Abstract
A spontaneous mutant isolated in the presence of a high concentration of puromycin acquired a multidrug-resistant phenotype. Expression of the bmr3 gene was dramatically increased. A base substitution, T to A at the +4 position, detected in the mutant resulted in the stabilization of bmr3 mRNA.
One of the mechanisms by which bacteria acquire multidrug resistance (MDR) is by increasing the expression of multidrug efflux transporters (MDTs). In a spontaneous mutant of Bacillus subtilis isolated in the presence of an inhibitory concentration of rhodamine 6G, Bmr is overexpressed via intrachromosomal amplification of the bmr locus (17). Increased transcription also results in overexpression of the MDT. Puromycin- and lincomycin-resistant (PLR) mutants, isolated by growing B. subtilis 168 in the presence of high concentrations of puromycin and lincomycin, display increased expression of LmrB because of the inactivation of a negative transcriptional regulator, LmrA, or because of mutations in the 5′ untranslated region (5′ UTR), which is a binding site for the LmrA protein (16, 25). Expression of Bmr and Blt is regulated by the product of adjacent genes encoding the transcriptional activators BmrR and BltR, respectively (1, 2). In Escherichia coli, overexpression of the transcriptional regulators of a two-component regulatory system up-regulates the expression of several MDTs, resulting in an MDR phenotype (7, 12, 13).
The steady-state level of mRNA in a cell is a function of its rate of synthesis and degradation. An increase in the stability of MDT mRNA may result in a higher level of mRNA. Therefore, it is reasonable to expect that cells may acquire an MDR phenotype by stabilization of MDT mRNA. However, such an example has not been reported to date. In the present report, we describe the first example of increased stability of MDT mRNA (bmr3 mRNA) resulting in an MDR phenotype in B. subtilis.
Spontaneous mutants (PR mutants) isolated in the presence of high concentrations of puromycin showed an MDR phenotype.
Spontaneous mutants isolated by growing B. subtilis 168 in Luria-Bertani (LB) medium containing high concentrations of puromycin (100 μg/ml) were found to be divided into two groups. One group of mutants showed high levels of resistance to lincomycin as well as puromycin (PLR mutants), whereas the other group (PR mutants) showed levels of lincomycin resistance similar to that of strain 168. Both groups of mutants expressed an MDR phenotype, although their drug specificities were somewhat different. It has previously been reported that PLR mutants have mutations in the lmrAB operon (16).
In the present study, a PR mutant was further characterized. Resistance to various drugs was assayed as described previously (20). As shown in Table 1, the PR mutant showed increased resistance to puromycin, norfloxacin, tosufloxacin, daunomycin, and ethidium bromide. Resistance levels to levofloxacin, lincomycin, tetraphenylphosphonium chloride, and rhodamine 6G were not significantly increased. The results indicated the possibility that expression of one of the MDTs was increased in the PR mutant.
TABLE 1.
MDR phenotypes of the PR mutant
| Drug | Relative resistancea
|
||
|---|---|---|---|
| PR mutant | PR::cat | TF-5′ UTR | |
| Puromycin | 54.1 | 0.9 | 52.6 |
| Norfloxacin | 8.7 | 1.1 | 9.0 |
| Tosufloxacin | 5.4 | 1.0 | 5.4 |
| Daunomycin | 2.7 | 0.9 | 2.9 |
| Ethidium bromide | 2.6 | 2.5 | |
| Levofloxacin | 1.3 | 1.4 | |
| Lincomycin | 1.4 | 1.4 | |
| TPPb | 1.2 | 1.2 | |
| Rhodamine 6G | 1.1 | 1.1 | |
Relative resistance was determined by dividing the 50% inhibitory concentration (IC50; the drug concentration required to inhibit growth by 50%) of various mutants by the IC50 of strain 168. PR::cat, PR mutant with cat inserted; TF-5′ UTR, 5′ UTR transformants (see text for details).
TPP, tetraphenylphosphonium chloride.
Identification of the gene responsible for the MDR phenotype.
In order to identify the gene responsible for the MDR phenotype, the PR mutant was transformed with pHV1248 (23). At a temperature inhibiting plasmid replication, a chloramphenicol resistance gene (cat) was inserted at random positions on the chromosome of the PR mutant by transposition. If insertion into the gene responsible for the MDR phenotype occurs, the cells become sensitive to puromycin. Therefore, we selected the chloramphenicol-resistant and puromycin-sensitive clones. One of the clones obtained (PR mutant with a cat insertion) showed wild-type levels of resistance to puromycin, norfloxacin, tosufloxacin, and daunomycin (Table 1), indicating that insertion of the cat gene inactivated the gene responsible for the MDR phenotype of the PR mutant.
