTC, citABTC, MMC, PQ (ilvG: Lrp) |
| wrbAS | Flavodoxin-like protein; binds the trp repressor | SSR | 364 | 5.0 ± 2.0 | NaSal, Lrp, BG, DHCP, OS |
| Iron uptake and storage | |||||
| bfr | Bacterioferrin, iron storage homoprotein | IUS | 905 | 5.1 ± 2.1 | DHCP, ryhB RNA |
| entD | Enterochelin synthetase, component D | IUS | 508 | 4.2 ± 0.5 | acrBTC |
| Carbohydrate metabolism | |||||
| crr | Phosphotransferase system, glucose-specific IIA component | CHM | 443 | 2.7 ± 0.6 | CD |
| fbaB | Fructose 1,6-bisphosphate aldolase (b2097) | CHM | 1,264 | 4.6 ± 1.4 | MMC, Lrp |
| gip | Glyoxylate-induced protein | 384 | 3.6 ± 1.0 | kdpABCDETC | |
| glcG | Putative oxidase, glycolate utilization (b2977) | CHM | 462 | 3.0 ± 1.7 | uvrYTC, arcATC, arcBTC |
| nagB | Glucosamine-6-phosphate deaminase | CHM | 860 | 2.6 ± 0.6 | ompR/envZTC, arcBTC |
| pfkB | Phosphofructokinase-2 | CHM | 512 | 2.5 ± 2 | MMC, CD, DHCP |
| talA | Transaldolase A | CHM | 1,097 | 2.9 ± 1.8 | rssBTC, MMC, DHCP, CreBC |
| Miscellaneous function or unknown function | |||||
| deoD | Purine nucleoside phosphorylase | NM | 349 | 4.8 ± 3.0 | |
| moeB | Molybdopterin biosynthesis | CB | 1,100 | 3.4 ± 1.2 | |
| msyB | Membrane protein, protein export (b1051) | MF | 516 | 6.1 ± 4.0 | CD, Lrp, DHCP |
| yafN | Hypothetical (adherence) protein (b0232) | (MF) | 354 | 2.6 ± 0.1 | MMC, GadX |
| yahOS | Hypothetical protein (b0329) | 933 | 4.5 ± 2.2 | Lrp | |
| ybgA | Hypothetical (inner membrane) protein (b0707) | 2,038 | 4.5 ± 1.7 | ||
| ybjP | Putative enzyme (b0865) | 1,317 | 4.2 ± 2.5 | rssBTC, baeSRTC, atoSCTC, Lrp | |
| yddX | Hypothetical protein, biofilm dependent (b1481) | (EF) | 376 | 4.2 ± 1.4 | |
| ydiZ | Hypothetical protein (b1724) | 707 | 3.1 ± 1.7 | rssBTC, OS | |
| yfeT | Hypothetical protein (b2427) | 341 | 3.5 ± 0.9 | ompR/envZTC | |
| ygaF | Putative dehydrogenase protein (b2660) | 453 | 2.8 ± 0.5 | rssBTC, NarXLTC | |
| yggE | Putative immunogenic protein (b2922) | (EF) | 813 | 4.8 ± 1.7 | rssBTC, MMC; Rcs, phoQP |
| ygiS | Putative transport periplasmic protein (b3020) | (MF) | 436 | 2.7 ± 0.8 | |
| ygiW | Hypothetical (outer membrane) protein (b3024) | (MF) | 892 | 4.0 ± 1.9 | rssBTC, citABTC, arcATC, NaSal |
| yiaG | Hypothetical (regulatory) protein (b3555) | 1,227 | 2.7 ± 0.5 | rssBTC, ntrBCTC, NaSal | |
| yjbJ | Hypothetical protein (b4045) | 601 | 4.5 ± 1.2 | ||
| ynfA | Hypothetical (membrane) protein (b1582) | (MF) | 3,334 | 3.5 ± 1.1 | |
| yqjC | Hypothetical (periplasmic) protein (b3097) | 1,121 | 5.9 ± 1.3 | ||
| ytfK | Hypothetical protein (b4217) | 556 | 3.9 ± 3.0 | PQ, Fur, BG |
SSR, starvation and stress response; PS, protein synthesis; AATM, amino acid transport and metabolism; IUS, iron uptake and storage; CHM, carbohydrate metabolism; NM, nucleoside metabolism; CB, cofactor biosynthesis; MF, membrane function; EF, extracellular function. Parentheses indicate a putative function.
Mean relative expression level in the wild-type (WT) strain; average values of the signal intensities obtained in the wild-type strain in four experiments without correction (i.e., without dye swapping).
Averages and standard deviations from four experiments are presented. The genes listed in this table presented a ratio of expression in the wild-type versus the rpoS strain (R) greater than 1.9 in at least three out of four experiments and never presented an R of <1. Genes with an average R lower than 2.5 were excluded from this table, except for ytfK and pfkB, which were confirmed by primer extension, and artI, located in a cluster with ybjP and artP, both of which are down-regulated in the rpoS strain.
Genes known to be regulated by RpoS are marked with a superscript capital S. Relevant references for these genes are given in parentheses dps (3), osmE (11, 17), osmY (8, 50, 101), sodC (27), rpsV (sra [42]), wrbA (100), and yahO (40). Growth conditions or regulatory pathways known (by either genomic or genetic analysis) to affect expression of the genes listed include the following: (i) disruption of known two- component (TC) regulatory systems (74); (ii) knockout of the specific regulatory gene(s) fur (10), rcs or phoQP (30), lrp (39, 89), gadX (64), creBC (7), or ryhB (63); (iii) specific growth conditions, including exposure to mitomycin C (MMC) (44), oxidative stress induced by either paraquat (PQ) or sodium salicylate (NaSal) (76), biofilm growth (BG) (86), 4,5-dihydroxy-2-cycloenten-1-one (DHCP) treatment (antimicrobial agent [75]), high-cell-density (CD) culture (102), osmotic stress (OS) response (supercoiling- dependent genes [16]), and minimal versus LB medium (MD) (97).
RT PCR.
For real-time PCR (RT-PCR), reverse transcription from RNA samples (extracted as for the microarray experiments) was carried out using 62.5 U of MultiScribe (Applied Biosystems) reverse transcriptase per μg of total RNA, in the presence of 1.25 μM random hexamers. Amplification reactions were carried out in an Applied Biosystems ABI Prism 7000 sequence detection system using the SYBR Green PCR master mix according to the manufacturer's standard protocol. cDNA produced from 20 ng of total RNA was used for each reaction. The internal-control 16S rRNA gene, used to normalize the values obtained for the genes analyzed, was amplified by primers rrsBfw (5′-GAATGCCACGGTGAATACGTT) and rrsBrev (5′-ACCCACTCCCATGGTGTGA). The PCR products of the genes of interest (shown in Fig. 1) were generated by using the primers listed in Table 1. All reactions were performed in triplicate, in addition to samples using DNase I-digested RNA as templates, to verify the lack of residual DNA. Real-time PCR data were analyzed by using ABI PRISM 7000 SDS software (version 1.0) and according to Relative Quantitation of Gene Expression (P/N 4303859), issued by the manufacturer.
