Abstract
The Escherichia coli energy-sensing Aer protein initiates aerotaxis towards environments supporting optimal cellular energy. The Aer sensor is an N-terminal, FAD-binding, PAS domain. The PAS domain is linked by an F1 region to a membrane anchor, and in the C-terminal half of Aer, a HAMP domain links the membrane anchor to the signaling domain. The F1 region, membrane anchor, and HAMP domain are required for FAD binding. Presumably, alterations in the redox potential of FAD induce conformational changes in the PAS domain that are transmitted to the HAMP and C-terminal signaling domains. In this study we used random mutagenesis and intragenic pseudoreversion analysis to examine functional interactions between the HAMP domain and the N-terminal half of Aer. Missense mutations in the HAMP domain clustered in the AS-2 α-helix and abolished FAD binding to Aer, as previously reported. Three amino acid replacements in the Aer-PAS domain, S28G, A65V, and A99V, restored FAD binding and aerotaxis to the HAMP mutants. These suppressors are predicted to surround a cleft in the PAS domain that may bind FAD. On the other hand, suppression of an Aer-C253R HAMP mutant was specific to an N34D substitution with a predicted location on the PAS surface, suggesting that residues C253 and N34 interact or are in close proximity. No suppressor mutations were identified in the F1 region or membrane anchor. We propose that functional interactions between the PAS domain and the HAMP AS-2 helix are required for FAD binding and aerotactic signaling by Aer.
Escherichia coli responds to its chemical environment by integrating signals from membrane-bound methyl-accepting chemoreceptors (MCPs) Tsr, Tar, Tap, and Trg and navigating bacteria towards nutrient-rich environments. E. coli also has an aerotaxis receptor, Aer, that guides bacteria towards oxygen and favorable environments for energy production (7, 33). The C-terminal signaling domains of Aer and the chemoreceptors are highly conserved (7, 33, 36), allowing specific interactions between receptors and the cytoplasmic CheA and CheW signaling proteins (8, 21). The MCPs are located in polar clusters in E. coli (25), organized as functional trimers of receptor dimers (2, 19, 20), where the trimers can include a mixture of MCP homodimer types (2, 37).
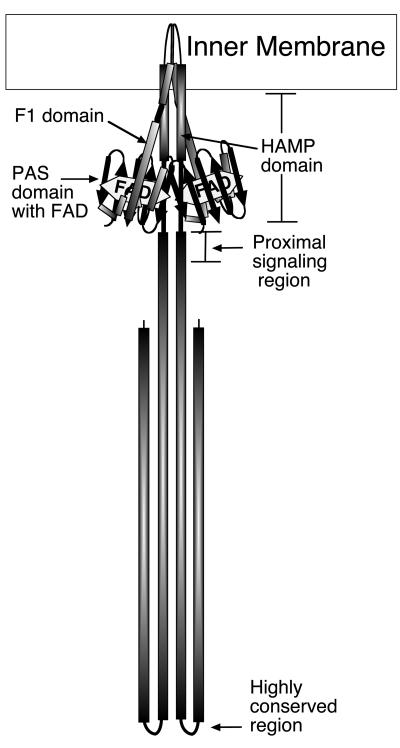
The N-terminal sensing domain of Aer differs from the periplasmic sensing domain of the other chemoreceptors because it consists of a cytoplasmic PAS domain that binds FAD cofactor (7, 33). The Aer-PAS domain is connected to an F1 domain and transmembrane (TM) anchor, followed by a cytoplasmic HAMP linker domain and the conserved signaling domain (Fig. 1). HAMP (histidine kinase, adenylyl cyclase, methyl-accepting chemotaxis protein, and phosphatase) domains are conserved in many chemoreceptors, sensory histidine kinases, and other sensory proteins in archaea, bacteria, and eukaryotes (5, 23, 35; http://smart.embl-heidelberg.de/). The HAMP domains of the MCPs and Aer consist of approximately 50 amino acids linking the signal input and output domains (5, 40). HAMP domains are predicted to consist of two amphipathic sequences arranged as α-helices (AS-1 and AS-2, residues 207 to 224 and 235 to 253, respectively, for Aer) connected by a region of undetermined structure (3, 4, 9, 22). The proposed secondary structure has been supported by cysteine scanning analysis of the Tar HAMP domain from Salmonella enterica serovar Typhimurium (9).
FIG. 1.
Proposed domain structure and working model of an Aer dimer.
Binding of aspartate to the periplasmic sensing domain of Tar results in a 1- to 2-Å inward movement of the transmembrane helix (10, 11, 15). HAMP domains have traditionally been thought of as simple linkers that transfer the same degree of sliding displacement from the sensing to signaling domains (15). However, it is also possible that the HAMP domain translates the size of the original helical displacement. A role for HAMP domains in signal transduction is supported by genetic studies of the HAMP domains from several bacterial sensory receptors (1, 4, 6, 9, 12, 18, 23a, 27, 29, 30, 39, 44). The majority of HAMP mutants from these genetic studies bias receptor signaling towards the ligand-bound state (4, 5, 40). A signal received by the HAMP domain is transmitted to the C-terminal signaling domain, where it modulates the phosphorylation of bound CheA histidine kinase. Attractant binding to the receptor inhibits CheA autophosphorylation. When the attractant dissociates, however, CheA autophosphorylation is reactivated. Phospho-CheA transphosphorylates the CheY protein, which can bind to the flagellum motor, changing the direction of flagellar rotation from counterclockwise to clockwise. This results in a brief bacterial tumbling response and a change in the direction of swimming.
PAS domains are known sensors of oxygen, redox, and light (38). Approximately 170 proteins are predicted to contain both a HAMP and a PAS domain, of which the majority are sensory proteins and/or histidine kinases (23, 35; http://smart.embl-heidelberg.de/). This suggests a role for PAS-HAMP combinations in bacterial signal transduction that may be common to many proteins and many bacteria. The Aer protein is anchored to the bacterial cytoplasmic membrane, and its PAS domain is thought to sense cellular energy changes through interactions with an undetermined component of the electron transport system. This is proposed to reduce the FAD redox center, which is bound to the PAS domain, initiating a conformational change that is transmitted to the C terminus of Aer. Whether the signal from the Aer-PAS domain travels to the HAMP domain through direct amino acid interactions or is transmitted via the F1 and TM regions to the HAMP domain is currently unknown. Possible PAS-HAMP interactions were suggested by studies showing that the Aer-HAMP domain is essential for FAD binding to the PAS domain (6, 23a). However, missense mutations in the F1 segment also abolished FAD binding to the Aer protein (6), and residues in the membrane anchor and the HAMP domain may be required for proper folding of the N-terminal region (16). In this study we employed an intragenic second-site suppressor analysis to investigate functional interactions between the N-terminal half of Aer and its HAMP domain. The results show that HAMP mutations that disrupt FAD binding to the Aer protein can be restored by nonspecific and specific PAS domain suppressor mutations that restore both FAD binding and Aer function.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All strains used in this work are derivatives of E. coli RP437 (31), which is wild-type (WT) for chemotaxis. Plasmids containing mutated aer genes were expressed in BT3388 (aer tsr tar tap trg [42]), BT3312 (aer tsr [34]), or BT3340 (aer tsr tar tap trg recA [23a]). Relevant strain properties are given in Table 1. Bacteria were grown at 30°C in Luria-Bertani (LB) medium (14) supplemented with 0.5 μg of thiamine · ml−1.