Southern hybridization analysis showed that a 5.0-kb HincII fragment of chromosomal DNA from the PR mutant with a cat insertion contained the cat gene and flanking regions. This fragment was cloned into pUC18, and the partial DNA sequence of the insert was determined. The sequences obtained matched those of the bmr3 gene which encodes a third MDT of B. subtilis belonging to the major facilitator superfamily (21). A 1.8-kb fragment containing the bmr3 locus was amplified with High Fidelity Platinum Taq (Invitrogen) with chromosomal DNA from the PR mutant used as a template and transformed into strain 168. The transformant selected by puromycin resistance showed the same MDR phenotype as the original PR mutant. These results indicate that the bmr3 gene was responsible for the MDR phenotype of the PR mutant.
Mutations found in the bmr3 gene.
The nucleotide sequence of a 1.8-kb fragment of the PR mutant containing the bmr3 gene was determined and compared to that of strain 168, which has been previously reported (21). As members of a European-Japanese cooperative B. subtilis genome sequencing project, Yamane et al. determined the nucleotide sequence for this region (accession no. D50453). In a comparison of the sequences of the 1.8-kb region containing the bmr3 gene determined by us (accession no. D50098) and by Yamane et al., many discrepancies (at more than 20 sites) were found. For example, Yamane et al. reported that the bmr3 gene encodes a protein containing 315 amino acids (24), whereas based on our data, it encodes a protein containing 536 amino acids (21). We confirmed that our sequence is correct at each discrepant site, and consequently our sequence data were adopted for this study.
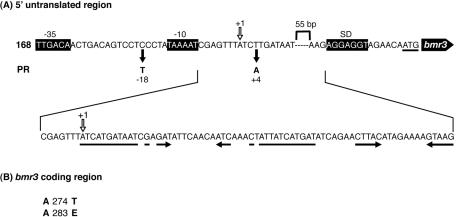
As shown in Fig. 1, two nucleotide changes were found in the 5′ UTR. One change was C to T at the −18 position and the other was T to A at the +4 position, both relative to the initiation site of transcription (see below). Two amino acid changes, A274T and A285E, which are located in the 9th of 14 transmembrane segments of the Bmr3 protein, were also found. In order to determine whether these two amino acid changes affect the MDR phenotype of the PR mutant, a 0.9-kb fragment containing the 5′ UTR and the coding region up to the 67th amino acid were amplified with High Fidelity Platinum Taq (Invitrogen), with chromosomal DNA from the PR mutant used as a template, and transformed into strain 168 cells. The transformants showed the same MDR phenotype as the original PR mutant (Table 1). No difference in drug specificity between the transformants and the original PR mutant was detected for the drugs listed in Table 1. From these results, it was concluded that the two base substitutions in the 5′ UTR of the bmr3 gene resulted in the MDR phenotype and that the two amino acid changes had no effect on the drug specificity of the Bmr3 efflux protein.
FIG. 1.
Mutations detected in the PR mutant. Putative promoter −35 and −10 consensus sequences and Shine-Dalgarno sequences are shown as boxes in black. The transcription initiation site is marked +1. The horizontal arrows indicate inverted repeat sequences. (A) Mutations in the 5′ UTR. Nucleotide changes detected in the PR mutant are indicated by filled arrows. (B) Amino acid changes in the bmr3 coding region. Both amino acids are located in the ninth transmembrane segment of Bmr3.
bmr3 mRNA increased more than 20-fold in the PR mutant.
In order to examine whether the expression of the bmr3 gene increased at a transcriptional level, Northern hybridization analysis was carried out with total RNA isolated from strain 168 and PR mutant cells as described previously (19). In strain 168, the bmr3 gene was transcribed as a 1.6-kb monocistronic mRNA whose expression level was dependent on growth phase: high level in early log phase and low level in late log phase (21). In the PR mutant, the cellular level of bmr3 mRNA increased about 24-fold in early-log-phase cells (Fig. 2). In late-log-phase cells, the cellular level of bmr3 mRNA decreased in the PR mutant. However, the difference between levels in early and late log phase was only 3.5-fold, compared to 20-fold in the wild-type strain. No experimental data have been obtained to explain this observation.
FIG. 2.