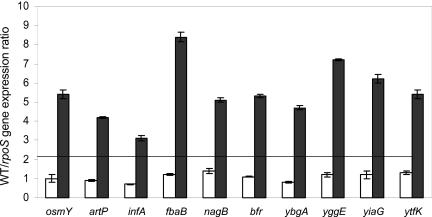
FIG. 1.
Real-time PCR experiments. Values are shown as induction factors for expression in the wild-type (WT) strain MG1655 versus the rpoS mutant strain EB1.3 in the exponential (white bars) and stationary (black bars) phases of growth. The genes tested are indicated in the figure. Values were normalized to those for the 16S rRNA gene, and results shown are averages of three experiments. The horizontal line shows the 2.5-fold threshold used to select rpoS-dependent genes in the microarray experiments.
TABLE 1.
Primers used in this study
| Name | Sequence |
|---|---|
| For RT-PCR | |
| osmYfw | 5′-GTCGATAGCTCTATG-3′ |
| osmYrev | 5′-GAAGGCGGCCCTGGG-3′ |
| artPfw | 5′-GAGTATTCAATTAAACGGC-3′ |
| artPrev | 5′-CCCTGTGGGCAATCCAGCGTG-3′ |
| infAfw | 5′-CGGTCACGTGGTTACTGCAC-3′ |
| infArev | 5′-GTCGCCCGTCAGGATGCG-3′ |
| fbaBfw | 5′-CGGGAGCATAGTAATGAC-3′ |
| fbaBrev | 5′-ACGGTGCTGTAAAAGGTTGTCG-3′ |
| nagBfw | 5′-CTCAACGGCAACGCCCCGG-3′ |
| nagBrev | 5′-GGCTCGTTACCTACACCGCCC-3′ |
| bfrfw | 5′-CTGGGGTCTCAAACGTCTC-3′ |
| bfrrev | 5′-CGGCGTGTTTCATCTCATC-3′ |
| ybgAfw | 5′-GATTTGATGACAGCACCCTAATC-3′ |
| ybgArev | 5′-AGCGGCTGTGATAATCGAGAAGC-3′ |
| yggEfw | 5′-GTGAAGTTCAAAGTTATCGCC-3′ |
| yggErev | 5′-TGACAATATGCGGTCCATCCG-3′ |
| yiaGfw | 5′-GATCCAATGCATGAGCTGTTGAG-3′ |
| yiaGrev | 5′-AGGACGTTGTTCTGTGCGTCAGG-3′ |
| ytfkfw | 5′-GGTACAACCCACTTCAGGTG-3′ |
| ytfKrev | 5′-TCCTTGATGTATAACCGTCC-3′ |
| For determination of transcription start sites | |
| ansP | 5′-AATGGCGATCATCTGCACCTGG-3′ |
| artP | 5′-CCCTGTGGGCAATCCAGCGTG-3′ |
| dps | 5′-AACTGGATAACCTGGCGATTCAG-3′ |
| fbaB | 5′-ACGGTGCTGTAAAAGGTTGTCG-3′ |
| mysB | 5′-TTTGGGCATTGAACTGTTGCAC-3′ |
| pfkB | 5′-AACACCGGTGCGGTACAGCG-3′ |
| rpsV | 5′-TGCCGGTTGGGTTATTTACTACG-3′ |
| talA | 5′-TGAGTAACAGCGAAGGATTGGTG-3′ |
| ybjP | 5′-TTCAACGCAAGGACCACTGCG-3′ |
| ybgA | 5′-AGCGGCTGTGATAATCGAGAAGC-3′ |
| yggE | 5′-TGACAATATGCGGTCCATCCG-3′ |
| yiaG | 5′-AGGACGTTGTTCTGTGCGTCAGG-3′ |
| ytfK | 5′-TCCTTGATGTATAACCGTCC-3′ |
Primer extension.
Samples were collected, and RNA was extracted, in the same way as for microarray hybridization (see above). Twelve micrograms of RNA was used by the primer extension assay with ImpromII reverse transcriptase (Promega) according to the manufacturer's instructions. RNA and labeled primers were annealed at 70°C (for 5 min), and the reaction proceeded at 42°C for 1.5 h. The labeled extended fragments were separated from mRNA by heating at 95°C and were placed on ice prior to addition of formamide dye (MWG); the samples were then loaded onto a 7% polyacrylamide-urea denaturing sequencing gel, and transcription start sites were determined by comparison with a known DNA sequence used as a molecular weight marker. Reaction products were quantified with ImageQuant (Molecular Dynamics) after background subtraction from the TIFF images generated by the laser scanner (Li-Cor 4200 long reader; MWG). Primers used in this assay are shown in Table 1.
Other genetic methods.
The tnaA gene was inactivated by the λ red gam method as described in reference 20, producing strain PL614. Insertion was verified by direct amplification of the tnaA gene. To measure tnaA gene expression, its promoter region, including the short leader peptide for the TnaA protein encoded by the tnaL gene, was amplified by PCR using primers tnaA1 (CTGAAGCTTGATTGTGATTCGATTC, annealing at positions −392 to −376 relative to the tnaA start codon) and tnaA2 (CGGAATTCCTCTTCACGATAAGC, annealing at positions +76 to +90 relative to the tnaA start codon). The primers introduced a HindIII and an EcoRI site (underlined in the primer sequences) into the amplified fragment, which was then subcloned into the multicopy plasmid pRS1274 (49) by using the corresponding restriction sites. β-Galactosidase activity was determined as described in reference 66.
Sequence analysis and gene function.
The pattern search tools of Colibri (http://genolist.pasteur.fr/Colibri/genome.cgi) (65) were used to search for putative −10 promoter elements upstream of the primer extension products. The Colibri, PromEC (38) (http://bioinfo.md.huji.ac.il/marg/promec/prom.seq.final.html), and RegulonDB (84) (http://www.cifn.unam.mx/Computational_Genomics/regulondb) databases, and J. E. Mitchell's collection of σ70-dependent promoters described in reference 67 (http://www.biosciences.bham.ac.uk/labs/minchin/mitchell2003), were used for analysis of promoter and operon structures. Finally, the Swiss-Prot (http://www.expasy.org/) and ProDom (http://prodes.toulouse.inra.fr/prodom/current/html/home.php) databases were used to gather more information about genes encoding hypothetical proteins.
RESULTS AND DISCUSSION
rpoS-dependent genes expressed at the onset of stationary phase.
Whole-genome expression in either E. coli MG1655 or its rpoS mutant derivative was determined at the onset of stationary phase (i.e., cells were harvested as soon as an increase in cell turbidity was no longer detectable) for cells grown in LB (rich) medium. At this stage of bacterial growth, we expected that the subset of rpoS-regulated genes mostly involved in the establishment of stationary-phase physiology would be maximally expressed. Through microarray experiments we detected 41 genes whose expression was significantly reduced in the rpoS strain (Table 2); only 7 of these had already been described as rpoS dependent (dps, osmE, osmY, rpsV, sodC, wrbA, and yahO). Known rpoS-dependent genes not detected in our experiment might be expressed only at later stages of stationary phase or in different growth media and conditions; however, lack of detection of some rpoS-dependent genes might be due to shortcomings of the microarray technique and to the criteria chosen for microarray analysis (see Materials and Methods). The known rpoS-dependent genes found in our experiments mainly encode stress response proteins with either regulatory or detoxification functions (35, 41; reviewed in reference 33). Among the newly identified σS-regulated genes, almost 50% have unknown functions. The others belong to four main functional classes: genes involved in amino acid transport and metabolism, iron uptake and storage, protein synthesis, and carbohydrate and nucleoside metabolism. It is noteworthy that the newly identified rpoS-dependent genes do not appear to be essential genes as defined by Gerdes et al. (26), except for infA (18), a finding consistent with the lack of major effects of the rpoS mutation on cell viability.