TABLE 1.
Strains, plasmids, and PCR primers
| Strain, plasmid, or primer | Relevant properties or primer sequence (5′ to 3′) | Reference |
|---|---|---|
| Strains | ||
| RP437 | Wild type for chemotaxis | 31 |
| BT3312 | Δaer-1 Δtsr-7028 | 34 |
| BT3340 | Δtsr-7021 Δtar-tap-5201 trg::Tn10 aer-2::kan recA::cat | 23a |
| BT3388 | Δaer::erm Δtsr-7021 Δtar-tap-5201 trg::Tn10 | 42 |
| XL1-Red | ΔmutD5 ΔmutS ΔmutT | Stratagene |
| Plasmids | ||
| pTrc99A | ptrc expression vector, ColE1 lacIq | Pharmacia |
| pGH1 | pTrc99A Aer+ [1-506] | 34 |
| pQH16 | pGH1 Aer-ΔHAMP domain, BstBI and SacI sites | 23a |
| pKW1 | pQH16 Aer-WT HAMP+, NheI site | This study |
| pKW75 | pKW1 Aer[M21R] | This study |
| pKW76 | pKW1 Aer[D35R] | This study |
| pKW77 | pKW1 Aer[Q39R] | This study |
| pKW78 | pGH1 Aer[N34R] | This study |
| pKW79 | pGH1 Aer[C253D] | This study |
| Plasmid templates for suppressor analysis | ||
| pKW2 | pKW1 Aer[D237G] | This study |
| pKW3 | pKW1 Aer[G240R] | This study |
| pKW4 | pKW1 Aer[L251P] | This study |
| pKW5 | pKW1 Aer[C253R] | This study |
| pKW6 | pKW1 Aer[V230D] | This study |
| pKW7 | pKW1 Aer[R235E] | This study |
| pKW8 | pKW1 Aer[L239Q] | This study |
| pKW9 | pKW1 Aer[L239R] | This study |
| pKW10 | pKW1 Aer[L241P] | This study |
| pKW11 | pKW1 Aer[Q248R] | This study |
| pKW12 | pKW1 Aer[L249P] | This study |
| pKW13 | pKW1 Aer[D259H] | This study |
| pKW14 | pKW1 Aer[V260A] | This study |
| pKW15 | pKW1 Aer[S265P] | This study |
| pKW74 | pKW1 Aer[N34D] | This study |
| Primers | ||
| BstBIF | GGTTTCGAATGGCAGATTGTG | This study |
| SacIR | GCTGAGCTCATCGGTGCCTTTCGCCAGCGT | This study |
| NheIF | ATACCCCGCTAGCGGACGATACC | This study |
| NheIR | GTATCGTCCGCTAGCGGGGTATT | This study |
| BstBIR | GTCTTCGAAACAGGCGCTTGC | This study |
| M21RF | GATACCACTCTGAGGTCCACTACCGATCTG | This study |
| M21RR | CAGATCGGTAGTGGACCTCAGAGTGGTATC | This study |
| N34RF | GCAAAGCTATATCACTCATGCTCGTGACACTTTTGTGCAGGT | This study |
| N34RR | CTCACCTGCACAAAAGTGTCACGAGCATGAGTGATATAGCTT | This study |
| D35RF | CTCATGCTAATCGCACTTTTGTGCAGGTGAGCG | This study |
| D35RR | CGCTCACCTGCACAAAAGTGCGATTAGCATGAG | This study |
| Q39RF | GACACTTTTGTGCGGGTGAGCGGCTATACC | This study |
| Q39RR | GGTATAGCCGCTCACCCGCACAAAAGTGTC | This study |
| C253DF | GGTAGGGCAACTTGGCCTGATGGACCGTTGGCTAATTAACG | This study |
| C253DR | CATCGTTAATTAGCCAACGGTCCATCAGGCCAAGTTGCCCTA | This study |
The parental plasmids used in this work were pTrc99A (Pharmacia), an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible ptrc expression vector, and pGH1 (33), a pTrc99A derivative expressing a WT aer gene under the control of the ptrc promoter. Silent mutations were engineered into the aer insert of pGH1 by PCR to introduce BstBI and SacI restriction sites at codons 205 and 280/281, respectively, creating pQH16 (23a). A fragment of the WT aer gene encoding residues 204 to 281 was amplified by PCR using PfuTurbo (Stratagene, La Jolla, Calif.) and BstBIF and SacIR primers (Table 1) and ligated into the BstBI and SacI sites of pQH16. An NheI restriction site (silent mutation) was also introduced into this plasmid at Aer codon 14 by PCR site-directed mutagenesis using PfuTurbo and NheIF and NheIR primers (Table 1) to create pKW1. Constructs containing individual HAMP or proximal signaling domain mutations were created by PCR using the BstBIF and SacIR primers and constructs with individual mutations (23a, or this study) and ligating into the BstBI/SacI sites of pKW1. Aer residues M21, N34, D35, and Q39 were converted to arginine, and C253 was converted to aspartic acid, by PCR site-directed mutagenesis of pGH1 or pKW1 using PfuTurbo and the primer pairs in Table 1. To place the mutation encoding C253D into Aer-N34R, the KpnI-PstI fragment was removed from pKW79 (corresponding to residues from Aer-100 to the multicloning site of pTrc99A) and ligated in place of the corresponding DNA fragment in pKW78. Relevant plasmid properties are given in Table 1. The sequences of all constructs were confirmed by DNA sequencing, and expression of full-length Aer protein was analyzed by Western blot analysis with anti-His6Aer2-166 antiserum (34). Strains transformed with plasmids were grown in LB-thiamine (0.5 μg · ml−1) supplemented with 100 μg of ampicillin · ml−1.
Random mutagenesis.
