Abstract
When an Escherichia coli culture changes from exponential growth to the stationary phase, expression of growth-related genes levels off, while a number of stationary-phase-specific genes are turned on. To gain insight into the growth phase-dependent global regulation of genome transcription, we analyzed the strength and specificity of promoters associated with the stationary-phase genes. For the in vivo assay of promoter activity, 300- to 500-bp DNA fragments upstream from the translation initiation codon were isolated and inserted into a newly constructed doubly fluorescent protein (DFP) vector. The activity of test promoters was determined by measuring the green fluorescent protein (GFP). To avoid the possible influence of plasmid copy number, the level of transcription of reference promoter lacUV5 on the same plasmid was determined by measuring the red fluorescent protein (RFP). Thus, the activities of test promoters could be easily and accurately determined by determining the GFP/RFP ratio. Analysis of the culture time-dependent variation of 100 test promoters indicated that (i) a major group of the stationary-phase promoters are up-regulated only in the presence of RpoS sigma; (ii) the phase-coupled increase in the activity of some promoters takes place even in the absence of RpoS; and (iii) the activity of some promoters increases in the absence of RpoS. This classification was confirmed by testing in vitro transcription by using reconstituted RpoD and RpoS holoenzymes.
The RNA polymerase of Escherichia coli is composed of the core enzyme (subunit composition, α2ββ′) with RNA polymerization catalytic activity and one of seven different species of the sigma subunit, each of which participates in transcription of a specific set of genes (10, 15). The intracellular concentration of RNA polymerase in the steady state of growing E. coli W3350 cells is maintained at a constant level characteristic of the rate of cell growth (16). The total number of core enzymes is not more than the total number of genes on the E. coli genome (1). Thus, the distribution pattern for RNA polymerase genes among about 4,000 genes in the genome should vary depending on the culture conditions (15). This finding accentuates the importance of the need for the RNA polymerase to choose which genes to transcribe and how often they are transcribed. The replacement of one core enzyme-associated sigma subunit by another sigma subunit is the most efficient way to alter the promoter recognition specificity of the transcription apparatus and is thus believed to be the major mechanism for switching of the transcription pattern. Thus, the competition between available sigma subunits should be a key determinant of which group genes are transcribed (8, 33). In addition to sigma subunit replacement, the activity and specificity of RNA polymerase is also modulated by interaction with about 300 molecular species of transcription factors (14, 15). Most of these accessory transcription factors are DNA-binding proteins and interact with RNA polymerase when both bind to their respective target sites.
When E. coli enters the stationary phase of growth, the majority of growth-related genes are turned off or their expression is leveled down, and a number of genes which are needed for morphological and physiological alteration of cells for adaptation to the dormant state are switched on (12, 15). For expression of the stationary-phase genes, both the transcription apparatus and the translation apparatus are modified. For modulation of the transcription apparatus, the RNA polymerase-associated sigma subunit is changed from RpoD (σ70) to RpoS (σ38) (11, 12). The intracellular levels of some transcription factors also fluctuate to various extents with the change in the cell growth phase (E. Koshio and A. Ishihama, unpublished data). On the other hand, one major change in the translation apparatus is the conversion of 70S ribosomes to 100S ribosome dimers after association of ribosome modulation factor (45). In addition, the protein composition and the configuration of the nucleoid change markedly upon entry into the stationary phase (41), which leads to changes in the genome expression pattern.
The synthesis and accumulation of the RpoS sigma subunit are controlled at multiple levels, including transcription, translation, protein turnover, and activity control (15, 32). Transcription control of rpoS involves a number of factors, including ppGpp (9, 29) and polyphosphate (polyP) as positive regulators and cAMP (12, 32) and UDP-glucose (2) as negative regulators. Translation of rpoS mRNA is stimulated under various stress conditions by several regulatory factors, including the RNA-binding Hfq (HF-1) protein (3, 36) and the small regulatory DsrA RNA (30, 40), and is repressed by the histone-like protein H-NS (24) and a regulatory oxyS RNA (48). The RpoS protein is subject to rapid turnover in exponential-phase E. coli cells. The increase in the RpoS level in stationary-phase E. coli results, at least in part, from a large increase in the stability of the RpoS protein (47).
The RNA polymerase holoenzyme containing the RpoS sigma subunit is essential for transcription of some, if not all, stationary-phase-specific genes. The promoter recognition specificity of the RpoS holoenzyme is not entirely understood, since the promoters of the stationary-phase-specific genes identified so far do not have a clear distinctive consensus sequence and for the most part are recognized in vitro by both the RpoD and RpoS holoenzymes (25, 42, 43). Furthermore, the intracellular concentration of RpoD is always the highest concentration in both the exponential and stationary phases even though RpoS becomes detectable in the stationary phase, as do two of the alternative sigma subunits, RpoN and RpoF (17, 21).
One critical factor for selective utilization of the RpoS sigma subunit is inactivation of sigma factors by the corresponding anti-sigma factors. Rsd (a regulator of sigma D) is involved in selective inhibition of the RpoD sigma subunit (19). Several lines of evidence indicate that some additional factors influence, in various ways, the activity and specificity of the two different forms (the RpoD and RpoS holoenzymes) of the RNA polymerase (15). Under stress conditions, for instance, the intracellular concentrations of compatible solutes, including stress protectants, such as trehalose and glycine betaine, and of some storage products, such as glycogen and polyphosphate, increase markedly (37). At least some of these compounds influence the activity and specificity of the two holoenzymes to different degrees. For instance, the activity of the RpoS holoenzyme is modulated by glutamate (7, 31, 38) and trehalose (27) at the steps for holoenzyme formation and holoenzyme binding to promoters.
RNA polymerase from stationary-phase cells of E. coli is associated with inorganic polyphosphate and shows altered promoter selectivity (28). Since mutants defective in the ppk gene encoding polyphosphate kinase are defective in survival in the stationary-phase function (39), polyP is now believed to play a role in bacterial adaptation to the stationary phase. At low salt concentrations, polyP inhibits transcription in vitro by both the RpoD and RpoS holoenzymes. When the concentration of potassium glutamate increases, however, polyP inhibition is relieved for the RpoS RNA polymerase but not for RpoD holoenzyme, suggesting that polyP may play a role in promoter selectivity control of RNA polymerase in E. coli growing under high-osmolarity conditions and in the stationary phase. Together, these observations indicate that the promoter selectivity of each holoenzyme is expressed only under defined conditions, and thus it is difficult to determine the promoter selectivity of RpoS until the specific optimum reaction conditions for in vitro transcription are established.
To gain further insight into the promoter selectivity control of the RNA polymerase holoenzyme containing the RpoS sigma subunit, our recent efforts have been directed towards identification of a complete list of the stationary-phase-specific gene promoters and towards functional characterization of these promoters in vivo, with a focus on growth phase-coupled variation of their strengths. In this study, we first performed a systematic search for the stationary-phase-specific promoters based on transcriptome analyses. The strength order of 80 representative stationary-phase gene-associated promoters from the set consisting of previously identified (12, 15) and newly identified stationary-phase promoters was examined in vivo by using the novel doubly fluorescent protein (DFP) vector pGRP for a promoter assay. The newly developed promoter assay vector was also effectively used for classification of the stationary-phase promoters on the basis of their dependence on the RpoS sigma subunit.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli KP7600 (W3110 lacI lacZ Δλ galK2 galT22) was used as the wild-type strain. By starting with the KP7600 strain, rpoS disruptant strain JD22323 was constructed by a transposon insertion method (T. Miki, unpublished data). The absence of the RpoS protein was confirmed by immunoblotting of whole-cell lysates. Cells were grown at 37°C with aeration in Luria broth (LB). An overnight culture in LB was diluted 1,000-fold into fresh LB, and incubation was carried out at 37°C with shaking at a constant rate (150 rpm). Cell growth was monitored by measuring the turbidity at 600 nm.