To confirm the results of the whole-genome expression analysis, we performed RT-PCR experiments on a selection of differentially expressed genes belonging to each functional category (Fig. 1). RT-PCR experiments confirmed the increased expression of all the genes tested in strain MG1655 compared to the rpoS mutant strain EB1.3. Increased gene expression in MG1655 was detectable only at the onset of stationary phase; samples taken in mid-exponential phase (OD600, 0.6 to 0.8) showed no significant differences in gene expression between the two strains (Fig. 1).
Genes involved in cell metabolism.
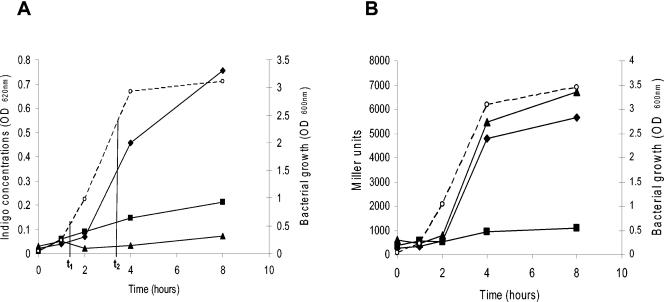
We found several genes involved in amino acid transport (artI, artP, and ansP) or metabolism (tnaA and ilvD) to be induced in a σS-dependent fashion at the onset of stationary phase. In particular, rpoS appears to promote tryptophan degradation by inducing the tryptophanase gene tnaA, which results in the production of indole, and by stimulating expression of the WrbA protein, which might strengthen negative regulation of the trp biosynthetic operon via binding to the TrpR repressor protein (100). Expression of the wrbA gene has already been shown to be rpoS dependent (41, 100). Interestingly, indole has been proposed to act as an extracellular signal in stationary cells of E. coli (95), and tnaA-mediated indole biosynthesis appears to stimulate biofilm formation (62). We investigated the possibility that indole production is negatively affected by rpoS mutation. Indole is converted into indigo (which is not further degraded in E. coli) by several monooxygenases, thus providing an easy method for its determination (71). We transformed both strains MG1655 and EB1.3 with pStyABB, a plasmid expressing styrene monooxygenase, and monitored indole production through its conversion into indigo (Fig. 2A). The production of indole closely follows the growth curve in MG1655, as revealed by indigo accumulation; in contrast, indole production is totally abolished in a tnaA-null mutant and severely affected by inactivation of the rpoS gene (Fig. 2A). To rule out the possibility that indigo production might be affected by different expression of styrene monooxygenase in the rpoS mutant strain, we exposed MG1655/pStyABB cells in early-exponential phase to filtered supernatants of either MG1655 or EB1.3 cultures. MG1655/pStyABB produced indigo only when treated with MG1655 conditioned medium, confirming that the rpoS mutant strain EB1.3 is deficient in indole production (data not shown). The results of the microarray experiments strongly suggest that lack of indole production in the rpoS mutant strain depends on reduced tnaA expression, which was further confirmed by tnaA gene expression measurement using the lacZ reporter gene (Fig. 2B).
FIG. 2.
(A) Indole production in strains MG1655 (wild type) (black diamonds), EB1.3 (rpoS) (black squares), and PL614 (tnaA) (black triangles), all carrying the pStyABB plasmid. The indole concentration was determined through its conversion into indigo by the StyAB monooxygenase as described in Materials and Methods. The growth curve in LB for MG1655 is shown by the dashed line and open circles. The growth rate of either the EB1.3 or the PL614 strain was not affected by the mutation. Samples for RT-PCR (Fig. 1) from exponential- and stationary-phase cells were collected at time points t1 (OD600, 0.6 to 0.8) and t2 (OD600, 2.5 to 3), respectively. (B) tnaA promoter activity from a multicopy plasmid in strains MG1655, EB1.3, and PL614. Symbols are as explained for panel A.
While tryptophan is degraded in stationary phase, accumulation of the amino acids arginine and asparagine appears to be stimulated by rpoS through activation of the artP and artI genes, whose products are components of the arginine transport system and of the asparagine transporter ansP. Thus, the rpoS gene seems to play an important role in determining intracellular amino acid concentrations in stationary phase. This function might be related to the synthesis of amino acid- or peptide-derived signal molecules, as for indole, or to the need to redirect general amino acid metabolism in stationary phase. It is noteworthy that both the artP and the wrbA promoter are controlled by the global regulator Lrp (leucine-responsive protein) (Table 2), the main regulator of amino acid biosynthesis and metabolism. The unknown ybjP gene, located immediately upstream of the art operon but controlled by its own promoter (see Fig. 4), is also regulated by lrp (39) as well as by rpoS. Concerted regulation by RpoS and Lrp has already been reported for a number of genes (osmC, osmY, aidB, and csiD [13, 48, 50, 59]), suggesting tight integration of Lrp- and RpoS-dependent regulation.
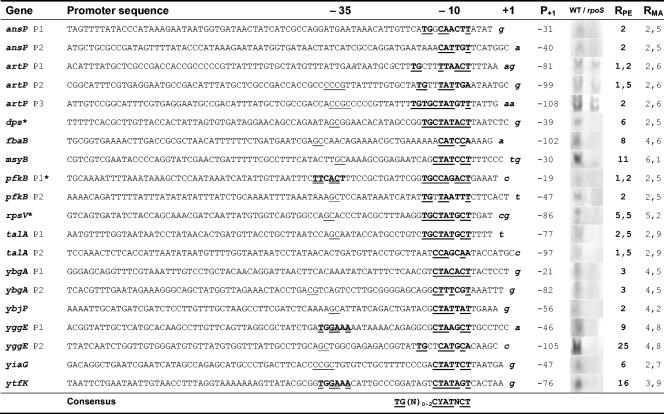
FIG. 4.
Promoter sequences of σS-regulated genes induced at the onset of stationary phase, characterized by primer extension. The −10 sequence of each promoter is boldfaced, as is the conserved −35 element when present. Nucleotides that match the proposed consensus for the −10 element [(TGN0-2)CYANNMT, where Y stands for C or T, M stands for A or C, and N stands for any nucleotide (47)] or the −35 element (TTGACA) at a functional location (15 to 17 nucleotides away from the −10 sequence) are underlined (for the −10 sequence) or double underlined (for the −35 sequence). The transcription start sites are lowercased, boldfaced, and italicized. Recurrent CG dinucleotides in the −35 region and the CCG motifs proposed by Wise et al. (98) are also underlined. Promoters marked with an asterisk are known promoters (3, 19, 42). P+1 indicates the position of the transcription start site relative to the start codon. WT and rpoS designate the products of primer extension obtained with total mRNA from the wild-type strain MG1655 and its rpoS derivative, respectively; the sizes of the primer extension products were determined by use of a DNA sequencing ladder of a known sequence run on the same gel. RPE indicates the WT/rpoS expression ratio as determined from the primer extension experiments. RMA indicates the WT/rpoS expression ratio as determined from the microarray experiments (Table 2).