The aer gene was mutagenized using several different methods. To obtain single amino acid substitutions in the Aer-HAMP and proximal signaling domains (codons 204 to 281), random PCR mutagenesis was performed on pGH1 using BstBIF and SacIR primers. To obtain single amino acid substitutions in the N terminus of Aer (codons 14 to 204), random PCR mutagenesis was performed on pGH1 using NheIF and BstBIR primers (Table 1). The PCR was performed using Taq DNA polymerase under conditions of reduced fidelity (17). Each reaction mixture contained 250 ng of pGH1 DNA, 2 U of Taq DNA polymerase (Fisher Scientific, Pittsburgh, Pa.), a 0.5 μM concentration of each primer, a 0.2 mM concentration of each deoxynucleoside triphosphate (dNTP), 0.05 mM MnCl2, 1.5 mM MgCl2, and 5 μg of bovine serum albumin · ml−1 in a total volume of 50 μl. PCR was performed for 10 cycles of 95°C (1 min), 50°C (2 min), and 75°C (3 min), following an initial denaturing step at 95°C (1 min). The randomly mutated DNA fragments were subsequently purified using a QIAquick PCR purification kit (QIAGEN Inc., Valencia, Calif.) and subjected to 20 further cycles of PCR amplification as before except without MnCl2 and bovine serum albumin. The N terminus of the aer gene was also mutagenized by low-fidelity PCR amplification under conditions in which the concentration of one dNTP in the reaction was increased by 40% or decreased by 40%. The PCR products were gel purified using a QIAquick gel extraction kit (QIAGEN) and then digested with either SacI and BstBI (for the HAMP signaling region fragments) or NheI and BstBI (for N-terminal coding fragments). The digested fragments were gel purified and cloned into their respective sites in pQH16 (for the HAMP signaling region fragments) or pKW1 derivatives with the corresponding NheI/BstBI piece removed (for the N-terminal fragments).
DNA mutagenesis with hydroxylamine was carried out using a modification of the method of Wolff and Parkinson (41), in which 400 ng of a pKW1 plasmid derivative containing an aer mutation was added to 0.4 M hydroxylamine in 0.5 M potassium phosphate buffer, pH 6.0, containing 5 mM EDTA and incubated at 37°C for 17 h. The chemically modified plasmid DNA was purified with the QIAquick PCR purification kit.
Plasmids were also randomly mutagenized by passage through XL1-Red (mutS mutD mutT [Stratagene]). Pinpoint-sized colonies were washed from LB agar plates with LB broth, and the plasmid DNA was reisolated using a QIAprep spin miniprep kit (QIAGEN).
Phenotypic screening for HAMP and proximal signaling domain mutants.
Mutagenized DNA fragments corresponding to Aer codons 204 to 281 were prepared as described above and ligated into pQH16 using the engineered BstBI and SacI sites. Plasmids were transformed into BT3388, and aerotaxis phenotypes were analyzed by transferring individual colonies from LB agar plates with a sterile toothpick to tryptone semisoft agar swarm plates containing 0.28% agar, 100 μg of ampicillin · ml−1, and 20 μM IPTG and incubating at 30°C. BT3388/pKW1 was used as a positive control, which swarmed out from the site of inoculation, producing an aerotactic ring at its edge. Cells with abnormal phenotypes were isolated and tested on swarm plates supplemented with 0 to 0.4 mM IPTG to distinguish transformants with low levels of protein expression from genuine Aer functional mutants. Cultures containing Aer mutants were then grown at 30°C to an optical density at 600 nm of 0.4, induced with 0.6 mM IPTG for 3 h, and checked for expression of full-length Aer protein by Western blot analysis using anti-His6Aer2-166 antiserum. Quantitation of Aer bands was performed using an Alpha Innotech IS-1000 digital imaging system. The mutation carried by each plasmid was then identified by DNA sequencing. Aer mutants that were nonaerotactic in a background lacking all chemoreceptors (BT3388) were retested in a strain expressing the Tar, Trg, and Tap chemoreceptors (BT3312) by plating onto 30 mM succinate semisoft agar swarm plates containing 0.28% agar and 100 μg of ampicillin · ml−1 and incubating at 30°C. Aer mutants also displaying a null phenotype in BT3312 were analyzed with a temporal assay to observe responses to increased and decreased oxygen concentrations as described previously (33).
Isolation of aerotactic pseudorevertants.
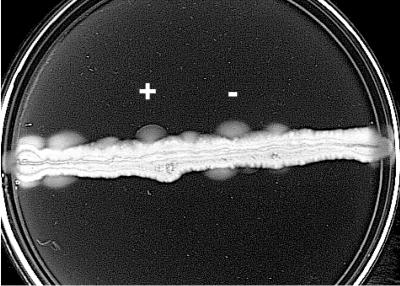
Mutagenized DNA fragments corresponding to Aer codons 14 to 204 were prepared as described above. The DNA fragments were ligated using NheI and BstBI in place of the corresponding DNA fragment in derivatives of the pKW1 plasmid that contained a primary mutation in the HAMP or proximal signaling domain (Table 1). Constructs were transformed into BT3340 or BT3388, and approximately 106 clones were spread in a line beneath the surface of a semisoft tryptone swarm plate containing 100 μg of ampicillin · ml−1 and 20 μM IPTG. Swarm plates were grown for 24 to 36 h at 30°C. Aerotactic blebs were selected from the nonaerotactic clones as they swam out from the inoculation site and formed a characteristic ring at the outer edge of the colony (Fig. 2). Potential aerotactic pseudorevertants were plated onto LB agar, and individual colonies were rescreened for an aerotactic phenotype on tryptone semisoft agar swarm plates. Plasmids isolated from mutants containing potential suppressor mutations were transferred into BT3312 and tested on 30 mM succinate semisoft agar swarm plates. In order to eliminate simple revertants, the N terminus of a potential suppressor was excised from the plasmid with NheI and BstBI and religated to a plasmid containing the primary mutation and their function was retested. Pseudorevertant clones passing this test were sequenced to confirm the presence of the primary HAMP mutation and to identify the suppressor mutation in the N terminus.
FIG. 2.
Pseudoreversion analysis. Approximately 106 mutagenized aer clones were spread in a line beneath the surface of a semisoft tryptone swarm plate containing 100 μg of ampicillin · ml−1 and 20 μM IPTG. Blebs showing an aerotactic ring (+) can be easily differentiated from blebs caused by mutations in loci other than aer, which do not show an aerotactic ring (-). Aerotaxis was observed in BT3340 (aer tsr tar tap trg recA) after incubating the plate for 24 h at 30°C.
The NheI-BstBI DNA fragments from plasmids with a pseudorevertant aer gene were then excised and cloned into pKW1 or pKW1 derivatives containing a range of primary mutations in the HAMP or proximal signaling regions (Table 1) to determine the specificity of the suppressor mutation. The resulting plasmids were transformed into BT3312 and screened for aerotactic phenotype on 30 mM succinate swarm plates.