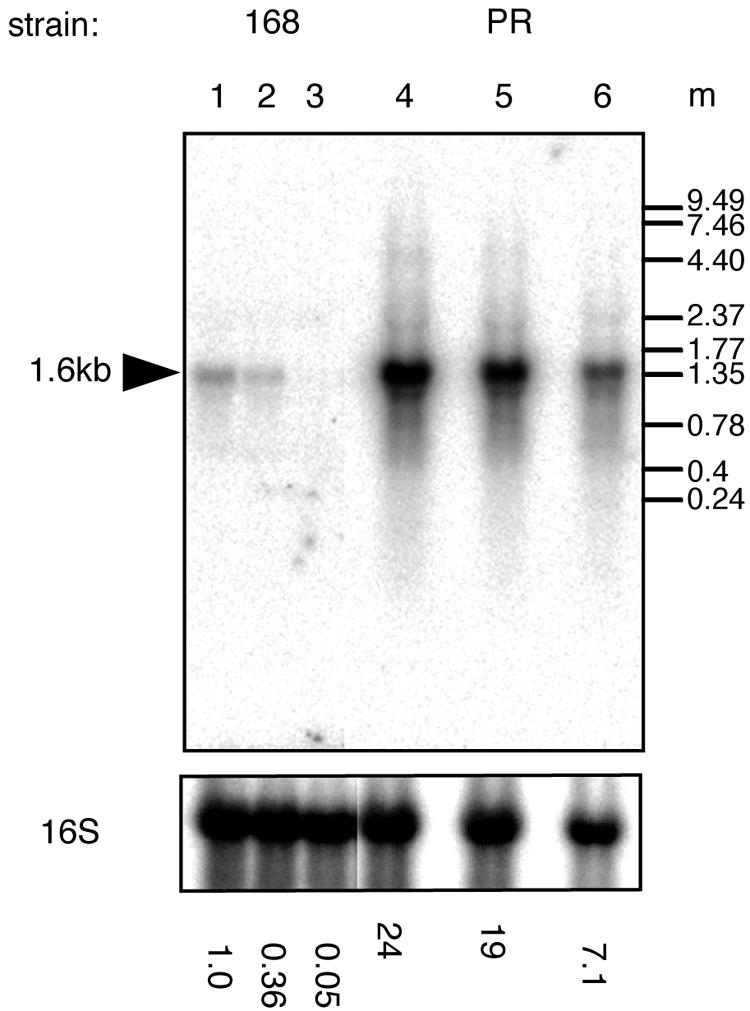
Northern hybridization analysis of RNA isolated from strain 168 and the PR mutant. Cells were grown in LB medium to early (optical density at 530 nm [OD530], 0.5) (lanes 1 and 4), middle (OD530, 1.2) (lanes 2 and 5), and late (OD530, 2.0) (lanes 3 and 6) log phases. Five micrograms of total RNA was loaded per lane. A 32P-labeled bmr3 probe was used. The same membrane was reprobed with a 32P-labeled 16S rRNA fragment, and the results are shown in the lower part of the figure. The radioactivity of each band was quantified with a BAS2000 imaging analyzer (Fuji). The relative amounts of bmr3 mRNA normalized to 16S rRNA are shown at the bottom of the figure. m, molecular size standard (Invitrogen RNA ladder, 0.24 to 9.5 kb and 0.16 to 1.77 kb).
Primer extension analysis was carried out to determine the start site of the transcription. Total RNA (40 μg) and 2 pmol of a rhodamine X-isothiocyanate (XRITC)-labeled primer (5′-XRITC-CAGGACCACAAATTTGGTGGAAGCC) complementary to the sequence located 48 to 23 bp downstream from the putative initiation codon of the bmr3 gene were mixed, made up to a total volume of 17 μl with distilled water, heated at 95°C for 5 min, and incubated at 40°C for 60 min. The first-strand cDNA was synthesized by use of an avian myeloblastosis virus reverse transcriptase first-strand cDNA synthesis kit (LIFE SCIENCE, Inc.) in accordance with the manufacturer's instruction. Extension products were subjected to gel electrophoresis (5% polyacrylamide sequencing gel), alongside sequence ladders obtained with the same primer, and analyzed with an FMBIO-100 Fluor Bio image analyzer (Hitachi Software Engineering Co., Ltd.). The results showed that transcription was initiated from an adenosine residue located 83 bp upstream from the initiation codon in both strain 168 and the PR mutant (Fig. 3). Typical σA-dependent −35 and −10 consensus sequences were found in the upstream region, as shown in Fig. 1.
FIG. 3.