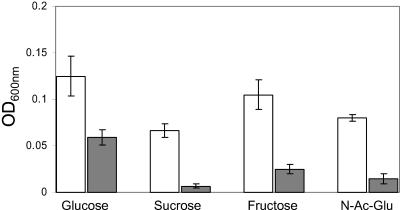
We also detected rpoS-dependent activation of genes encoding carbohydrate metabolism proteins such as the carbohydrate phosphotransferase systems (crr) or glycolysis enzymatic proteins (fbaB and pfkB). Interestingly, both the fbaB and talA genes encode isoenzymes of fbaA and talB, genes that do not appear to be regulated by rpoS, suggesting that σS could up-regulate the expression of isoenzymes that are possibly more functional during the stationary phase of growth. The rpoS gene also appears to be involved in alternative metabolic pathways for carbohydrate utilization such as the pentose phosphate or the glyoxylate shunt to the carboxylic acid pathway (glcG or gip). In addition, rpoS activates the catabolism of N-acetylglucosamine through induction of nagB. We directly tested the possible role of rpoS in the expression of carbohydrate metabolism genes by growing strains MG1655 and EB1.3 on limiting amounts of different sugars as sole carbon sources. Both the parental and rpoS strains were able to grow to an OD600 of >1.0 on 10 mM concentrations of glucose, fructose, N-acetylglucosamine, or sucrose, with similar growth rates (data not shown). However, in the presence of a growth-limiting sugar concentration (0.1 mM), the rpoS mutant showed significantly impaired ability to grow, ranging from a twofold reduction for growth on glucose to an eightfold reduction for growth on sucrose (Fig. 3), despite the fact that the inocula contained similar numbers of viable cells (data not shown) (see Materials and Methods). Although rpoS dependence of sugar uptake or catabolism genes would be consistent with their induction in response to starvation, it is in contrast with previous observations suggesting that rpoS might negatively regulate the nagB and crr genes in continuous cultures grown under sugar-limited conditions (92). It is possible that rpoS might be involved in feedback regulation of carbohydrate metabolism genes. rpoS could activate the expression of sugar uptake and metabolism genes at the onset of stationary phase, to scavenge for the presence of low concentrations of sugars. However, in the absence of the specific inducers, expression would be shut down by rpoS itself through indirect regulation at a later stage of the stationary phase. An example of feedback loop regulation by rpoS has already been described for the curli-encoding csg operon (6, 78).
FIG. 3.
Growth of MG1655 (wild type) (white bars) or EB1.3 (rpoS) (grey bars) on limiting concentrations (0.1 mM) of either glucose, sucrose, fructose, or N-acetylglucosamine (N-Ac-Glu). The growth experiments were performed as described in Materials and Methods. Values represent the final OD600 reached at the end of the 24-h incubation and are averages from three experiments.
Iron-dependent genes.
Iron is an essential cofactor of many enzymes, and bacteria have evolved a range of strategies to acquire and store iron, which is often not bioavailable in the environment. E. coli possesses two main iron storage proteins: bacterioferritin and ferritin (4). Bacterioferritin, whose precise function is still unclear (1), is induced during slow growth or at the transition to stationary phase, consistent with its regulation by rpoS. The entD gene encodes enterochelin, a synthase of a catechol siderophore from chorismic acid; the EntD protein is located in the inner membrane, but its function is not fully understood (5, 29). The gene is located downstream of the fepA gene, apart from the rest of the ent cluster, suggesting possible differential regulation. Regulation of ferritins and iron-dependent genes by rpoS is not entirely surprising, since rpoS is already known as the main regulator of dps expression. Although the Dps protein was originally described as a nonspecific DNA binding protein involved in resistance to oxidative stress (60), it is actually a bacterial ferritin (28), whose main function could be to chelate intracellular iron and thus to prevent DNA damage through Fenton reaction-catalyzed oxyradical formation (31). Many iron-dependent genes are regulated by Fur (ferric uptake regulator), which, in the presence of iron, represses the transcription of iron-responsive genes (25, 63, 80). A recent report proposes that Fur accumulation would be modulated by rpoS in Vibrio vulnificus (52), pointing to a possible overlap of fur- and rpoS-dependent gene regulation in enterobacteria.
Protein synthesis.
We found two genes involved in protein synthesis to be activated in an rpoS-dependent fashion under the conditions tested. Interestingly, in addition to the already known rpsV gene, we could show that transcription of the infA gene is also stimulated by rpoS (Table 2; Fig. 1). The infA gene encodes the initiation factor IF-1, whose precise role has not yet been assigned (77). Similarly, no precise function has yet been assigned to the product of rpsV, a small, stationary-phase-induced, ribosome-associated protein (SRA) which appears to be specific to enterobacteria (42). The RpsV protein might play a role similar to that of the ribosome-modulating factor (RMF), i.e., to stabilize ribosomes in stationary phase. The RMF protein is also produced in stationary phase, although not in an rpoS-dependent fashion (94), and E. coli mutants with defects in rmf cannot survive long periods of starvation (99). Recent findings point to a close relationship between protein synthesis and entrance into stationary phase: aberrant proteins result from increased mistranslation in nonproliferating bacteria and are likely to accumulate in stationary phase (70). We propose that σS-dependent induction of IF-1 and SRA might either prevent translation errors or modulate protein synthesis by blocking ribosomes in an inactive form.
Genes of unknown function.
The rpoS mutation affects the expression of 16 genes encoding hypothetical proteins of unknown function, several of which might have functions associated with membrane trafficking (ybgA, ygiS, ygiW, ynfA, and yqjC) or might be located at the cell surface (yafN, yddX, and yggE). Some of these proteins are common to a wide number of enterobacteria or, in the case of ynfA and yiaG, even to a larger group including gram-positive bacteria. Interestingly, the yahO gene has previously been reported to be regulated by RpoS in Salmonella enterica (40); the YahO protein shares 66% identity and 79% similarity with its E. coli homologue. The yggE gene, conserved in other enterobacteria such as Shigella flexneri and Edwardsiella ictaluri at an identical locus (68, 96), encodes a putative extracellular protein, considered a potential antigenic protein for development of a vaccine against E. ictaluri in catfish (68). Two rpoS-dependent genes encode either a potential adhesion factor (yafN) or a factor involved in biofilm formation (yddX [79]). Another gene, ytfK, is up-regulated in E. coli cells growing as a biofilm (86). These observations would support a general positive role of rpoS in biofilm formation (2), possibly mediated by indole production (Fig. 2) (62), consistent with the dependence on rpoS of known adhesion and biofilm formation determinants such as the csg genes in Salmonella and in pathogenic E. coli strains (73).
Promoter sequences of σS-regulated genes.