Measurement of total FAD content in the cell membrane.
HAMP mutants and mutants containing both HAMP and suppressor mutations were tested for FAD binding by determining total FAD in membrane fractions of BT3312 cells in which Aer expression was induced with 1 mM IPTG, compared with that in uninduced cells. This was carried out using a modification (34) of the method of Bibikov et al. (6).
RESULTS
Random mutagenesis of the HAMP and proximal signaling domains.
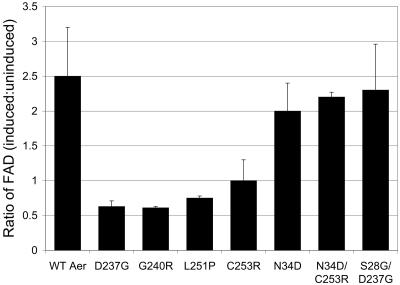
Previously, we identified 12 mutations in the Aer-HAMP and proximal signaling regions that disrupted aerotaxis (23a). Here we extended this analysis by performing random PCR mutagenesis on the DNA fragment corresponding to Aer codons 204 to 281 and ligating the BstBI-SacI fragments with the remaining aer gene in pQH16. A pool of plasmids containing the recombinant aer genes were transformed into an E. coli background that was deficient in Aer and all chemoreceptors (BT3388), and 455 colonies were screened for a null aerotaxis phenotype on tryptone semisoft agar swarm plates. Colonies showing an aerotactic phenotype formed a dome-shaped swarm in the semisoft agar, with a focused ring at the outer edge of the colony. Thirteen missense mutations in the Aer-HAMP and proximal signaling domains eliminated aerotaxis in the chemoreceptor-deficient background of BT3388, resulting in cylinder-shaped colonies lacking an aerotactic ring at their edge. This included the amino acid substitutions V215A, K221E/Q, N228D, D237G, G240E/R, A245V, L251P, C253R, S265Y, V267I, and R268G. Overall, transition mutations were more commonly detected (10 of 13; 77%) in the DNA of these mutants than transversion mutations (3 of 13; 23%), as expected for PCR mutagenesis. Nine of the Aer mutants recovered aerotactic behavior when the Tar, Trg, and Tap chemoreceptors were present in the background (in BT3312), perhaps by their ability to alleviate the bias of the flagellar motors. However, Aer-D237G, Aer-G240R, Aer-L251P, and Aer-C253R retained a null aerotactic phenotype in BT3312 (for example, Aer-D237G in Fig. 3A). In a temporal assay, Aer-D237G, Aer-G240R, Aer-L251P, and Aer-C253R did not respond to increased or decreased oxygen concentrations when observed under a dark-field microscope. Motility was random for all mutants except Aer-D237G, which had a slightly tumbly bias. This indicated that the nonaerotactic phenotype of these mutants was not caused by a strong bias of the flagellar motors. Expression of the Aer-C253R protein was approximately 65% of Aer expression from pKW1, which was approximately two- to fourfold above chromosomal expression levels. However, Aer proteins containing the D237G, G240R, or L251P substitutions expressed only 15 to 35% of the amount of Aer expressed from pKW1. Aerotaxis was not restored to any of the four BT3312 mutants when expression was induced in succinate swarm plates by the presence of IPTG at various concentrations. Unlike BT3312 expressing the WT Aer protein from pKW1, membrane FAD levels were not increased when Aer-D237G, Aer-G240R, Aer-L251P, or Aer-C253R was overexpressed by IPTG induction in BT3312 (Fig. 4), suggesting that the aerotaxis defect was associated with impaired FAD binding.
FIG. 3.
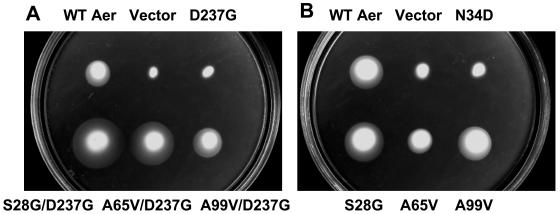
Restoration of aerotaxis by second-site suppression of the Aer-HAMP mutant D237G. (A) Upper row: WT Aer (expressed from pKW1), plasmid vector (pTrc99A), and Aer-D237G. Bottom row: Aer-S28G/D237G, Aer-A65V/D237G, and Aer-A99V/D237G. (B) Individual effects of PAS suppressors on aerotaxis by Aer. Upper row: WT Aer (pKW1), plasmid vector (pTrc99A), and Aer-N34D. Bottom row: Aer-S28G, Aer-A65V, and Aer-A99V. Plasmids containing a cloned aer gene were expressed in BT3312 (aer tsr), and aerotaxis was observed on a 30 mM succinate semisoft agar plate after incubation at 30°C for 20 h (A) or 24 h (B).
FIG. 4.
Relative FAD content in membranes after Aer induction with 1 mM IPTG. Total membrane-bound FAD was measured with and without induction in BT3312 (aer tsr) as described elsewhere (34). A ratio of 1.2 or less indicates the mutant Aer protein did not bind FAD. FAD ratios are shown for WT Aer (pKW1), the Aer missense mutants D237G, G240R, L251P, C253R, and N34D, and the Aer pseudorevertants N34D/C253R and S28G/D237G.
Pseudoreversion analysis.
Pseudorevertants maintain the original mutation but recover function by a secondary mutation(s) within the same or a different gene (28). Pseudoreversion analysis and the identification of allele-specific suppressor mutations in trans previously suggested protein-protein interactions in the E. coli chemotaxis pathway (32). We adapted second-site suppressor analysis to investigate interactions between domains within the Aer protein. Initially, BT3340 (aer tsr tar tap trg recA) was transformed with a plasmid expressing an Aer-HAMP or proximal signaling domain mutant. Transformants were spread through tryptone semisoft agar plates and incubated at 30°C to select for spontaneous aerotactic revertants. However, cells picked from aerotactic blebs were predominantly revertants with back-reversion of the original mutation (Fig. 2) rather than pseudorevertants. To select for pseudorevertants, the N terminus of Aer (codons 14 to 204), which might interact with the HAMP domain, was mutagenized by random PCR mutagenesis, chemical mutagenesis with hydroxylamine, and passage through the mutator strain XL1-Red. Plasmids derived from pKW1 (Table 1) encoding substitutions in the Aer-HAMP domain (V230D, R235E, L239Q, Q248R, or C253R) or proximal signaling domain (D259H) were digested with NheI and BstBI and ligated to randomly mutated N-terminal fragments. The HAMP and proximal signaling domain mutants inhibit aerotaxis and were identified either in this study or in a recent study (23a). The recombinant plasmids were then transformed into BT3388 and selected for recovery of aerotaxis on tryptone semisoft agar plates as previously described. Revertants that recovered an aerotactic phenotype were identified as blebs that swam out from the inoculation site and formed a characteristic ring at the edge of the colony (Fig. 2).