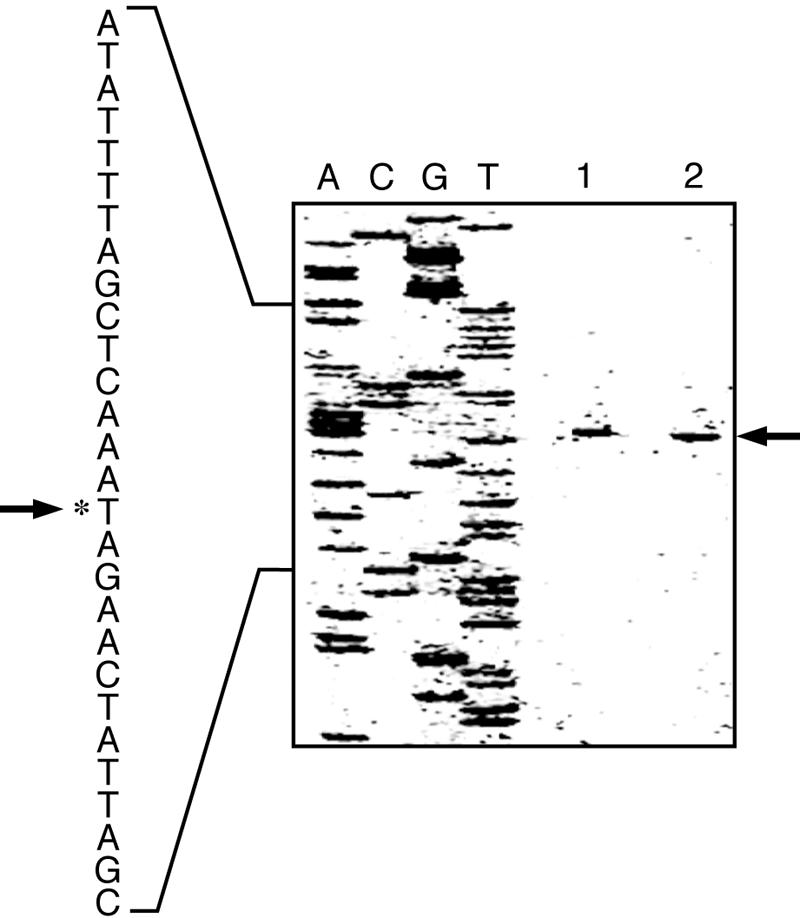
Mapping of the 5′-end of bmr3 mRNA by primer extension analysis. Total RNA was isolated from an early-log-phase (OD530, 0.5) culture of strain 168 and the PR mutant grown in LB medium. Forty micrograms of total RNA was used for primer extension. The amount of sample loaded on lane 2 was 1/10 of that loaded on lane 1. The potential transcription start site is marked with arrows. Lanes: 1, strain 168; 2, PR mutant; A, C, G, and T, dideoxy sequencing ladder obtained with the same primer used for primer extension.
Expression of pbmr3-lacZ transcriptional fusion genes.
To determine whether only one or both of the C-to-T(−18) and T-to-A(+4) changes in the PR mutant were responsible for the increased expression of the bmr3 gene, a 386-bp fragment encompassing positions −103 to +283 of the bmr3 gene was transcriptionally fused to the lacZ gene. In order to construct 386-bp fragments having a base change of C to T at the −18 position, T to A at the +4 position, or both, in vitro mutagenesis was carried out with a Quick Change site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) in accordance with the method described by the manufacturers. The primers used were as follows: for the C-to-T change at the −18 position, 5′-CTTGTTGACAACTGACAGTCCTTCCTATAAAATCGAG and 5′-CTCGATTTTATAGGAAGGACTGTCAGTTGTCAACAAG, and for the T-to-A change at the +4 position, 5′-CTGACAGTCCTCCCTATAAAATCGAGTTTATCATGATAATCGAG and 5′-TCGATTATCATGATAAACTCGATTTTATAGGGAGGACTGTCAG. Nucleotide changes were confirmed by DNA sequencing with an ABI Prism 310 genetic analyzer (PE Biosystems). The 386-bp fragments obtained by digestion with EcoRI and BamHI were cloned into pDL2 (8), and the resulting plasmids were linearized by digestion with PstI and transformed into B. subtilis 168. Chloramphenicol-resistant colonies were selected. The integration of pbmr3-lacZ fusion genes at the amyE locus was confirmed by PCR. The resulting strains obtained were described as 168 amyE::pbmr3(C→T)-lacZ, 168 amyE::pbmr3(T→A)-lacZ, 168 amyE::pbmr3(PR mutant)-lacZ, and 168 amyE::pbmr3(wild type)-lacZ.