Primer extension assays were performed to determine the promoter sequences of the genes identified and to confirm the results of the gene array experiment. We selected 11 genes belonging to different functional classes and showing variable expression levels and wild-type/rpoS mutant ratios in the gene array experiment. In addition, we performed primer extension assays on the dps and rpsV genes, whose σS-dependent promoters have already been characterized, as a control for our experimental conditions. The promoter sequences found for both dps and rpsV correspond to those previously described (3, 42); this was also the case for the known pfkB promoter, which, however, had not yet been identified as a σS-dependent promoter (19). Primer extension was performed from total RNAs of both wild-type and rpoS strains, and transcription start sites were determined by comparison with a DNA sequencing reaction ladder (see Fig. S1 in the supplemental material). Quantification of primer extension products consistently showed lower levels of expression in the rpoS mutant, confirming the results of the gene array experiments (Fig. 4). For several rpoS-dependent genes, multiple signals were detected in the primer extension experiments (ansP, artP, talA, pfkB, ybgA, and yggE), which might depend either on secondary RNA structures resulting in pausing of reverse transcriptase, or on the presence of multiple promoters. Indeed, well-conserved promoter-like sequences could be detected upstream of most reverse transcriptase extension products, suggesting that the presence of multiple promoters might be a rather common feature for rpoS-dependent genes.
Our results do not allow us to ascertain that all rpoS-dependent genes identified are directly transcribed by EσS. However, we expect that most promoters induced at the onset of stationary phase should belong to the class of promoters recognized with high affinity by the σS subunit of RNA polymerase and thus should display promoter features favorable for EσS. Indeed, the sequences of the −10 regions of the promoters tested are in good agreement with previous compilations of σS-dependent promoters (23, 47, 54) and reflect the recently proposed motif TGN0-2CYATAMT (47). The −10 sequence of around 70% (12 of 18) of the newly characterized σS-dependent promoters is immediately preceded by a C at the position conventionally indicated as −13; −13C is a widely conserved feature for promoter recognition by EσS (9, 15). Six (30%) promoters (ansPp1, ansPp2, fbaB, talAp2, pfkBp1, and yggEp2) display a C as the first nucleotide of the −10 promoter element (conventionally −12C), a feature that might prevent their transcription by Eσ70 (47). A TG motif is present upstream of the −10 element in 10 (50%) of the newly determined promoters, at positions from −17 to −14. In a previous study (47), we proposed that σS can recognize the TG motif at different positions, in contrast to σ70, which can interact only with a TG placed at −15/−14 (14, 67). In the novel σS-dependent promoters identified in this report, the distribution of the TG motif is not as wide as that which we had found at the 56 previously characterized σS-dependent promoters (47), as it is more restricted to the −15/−14 position. Interestingly, the artPp3 promoter, one of the promoters with the lowest σS/σ70 activity ratio, harbors a double TG, which is supposed to further enhance promoter recognition and transcription initiation by both forms of polymerase (24, 67).
The conservation of the amino acids of region 4.2 of both σ factors and the results obtained by the SELEX approach (24) suggest that EσS might be able to recognize a −35 element and would have the same optimal sequence as σ70 (TTGACA). At some rpoS-dependent promoters, such as osmE (12), disruption of the −35 sequence (TTGAAA) has a negative effect on σS-mediated transcription. However, data on the importance of a −35 sequence for EσS are controversial, and very few known σS-dependent promoters possess a −35 sequence similar to that recognized by Eσ70 (12, 72, 87, 98). Among the newly identified promoters, only three (pfkB, ytfK, and yggE) have a discernible −35 element with four of six matches to the σ70 consensus sequence, possibly suggesting that EσS-specific −35 sequences might strongly diverge from the Eσ70 consensus (Fig. 4). It has been proposed that DNA bending upstream of the promoter region as well as motifs rich in C/G in the −35 region would favor σS selectivity (45, 98). Although our results do not allow us to confirm this hypothesis, we did find fairly high occurrence of GC or CG dinucleotides in the −35 area (Fig. 4). Indeed, the artPp1, artPp2, and yiaG promoters carry a CCG sequence, proposed to be a σS-specific −35 promoter sequence by Wise et al. (98), while dps and ytfK display the GCGG motif recently proposed as an osmotic shock-responding module in a subset of σS-dependent promoters (53).
Finally, five promoters (ansPp1, fbaBp1, pfkB, talA, and ytfK) possess UP-like elements, i.e., putative binding sites for the α subunit of the RNAP (82). The role of the UP element in promoter recognition by EσS has not yet been addressed in detail; at the aidB promoter, an UP-like element functional for Eσ70 also contributes to EσS-dependent transcription in vitro, suggesting that UP elements might indeed play a role in EσS-promoter interactions (S. Lacour and P. Landini, unpublished data).
Conclusions.
In this report, we compared the expression profiles of strain MG1655 and its rpoS derivative in LB (rich) medium. Since we tested rpoS-dependent expression at only one time point (the onset of stationary phase) and in one growth medium, our results give only an incomplete picture of the rpoS regulon; however, they provide information on genetic and physiological adaptation to stationary phase under the conditions tested, as well as on the structure of rpoS-dependent promoters. Indeed, in addition to known stress response genes, we found genes involved in a variety of metabolic functions to be induced in an rpoS-dependent fashion. The rpoS gene appears to be directly involved in the biosynthesis of signal molecules such as indole, which plays a role in biofilm formation in some E. coli strains (62) and might regulate gene expression in coordination with other cell-signaling systems directly related to quorum sensing and cell density (81). Our results also point to a direct role of σS in response to nutrient starvation, suggesting that the σS regulon overlaps with other nutrient-specific regulatory pathways such as cAMP/CRP, NtrB/NrtC/σ54, and PhoB/PhoR. We could confirm with primer extension experiments that, for a large number of the newly identified σS-dependent promoters, the −10 promoter element matches the proposed consensus TGN0-2CYATAMT. In contrast to strong conservation in the −10 region, little similarity to the −35 sequence for Eσ70 and no significant presence of UP element sequences could be detected.
Supplementary Material
Acknowledgments
We thank Eva Brombacher and Luciana Gualdi for assistance and Annie Kolb for encouragement and useful discussions.