Missense mutations that suppress defects in the HAMP and proximal signaling domains.
Four second-site suppressors were identified by different mutagenesis methods: Aer-S28G (identified as a suppressor for V230D, R235E, L239Q, and Q248R), Aer-A65V (identified for V230D and R235E), Aer-A99V (identified for Q248R), and Aer-N34D (identified for C253R). These suppressor substitutions were located exclusively in the PAS domain (Table 2), rather than the F1 or transmembrane regions, which were also mutated during the analysis. The phenotype of the pseudorevertants was similar when the plasmids were transformed into a background with (BT3312) or without (BT3388) other chemoreceptors.
TABLE 2.
Second-site suppressor mutations
| Specificity of suppression | Second-site suppressor | Corresponding codon change | Location in PAS domain | Frequency of amino acid substitution in other Aer-like PAS domainsa |
|---|---|---|---|---|
| Nonspecific | S28G | AGC → GGC | PAS core, Bβ | G conserved at this position in many other PAS domains |
| A65V | GCG → GTG | EF loop | A and V occur in other PAS domains, but this is not a conserved position in the PAS alignment | |
| A99V | GCG → GTG | β-Scaffold, Hβ | Type of amino acid not conserved at this position | |
| Site specific | N34D | AAT → GAT | PAS core, Cα | N is conserved at this position in many other PAS domains; no PAS domains contain D at this position |
Alignment from reference 38.
Nonspecific second-site suppressor mutations.
The amino acid replacement identified most frequently as a suppressor by PCR mutagenesis was Aer-S28G, and its reoccurrence limited the search for other suppressors. Since the S28G substitution resulted from an adenine-to-guanine transition in codon 28 of Aer (Table 2), we altered the PCR cocktail by decreasing dGTP by 40% or by increasing dATP, dCTP, or dTTP by 40%. This method was successful in preventing the formation of S28G. Under these conditions, A65V, the second most common PAS suppressor from PCR mutagenesis experiments, then dominated the selection process with Aer-V230D and Aer-R235E as the parental mutants. This was also true for hydroxylamine mutagenesis. Hydroxylamine deaminates cytosine and does not create the transition mutation responsible for Aer-S28G. Mutating the plasmid encoding Aer-V230D with hydroxylamine resulted in aerotactic pseudorevertant colonies that contained the A65V suppressor. Passage of the plasmid encoding Aer-R235E through the mutator strain XL1-Red yielded exclusively the S28G suppressor.
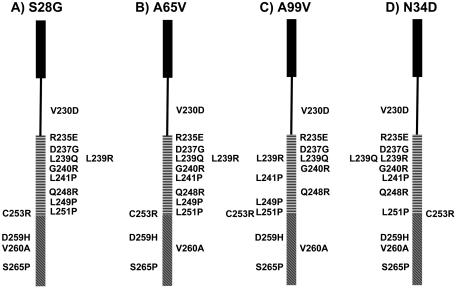
The specificity of each suppressor was then tested by excising the NheI-BstBI fragment containing the suppressor mutation from the pseudorevertant plasmids and replacing the comparable fragment in a series of 14 plasmids containing representative mutations in the HAMP and proximal signaling domains (Table 1). Recombinant BT3312 transformants were each tested for aerotactic phenotype on succinate semisoft agar in order to determine which original mutants were functionally compensated by the suppressor substitutions. The PAS suppressors Aer-S28G, Aer-A65V, and Aer-A99V were each capable of complementing a large spectrum of the mutants tested (Fig. 5A, B, and C). Typical results are shown in Fig. 3A for the D237G mutant, whereby aerotaxis was recovered with all three nonspecific suppressors. Although Aer-S28G was identified more often than Aer-A65V by random DNA mutagenesis, S28G and A65V both suppressed comparable numbers of HAMP mutants (Fig. 5A and B). Conversely, Aer-A99V complemented fewer mutants (Fig. 5C). Most of the functionally corrected mutants contained primary mutations in the second amphipathic helix (AS-2) of the HAMP domain that prevented FAD binding (23a). Amino acid changes in the proximal signaling domain were generally not compensated by the suppressors, with the exception of Aer-V260A, which recovered aerotaxis with both A65V and A99V (Fig. 5B and C). The addition of a suppressor mutation to a construct containing a primary HAMP or proximal signaling domain mutation generally increased Aer expression up to 2.5-fold, regardless of whether the mutation complemented the parental mutation or not. Site-specific replacement of S28G, A65V, and A99V in WT Aer did not affect Aer function (Fig. 3B).
FIG. 5.
Complementation of Aer-HAMP and proximal signaling domain mutants by amino acid substitutions in the PAS domain S28G (A), A65V (B), A99V (C), or N34D (D). The distal end of the Aer-HAMP domain is placed at residue C253. The areas of the HAMP domain corresponding to AS-1 and AS-2 are indicated by the filled black rectangle and horizontal bars, respectively. The proximal signaling region is indicated with diagonal bars, and the HAMP connector region is indicated with a line. Amino acid substitutions in Aer shown to the right of each bar were suppressed by the amino acid substitution in the PAS domain shown at the top and recovered aerotaxis, while amino acid substitutions shown to the left of the bar were not suppressed.
Site-specific suppressor mutation.
A common property of the Aer-HAMP mutant proteins that were suppressed by the nonspecific repressors was their impairment for FAD binding (Fig. 4) (23a). Aer-C253R was unique in that it did not bind FAD but was not complemented by the nonspecific suppressors (Fig. 5). Lack of suppression made Aer-C253R a suitable candidate for studies to identify a specific suppressor and possible direct interaction between residues of the Aer-HAMP and PAS domains.
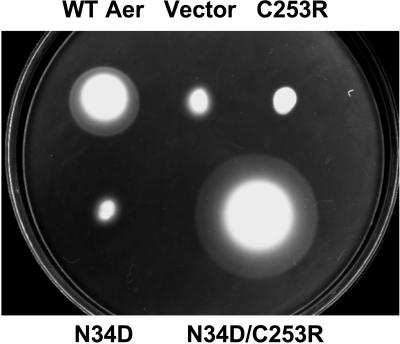
The Aer PAS substitution N34D was identified as a second-site suppressor for Aer-C253R (Fig. 6 and Table 2) from five experiments using the altered dNTP PCR mutagenesis protocol with either the dCTP or dTTP concentration in excess. The Aer-N34D/C253R mutant exhibited a large swarm diameter in BT3312, with a colony diameter almost twofold larger than that with WT Aer (Fig. 6). The NheI-BstBI fragment encoding the N34D mutation was ligated to 12 other mutants; however, none regained aerotaxis on swarm plates (Fig. 5D), implying that Aer-N34D is a specific second-site suppressor for Aer-C253R. Aer-N34D also disrupted WT Aer function (Fig. 6). Expression levels of the noncomplemented Aer mutants also containing the N34D substitution were approximately 30 to 200% of the expression level of Aer-N34D, while expression of Aer-N34D/C253R was approximately 110% that of Aer-N34D.