The β-galactosidase activity of 168 amyE::pbmr3(PR mutant)-lacZ was about 200-fold higher than that of 168 amyE::pbmr3(wild type)-lacZ. The β-galactosidase activity of 168 amyE::pbmr3(T→A)-lacZ was about 50-fold higher than that of 168 amyE::pbmr3(wild type)-lacZ, whereas the β-galactosidase activity of 168 amyE::pbmr3(C→T)-lacZ was only twice that of the wild-type fusion strain (Table 2). These results indicate that the T-to-A(+4) nucleotide change in the PR mutant was mainly responsible for increased expression of the bmr3 gene at a transcriptional level, although the C-to-T(−18) change together with T-to-A(+4) change resulted in about a fourfold increase in expression.
TABLE 2.
Expression of pbmr3-lacZ transcriptional fusion genes
| Strain | β-Galactosidase activity (U)a |
|---|---|
| 168 amyE::pbmr3(wild type)-lacZ | 1.05 |
| 168 amyE::pbmr3(PR)-lacZ | 198 |
| 168 amyE::pbmr3(T→A)-lacZ | 49.0 |
| 168 amyE::pbmr3(C→T)-lacZ | 2.37 |
β-Galactosidase activity was measured in cells at mid-log phase. One unit of β-galactosidase activity was defined as the amount of enzyme that catalyzed the production of 1 μmol of o-nitrophenol/h/optical density unit at 530 nm.
Increased stability of bmr3 mRNA in the PR mutant.
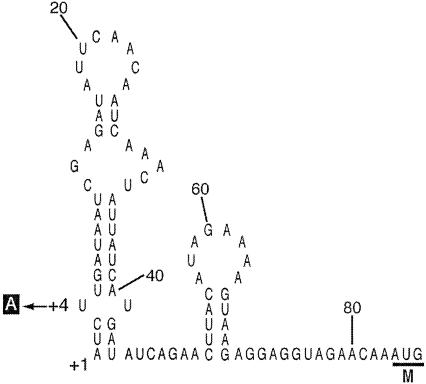
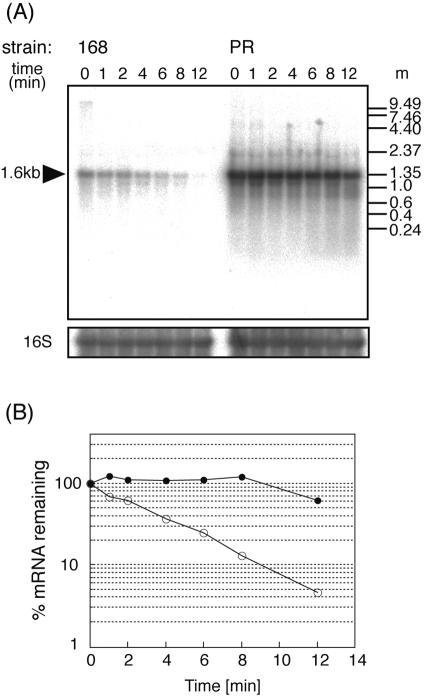
We examined potential secondary structures of bmr3 mRNA in the 83-bp 5′ UTR and found that the 5′-proximal region of bmr3 mRNA can fold to form stem-loop structures without any unpaired nucleotides at the 5′ end. The nucleotide change of T to A at the +4 position in the PR mutant changed one mismatched T-T pair to a matched A-T pair, resulting in a perfectly matched 11-bp stem (Fig. 4). It is well known that the 5′-terminal stem-loop is a determinant of mRNA stabilization (9, 10). Therefore, we compared the stabilities of bmr3 mRNA for strain 168 and the PR mutant. To determine the stability of mRNA, rifampin was added to the early-log-phase culture growing in LB medium at a final concentration of 500 μg/ml, and samples were removed at set time intervals as described previously (3). In strain 168 cells, the half-life of bmr3 mRNA was 3.0 min, whereas in the PR mutant, it was more than 12 min (Fig. 5). As a control, we also measured the half-life of the bcrC mRNA, which encodes a membrane protein involved in bacitracin resistance (20). The half-life of bcrC mRNA (1.8 min) in the PR mutant was found to be the same as that of strain 168 (data not shown).
FIG. 4.
mRNA secondary structure predicted for the 5′ UTR of bmr3. A base change, T to A at position +4, is shown by an arrow. +1, transcriptional start site; M, initiation codon.
FIG. 5.
Increased stability of bmr3 mRNA in the PR mutant. Cells were grown in LB medium to early log phase (OD530, 0.5), and rifampin (500 μg/ml) was added to inhibit transcription. Portions of the culture were removed at the indicated times (in minutes) after the addition of rifampin (shown at the top of the each lane). m, molecular size standard (Invitrogen RNA ladder, 0.24 to 9.5 kb; Novagen Perfect RNA markers, 0.1 to 1 kb). (A) Hybridization was carried out with probes specific for bmr3 and 16S rRNA. (B) The relative amounts of mRNAs remaining after the addition of rifampin were calculated from the results shown in panel A and plotted against time. Open circles, strain 168; closed circles, PR mutant.