Financial support for this work was provided by the Swiss National Science Foundation (SNF grant 3100-056742.99).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abdul-Tehrani, H., A. J. Hudson, Y. S. Chang, A. R. Timms, C. Hawkins, J. M. Williams, P. M. Harrison, J. R. Guest, and S. C. Andrews. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 181:1415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, J. L., and R. J. McLean. 1999. Impact of rpoS deletion on Escherichia coli biofilms. Appl. Environ. Microbiol. 65:4285-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altuvia, S., M. Almiron, G. Huisman, R. Kolter, and G. Storz. 1994. The dps promoter is activated by OxyR during growth and by IHF and σS in stationary phase. Mol. Microbiol. 13:265-272. [DOI] [PubMed] [Google Scholar]
- 4.Andrews, S. C., J. M. Smith, C. Hawkins, J. M. Williams, P. M. Harrison, and J. R. Guest. 1993. Overproduction, purification and characterization of the bacterioferritin of Escherichia coli and a C-terminally extended variant. Eur. J. Biochem. 213:329-338. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong, S. K., G. S. Pettis, L. J. Forrester, and M. A. McIntosh. 1989. The Escherichia coli enterobactin biosynthesis gene, entD: nucleotide sequence and membrane localization of its protein product. Mol. Microbiol. 3:757-766. [DOI] [PubMed] [Google Scholar]
- 6.Arnqvist, A., A. Olsen, and S. Normark. 1994. σS-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by σ70 in the absence of the nucleoid-associated protein H-NS. Mol. Microbiol. 13:1021-1032. [DOI] [PubMed] [Google Scholar]
- 7.Avison, M. B., R. E. Horton, T. R. Walsh, and P. M. Bennett. 2001. Escherichia coli CreBC is a global regulator of gene expression that responds to growth in minimal media. J. Biol. Chem. 276:26955-26961. [DOI] [PubMed] [Google Scholar]
- 8.Barth, M., C. Marschall, A. Muffler, D. Fischer, and R. Hengge-Aronis. 1995. Role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation of σS and many σS-dependent genes in Escherichia coli. J. Bacteriol. 177:3455-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker, G., and R. Hengge-Aronis. 2001. What makes an Escherichia coli promoter σS-dependent? Role of the −13/−14 nucleotide promoter positions and region 2.5 of σ S. Mol. Microbiol. 39:1153-1165. [DOI] [PubMed] [Google Scholar]
- 10.Bjarnason, J., C. M. Southward, and M. G. Surette. 2003. Genomic profiling of iron-responsive genes in Salmonella enterica serovar Typhimurium by high-throughput screening of a random promoter library. J. Bacteriol. 185:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bordes, P., J. Bouvier, A. Conter, A. Kolb, and C. Gutierrez. 2002. Transient repressor effect of Fis on the growth phase-regulated osmE promoter of Escherichia coli K12. Mol. Genet. Genomics 268:206-213. [DOI] [PubMed] [Google Scholar]
- 12.Bordes, P., F. Repoila, A. Kolb, and C. Gutierrez. 2000. Involvement of differential efficiency of transcription by EσS and Eσ70 RNA polymerase holoenzymes in growth phase regulation of the Escherichia coli osmE promoter. Mol. Microbiol. 35:845-853. [DOI] [PubMed] [Google Scholar]
- 13.Bouvier, J., S. Gordia, G. Kampmann, R. Lange, R. Hengge-Aronis, and C. Gutierrez. 1998. Interplay between global regulators of Escherichia coli: effect of RpoS, Lrp and H-NS on transcription of the gene osmC. Mol. Microbiol. 28:971-980. [DOI] [PubMed] [Google Scholar]
- 14.Burr, T., J. Mitchell, A. Kolb, S. Minchin, and S. Busby. 2000. DNA sequence elements located immediately upstream of the −10 hexamer in Escherichia coli promoters: a systematic study. Nucleic Acids Res. 28:1864-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Checroun, C., P. Bordes, O. Leroy, A. Kolb, and C. Gutierrez. 2004. Interactions between the 2.4 and 4.2 regions of σS, the stress-specific sigma factor of Escherichia coli, and the −10 and −35 promoter elements. Nucleic Acids Res. 32:45-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung, K. J., V. Badarinarayana, D. W. Selinger, D. Janse, and G. M. Church. 2003. A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli. Genome Res. 13:206-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conter, A., C. Menchon, and C. Gutierrez. 1997. Role of DNA supercoiling and RpoS sigma factor in the osmotic and growth phase-dependent induction of the gene osmE of Escherichia coli K12. J. Mol. Biol. 273:75-83. [DOI] [PubMed] [Google Scholar]
- 18.Cummings, H. S., and J. W. Hershey. 1994. Translation initiation factor IF1 is essential for cell viability in Escherichia coli. J. Bacteriol. 176:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daldal, F. 1983. Molecular cloning of the gene for phosphofructokinase-2 of Escherichia coli and the nature of a mutation, pfkB1, causing a high level of the enzyme. J. Mol. Biol. 168:285-305. [DOI] [PubMed] [Google Scholar]
- 20.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Biase, D., A. Tramonti, F. Bossa, and P. Visca. 1999. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 32:1198-1211. [DOI] [PubMed] [Google Scholar]
- 22.Ding, Q., S. Kusano, M. Villarejo, and A. Ishihama. 1995. Promoter selectivity control of Escherichia coli RNA polymerase by ionic strength: differential recognition of osmoregulated promoters by EσD and EσS holoenzymes. Mol. Microbiol. 16:649-656. [DOI] [PubMed] [Google Scholar]
- 23.Espinosa-Urgel, M., C. Chamizo, and A. Tormo. 1996. A consensus structure for σS-dependent promoters. Mol. Microbiol. 21:657-659. (Letter.) [DOI] [PubMed] [Google Scholar]
- 24.Gaal, T., W. Ross, S. T. Estrem, L. H. Nguyen, R. R. Burgess, and R. L. Gourse. 2001. Promoter recognition and discrimination by EσS RNA polymerase. Mol. Microbiol. 42:939-954. [DOI] [PubMed] [Google Scholar]
- 25.Garg, R. P., C. J. Vargo, X. Cui, and D. M. Kurtz, Jr. 1996. A [2Fe-2S] protein encoded by an open reading frame upstream of the Escherichia coli bacterioferritin gene. Biochemistry 35:6297-6301. [DOI] [PubMed] [Google Scholar]
- 26.Gerdes, S. Y., M. D. Scholle, J. W. Campbell, G. Balazsi, E. Ravasz, M. D. Daugherty, A. L. Somera, N. C. Kyrpides, I. Anderson, M. S. Gelfand, A. Bhattacharya, V. Kapatral, M. D'Souza, M. V. Baev, Y. Grechkin, F. Mseeh, M. Y. Fonstein, R. Overbeek, A. L. Barabasi, Z. N. Oltvai, and A. L. Osterman. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185:5673-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gort, A. S., D. M. Ferber, and J. A. Imlay. 1999. The regulation and role of the periplasmic copper, zinc superoxide dismutase of Escherichia coli. Mol. Microbiol. 32:179-191. [DOI] [PubMed] [Google Scholar]
- 28.Grant, R. A., D. J. Filman, S. E. Finkel, R. Kolter, and J. M. Hogle. 1998. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat. Struct. Biol. 5:294-303. [DOI] [PubMed] [Google Scholar]
- 29.Grossman, T. H., M. Tuckman, S. Ellestad, and M. S. Osburne. 1993. Isolation and characterization of Bacillus subtilis genes involved in siderophore biosynthesis: relationship between B. subtilis sfpo and Escherichia coli entD genes. J. Bacteriol. 175:6203-6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagiwara, D., M. Sugiura, T. Oshima, H. Mori, H. Aiba, T. Yamashino, and T. Mizuno. 2003. Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J. Bacteriol. 185:5735-5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halsey, T. A., A. Vazquez-Torres, D. J. Gravdahl, F. C. Fang, and S. J. Libby. 2004. The ferritin-like Dps protein is required for Salmonella enterica serovar Typhimurium oxidative stress resistance and virulence. Infect. Immun. 72:1155-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hengge-Aronis, R. 1996. Back to log phase: σS as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol. Microbiol. 21:887-893. [DOI] [PubMed] [Google Scholar]