Microarray assay of the transcriptome with DNA chips.
Total RNA was isolated from fresh cells with an RNeasy kit (QIAGEN, Chatsworth, Calif.) or by phenol-chloroform extraction. To remove genomic DNA, samples were treated with RNase-free DNase I (Takara), followed by phenol-chloroform extraction and precipitation with ethanol. The purity of RNA samples was checked by polyacrylamide gel electrophoresis in the presence of urea. A fluorescently labeled cDNA library was prepared in a 40-μl reaction mixture, which contained standard AMV-RT-XL buffer (Takara), 0.5 mM dATP, 0.5 mM dGTP, 0.5 mM dCTP, 0.2 mM dTTP, 10 μg of random primers (Takara), 0.4 nmol of Cy3-dUTP or Cy5-dUTP, and 20 μg of RNA. Cy3-dUTP was used to label control samples (log-phase samples for the time course experiments and wild-type samples for the comparative studies with the rpoS mutant), and Cy5-dUTP was used to prepare a test cDNA set (stationary-phase samples or rpoS mutant samples). Each reaction mixture was heated for 5 min at 65°C and cooled at room temperature. After addition of 50 U of reverse transcriptase (AMV-RT-XL; Takara), cDNA synthesis was carried out at 42°C for 1 h and then, after addition of another 50 U of AMV-RT-XL, was continued for additional 1 h. Synthesized cDNA was purified by using Centri-Sep spin columns preequilibrated with 0.1 M NaCl, extracted with phenol-chloroform, precipitated with ethanol, and dissolved in water (8 μl).
IntelliGene EcoliCHIP (version 1.0; Takara) containing 4,028 spots for the full-length coding frame DNA from 4,390 open reading frames of the E. coli W3110 genome was used for microarray analysis of the expression profile. Before hybridization, DNA-spotted glass slides were incubated at 65°C for 1 h with 20 μl of prehybridization buffer containing 6× SSC, 0.2% sodium dodecyl sulfate (SDS), 5× Denhardt's solution, and 1 mg of denatured salmon sperm DNA per ml and washed with 2× SSC at 65°C (1× SSC is 0.15 M NaCl plus 0.015 sodium citrate). cDNA samples were heated to 98°C for 2 min. Hybridization was performed for 16 h at 65°C by using 20 μl of reaction buffer (4.2× SSC, 0.14% SDS, 3.5× Denhardt's solution, 0.7 mg of denatured salmon sperm DNA per ml) containing both Cy3- and Cy5-labeled cDNA samples. Chips were washed once with 2× SSC for 5 min at 65°C, once with 0.2× SSC-0.1% SDS for 5 min at 65°C, and four times with 0.2× SSC for 5 min at room temperature. The microarrays were scanned with an Affimetrix laser scanner, and the intensities of hybridized Cy3 and Cy5 were independently quantified by using ImageQuant (v.4.0; Molecular Dynamics). Background correction was performed by measuring the fluorescence intensity of the chip regions outside the DNA spots. Averaged signals detected for the spots containing non-E. coli DNA samples (i.e., calf thymus DNA, human transferrin receptor [TFR], or human β-actin) were used as negative controls.
Construction of promoter assay vectors and measurement of the promoter activity in vivo.
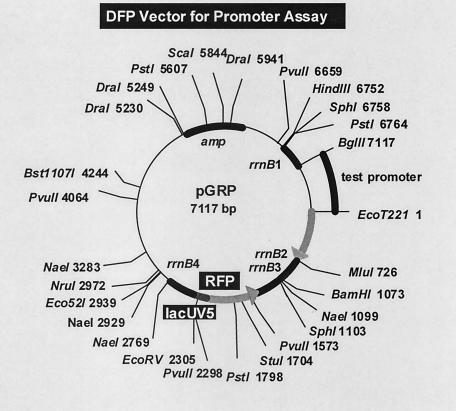
For quantitative measurement of the promoter activity in vivo, two types of fluorescent protein genes, one for the red fluorescent protein dsRed (Clontech) (referred to as RFP in this paper) and the other for the green fluorescent protein eGFP (Clontech) (referred to as GFP in this paper), wereinserted into a single vector. In most experiments described below the RFP gene was under control of reference promoter lacUV5, and the GFP gene was under control of a test promoter. The test promoter sequences upstream from the corresponding translation initiation codons up to about 500 bp were PCR amplified and inserted into pGRP (the second version of the DFP vector) between BglII and EcoT221sites (the sites for BglII and EcoT221 were included in the PCR primers) (see Fig. 2 for the physical map). The promoter assay vectors constructed in this way were transformed into the appropriate host strains.
FIG. 2.
aCategory 1, stationary-phase promoters selected based on the transcriptome analysis (this study); category 2, stationary-phase promoters selected based on the proteome analysis (Wada et al., unpublished); category 3, stationary-phase promoters identified previously (15); category C, reference promoters associated with the growth-related genes; category H, promoters associated with the heat shock-induced genes. DFP vector for the assay of in vivo promoter activity. The test promoter fragment upstream from the initiation codon with BglII and EcoT221 sites at the termini can be inserted in one step into pGRP (the second version of the DFP vector) between BglII and EcoT221 sites. The initiation codon for GFP is regenerated after ligation of the EcoT221 site. Major restriction enzyme sites and the distances from the EcoT221 site are shown.
LB contains unidentified natural substances that are fluorescent. For measurement of the fluorescence intensity of RFP or GRP expressed in E. coli, cells grown in LB for various times were harvested by centrifugation, resuspended in phosphate-buffered saline,and diluted with phosphate-buffered saline to obtain approximately the same cell density (0.6 A600 unit) for all samples. For measurement of bulk fluorescence, aliquots of a 0.3-ml cell suspension were added to 0.4 × 96 flat-bottom wells, and the fluorescence was measured with a FL600 Bio-Tek microplate reader (Bio-Tek Instruments, Winooski, Vt.). The net fluorescence value was measured after subtraction of the background fluorescence, which was determined by using E. coli cultures with the pDFP vector without promoter insertions.
Measurement of the promoter activity by an in vitro transcription assay.
PCR-amplified promoter fragments, which were used for construction of the promoter assay vectors (see above), were also used as truncated DNA templates for in vitro transcription. RNA polymerase core enzyme was purified from E. coli W3350 by passage of purified RNA polymerase at least three times through phosphocellulose columns (26). The RpoD sigma subunit was expressed by using pGEMD and was purified as described by Igarashi and Ishihama (13), while RpoS was expressed by using pEFT and was purified as described previously (42). Holoenzymes were reconstituted by mixing the core enzyme and a fourfold molar excess of each sigma subunit. Transcription by the reconstituted holoenzymes was carried out under standard reaction conditions described previously (23).
RESULTS AND DISCUSSION
Systematic search for the stationary-phase genes.
So far, more than 100 genes which are expressed when an E. coli culture enters the stationary phase after exponential growth have been identified (12, 15). For identification of the whole set of stationary-phase genes, we carried out systematic analyses of both the transcriptome and the proteome of E. coli wild-type strain W3110. Microarray-based comprehensive analyses of the gene expression profile during the early phase of the transition from exponential growth to the stationary phase have been performed with E. coli grown in poor media (44, 46). In this study, we performed the transcriptome analysis with E. coli cultures grown in a rich medium at various stages of the transition to the stationary phase.
E. coli KP7600, a derivative of A-type W3110, which contains the full set of all seven species of RNA polymerase sigma subunits in intact forms (18), was grown in LB (Fig. 1A). Total RNA was prepared at various times from both exponential-phase and stationary-phase cultures; exponential-phase RNA was labeled with Cy5, while stationary-phase cDNA was labeled with Cy5. Mixtures of Cy3- and Cy5-labeled RNA were subjected to microarray analysis by using an E. coli DNA chip containing 4,028 DNA spots, each of which corresponded to one open reading frame (Takara version 1). The experiment was repeated with the same RNA samples, but exponential-phase cDNA was labeled with Cy5 and stationary-phase cDNA was labeled with Cy3. For each time point of the culture, the same experiments were repeated with a different batch of mRNA.