FIG. 6.
Complementarity of the N34D and C253R substitutions in Aer. Experimental conditions were the same as for Fig. 3, and the plate was incubated for 24 h. Upper row: WT Aer (pKW1), plasmid vector (pTrc99A), and Aer-C253R. Bottom row: Aer-N34D and Aer-N34D/C253R.
We then used a reversion analysis of Aer-N34D to confirm the allele-specific nature of the N34D-C253R interaction. The construct encoding Aer-N34D was ligated to randomly mutated PCR fragments using the BstBI and SacI sites (codons 204 to 281), encompassing the HAMP and proximal signaling domains. Using Aer-N34D as the parental mutant, C253R was identified as a suppressor for Aer-N34D from each of four independent experiments.
Attempts to identify other interacting Aer residues in proximity to N34D-C253R were not successful. Using a model of the Aer PAS domain (34), the side chains of M21, D35, and Q39 were projected to be in close proximity to N34 on the PAS surface. Each residue was mutated to arginine by PCR site-directed mutagenesis, and its effect on aerotaxis in BT3312 was tested on succinate swarm plates. We hypothesized that the long, charged side chain of arginine might disrupt Aer function and that suppressor analysis might be useful for finding an interacting partner residue. On succinate swarm plates, Aer-D35R and Aer-Q39R displayed a normal aerotactic phenotype, while Aer-M21R was nonaerotactic (data not shown). Aer-M21R expression was approximately 30% of that for Aer expressed from pKW1. The Aer-M21R construct was ligated to randomly mutated BstBI-SacI fragments to screen for suppressors; however, none was identified by this approach. Aer-N34 was also mutated to arginine and Aer-C253 to aspartic acid, and their effects on BT3312 aerotaxis were tested on succinate swarm plates. Aer-N34R was nonaerotactic on swarm plates, while Aer-C253D did not appear to have an aerotaxis defect (data not shown). Aer-N34R/C253D, which expressed Aer protein at approximately 10% the concentration of Aer expressed from pKW1, did not recover aerotactic function in BT3312 on swarm plates nor on plates containing various concentrations of IPTG.
Binding of FAD by mutant Aer proteins.
The level of FAD bound to mutant Aer proteins was estimated by using a method that compared total membrane-bound FAD before and after inducing overexpression of the Aer protein with IPTG. Consistent with previous findings (23a), Aer proteins with a primary missense mutation in the HAMP domain (D237G, G240R, L251P, and C253R) did not bind FAD (Fig. 4). The PAS suppressor mutations, which restored an aerotactic phenotype to Aer containing a primary HAMP mutation (Fig. 3, 5, and 6), also restored FAD binding, as shown for Aer-S28G/D237G and Aer-N34D/C253R in Fig. 4. Individually, the nonspecific Aer-PAS suppressors S28G, A65V, and A99V did not impair aerotaxis by Aer (Fig. 3B), implying that these proteins bind FAD. The Aer protein containing the specific suppressor N34D also bound FAD (Fig. 4), although aerotaxis was inhibited by the amino acid substitution (Fig. 3B).
Relationship between suppressors and Aer structure.
Table 2 gives the nucleotide changes of each suppressor mutation and their putative locations in the Aer PAS domain. Based on PAS domain sequence alignments (38), a glycine at the equivalent position to Aer-28 is conserved in many other PAS domains. Similarly, an asparagine at residues equivalent to Aer-34 is conserved in other PAS domains. On the other hand, many different residues occur at the position equivalent to Aer-99, and a position equivalent to Aer-65 is not conserved in other PAS domains.
DISCUSSION
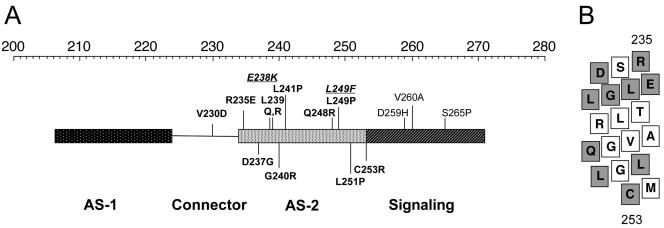
In this study, 13 missense mutations in the Aer-HAMP and proximal signaling domains prevented aerotaxis in the chemoreceptor-deficient background of BT3388. Nine of the mutant Aer proteins recovered aerotactic function in BT3312, which expresses the Tar, Trg, and Tap chemoreceptors. The most probable explanation for the complementary effects by the MCPs is that they decrease the counterclockwise bias of the flagellar motors; however, the possibility of physical interactions between the MCPs and Aer cannot be eliminated. The four Aer mutants that remained nonaerotactic in BT3312 (D237G, G240R, L251P, and C253R) did not bind FAD (Fig. 4). FAD binding to the E. coli Aer protein can be abolished by amino acid substitutions in the PAS, F1, or HAMP domains (6, 23a). Previously identified Aer-HAMP mutants that do not bind FAD are predominantly clustered in the predicted HAMP AS-2 helix, which was also the case for Aer-D237G, Aer-G240R, Aer-L251P, and Aer-C253R (Fig. 7A). When all Aer-HAMP substitutions that disrupted aerotaxis were represented on a putative AS-2 α-helix (Fig. 7B), the mutated residues were distributed over the helical surface rather than focused on one particular face. Disruption of the packing of AS-2 residues, including the putative HAMP-HAMP′ homodimeric interface between Aer monomers, may be sufficient to disrupt FAD binding to Aer. In this study we applied intragenic suppression in the aer gene to investigate interactions between the HAMP domain and the N terminus of Aer. The method used is analogous to using pseudoreversion analysis to identify second-site suppressors in other loci (32).
FIG. 7.