- 33.Hengge-Aronis, R. 2000. The general stress response in E. coli, p. 161-178. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 34.Hengge-Aronis, R. 1999. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr. Opin. Microbiol. 2:148-152. [DOI] [PubMed] [Google Scholar]
- 35.Hengge-Aronis, R. 2002. Recent insights into the general stress response regulatory network in Escherichia coli. J. Mol. Microbiol. Biotechnol. 4:341-346. [PubMed] [Google Scholar]
- 36.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hengge-Aronis, R. 2002. Stationary phase gene regulation: what makes an Escherichia coli promoter σS-selective? Curr. Opin. Microbiol. 5:591-595. [DOI] [PubMed] [Google Scholar]
- 38.Hershberg, R., G. Bejerano, A. Santos-Zavaleta, and H. Margalit. 2001. PromEC: an updated database of Escherichia coli mRNA promoters with experimentally identified transcriptional start sites. Nucleic Acids Res. 29:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hung, S. P., P. Baldi, and G. W. Hatfield. 2002. Global gene expression profiling in Escherichia coli K12. The effects of leucine-responsive regulatory protein. J. Biol. Chem. 277:40309-40323. [DOI] [PubMed] [Google Scholar]
- 40.Ibanez-Ruiz, M., V. Robbe-Saule, D. Hermant, S. Labrude, and F. Norel. 2000. Identification of RpoS (σS)-regulated genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:5749-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishihama, A. 2000. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 54:499-518. [DOI] [PubMed] [Google Scholar]
- 42.Izutsu, K., C. Wada, Y. Komine, T. Sako, C. Ueguchi, S. Nakura, and A. Wada. 2001. Escherichia coli ribosome-associated protein SRA, whose copy number increases during stationary phase. J. Bacteriol. 183:2765-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jishage, M., K. Kvint, V. Shingler, and T. Nystrom. 2002. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 16:1260-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khil, P. P., and R. D. Camerini-Otero. 2002. Over 1000 genes are involved in the DNA damage response of Escherichia coli. Mol. Microbiol. 44:89-105. [DOI] [PubMed] [Google Scholar]
- 45.Kusano, S., Q. Ding, N. Fujita, and A. Ishihama. 1996. Promoter selectivity of Escherichia coli RNA polymerase Eσ70 and Eσ38 holoenzymes. Effect of DNA supercoiling. J. Biol. Chem. 271:1998-2004. [DOI] [PubMed] [Google Scholar]
- 46.Kvint, K., A. Farewell, and T. Nystrom. 2000. RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of σS. J. Biol. Chem. 275:14795-14798. [DOI] [PubMed] [Google Scholar]
- 47.Lacour, S., A. Kolb, and P. Landini. 2003. Nucleotides from −16 to −12 determine specific promoter recognition by bacterial σS-RNA polymerase. J. Biol. Chem. 278:37160-37168. [DOI] [PubMed] [Google Scholar]
- 48.Landini, P., L. I. Hajec, L. H. Nguyen, R. R. Burgess, and M. R. Volkert. 1996. The leucine-responsive regulatory protein (Lrp) acts as a specific repressor for σS-dependent transcription of the Escherichia coli aidB gene. Mol. Microbiol. 20:947-955. [DOI] [PubMed] [Google Scholar]
- 49.Landini, P., L. I. Hajec, and M. R. Volkert. 1994. Structure and transcriptional regulation of the Escherichia coli adaptive response gene aidB. J. Bacteriol. 176:6583-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lange, R., M. Barth, and R. Hengge-Aronis. 1993. Complex transcriptional control of the σS-dependent stationary-phase-induced and osmotically regulated osmY (csi-5) gene suggests novel roles for Lrp, cyclic AMP (cAMP) receptor protein-cAMP complex, and integration host factor in the stationary-phase response of Escherichia coli. J. Bacteriol. 175:7910-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lange, R., and R. Hengge-Aronis. 1991. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor σS. J. Bacteriol. 173:4474-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee, H. J., K. J. Park, A. Y. Lee, S. G. Park, B. C. Park, K. H. Lee, and S. J. Park. 2003. Regulation of fur expression by RpoS and Fur in Vibrio vulnificus. J. Bacteriol. 185:5891-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee, S. J., and J. D. Gralla. 2004. Osmo-regulation of bacterial transcription via poised RNA polymerase. Mol. Cell 14:153-162. [DOI] [PubMed] [Google Scholar]
- 54.Lee, S. J., and J. D. Gralla. 2001. σ38 (RpoS)-RNA polymerase promoter engagement via −10 region nucleotides. J. Biol. Chem. 276:30064-30071. [DOI] [PubMed] [Google Scholar]
- 55.Lisser, S., and H. Margalit. 1993. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 21:1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loewen, P. C., B. Hu, J. Strutinsky, and R. Sparling. 1998. Regulation in the rpoS regulon of Escherichia coli. Can. J. Microbiol. 44:707-717. [DOI] [PubMed] [Google Scholar]
- 57.Lonetto, M., M. Gribskov, and C. A. Gross. 1992. The σ70 family: sequence conservation and evolutionary relationships. J. Bacteriol. 174:3843-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maeda, H., N. Fujita, and A. Ishihama. 2000. Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 28:3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marschall, C., V. Labrousse, M. Kreimer, D. Weichart, A. Kolb, and R. Hengge-Aronis. 1998. Molecular analysis of the regulation of csiD, a carbon starvation-inducible gene in Escherichia coli that is exclusively dependent on σS and requires activation by cAMP-CRP. J. Mol. Biol. 276:339-353. [DOI] [PubMed] [Google Scholar]
- 60.Martinez, A., and R. Kolter. 1997. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 179:5188-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martinez-Garcia, E., A. Tormo, and J. M. Navarro-Llorens. 2001. Further studies on RpoS in enterobacteria: identification of rpoS in Enterobacter cloacae and Kluyvera cryocrescens. Arch. Microbiol. 175:395-404. [DOI] [PubMed] [Google Scholar]
- 62.Martino, P. D., R. Fursy, L. Bret, B. Sundararaju, and R. S. Phillips. 2003. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can. J. Microbiol. 49:443-449. [DOI] [PubMed] [Google Scholar]
- 63.Masse, E., F. E. Escorcia, and S. Gottesman. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17:2374-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Masuda, N., and G. M. Church. 2003. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48:699-712. [DOI] [PubMed] [Google Scholar]
- 65.Medigue, C., A. Viari, A. Henaut, and A. Danchin. 1993. Colibri: a functional database for the Escherichia coli genome. Microbiol. Rev. 57:623-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 67.Mitchell, J. E., D. Zheng, S. J. Busby, and S. D. Minchin. 2003. Identification and analysis of ′extended −10′ promoters in Escherichia coli. Nucleic Acids Res. 31:4689-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moore, M. M., D. L. Fernandez, and R. L. Thune. 2002. Cloning and characterization of Edwardsiella ictaluri proteins expressed and recognized by the channel catfish Ictalurus punctatus immune response during infection. Dis. Aquat. Organ. 52:93-107. [DOI] [PubMed] [Google Scholar]
- 69.Mulvey, M. R., and P. C. Loewen. 1989. Nucleotide sequence of katF of Escherichia coli suggests KatF protein is a novel sigma transcription factor. Nucleic Acids Res. 17:9979-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nystrom, T. 2003. Conditional senescence in bacteria: death of the immortals. Mol. Microbiol. 48:17-23. [DOI] [PubMed] [Google Scholar]