The T-to-A mutation at the +4 position resulted in increased mRNA stability of the pbmr3-lacZ fusion gene.
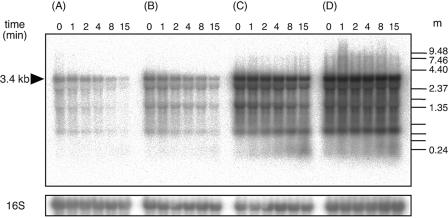
In order to examine whether the 5′ UTR of bmr3 mRNA functioned as a determinant of mRNA stability when it was fused to a heterologous gene, we also determined the mRNA stability of pbmr3-lacZ fusion genes. Relative values for steady-state mRNA levels of fusion genes were calculated from the intensities of the bands at time zero (Fig. 6) and were found to be 1:20:2:10 for 168 amyE::pbmr3(wild type)-lacZ, 168 amyE::pbmr3(PR mutant), 168 amyE::pbmr3(C→T), and 168 amyE::pbmr3 (T→A)-lacZ, respectively. The relative values of β-galactosidase activity in these strains, shown in Table 2, reflect the mRNA levels of the fusion genes.
FIG. 6.
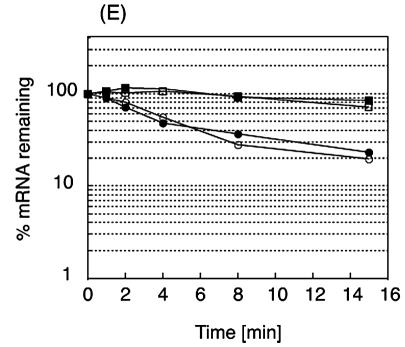
The 5′ UTR of the bmr3 transcript is a determinant of mRNA stability. Strain 168 clones containing various pbmr3-lacZ fusion genes at the amyE locus were grown in LB medium to early log phase (OD530, 0.5), and 500 μg of rifampin/ml was added to inhibit transcription. Portions of the culture were removed at the indicated times (in minutes) after the addition of rifampin (shown at the top of each lane). Five micrograms of total RNA was loaded per lane. m, molecular size standard (Invitrogen RNA ladder, 0.24 to 9.5 kb and 0.16 to 1.77 kb). A 32P-labeled lacZ probe was used for hybridization. An arrow indicates the mRNA of the pbmr3-lacZ fusion gene. (A) 168 amyE::pbmr3(wild type)-lacZ; (B) 168 amyE::pbmr3(C→T)-lacZ; (C) 168 amyE::pbmr3(T→A)-lacZ; (D) 168 amyE::pbmr3(PR)-lacZ. (E) The relative amounts of mRNA remaining after the addition of rifampin were calculated from the results shown in panels A through D and plotted against time. Open circles, 168 amyE::pbmr3(wild type)-lacZ; closed circles, 168 amyE::pbmr3(C→T)-lacZ; open squares, 168 amyE::pbmr3(T→A)-lacZ; closed squares, 168 amyE::pbmr3(PR mutant)-lacZ.
The half-lives of the pbmr3(wild type)-lacZ and pbmr3(C→T)-lacZ fusion gene mRNAs were about 4 min, whereas those of the pbmr3(PR mutant)-lacZ and pbmr3(T→A)-lacZ mRNAs were more than 15 min (Fig. 6). These results indicate that the 5′ UTR sequence of the bmr3 gene is a determinant of mRNA stability and that the base change of T to A at the +4 position resulted in increased stability of bmr3 mRNA. The base change of C to T at the −18 position had no effect on mRNA stability. Therefore, the twofold increase of pbmr3(C→T)-lacZ fusion gene mRNA compared to that of wild-type fusion gene mRNA may be due to an increase in the rate of mRNA synthesis.
Endonucleases RNase III and RNase M5 are not involved in the degradation of bmr3 mRNA.