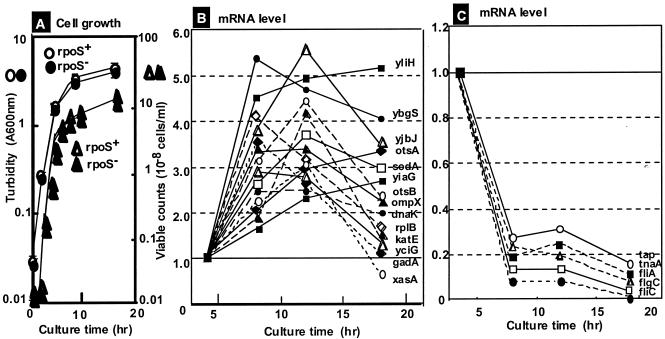
FIG. 1.
Growth phase-coupled variation of mRNA levels. (A) E. coli W3110 was grown in LB. Cell growth was monitored by measuring the turbidity at 600 nm, and the number of viable cells was determined by measuring colonies on LB agar plates. (B and C) Microarray assays were carried out as described in Materials and Methods, first for mixtures of Cy3-labeled cDNAs for exponential-phase RNAs and Cy5-labeled cDNAs for stationary-phase RNAs and second with the opposite combination. The assays were repeated twice with independent cultures. The level of each mRNA at each point during the stationary phase, relative to the exponential-phase level, was calculated. For the mRNAs whose levels increased (B) or decreased (C) in the stationary phase, the culture time-dependent variation is shown for some representative species (for details see Tables 1 and 3).
In a separate experiment, we recognized that the intracellular level of a population of mRNAs fluctuated greatly from experiment to experiment even though the experiments were carried out under essentially the same conditions (O. Ozoline and A. Ishihama, unpublished data). This group of mRNAs was excluded from the data analysis. Table 1 shows the 20 highly activated genes (more than 2.23-fold up-regulated) in the early stationary-phase (8 h of growth). Except for the unidentified genes, most of these genes have been identified previously as stationary-phase genes, including otsB, katE, sodA, cfa, cspE, bolA, and otsA (12, 15). The otsA and otsB genes form a single operon and are involved in trehalose metabolism, while gadB encoding glutamate decarboxylase and xasA for amino acid transport are organized in a single operon. The genes encoding ribosomal proteins are highly expressed in growing cells, and expression levels off upon entry into the stationary phase. The microarray assay, however, indicated that the levels of expression of some ribosomal protein operons were still high compared to those of other genes, at least during the transition from exponential growth to the stationary phase (46). The levels of mRNA for some ribosomal proteins, including L2 and S3, increased in the early stationary phase. The rplB and rpsC genes encoding these ribosomal proteins are organized in the same S10 operon. The apparent increase in S10 operon mRNA levels might have been due to the delay in the decline in transcription of the S10 operon compared to other ribosomal protein operons (Fig. 1B shows the growth phase-coupled change in mRNA level). Other up-regulated genes in the stationary phase, listed in Table 1, form single-gene operons. Some genes with unidentified functions, such as yciG, yliH, ybgS, yjbJ, yiaG, and ybaS, are highly induced and may be involved in the adaptation to the stationary phase. Among the top 20 highly transcribed genes, at least 11 are under the control of RpoS (see below), and at least 8 genes (yciG, otsB, katE, sodA, cfa, bolA, dagA, and otsA) have been found to be under control of a global regulator, Lrp, the leucine response regulatory protein (44).
TABLE 1.
Genes up-regulated in the stationary phase in wild-type E. coli W3110a
| Gene | Map position | Function | Transcription levelb |
|---|---|---|---|
| yciG | 28.32 | Unidentified | 5.57 |
| yliH | 18.91 | Unidentified | 4.94 |
| ybgS | 16.90 | Unidentified | 4.63 |
| otsB | 42.67 | Trehalose-6-phosphate phosphatase | 4.45 |
| katE | 39.06 | Catalase HPII; hydroperoxidase II | 4.39 |
| sodA | 88.34 | Superoxide dismutase (Mn) | 3.66 |
| cfa | 37.49 | Cyclopropane fatty acid synthase | 3.53 |
| ompX | 18.31 | Outer membrane protein X | 3.22 |
| rplB | 74.33 | 50S ribosomal subunit protein L2 | 3.14 |
| xasA | 33.78 | Acid-resistant amino acid transporter | 3.06 |
| yjbJ | 91.76 | Unidentified | 2.94 |
| cspE | 14.15 | Cold shock protein CspA homologue | 2.77 |
| bolA | 9.78 | Morphogenesis; PBP6 synthesis control | 2.63 |
| gadA | 78.97 | Glutamate decarboxylase alpha subunit | 2.56 |
| dnaK | 0.26 | Heat shock protein; Hsp70 chaperone | 2.56 |
| yiaG | 80.12 | Unidentified | 2.33 |
| otsA | 42.64 | Trehalose-6-phosphate synthase | 2.29 |
| ybaS | 11.01 | Unidentified (putative glutaminase) | 2.26 |
| guaB | 56.70 | IMP dehydrogenase; GMP synthesis | 2.23 |
| rpsC | 74.30 | 30S ribosomal subunit protein S3 | 2.23 |
The transcription patterns in wild-type E. coli W3110 were determined for both exponential growth and the stationary phase by a Microarray assay by using an E. coli DNA chip (Takara version 1).
Increase in transcription from exponential growth to the stationary phase.
The microarray assay was repeated twice for three times (8, 12, and 18 h) in the stationary phase. Figure 1B shows the culture time-dependent variation for some representative mRNAs from the highly activated genes in the stationary phase. The levels of transcripts of some stationary-phase genes, such as yliH, ybgS, sodA, otsA, and yiaG, remained high, while those of some other genes, such as yjbJ, otsB, ompX, katE, gadA, and xasA, showed maximum peaks at either 8 or 12 h but decreased thereafter. Although the mRNA levels for three genes, yliH, ybgS, and yjbJ, were always very high during the stationary phase, their physiological roles are not known yet.
The majority of stationary-phase genes are transcribed by RNA polymerase containing the RpoS sigma subunit (15). To identify the up-regulated genes, which are under the control of RpoS, we next carried out microarray assays for a mutant lacking RpoS. Table 2 shows 25 genes whose expression was marked reduced in the rpoS mutant. Of the top 20 genes that were highly up-regulated in the stationary phase, which are listed in Table 1, about one-half are included in this group of RpoS-dependent genes. For instance, the transcription level in the rpoS mutant decreased to less than 10% for ybgS, xasA, dps, yliH, yodT, osmC, hdeA, and osmB and to less than 20% for otsA, ompX, cacD, otsB, and cspC. Transcription of this group of genes must be under strict control of the RpoS sigma subunit, even though promoters of many stationary-phase genes are recognized and transcribed in vitro by both RpoD and RpoS holoenzymes (25, 42, 43).
TABLE 2.