Summary of amino acid substitutions altering Aer function mapped onto the HAMP and proximal signaling domains. (A) Segment of Aer representing helical (rectangle) and connector (line) regions of the HAMP and contiguous signaling regions. The top scale represents residue number. Substitutions that abolished FAD binding are shown in bold, while those that retained FAD binding are shown in nonbolded text. Mutants from a recent study (23a), (plain text) and those of Bibikov et al. (6) (underlined italics) are located on the upper part of the figure. Mutants that disrupted aerotaxis, identified in the present study from the background of BT3312, are indicated on the lower part of the figure. (B) Aer residues 235 to 253, which constitute HAMP AS-2, mapped onto a flattened α-helix viewed from the side. All residues for which missense mutations are known to abolish FAD binding to Aer in panel A are shaded in grey. These residues do not fall on a common face of the helix.
The finding that all HAMP residue substitutions that abolished FAD binding to Aer were compensated by a suppressor substitution in the PAS domain (either S28G, N34D, A65V, or A99V) is presumptive evidence that none of those residues per se are critical for FAD binding. Furthermore, none of the suppressors were found in the F1 domain, as would be expected if there were a direct interaction between the F1 and HAMP domains. The possibility of interaction between the F1 and PAS domains was not addressed in this study. Considered as a whole, the findings of this study support a direct interaction between the HAMP and PAS domains that stabilizes FAD binding to the PAS domain.
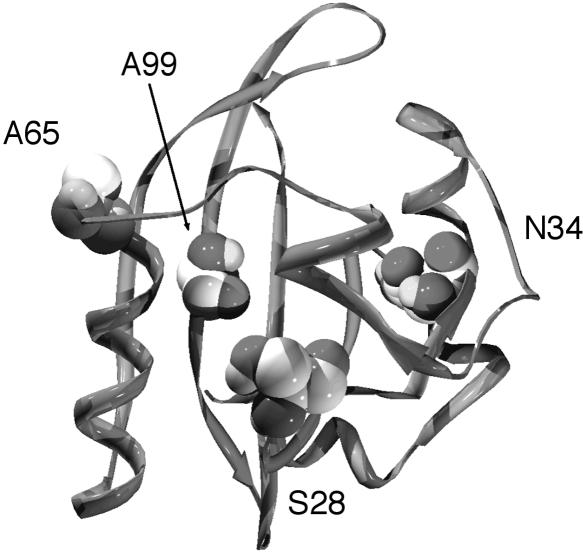
The primary Aer-HAMP mutations used to select pseudorevertants abolished FAD binding to the Aer protein (Fig. 4) (23a), resulting in a loss of aerotaxis. The suppressor substitutions (S28G, A65V, and A99V) that complemented multiple Aer-HAMP mutants restored the aerotactic phenotype of Aer, for which FAD binding is required. Restoration of FAD binding was confirmed for the Aer-S28G/D237G and Aer-N34D/C253R mutants (Fig. 4). The PAS suppressors may increase the affinity of the PAS domain for FAD through nonspecific compensatory structural changes, overcoming the negative effect of the HAMP substitutions on FAD binding. Aer-S28 has a predicted location in the PAS core Bβ (Table 2 and Fig. 8), where a glycine is conserved in many other PAS domains at the analogous position (Table 2) (38). Aer-A65 has a predicted location on the PAS EF loop (Table 2 and Fig. 8), which is a known active site for PAS signaling in Aer and other PAS domains (34, 43). By analogy to the crystal structure of the FMN-binding LOV2 PAS domain in Phy3 from Adiantum (13), S28G, A65V, and A99V are adjacent to a pocket in which the isoalloxazine ring of the FAD cofactor may bind in Aer (Fig. 8). FMN is bound noncovalently to the interior pocket of the LOV2 PAS domain, and the flavin-binding pocket is primarily polar on the pyrimidine side of the isoalloxazine ring (where Aer-S28G is analogously located) and nonpolar around the dimethylbenzene moiety (where Aer-A99V is analogously located) (13).
FIG. 8.
Suppressor residues mapped onto a putative Aer-PAS model (34). Residues S28, A65, and A99 surround a cleft proposed to bind FAD, while residue N34 is displaced from this region.
The allele-specific suppression of C253R by N34D provides evidence for an interaction between the Aer-HAMP and PAS domains. Isolation of a pseudorevertant to either mutation produced only the other mutation, indicating that the mutated residues may be in contact or in close proximity. Unlike the substitution at C253 to arginine, mutations encoding alanine (6) or aspartic acid at residue 253 do not affect Aer function. It is possible the negatively charged aspartic acid at Aer-34 shields the arginine at mutated residue 253 to restore a functional conformation. Site-specific replacement of N34 with aspartic acid, arginine, or cysteine (34) abolishes Aer function. Similarly, amino acid replacement in the E. coli ArcB PAS domain (N181A), corresponding to Aer-N34 (38), abolishes ArcB activity by preventing the ArcB transmitter module from autophosphorylating (26). Interestingly, oxidized Aer-C253 and Aer-C253′ can be cross-linked in vivo (24), suggesting close proximity between these residues in the WT Aer homodimer. An attractive model is one whereby residue 253 is located near the HAMP-HAMP′ interface as well as a HAMP-PAS interface. The identification of other specific suppressor pairs in the vicinity of N34D and C253R would strengthen the hypothesis of direct interaction between these regions of the PAS and HAMP domains. However, the search for other specific suppressors was hampered by the frequent identification of the nonspecific suppressors, in particular S28G.
During this study, Aer-PAS suppressor mutations were identified for a series of Aer-HAMP domain mutations, providing evidence for functional interactions between these domains in Aer. Cumulative findings support a model in which the AS-2 helix of the Aer-HAMP domain interacts with the PAS domain to stabilize a conformation that binds FAD. This is consistent with previous evidence that the membrane anchor and HAMP domain of Aer are necessary for native folding of the PAS domain (16). Altering the redox state of FAD bound to the Aer-PAS domain is thought to initiate a conformational signal, although it is uncertain how that signal is transmitted to the HAMP domain. The proposed interaction between the PAS and HAMP domains could provide a mechanism for direct signaling between these regions.
Acknowledgments
We are grateful to Debbie Thomas, Jack Scarbrough, Sheena Fry, and Nathan Abraham for technical assistance.
This work was supported by grants from the National Institute of General Medical Sciences (GM29481) and Loma Linda University to B. L. Taylor.