- 71.O'Connor, K. E., A. D. Dobson, and S. Hartmans. 1997. Indigo formation by microorganisms expressing styrene monooxygenase activity. Appl. Environ. Microbiol. 63:4287-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ojangu, E. L., A. Tover, R. Teras, and M. Kivisaar. 2000. Effects of combination of different −10 hexamers and downstream sequences on stationary-phase-specific sigma factor σS-dependent transcription in Pseudomonas putida. J. Bacteriol. 182:6707-6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Olsen, A., A. Arnqvist, M. Hammar, S. Sukupolvi, and S. Normark. 1993. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol. Microbiol. 7:523-536. [DOI] [PubMed] [Google Scholar]
- 74.Oshima, T., H. Aiba, Y. Masuda, S. Kanaya, M. Sugiura, B. L. Wanner, H. Mori, and T. Mizuno. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 46:281-291. [DOI] [PubMed] [Google Scholar]
- 75.Phadtare, S., I. Kato, and M. Inouye. 2002. DNA microarray analysis of the expression profile of Escherichia coli in response to treatment with 4,5-dihydroxy-2-cyclopenten-1-one. J. Bacteriol. 184:6725-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pomposiello, P. J., M. H. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pon, C. L., B. Wittmann-Liebold, and C. Gualerzi. 1979. Structure-function relationships in Escherichia coli initiation factors. II. Elucidation of the primary structure of initiation factor IF-1. FEBS Lett. 101:157-160. [DOI] [PubMed] [Google Scholar]
- 78.Prigent-Combaret, C., E. Brombacher, O. Vidal, A. Ambert, P. Lejeune, P. Landini, and C. Dorel. 2001. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol. 183:7213-7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prigent-Combaret, C., O. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Quail, M. A., P. Jordan, J. M. Grogan, J. N. Butt, M. Lutz, A. J. Thomson, S. C. Andrews, and J. R. Guest. 1996. Spectroscopic and voltametric characterisation of the bacterioferritin-associated ferredoxin of Escherichia coli. Biochem. Biophys. Res. Commun. 229:635-642. [DOI] [PubMed] [Google Scholar]
- 81.Ren, D., L. A. Bedzyk, R. W. Ye, S. M. Thomas, and T. K. Wood. 2004. Stationary-phase quorum-sensing signals affect autoinducer-2 and gene expression in Escherichia coli. Appl. Environ. Microbiol. 70:2038-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ross, W., K. K. Gosink, J. Salomon, K. Igarashi, C. Zou, A. Ishihama, K. Severinov, and R. L. Gourse. 1993. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 262:1407-1413. [DOI] [PubMed] [Google Scholar]
- 83.Sak, B. D., A. Eisenstark, and D. Touati. 1989. Exonuclease III and the catalase hydroperoxidase II in Escherichia coli are both regulated by the katF gene product. Proc. Natl. Acad. Sci. USA 86:3271-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Salgado, H., A. Santos-Zavaleta, S. Gama-Castro, D. Millan-Zarate, E. Diaz-Peredo, F. Sanchez-Solano, E. Perez-Rueda, C. Bonavides-Martinez, and J. Collado-Vides. 2001. RegulonDB (version 3.2): transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res. 29:72-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schellhorn, H. E., J. P. Audia, L. I. Wei, and L. Chang. 1998. Identification of conserved, RpoS-dependent stationary-phase genes of Escherichia coli. J. Bacteriol. 180:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schembri, M. A., K. Kjaergaard, and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48:253-267. [DOI] [PubMed] [Google Scholar]
- 87.Tanaka, K., S. Kusano, N. Fujita, A. Ishihama, and H. Takahashi. 1995. Promoter determinants for Escherichia coli RNA polymerase holoenzyme containing σ38 (the rpoS gene product). Nucleic Acids Res. 23:827-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tanaka, K., Y. Takayanagi, N. Fujita, A. Ishihama, and H. Takahashi. 1993. Heterogeneity of the principal sigma factor in Escherichia coli: the rpoS gene product, σ38, is a second principal sigma factor of RNA polymerase in stationary-phase Escherichia coli. Proc. Natl. Acad. Sci. USA 90:3511-3515. (Erratum, 90:8303.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tani, T. H., A. Khodursky, R. M. Blumenthal, P. O. Brown, and R. G. Matthews. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Taverna, P., and B. Sedgwick. 1996. Generation of an endogenous DNA-methylating agent by nitrosation in Escherichia coli. J. Bacteriol. 178:5105-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Touati, E., E. Dassa, and P. L. Boquet. 1986. Pleiotropic mutations in appR reduce pH 2.5 acid phosphatase expression and restore succinate utilisation in CRP-deficient strains of Escherichia coli. Mol. Gen. Genet. 202:257-264. [DOI] [PubMed] [Google Scholar]
- 92.Ueguchi, C., N. Misonou, and T. Mizuno. 2001. Negative control of rpoS expression by phosphoenolpyruvate: carbohydrate phosphotransferase system in Escherichia coli. J. Bacteriol. 183:520-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Volkert, M. R., L. I. Hajec, Z. Matijasevic, F. C. Fang, and R. Prince. 1994. Induction of the Escherichia coli aidB gene under oxygen-limiting conditions requires a functional rpoS (katF) gene. J. Bacteriol. 176:7638-7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wada, A. 1998. Growth phase coupled modulation of Escherichia coli ribosomes. Genes Cells 3:203-208. [DOI] [PubMed] [Google Scholar]
- 95.Wang, D., X. Ding, and P. N. Rather. 2001. Indole can act as an extracellular signal in Escherichia coli. J. Bacteriol. 183:4210-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wei, J., M. B. Goldberg, V. Burland, M. M. Venkatesan, W. Deng, G. Fournier, G. F. Mayhew, G. Plunkett III, D. J. Rose, A. Darling, B. Mau, N. T. Perna, S. M. Payne, L. J. Runyen-Janecky, S. Zhou, D. C. Schwartz, and F. R. Blattner. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 71:2775-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wei, Y., J. M. Lee, C. Richmond, F. R. Blattner, J. A. Rafalski, and R. A. LaRossa. 2001. High-density microarray-mediated gene expression profiling of Escherichia coli. J. Bacteriol. 183:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wise, A., R. Brems, V. Ramakrishnan, and M. Villarejo. 1996. Sequences in the −35 region of Escherichia coli rpoS-dependent genes promote transcription by EσS. J. Bacteriol. 178:2785-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yamagishi, M., H. Matsushima, A. Wada, M. Sakagami, N. Fujita, and A. Ishihama. 1993. Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: growth phase- and growth rate-dependent control. EMBO J. 12:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang, W., L. Ni, and R. L. Somerville. 1993. A stationary-phase protein of Escherichia coli that affects the mode of association between the Trp repressor protein and operator-bearing DNA. Proc. Natl. Acad. Sci. USA 90:5796-5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yim, H. H., R. L. Brems, and M. Villarejo. 1994. Molecular characterization of the promoter of osmY, an rpoS-dependent gene. J. Bacteriol. 176:100-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yoon, S. H., M. J. Han, S. Y. Lee, K. J. Jeong, and J. S. Yoo. 2003. Combined transcriptome and proteome analysis of Escherichia coli during high cell density culture. Biotechnol. Bioeng. 81:753-767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.