Stationary-phase genes that were not activated in an RpoS sigma-defective mutanta
| Gene | Map position | Function | Activity ratiob |
|---|---|---|---|
| ybgS | 16.90 | Unidentified | 0.033 |
| xasA | 33.78 | Acid-resistant amino acid transporter | 0.036 |
| dps | 18.27 | DNA-binding protein for starvation | 0.039 |
| yliH | 18.91 | Unidentified | 0.047 |
| ycdT | 23.54 | Unidentified | 0.063 |
| osmC | 33.51 | Osmotically inducible protein C | 0.074 |
| hdeA | 78.76 | Periplasmic protein A | 0.084 |
| osmB | 28.91 | Osmotically inducible protein B | 0.091 |
| otsA | 42.64 | Trehalose-6-phosphate synthase | 0.104 |
| ompX | 18.31 | Outer membrane protein X | 0.122 |
| zitB | 16.88 | Zinc transporter | 0.140 |
| otsB | 42.67 | Trehalose-6-phosphate phosphatase | 0.155 |
| cspC | 41.07 | Cold shock protein CspA homologue | 0.168 |
| sodC | 37.12 | Superoxide dismutase (Cu/Zn) | 0.209 |
| yjbJ | 91.76 | Unidentified | 0.213 |
| osmY | 99.35 | Osmotically inducible protein Y | 0.216 |
| hdeB | 78.75 | Periplasmic protein B | 0.246 |
| hdeD | 78.78 | Periplasmic protein D | 0.250 |
| slp | 78.71 | C starvation; outer membrane protein | 0.272 |
| gadA | 78.97 | Glutamate decarboxylase alpha subunit | 0.297 |
| wrbA | 22.99 | TrpR repressor-binding protein | 0.328 |
| osmE | 39.23 | Osmotically inducible protein E | 0.364 |
| adhE | 27.91 | Alcohol dehydrogenase | 0.422 |
| bolA | 9.78 | Morphogenesis; PBP6 control | 0.424 |
| rpoS | 61.75 | RNA polymerase RpoS sigma subunit | 0.453 |
The transcription patterns in stationary-phase cells of wild-type and rpoS mutant E. coli W3110 were determined by a microarray assay by using an E. coli DNA chip (Takara version 1).
Relative level of transcription for the rpoS mutant compared with wild-type E. coli.
Microarray assays also identified a set of down-regulated genes in the stationary phase (Table 3 and Fig. 1C). Most of the genes that showed marked reductions in the stationary phase are genes involved in the formation and function control of flagella, including fliC (flagellin), flgD (basal body modification), flgE (hook protein), flgB (basal body rod), flgC (hook cap), flgF (basal body rod), cheA (CheA/CheY kinase), flgA (flagellum synthesis), fliZ (flagellum synthesis), fliA (RpoF sigma subunit), and flgM (anti-RpoF). These genes are organized into three operons, flgBCDEFGHIJKL, flgAMN, and fliDCAZ. This finding agrees well with our previous observation that the flagellum density on the cell surface decreases in stationary-phase E. coli (34, 35). The down-regulation of flagellum genes was not observed in the mutant lacking the RpoS sigma subunit, supporting the sigma subunit competition model (35).
TABLE 3.
Genes down-regulated in the stationary phase in wild-type E. coli W3110a
| Gene | Map position | Function | Transcription levelb |
|---|---|---|---|
| fliC | 43.11 | Flagellin | 0.067 |
| flgD | 24.38 | Flagellum basal body rod modification | 0.114 |
| gatC | 46.80 | Galactitol-specific PTS systemc | 0.124 |
| gatD | 46.77 | Galactitol-1-phosphate dehydrogenase | 0.128 |
| flgE | 24.40 | Flagellum hook | 0.134 |
| flgB | 24.36 | Flagellum basal body rod | 0.151 |
| flgC | 24.37 | Flagellum hook cap | 0.157 |
| flgF | 24.42 | Flagellum basal body rod | 0.188 |
| sucB | 16.40 | 2-Oxoglutarate dehydrogenase | 0.204 |
| tnaA | 83.77 | Tryptophanase | 0.233 |
| cheA | 42.49 | CheY and CheB kinase | 0.249 |
| flgA | 24.35 | Flagellar synthesis | 0.258 |
| fliZ | 43.08 | Unidentified (putative RpoF regulator) | 0.269 |
| fliA | 43.09 | RpoF sigma factor | 0.277 |
| ompC | 49.79 | Outer membrane protein C | 0.290 |
| sucC | 16.43 | Succinyl coenzyme A synthetase | 0.301 |
| rbsB | 84.80 | Periplasmic d-ribose-binding protein | 0.316 |
| flgM | 23.34 | Anti-sigma F (RpoF) factor | 0.325 |
| gatZ | 46.84 | (Tagatose-6-phosphate kinase) | 0.345 |
| tap | 42.41 | Dipeptide chemoreceptor | 0.347 |
The transcription patterns in wild-type E. coli W3110 were determined for both the exponential and stationary phases by a microarray assay by using an E. coli DNA chip (Takara version 1).
Decrease in transcription from exponential growth to the stationary phase.
PTS, phosphotransferase.
In parallel with the transcriptome analysis, a proteome analysis was performed for 8 days with the same E. coli W3110 strain by using the newly developed radical-free and highly reducing method of two-dimensional gel electrophoresis (A. Wada et al., unpublished data). The major species of stationary-phase genes were the same in the transcriptome and proteome analyses.
Isolation of promoters from the stationary-phase genes and construction of promoter assay vectors.
So far, a number of stationary-phase genes have been identified, including genes identified in this study, but the nature of the promoters associated with the stationary-phase genes is poorly understood. Here we performed a systematic and comprehensive analysis of 80 promoters for the stationary-phase genes (50 known stationary-phase genes [category 3 in Table 4 ] and 30 newly identified stationary-phase genes [categories 1 and 2 in Table 4]). The promoters examined in this study, listed in Table 4, were selected on the basis of (i) the stationary-phase genes newly identified by the transcriptome analysis (this study) (category 1), (ii) the stationary-phase genes newly identified by the proteome analysis (A. Wada, unpublished data) (category 2), and (iii) the stationary-phase genes previously identified mostly by genetic analyses (listed in reference 15) (category 3). In addition, we included in the promoter analysis some reference promoters for the genes expressed in the exponential phase of cell growth (nine control promoters) and for the genes up-regulated upon exposure to heat shock (11 heat shock gene promoters). Transcription of these growth-related genes and heat shock genes is under control of RpoD and RpoH, respectively.
TABLE 4.