REFERENCES
- 1.Ames, P., and J. S. Parkinson. 1988. Transmembrane signaling by bacterial chemoreceptors: E. coli transducers with locked signal output. Cell 55:817-826. [DOI] [PubMed] [Google Scholar]
- 2.Ames, P., C. A. Studdert, R. H. Reiser, and J. S. Parkinson. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:7060-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appleman, J. A., L. L. Chen, and V. Stewart. 2003. Probing conservation of HAMP linker structure and signal transduction mechanism through analysis of hybrid sensor kinases. J. Bacteriol. 185:4872-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appleman, J. A., and V. Stewart. 2003. Mutational analysis of a conserved signal-transducing element: the HAMP linker of the Escherichia coli nitrate sensor NarX. J. Bacteriol. 185:89-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 6.Bibikov, S. I., L. A. Barnes, Y. Gitin, and J. S. Parkinson. 2000. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5830-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibikov, S. I., R. Biran, K. E. Rudd, and J. S. Parkinson. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourret, R. B., K. A. Borkovich, and M. I. Simon. 1991. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu. Rev. Biochem. 60:401-441. [DOI] [PubMed] [Google Scholar]
- 9.Butler, S. L., and J. J. Falke. 1998. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry 37:10746-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chervitz, S. A., and J. J. Falke. 1995. Lock on/off disulfides identify the transmembrane signaling helix of the aspartate receptor. J. Biol. Chem. 270:24043-24053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chervitz, S. A., and J. J. Falke. 1996. Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc. Natl. Acad. Sci. USA 93:2545-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, L. A., S. M. Egan, and V. Stewart. 1992. Mutational analysis reveals functional similarity between NARX, a nitrate sensor in Escherichia coli K-12, and the methyl-accepting chemotaxis proteins. J. Bacteriol. 174:3667-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crosson, S., and K. Moffat. 2001. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. USA 98:2995-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Falke, J. J., and G. L. Hazelbauer. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrmann, S., Q. Ma, M. S. Johnson, A. V. Repik, and B. L. Taylor. PAS domain of the Aer redox sensor requires C-terminal residues for native fold and FAD binding. J Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 17.Jung, K. H., and J. L. Spudich. 1998. Suppressor mutation analysis of the sensory rhodopsin I-transducer complex: insights into the color-sensing mechanism. J. Bacteriol. 180:2033-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalman, L. V., and R. P. Gunsalus. 1990. Nitrate- and molybdenum-independent signal transduction mutations in narX that alter regulation of anaerobic respiratory genes in Escherichia coli. J. Bacteriol. 172:7049-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, K. K., H. Yokota, and S. H. Kim. 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400:787-792. [DOI] [PubMed] [Google Scholar]
- 20.Kim, S. H., W. Wang, and K. K. Kim. 2002. Dynamic and clustering model of bacterial chemotaxis receptors: structural basis for signaling and high sensitivity. Proc. Natl. Acad. Sci. USA 99:11611-11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kofoid, E. C., and J. S. Parkinson. 1988. Transmitter and receiver modules in bacterial signaling proteins. Proc. Natl. Acad. Sci. USA 85:4981-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Moual, H., and D. E. Koshland, Jr. 1996. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J. Mol. Biol. 261:568-585. [DOI] [PubMed] [Google Scholar]
- 23.Letunic, I., L. Goodstadt, N. J. Dickens, T. Doerks, J. Schultz, R. Mott, F. Ciccarelli, R. R. Copley, C. P. Ponting, and P. Bork. 2002. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30:242-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Ma, Q., M. S. Johnson, and B. L. Taylor. Genetic analysis of the HAMP domain of the Aer aerotaxis sensor localizes FAD binding determinants to the AS-2 helix. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 24.Ma, Q., F. Roy, S. Herrmann, B. L. Taylor, and M. S. Johnson. The Aer protein of Escherichia coli forms a homodimer independent of the signaling domain and FAD binding. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 25.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717-1723. [DOI] [PubMed] [Google Scholar]
- 26.Matsushika, A., and T. Mizuno. 2000. Characterization of three putative sub-domains in the signal-input domain of the ArcB hybrid sensor in Escherichia coli. J. Biochem. (Tokyo) 127:855-860. [DOI] [PubMed] [Google Scholar]
- 27.Mehan, R. S., N. C. White, and J. J. Falke. 2003. Mapping out regions on the surface of the aspartate receptor that are essential for kinase activation. Biochemistry 42:2952-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Norwood, W. I., and J. R. Sadler. 1977. Pseudoreversion of lactose operator-constitutive mutants. J. Bacteriol. 130:100-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park, H., and M. Inouye. 1997. Mutational analysis of the linker region of EnvZ, an osmosensor in Escherichia coli. J. Bacteriol. 179:4382-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park, H., S. K. Saha, and M. Inouye. 1998. Two-domain reconstitution of a functional protein histidine kinase. Proc. Natl. Acad. Sci. USA 95:6728-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkinson, J. S., S. R. Parker, P. B. Talbert, and S. E. Houts. 1983. Interactions between chemotaxis genes and flagellar genes in Escherichia coli. J. Bacteriol. 155:265-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rebbapragada, A., M. S. Johnson, G. P. Harding, A. J. Zuccarelli, H. M. Fletcher, I. B. Zhulin, and B. L. Taylor. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. USA 94:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Repik, A., A. Rebbapragada, M. S. Johnson, J. O. Haznedar, I. B. Zhulin, and B. L. Taylor. 2000. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 36:806-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stock, J. B., and M. G. Surette. 1996. Chemotaxis, p. 1103-1148. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 37.Studdert, C. A., and J. S. Parkinson. 2004. Crosslinking snapshots of bacterial chemoreceptor squads. Proc. Natl. Acad. Sci. USA 101:2117-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tokishita, S., A. Kojima, and T. Mizuno. 1992. Transmembrane signal transduction and osmoregulation in Escherichia coli: functional importance of the transmembrane regions of membrane-located protein kinase, EnvZ. J. Biochem. (Tokyo) 111:707-713. [DOI] [PubMed] [Google Scholar]
- 40.Williams, S. B., and V. Stewart. 1999. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol. Microbiol. 33:1093-1102. [DOI] [PubMed] [Google Scholar]
- 41.Wolff, C., and J. S. Parkinson. 1988. Aspartate taxis mutants of the Escherichia coli Tar chemoreceptor. J. Bacteriol. 170:4509-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, H. S., J. H. Saw, S. Hou, R. W. Larsen, K. J. Watts, M. S. Johnson, M. A. Zimmer, G. W. Ordal, B. L. Taylor, and M. Alam. 2002. Aerotactic responses in bacteria to photoreleased oxygen. FEMS Microbiol. Lett. 217:237-242. [DOI] [PubMed] [Google Scholar]
- 43.Zhong, X., B. Hao, and M. K. Chan. 2003. Structure of the PAS fold and signal transduction mechanisms, p. 1-16. In S. T. Crews (ed.), PAS proteins: regulators and sensors of development and physiology. Kluwer Academic Publishers, Boston, Mass.
- 44.Zhu, Y., and M. Inouye. 2003. Analysis of the role of the EnvZ linker region in signal transduction using a chimeric Tar/EnvZ receptor protein, Tez1. J. Biol. Chem. 278:22812-22819. [DOI] [PubMed] [Google Scholar]