A predicted ρ-independent transcriptional terminator, which includes 17-bp perfectly matched inverted repeats, is located immediately downstream of the stop codon of the bmr3 gene. Therefore, bmr3 mRNA is predicted to have stem-loop structures at both the 3′ and 5′ ends. We attempted to determine whether endonucleases are involved in degradation of bmr3 mRNA. It is expected that the deletion of an RNase, which is involved in the critical step of bmr3 mRNA degradation in strain 168, may result in increased drug resistance as well as an increase in the expression of the bmr3(wild type)-lacZ fusion gene. An endoribonuclease, RNase III, has been reported to be involved in the degradation of certain mRNAs in B. subtilis. The mRNA of the early genes of B. subtilis phage SP82 is cleaved by RNase III at the loop of a hairpin structure (22), and it also cleaves the small cytoplasmic RNA precursor of the signal recognition particle at the bulge in the stem structure (18). Drug resistance levels of an rncS deletion mutant (BG322) and a control strain (BG324) were compared (11). No increase in resistance to puromycin was observed in BG322. The pbmr3(wild type)-lacZ fusion gene was also introduced into the amyE locus of BG322 and BG324. β-Galactosidase activity was not increased by the rncS deletion. rnmV encodes RNase, which has been reported to be responsible for 5S rRNA maturation (5). We compared puromycin resistance levels of strain 168 and the pMutin insertional disruption mutant of the rnmV gene (YABFd). No difference in resistance was observed between the two strains. These results indicate that endoribonucleases RNase III and RNase M5 are not involved in the degradation of bmr3 mRNA. Based on a gene array analysis, Condon et al. reported that RNase M5 has few, if any, mRNA substrates in B. subtilis (6).
Several mRNAs with extreme stability have been reported for B. subtilis. Individual factors contributing to the stabilization of these mRNAs have been discussed in each case. The half-life of an ermC transcript increases about 20-fold upon exposure to erythromycin. The erythromycin-bound ribosome stalls while translating a leader peptide preceding the coding region of ErmC. The stalled ribosome protects the transcript from degradation (4, 14). The gsiB mRNA, which encodes a σB-dependent general stress protein, has a remarkably long half-life (∼20 min) (15). It was found that a strong ribosome binding site was crucial for the increased stability of the gsiB mRNA. The mRNA of the aprE gene, which encodes subtilisin, is stable, with a half-life exceeding 20 min (9). aprE-5′ UTR-lacZ fusion mRNA has a similar half-life, indicating that the determinants for aprE mRNA stability are located in the 5′ UTR, which is predicted to fold into a stem-loop structure at the 5′ end. Our results indicate that endonucleases RNase III and RNase M5 are not involved in the degradation of bmr3 mRNA. The possibility that a protein factor other than RNase is involved in the regulation of mRNA stability has not been excluded. Further work is required to elucidate the precise mechanism of the bmr3 mRNA degradation process.
The results obtained in the present study indicate the possibility that MDR clinical isolates have a mutation which results in the stabilization of MDT mRNA. This finding provides an additional target for potential drugs designed to overcome MDR pathogens.
Acknowledgments
We are grateful to D. H. Bechhofer for providing strains BG322 and BG324. We also appreciate the members of the Japan and EU consortia of B. subtilis functional genomics for providing the pMutin insertional disruption mutant of rnmV.
This work was partly supported by a grant in aid from the Scientific Research Promotion Fund of the Japan Private School Promotion Foundation.
REFERENCES
- 1.Ahmed, M., C. M. Borsch, S. S. Taylor, N. Vázquez-Laslop, and A. A. Neyfakh. 1994. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J. Biol. Chem. 269:28506-28513. [PubMed] [Google Scholar]
- 2.Ahmed, M., L. Lyass, P. N. Markham, S. S. Taylor, N. Vázquez-Laslop, and A. A. Neyfakh. 1995. Two highly similar multidrug transporters of Bacillus subtilis whose expression is differentially regulated. J. Bacteriol. 177:3904-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aiso, T., and R. Ohki. 2003. Instability of sensory histidine kinase mRNAs in Escherichia coli. Genes Cells 8:179-187. [DOI] [PubMed] [Google Scholar]