Promoters analyzed with the DFP vector
| Gene | Category(ies)a | Function | Gene | Category(ies)a | Function | |
|---|---|---|---|---|---|---|
| acnA | 3 | Aconitate hydrase | ||||
| adhE | 1, 2 | Alcohol dehydrogenase | ||||
| ahpC | 3 | Alkyl hydroperoxide reductase small subunit | ||||
| aldB | 3 | Aldehyde dehydrogenase | ||||
| appY | 3 | Transcription factor (stationary-phase genes) | ||||
| bolA | 1, 2, 3 | Morphogenesis; control of PBP6 synthesis | ||||
| cbpA | 3 | Curved DNA-binding nucleoid protein | ||||
| cfa | 1, 3 | Cyclopropane fatty acid synthase | ||||
| clpA | 3 (H) | ATP-dependent ClpAP protease A subunit | ||||
| clpP | H | ATP-dependent ClpAP protease P subunit | ||||
| cpxP | 2 | Periplasmic protein | ||||
| csgB | 3 | Minor curlin subunit precursor | ||||
| csiD | 3 | Carbon starvation-inducible gene D | ||||
| csiE | 3 | Stationary-phase inducible protein E | ||||
| cspC | 1, 2 | Cold shock protein C; mukB suppression | ||||
| cspD | 2 | Cold shock protein D | ||||
| cspE | 1 | Cold shock protein E; chromosome condensation | ||||
| dnaK | 1 (H) | Heat shock protein; chaperone Hsp70 | ||||
| dps | 1, 2, 3 | DNA-binding protein in starved cells | ||||
| fic | 3 | Cell filamentation | ||||
| ftsQ | 3 | Cell division protein; septum formation | ||||
| fxsA | H | F exclusion suppressor | ||||
| gabP | 3 | γ-Aminobutyric acid transport permease | ||||
| gadA | 1, 2, 3 | Glutamate decarboxylase alpha subunit | ||||
| gadB | 3 | Glutamate decarboxylase beta subunit | ||||
| galE | 3 | ODP-galactose 4-epimerase | ||||
| galM | H | Mutarotase; alpha- to beta-aldose conversion | ||||
| gatY | 2 | d-Tagatose-1,6-bisphosphate aldolase | ||||
| ggt | C | γ-Glutamyltranspeptidase | ||||
| glgC | 3 | Glucose-1-phosphate adenylyltransferase | ||||
| glgS | 3 | Priming of glycogen synthesis | ||||
| glpD | 2 | sn-Glycerol-3-phosphate dehydrogenase | ||||
| gor | 3 | Glutathione oxidoreductase | ||||
| guaB | 1 | IMP dehydrogenase | ||||
| hdeA | 1, 2, 3 | Periplasmic protein A | ||||
| hdeB | 1, 2, 3 | Periplasmic protein B | ||||
| hdeD | 1, 2 | Periplasmic protein D | ||||
| hfq | C | RNA-binding protein; RNA function control | ||||
| hlpA | C | Nucleoid histone-like protein HLP-I | ||||
| hns | C | Nucleoid curved DNA-binding protein HLP-II | ||||
| hscB | H | Cochaperone protein HscB | ||||
| htpG | H | Heat shock chaperone Hsp90 | ||||
| htrE | 3 | Outer membrane porin protein; fimbrial assembly | ||||
| hupA | C | DNA-binding protein HU-alpha | ||||
| hyaA | 3 | Hydrogenase 1 small subunit | ||||
| ihfA | C | Nucleoid protein; integration host factor | ||||
| katE | 1 | Catalase HPII; hydroperoxidase II | ||||
| katG | 1, 3 | Catalase HPI; hydroperoxidase I | ||||
| Ion | H | Heat shock-induced ATP-dependent protease La | ||||
| mdh | H | Malate dehydrogenase | ||||
| modA | 2 | Molybdate-binding periplasmic protein | ||||
| nlpD | 2 | Inner membrane lipoprotein | ||||
| ompC | 1 | Outer membrane protein C (1b) | ||||
| ompX | 1, 2 | Outer membrane protein X | ||||
| oppA | 2 | Oligopeptide transport; periplasmic protein | ||||
| osmB | 1, 2, 3 | Osmotically inducible protein B | ||||
| osmC | 1, 2, 3 | Osmolarity-inducible protein C | ||||
| osmE | 1, 2, 3 | Osmolarity-inducible protein E | ||||
| osmY | 1, 2, 3 | Osmolarity-inducible protein Y | ||||
| otsA | 1, 2, 3 | Trehalose-6-phosphate synthase | ||||
| otsB | 1, 2, 3 | Trehalose-6-phosphate phosphatase | ||||
| oxyR | 3 | Transcription factor (H2O2-inducible genes) | ||||
| pcm | 3 | l-Isoaspartate protein carboxylmethyltransferase | ||||
| phoA | H | Alkaline phosphatase | ||||
| poxB | 3 | Pyruvate oxidase | ||||
| proP | 3 | Glycine betaine/proline transport permease II | ||||
| proV | 2 | Glycine betaine/proline transport permease I | ||||
| pspA | 3 | Phage shock protein | ||||
| rbsB | 1 | Periplasmic d-ribose-binding protein | ||||
| rbsD | 2 | Periplasmic d-ribose high-affinity transport system | ||||
| rmf | 3 | Ribosome modulation factor | ||||
| rob | C | Right origin-binding protein | ||||
| rpmE | C | 50S ribosomal protein L31 | ||||
| rpoS | 1, 2, 3 | RNA polymerase RpoS sigma subunit | ||||
| rsd | 3 | Regulator for RpoD (anti-RpoD) | ||||
| slp | 1, 2 | Carbon starvation; outer membrane lipoprotein | ||||
| slyD | 3 | FKBP-type peptidyl-prolyl cis-trans-isomerase | ||||
| sodA | 1 | Superoxide dismutase (Mn) | ||||
| sodB | 2 | Superoxide dismutase (Fe) | ||||
| sodC | 1, 2, 3 | Superoxide dismutase (Cu-Zn) | ||||
| ssnA | 2 | Putative proteoglycan | ||||
| stpA | C | H-NS-like nucleoid protein | ||||
| treA | 3 | Periplasmic trehalase | ||||
| treF | 3 | Cytoplasmic trehalase | ||||
| uspB | 3 | Universal stress protein B | ||||
| uxuA | H | Mannonate dehydratase | ||||
| wrbA | 1, 2, 3 | Trp repressor-binding protein | ||||
| xasA | 1, 2 | Acid-resistant amino acid transporter | ||||
| xthA | 1, 3 | Exonuclease III | ||||
| ybaS | 1 | Unidentified (putative glutaminase) | ||||
| ybgS | 1,2 | Unidentified | ||||
| yceP | H | Unidentified | ||||
| ycgZ | 2 | Unidentified | ||||
| yciF | 2 | Unidentified | ||||
| yciG | 1, 2 | Unidentified | ||||
| ygaU | 2 | Unidentified | ||||
| yiaG | 1 | Unidentified | ||||
| yifE | 2 | Unidentified | ||||
| yjbJ | 1, 2 | Unidentified | ||||
| yliH | 1, 2 | Unidentified |
Category 1, stationary-phase promoters selected based on the transcriptome analysis (this study); category 2, stationary-phase promoters selected based on the proteome analysis (Wada et al., unpublished); category 3, stationary-phase promoters identified previously (15); category C, reference promoters associated with the growth-related genes; category H, promoters associated with the heat shock-induced genes.
For determination of the activity and regulation of 100 promoters selected as described above, DNA segments containing the corresponding promoters were isolated by PCR. Each promoter segment covered the sequence from the translation initiation codon to approximately 300 bp upstream of the transcription initiation site or to about 500 bp upstream of the initiation codon (when the transcription initiation site has not been identified). In some cases, 500-bp fragments included terminal segments of flanking genes. PCR-amplified fragments were inserted into the second version of the DFP vector pGRP for the promoter assay (Fig. 2). In the case of the original DFP vector, pGRMI, two long segments, one containing the GFP reading frame and the other containing the RFP coding frame, had to be ligated (34, 35), but the ligation of two large fragments was inefficient. By using the second version, PCR-amplified fragments could be directly inserted into the DFP vector in one step between BglII and EcoT221 sites. The promoter activity in vivo was determined by determining the GFP/RFP ratio, thereby avoiding the fluctuation of reporter gene expression resulting from changes in plasmid copy number.
Promoter activity in vivo and growth phase-dependent variation.
One useful application of the newly developed promoter assay system with the DFP vector is to determine the promoter activity in vivo and to compare the relative levels for various test promoters. In this study, we initially determined the promoter activity for 100 promoters in wild-type E. coli KP7600 (a W3110 derivative) grown in a rich LB. The promoter activity was measured for nine times from the exponential growth phase (3 h after inoculation of an overnight culture into fresh medium) to the stationary phase (36 h). The activity of reference promoter lacUV5 was almost the same throughout the culture period (35). A culture time-dependent increase in the promoter activity was observed, as expected, for most of the stationary-phase promoters tested (Fig. 3), while the variation in activity was low or nonexistent for the control reference promoters for the constitutively expressed growth-related genes.
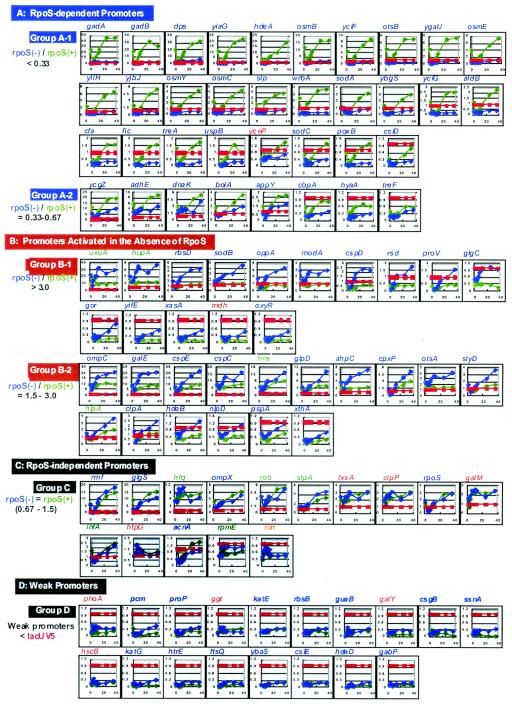
FIG. 3.
Growth phase-dependent variation of promoter activity. A total of 100 promoters, listed in Table 4, were inserted into pGRP, and each of the resultant promoter assay plasmids was transformed into both wild-type E. coli KP7600 and its rpoS mutant JD22323. An overnight culture of each transformant grown in LB was transferred into fresh LB, and the culture was incubated at 37°C with shaking. The promoter activity was measured at nine times (0, 3, 4, 6, 8, 10, 12, 24, and 36 h). The activity of the test promoter relative to that of reference promoter lacUV5 was determined by determining the GFP/RFP ratio (for details see Materials and Methods). (A) RpoS-dependent promoters (group A); (B) RpoS-independent promoters (group B); (C) RpoS-independent promoters (group C); (D) weak promoters (group D).
In the stationary phase, three promoters, gadA, gadB, and dps, showed more than 30-fold-higher activities than the reference promoter lacUV5 (Table 5). Glutamate decarboxylation contributes to pH homeostasis by consuming intracellular glutamic acid (6). E. coli contains two genes, gadA and gadB, encoding two isoforms of glutamate decarboxylase, which differ in five amino acid residues. In agreement with the previous finding that acid-induced stationary-phase expression of gadA and gadB is controlled at the level of transcription (5), the gadA and gadB promoters were among the group of promoters with the highest activity in the stationary phase under the culture conditions employed. The level of gadA mRNA was included in the group of the top 20 mRNA levels as measured by the microarray assay (Table 1). The level of gadB mRNA was also elevated 1.5- to 2.0-fold in the stationary phase (data not shown). It is worth noting that selective activation in vitro of the RpoS sigma subunit takes place in the presence of high concentrations of glutamate (7, 31, 38), and therefore the activity control of the RpoS sigma subunit and the elevation of the GAD (glutamate decarboxylase) system must be tightly correlated during the growth transition for adaptation to the stationary phase.
TABLE 5.
Promoters highly expressed in the stationary phase
| Expression level | Promoter | Function |
|---|---|---|
| >30 | gadA | Glutamate decarboxylase |
| gadB | Glutamate decarboxylase | |
| dps | DNA-binding protein in starved cells | |
| 10-30 | yiaG | Unidentified |
| hdeA | Periplasmic protein A | |
| ycgZ | Unidentified | |
| osmB | Osmotic stress-induced protein B | |
| uxuA | Mannonate dehydratase | |
| ompC | Outer membrane protein C | |
| rmf | Ribosome modulation factor | |
| glgS | Glycogen synthetase | |
| 5-10 | galE | UDP-galactose 4-epimerase |
| cspE | Cold shock protein; chromosome condensation | |
| yciF | Unidentified | |
| hfq | RNA-binding protein; RNA function control | |
| otsB | Trehalose-6-phosphate phophatase | |
| ygaU | Unidentified | |
| osmE | Osmotic stress-induced protein E | |
| adhE | Acetaldehyde dehydrogenase | |
| cspC | Cold shock protein; mukB suppression | |
| hns | DNA-binding protein HLP-II | |
| yliH | Unidentified | |
| dnaK | Heat shock Hsp70 chaperone | |
| ompX | Outer membrane protein X | |
| rob | Replication right-origin-binding protein | |
| 3-5 | osmY | Osmotic stress-induced protein Y |
| osmC | Osmotic stress-induced protein C | |
| slp | Carbon starvation-induced outer membrane protein | |
| wrbA | Trp repressor-binding protein | |
| glpD | sn-Glycerol-3-phosphate dehydrogenase | |
| bolA | Morphogenesis; control of PBP6 synthesis | |
| galM | Mutarotase; alpha-aldose to beta anomer conversion | |
| fxsA | F exclusion suppressor | |
| sodA | Superoxide dismutase (Mn) | |
| ybgS | Unidentified | |
| appY | Transcription factor for stationary-phase genes | |
| stpA | H-NS-like curved DNA-binding protein | |
| clpP | Proteosome-like ATP-dependent protease |
Activation of the dps promoter during the transition from exponential growth to the stationary phase is required for high-level accumulation of Dps to cover approximately two-thirds of the stationary-phase nucleoid surface (41). Five promoters, yiaG, hdeA, ycgZ, osmB, and osmC, exhibited 10- to 30-fold activation (Table 5), and three of these genes, hdeA, osmB, and ompC, are all involved in the modulation of the cell membrane for adaptation to the stationary phase.
Fourteen promoters (11 known genes [galE, cspE, hfq, otsB, osmE, adhE, cspC, hns, dnaK, ompX, and rob] and three unidentified genes [yciF, ygaU, and yliH]) were activated 5- to 10-fold compared with the reference promoter lacUV5 in the stationary phase, and 11 promoters (10 known genes and 1 unidentified gene) were activated 3- to 5-fold (Table 5). Besides the osmB promoter, which was activated 10- to 30-fold, the promoters for three high-osmolarity-induced proteins, OsmE, OsmC, and OsmY, were in the group of promoters that were activated 5- to 10-fold. In addition, the promoters for the slp and bolA genes, both of which are involved in the modulation of the membrane in the stationary phase, were also in this group. The promoters for two cold shock-inducible proteins, CspE and CspC, and one heat shock-inducible chaperone, DnaK, were also highly activated.
Drastic changes in metabolism take place in E. coli during the growth transition from the exponential phase to the stationary phase. Promoters associated with some genes, such as galE, otsB, adhE, and glpD, which participate in the modulation of metabolism during the growth transition, are also activated to high levels. Most of these highly activated promoters are associated with genes which have previously been found to be expressed at high levels in the stationary phase (15).
Identification of the RpoS sigma-dependent promoters (group A promoters).
Another useful application of the newly developed promoter assay system with use of the DFP vector is to identify the set of promoters under the control of each sigma or transcription factor. To identify the stationary-phase promoters, which are under the control of the RpoS sigma subunit, the culture time-dependent changes in promoter activity were measured in parallel for both wild-type E. coli KP7600 (a W3110 derivative) and its rpoS deletion mutant JD22323. The patterns of promoter activity variation are summarized in Fig. 3. Based on the dependence on the RpoS sigma subunit, the test promoters could be classified into two groups: RpoS-dependent promoters (groups A1 and A2) (Fig. 3A) and RpoS-independent promoters (groups B1, B2, and C) (Fig. 3B and C). The group A1 promoters (rpoS/rpoS+, <1/3; 28 species) absolutely required RpoS for activity expression, and virtually no activity was detected in the absence of RpoS (Fig. 3A). More than 10-fold activation was observed in the stationary phase for six promoters in this group, gadA, gadB, dps, yiaG, hdeA, and osmB (note that the time-dependent patterns of promoter activity in Fig. 3A are aligned in order of promoter activity). Transcription regulation of the highly expressed gadA and gadB genes is extremely complex, but the results described here agree well with the notion that RpoS plays a major role in transcription of the gad genes in cells grown in rich medium (4).
The group A2 promoters (rpoS/rpoS+, 1/3 to 2/3; eight species) also showed RpoS dependence for activity expression, but about one-half of the activity was detected in the absence of RpoS even though the activity was generally low for this group of promoters. Except for yceP (a heat shock gene), the majority of group A promoters (35 of 36 promoters) are associated with the stationary-phase genes (Table 4), including eight unidentified open reading frames, which were identified in this study as stationary-phase genes after transcriptome and proteome analyses (see above).
Promoters activated in the absence of RpoS (group B promoters).