- 4.Bechhofer, D. H., and K. H. Zen. 1989. Mechanism of erythromycin-induced ermC mRNA stability in Bacillus subtilis. J. Bacteriol. 171:5803-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condon, C., D. Brechemier-Baey, B. Beltchev, M. Grunberg-Manago, and H. Putzer. 2001. Identification of the gene encoding the 5S ribosomal RNA maturase in Bacillus subtilis: mature 5S rRNA is dispensable for ribosome function. RNA 7:242-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Condon, C., J. Rourera, D. Brechemier-Baey, and H. Putzer. 2002. Ribonuclease M5 has few, if any, mRNA substrates in Bacillus subtilis. J. Bacteriol. 184:2845-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eguchi, Y., T. Oshima, H. Mori, R. Aono, K. Yamamoto, A. Ishihama, and R. Utsumi. 2003. Transcriptional regulation of drug efflux genes by EvgAS, a two-component system in Escherichia coli. Microbiology 149:2819-2828. [DOI] [PubMed] [Google Scholar]
- 8.Fukuchi, K., Y. Kasahara, K. Asai, K. Kobayashi, S. Moriya, and N. Ogasawara. 2000. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 146:1573-1583. [DOI] [PubMed] [Google Scholar]
- 9.Hambraeus, G., M. Persson, and B. Rutberg. 2000. The aprE leader is determinant of extreme mRNA stability in Bacillus subtilis. Microbiology 146:3051-3059. [DOI] [PubMed] [Google Scholar]
- 10.Hansen, M. J., L.-H. Chen, M. L. S. Fejzo, and J. G. Belasco. 1994. The ompA 5′ untranslated region impedes a major pathway for mRNA degradation in Escherichia coli. Mol. Microbiol. 12:707-716. [DOI] [PubMed] [Google Scholar]
- 11.Herskovitz, M. A., and D. H. Bechhofer. 2000. Endoribonuclease RNase III is essential in Bacillus subtilis. Mol. Microbiol. 38:1027-1033. [DOI] [PubMed] [Google Scholar]
- 12.Hirakawa, H., K. Nishino, J. Yamada, T. Hirata, and A. Yamaguchi. 2003. β-Lactam resistance modulated by the overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J. Antimicrob. Chemother. 52:576-582. [DOI] [PubMed] [Google Scholar]
- 13.Hirakawa, H., K. Nishino, T. Hirata, and A. Yamaguchi. 2003. Comprehensive studies of drug resistance mediated by overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J. Bacteriol. 185:1851-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hue, K. K., and D. H. Bechhofer. 1991. Effect of ermC leader region mutations on induced mRNA stability. J. Bacteriol. 173:3732-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jürgen, B., T. Schweder, and M. Hecker. 1998. The stability of mRNA from the gsiB gene of Bacillus subtilis is dependent on the presence of a strong ribosome binding site. Mol. Gen. Genet. 258:538-545. [DOI] [PubMed] [Google Scholar]
- 16.Murata, M., S. Ohno, M. Kumano, K. Yamane, and R. Ohki. 2003. Multidrug resistant phenotype of Bacillus subtilis spontaneous mutants isolated in the presence of puromycin and lincomycin. Can. J. Microbiol. 49:71-77. [DOI] [PubMed] [Google Scholar]
- 17.Neyfakh, A. A., V. E. Bidnenko, and L. B. Chen. 1991. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc. Natl. Acad. Sci. USA. 88:4781-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oguro, A., H. Kakeshita, K. Nakamura, K. Yamane, W. Wang, and D. H. Bechhofer. 1998. Bacillus subtilis RNase III cleaves both 5′- and 3′-sites of the small cytoplasmic RNA precursor. J. Biol. Chem. 273:19542-19547. [DOI] [PubMed] [Google Scholar]
- 19.Ohki, R., Giyanto, K. Tateno, W. Masuyama, S. Moriya, K. Kobayashi, and N. Ogasawara. 2003. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49:1135-1144. [DOI] [PubMed] [Google Scholar]
- 20.Ohki, R., K. Tateno, Y. Okada, H. Okajima, K. Asai, Y. Sadaie, M. Murata, and T. Aiso. 2003. A bacitracin-resistant Bacillus subtilis gene encodes a homologue of the membrane-spanning subunit of the Bacillus licheniformis ABC transporter. J. Bacteriol. 185:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohki, R., and M. Murata. 1997. bmr3, a third multidrug transporter gene of Bacillus subtilis. J. Bacteriol. 179:1423-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panganiban, A. T., and H. R. Whiteley. 1983. Bacillus subtilis RNase III cleavage sites in phage SP82 early mRNA. Cell 33:907-913. [DOI] [PubMed] [Google Scholar]
- 23.Petit, M.-A., C. Bruand, L. Jannière, and S. D. Ehrlich. 1990. Tn10-derived transposons active in Bacillus subtilis. J. Bacteriol. 172:6736-6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamane, K., M. Kumano, and K. Kurita. 1996. The 25o-36o region of the Bacillus subtilis chromosome: determination of the sequence of a 146 kb segment and identification of 113 genes. Microbiology 142:3047-3056. [DOI] [PubMed] [Google Scholar]
- 25.Yoshida, K.-I., Y.-H. Ohki, M. Murata, M. Kinahara, H. Matsuoka, H. Yamaguchi, R. Ohki, M. Kumano, K. Yamane, and Y. Fujita. 2004. Bacillus subtilis LmrA is a repressor for the lmrAB and yxaGH operons: identification of its binding sites and functional analysis of lmrB and yxaGH. J. Bacteriol. 186:5640-5648. [DOI] [PMC free article] [PubMed] [Google Scholar]