Promoters with weak affinity to the RpoD holoenzyme are activated in the absence of other minor sigma factors (15). As predicted from this sigma competition model, some promoters showed higher activity in the absence of the RpoS sigma subunit (Fig. 3B). Most of the group B1 promoters (rpoS/rpoS+, >3) in wild-type E. coli were weak, showing activities that were not higher than that of the lacUV5 promoter in the growth phase, but in the stationary phase, the activities increased to levels that were higher than the level of lacUV5. The activities of some promoters (gor, yifE, xasA, mdh and oxyR) were virtually undetectable in the exponential growth phase but increased to detectable levels in the stationary phase.
The stationary-phase genes, which are classified in group B1, are expressed in the early phase of the growth transition from the exponential phase to the stationary phase and must be transcribed by the holoenzyme containing the RpoD sigma subunit. Thus, it is reasonable that the transcription levels of this group of genes increased in the absence of sigma competition with RpoS (33). The rsd gene coding for Rsd (regulator for sigma D), an anti-sigma for RpoD (16), is a member of this group of genes. The rsd promoter P1 is transcribed by the RpoD holoenzyme (20) and is activated by the alarmone ppGpp (22), but the downstream P2 promoter is recognized by RpoS (20). The rsd promoter analyzed here included both P1 and P2, but the promoter activity was markedly enhanced in the absence of RpoS, implying that the RpoS-dependent downstream P2 promoter is weaker than P1.
A set of promoters (group C) was not affected in the presence or absence of RpoS (Fig. 3). The promoters for the genes encoding two main factors, RpoS and Rmf (ribosome modulation factor), for modulation of the transcription and translation apparatus are group C promoters. The affinity of these promoters for the RpoD holoenzyme is high, and the absence of RpoS might not lead to an increase in the promoter activity.
Stress response promoters (group D promoters).
A considerable number of promoters showed low levels of activity in both wild-type and rpoS mutant cells (group D in Fig. 3C). This group of promoters must require specific transcription factors or specific conditions for expression of activity. The holoenzyme alone is able to recognize the simple promoters of constitutive genes and to initiate transcription at constant rates. Most of the genes in bacteria are, however, subject to regulation in response to changes in environmental conditions. For transcription of the majority of E. coli genes, therefore, one or more additional accessory factors are involved in regulated transcription (14, 15). Even within the same group of genes under control of a single sigma species, the order of transcription levels depends on the presence or absence of respective transcription factors. The collection of stationary-phase promoters constructed in this study may be useful for identification of accessory factors for promoter activation and classification of these promoters depending on the regulatory factors.
The lack of an apparent consensus sequence for RpoS-dependent promoters may reflect the fact that each promoter requires a specific factor(s) or condition(s) for expression of activity. A systematic search for putative transcription factors involved in activation of the group D promoters is in progress. The consensus promoter sequence could be found once the promoters are classified based on the conditions that are needed for expression of activity.
Transcription in vitro of some stationary-phase promoters.
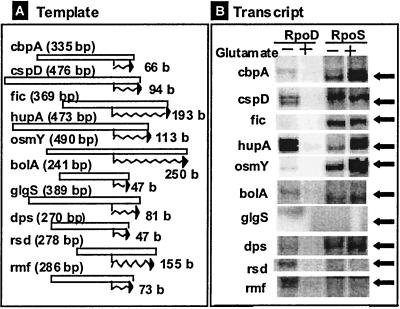
To confirm the dependence of some stationary-phase promoters on the RpoS sigma subunit, we carried out in vitro transcription assays directed by the promoter fragments, which were used for construction of the DFP promoter assay vectors. Since the RpoS holoenzyme is activated in the presence of high concentrations of glutamate (7, 31, 38, 43), transcription by both the RpoD and RpoS holoenzymes was carried out in the presence and absence of 0.3 M potassium glutamate. Some representative promoters belonging to group A (cbpA, fic, osmY, bolA, and dps) showed high activities with the RpoS holoenzyme in the presence and absence of glutamate (Fig. 4). The levels of transcription of these promoters by the RpoD holoenzyme were negligible or low in the absence of glutamate, and transcription was undetectable in the presence of glutamate. On the other hand, group B promoters, including cspD, hupA, and rsd, exhibited activities with both the RpoD and RpoS holoenzymes in the absence of glutamate. As expected, transcription of hupA coding for HU was much higher with the RpoD holoenzyme than with the RpoS enzyme.
FIG. 4.
Promoter specificity test with the in vitro transcription assay. Transcription in vitro was carried out by using two forms of RNA polymerase (RpoD and RpoS holoenzymes) and the promoter fragments which were used for cloning the promoter assay vectors (see Fig. 3). The holoenzymes were reconstituted from sigma-free core enzyme and purified RpoD or RpoS. Multiple-round transcription was carried out under the standard reaction conditions as described in Materials and Methods.
Overall, the results of in vitro transcription assays agreed well with the in vivo observations. However, two promoters, glgS and rmf, which were virtually inactive in the promoter assay in vivo (Fig. 3C), were transcribed in vitro with the purified RNA polymerase alone. The glgS promoter was transcribed in vitro by both the RpoD and RpoS holoenzymes, and the rmf promoter was transcribed by the RpoD holoenzyme. These promoters must be repressed in vivo by unidentified specific transcription factors. Again, the DFP vectors could be useful, in conjunction with the collection of E. coli mutants defective in each transcription factor, for identification of repressors for the RpoS-dependent promoters.
Acknowledgments
A collection of E. coli promoters is being constructed by the promoter project team, which includes the Ishihama lab (Nippon Institute for Biological Science, Ome, Tokyo, Japan), the Utsumi lab (Kinki University, Nara, Japan), and the Maeda lab (Meiji University, Kanagawa, Japan).
This work was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, Y. Burland, M. Rilsy, J. Collado-Vides, J. D. Glasner, C. K. Rods, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1432-1474. [DOI] [PubMed] [Google Scholar]
- 2.Böhringer, J., D. Fischer, G. Mosler, and R. Hengge-Aroni. 1995. UDP-glucose is a potential intracellular signal molecule in the control of expression of σS and σS-dependent genes in Escherichia coli. J. Bacteriol. 177:413-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, L., and T. Elliott. 1996. Efficient translation of the RpoS sigma factor in Salmonella typhimurium requires host factor I, an RNA-binding protein encoded by the hfq gene. J. Bacteriol. 178:3763-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costanie-Conrat, M., and J. W. Foster. 2001. Escherichia coli resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709-715. [DOI] [PubMed] [Google Scholar]
- 5.Costanie-Cornet, M., T. A. Penfound, D. Smith, J. F. Elliot, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Biase, D., A. Tramonti, R. A. John, and F. Bossa. 1996. Isolation, over-expression and biochemical characterization of the two isoforms of glutamic acid decarboxylase from Escherichia coli. Protein Expr. Purif. 8:430-438. [DOI] [PubMed] [Google Scholar]
- 7.Ding, Q., S. Kusano, M. Villarejo, and A. Ishihama. 1995. Promoter selectivity control of Escherichia coli RNA polymerase by ionic strength: differential recognition of osmo-regulated promoters by EσD and EσS holoenzymes. Mol. Microbiol. 16:649-656. [DOI] [PubMed] [Google Scholar]
- 8.Farewell, A., K. Kvint, and T. Nyström. 1998. Negative regulation by RpoS: a case of sigma factor competition. Mol. Microbiol. 29:1039-1051. [DOI] [PubMed] [Google Scholar]
- 9.Gentry, D. R., V. J. Hernandez, L. H. Nguyen, D. B. Jensen, and M. Cashel. 1993. Synthesis of the stationary-phase sigma factor σS is positively regulated by ppGpp. J. Bacteriol. 175:7982-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross, C. A., M. Lonetto, and R. Losick. 1992. Bacterial sigma factors, p. 129-176. In K Yamamoto and S. McKnight (ed.), Transcriptional regulation. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 11.Hengge-Aronis, R. 1996. Back to log phase: sigma S as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol. Microbiol. 21:887-893. [DOI] [PubMed] [Google Scholar]
- 12.Hengge-Aronis, R. 1996. Regulation of gene expression during entry into stationary phase, p. 1497-1512. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 13.Igarashi, K., and A. Ishihama. 1991. Bipartite functional map of the E. coli RNA polymerase α subunit: involvement of the C-terminal region in transcription activation by cAMP-CRP. Cell 65:1015-1022. [DOI] [PubMed] [Google Scholar]
- 14.Ishihama, A. 1988. Promoter selectivity of prokaryotic RNA polymerases. Trends Genet. 4:282-286. [DOI] [PubMed] [Google Scholar]
- 15.Ishihama, A. 2000. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 54:499-518. [DOI] [PubMed] [Google Scholar]
- 16.Ishihama, A., T. Saitoh, M. Taketo, and R. Fukuda. 1976. Formation of RNA polymerase in Escherichia coli, p. 475-485. In R. Losick and M. Chamberlin (ed.), RNA polymerase. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 17.Jishage, M., and A. Ishihama. 1995. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of σ70 and σ38. J. Bacteriol. 177:6832-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jishage, M., and A. Ishihama. 1997. Variation in RNA polymerase sigma subunit composition within different stocks of Escherichia coli W3110. J. Bacteriol. 179:959-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jishage, M., and A. Ishihama. 1998. A stationary phase protein in Escherichia coli with binding activity to the major σ subunit of RNA polymerase. Proc. Natl. Acad. Sci. USA 95:4953-4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jishage, M., and A. Ishihama. 1999. Transcriptional organization and in vitro role of the Escherichia coli rsd gene, encoding the regulator of RNA polymerase sigma D. J. Bacteriol. 181:3738-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jishage, M., A. Iwata, S. Ueda, and A. Ishihama. 1996. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol. 178:5447-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jishage, M., K. Kvint, V. Shingler, and T. Nystrom. 2002. Regulation of factor competition by the alamone ppGpp. Genes Dev. 16:1260-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kajitani, M., and A. Ishihama. 1983. Determination of the promoter strength in the mixed transcription system: promoters of lactose, tryptophan and ribosomal protein L10 operons from Escherichia coli. Nucleic Acids Res. 11:671-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klauck, E., J. Bohringer, and R. Hengge-Aronis. 1997. The LysR-like regulator LeuO in Escherichia coli is involved in the translational regulation of rpoS by affecting the expression of the small regulatory DsrA-RNA. Mol. Microbiol. 25:559-569. [DOI] [PubMed] [Google Scholar]
- 25.Kolb, A., D. Kotlarz, S. Kusano, and A. Ishihama. 1995. Selectivity of the Escherichia coli RNA polymerase Eσ38 for overlapping promoters and ability to support CRP activation. Nucleic Acids Res. 23:819-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kusano, S., Q. Ding, N. Fujita, and A. Ishihama. 1996. Promoter selectivity of Escherichia coli core RNA polymerase Eσ70 and Eσ38 holoenzyme: effect of DNA supercoiling. J. Biol. Chem. 271:1998-2004. [DOI] [PubMed] [Google Scholar]
- 27.Kusano, S., and A. Ishihama. 1997. Stimulatory effect of trehalose on the formation and activity of Escherichia coli RNA polymerase Eσ38 holoenzyme. J. Bacteriol. 179:3649-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kusano, S., and A. Ishihama. 1997. Functional interaction of Escherichia coli RNA polymerase with inorganic polyphosphate. Genes Cells 2:433-441. [DOI] [PubMed] [Google Scholar]
- 29.Lange, R., D. Fischer, and R. Hengge-Aronis. 1995. Identification of the transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the σS subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 177:4676-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lease, R. A., M. E. Cusick, and M. Belfort. 1998. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl. Acad. Sci. USA 95:12456-12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, S. J., and J. D. Gralla. 2004. Osmo-regulation of bacterial transcription via poised RNA polymerase. Mol. Cell 23:153-162. [DOI] [PubMed] [Google Scholar]
- 32.Loewen, P. C., and R. Hengge-Aronis. 1994. The role of the sigma-factor sigma S (KatF) in bacterial global regulation. Annu. Rev. Microbiol. 48:53-80. [DOI] [PubMed] [Google Scholar]
- 33.Maeda, H., M. Jishage, T. Nomura, N. Fujita, and A. Ishihama. 2000. Promoter selectivity of the RNA polymerase holoenzyme containing the extracytoplasmic function (ECF) sigma subunit σE or σFecI. J. Bacteriol. 182:1181-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makinoshima, H., S. Aizawa, H. Hayashi, T. Miki, A. Nishimura, and A. Ishihama. 2003. Growth phase-coupled alterations in cell structure and function of Escherichia coli. J. Bacteriol. 185:1338-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makinoshima, H., A. Nishimura, and A. Ishihama. 2002. Fractionation of Escherichia coli cell populations at different stages during growth transition to stationary phase. Mol. Microbiol. 43:269-279. [DOI] [PubMed] [Google Scholar]
- 36.Muffler, A., D. Fischer, S. Altuvia, G. Storz, and R. Hengge-Aronis. 1996. The response regulator RssB controls stability of the σS subunit of RNA polymerase in Escherichia coli. EMBO J. 15:1333-1339. [PMC free article] [PubMed] [Google Scholar]
- 37.Poolman, B., and E. Glassker. 1998. Regulation of compatible solute accumulation in bacteria. Mol. Microbiol. 29:397-407. [DOI] [PubMed] [Google Scholar]
- 38.Rajkumari, K., S. Kusano, A. Ishihama, T. Mizuno, and J. Gowrishankar. 1996. Effects of H-NS and potassium glutamate on σS- and σ70-directed transcription in vitro from osmotically regulated P1 and P2 promoters of proU in Escherichia coli. J. Bacteriol. 178:4176-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao, N. N., and A. Kornberg. 1996. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 178:1394-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sledjeski, D. D., A. Gupta, and S. Gottesman. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15:3993-4000. [PMC free article] [PubMed] [Google Scholar]
- 41.Talukder, A. Z., A. Iwata, S. Ueda, and A. Ishihama. 1999. Growth phase-dependent variation in the protein composition of Escherichia coli nucleoid. J. Bacteriol. 181:6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka, K., N. Fujita, A. Ishihama, and H. Takahashi. 1993. Heterogeneity of the principal sigma factor in Escherichia coli: the rpoS gene product, σ38, is a principal sigma factor of RNA polymerase in stationary phase Escherichia coli. Proc. Natl. Acad. Sci. USA 90:3511-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka, K., S. Kusano, N. Fujita, A. Ishihama, and H. Takahashi. 1995. Promoter determinants for Escherichia coli RNA polymerase holoenzyme containing σ38 (the rpoS gene product). Nucleic Acids Res. 23:827-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tani, T. H., A. Khodursky, R. M. Blumenthal, P. O. Brown, and R. G. Mattews. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wada, A. 1998. Growth phase coupled modulation of Escherichia coli ribosomes. Genes Cells 3:203-208. [DOI] [PubMed] [Google Scholar]
- 46.Wei, Y., J.-M. Lee, C. Richmond, F. R. Blattner, J. A. Rafalski, and R. A. LaRossa. 2001. High-density microarray-mediated gene expression profiling of Escherichia coli. J. Bacteriol. 183:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zgurskaya, H. I., M. Keyhan, and S. Matin. 1997. The sigma S level in starving Escherichia coli cells increases solely as a result of its increased stability, despite decreased synthesis. Mol. Microbiol. 24:643-651. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, A., S. Altuvia, A. Tiwari, L. Argaman, R. Hengge-Aronis, and G. Storz. 1998. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 17:6061-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]