Abstract
Coxiella burnetii undergoes a poorly defined developmental cycle that generates morphologically distinct small-cell variants (SCV) and large-cell variants (LCV). We developed a model to study C. burnetii morphogenesis that uses Vero cells synchronously infected with homogeneous SCV (Nine Mile strain in phase II) harvested from aged infected cell cultures. A time course transmission electron microscopic analysis over 8 days of intracellular growth was evaluated in conjunction with one-step growth curves to correlate morphological differentiations with growth cycle phase. Lag phase occurred during the first 2 days postinfection (p.i.) and was primarily composed of SCV-to-LCV morphogenesis. LCV forms predominated over the next 4 days, during which exponential growth was observed. Calculated generation times during exponential phase were 10.2 h (by quantitative PCR assay) and 11.7 h (by replating fluorescent focus-forming unit assay). Stationary phase began at approximately 6 days p.i. and coincided with the reappearance of SCV, which increased in number at 8 days p.i. Quantitative reverse transcriptase-PCR demonstrated maximal expression of scvA, which encodes an SCV-specific protein, at 8 days p.i., while immunogold transmission electron microscopy revealed degradation of ScvA throughout lag and exponential phases, with increased expression observed at the onset of stationary phase. Collectively, these results indicate that the overall growth cycle of C. burnetii is characteristic of a closed bacterial system and that the replicative form of the organism is the LCV. The experimental model described in this report will allow a global transcriptome and proteome analysis of C. burnetii developmental forms.
Coxiella burnetii is a bacterial obligate intracellular parasite that is the etiologic agent of human Q fever. The organism is phylogenetically related to Legionella spp. and resides in an acidic (pH ≈ 5) parasitophorous vacuole (PV) with lysosomal characteristics. Here the acid pH activates C. burnetii metabolism and initiates replication of the organism. Q fever is a zoonosis that typically manifests as an acute, debilitating, influenza-like illness. Rare chronic disease can occur, usually in the form of hepatitis or endocarditis. Humans become infected primarily by inhaling contaminated aerosols that are generated by domestic livestock operations (reviewed in reference 26). C. burnetii is a recognized potential agent of bioterrorism and was recently designated by the Centers for Disease Control and Prevention as a category B agent. Characteristics of C. burnetii that warrant this classification include aerosolic infection, low infectious dose (fewer than 10 organisms), and the ability to survive for prolonged periods in the environment. Indeed, C. burnetii is highly resistant to osmotic shock, elevated temperature, desiccation, UV light, and various chemical disinfectants (29, 41).
C. burnetii's impressive stability and resistance are thought to be characteristics of a small-cell form that is part of a biphasic developmental cycle. Davis and Cox (10) noted pleomorphic small- and large-cell forms of C. burnetii in their original description of the organism. McCaul and Williams (30) termed these cells small-cell variants (SCV) and large-cell variants (LCV) in their proposed model of C. burnetii differentiation that includes both vegetative and sporogenic differentiations. By electron microscopy, SCV are typically rod shaped and 0.2 to 0.5 μm in length, with a characteristic electron-dense chromatin and an array of intracytoplasmic membranes. A subpopulation of the SCV that displays extreme tolerance to breakage by high pressure (20,000 lb/in2), termed the “small dense cell,” has been described (28). LCV can exceed 1 μm in length and are more pleomorphic, with a dispersed, filamentous chromatin (17, 30). The idea of C. burnetii sporogenesis is controversial. While an electron-dense polar body resembling an endospore is occasionally observed in LCV, these spore-like forms have not been demonstrated to be infectious, and the genome lacks homologues of sporulation genes (17, 30, 34). Moreover, the physical properties of the SCV are sufficient to account for the extracellular stability of the agent and environmental transmission of Q fever (29).
A few proteins that are differentially expressed by SCV and LCV have been identified. Two highly basic proteins, ScvA and Hq1, are DNA binding proteins specific to the SCV that likely play roles in chromatin condensation (18, 20). Elongation factors EF-Tu and EF-Ts, the stationary-phase sigma factor RpoS, and a protein with porin activity termed P1 are preferentially expressed by LCV (33, 35, 39).
The cellular and molecular biology of C. burnetii morphological development within the host PV are largely undefined. Studies have been hampered because of experimental constraints inherent in working with an obligate intracellular bacterium and the absence of an infection model that employs synchronous infection with purified cell forms. In this study, we conducted a systematic evaluation of C. burnetii developmental kinetics and gene expression using Vero cells synchronously infected with homogenous SCV purified from aged infected cell cultures. The kinetics of LCV and SCV morphological development, growth cycle characteristics, and developmental form transcriptional and translational properties are discussed.
MATERIALS AND METHODS
Cultivation and purification of C. burnetii.
C. burnetii (Nine Mile strain in phase II) was propagated in African green monkey kidney (Vero) fibroblasts (CCL-81; American Type Culture Collection) grown in RPMI medium (Invitrogen, Carlsbad, Calif.) supplemented with 2% fetal bovine serum. To induce production of SCV, infected cells in 150-cm2 tissue culture flasks were incubated for 4 weeks without replenishment of the growth medium: the first week at 37°C in 5% CO2 followed by 3 weeks at room temperature with flask caps tightened. Organisms were purified from infected cells by Renografin density gradient centrifugation (14). Purified SCV were resuspended in K-36 buffer (0.1 M KCl, 0.015 M NaCl, 0.05 M potassium phosphate, pH 7.0) and stored at −80°C. The homogeneity of SCV was assessed by transmission electron microscopy (TEM) as previously described (20).
Replating FFU and TaqMan QPCR assays.
C. burnetii replication during its developmental cycle was quantified by using a replating fluorescent focus-forming unit (FFU) assay and TaqMan quantitative PCR (QPCR) of genome equivalents. For both procedures, confluent Vero cells in individual wells of a 6-well tissue culture plate were incubated with the SCV inoculum at a multiplicity of infection of 5 for 1 h at room temperature to allow internalization. The inoculum was then removed and replaced with fresh RPMI medium supplemented with 2% fetal bovine serum. (This point was considered 0 h postinfection [p.i.] for all time course experiments, which were conducted three times.) For replating FFU assays, infected cells were harvested at specified time points by scraping and disrupted by gentle sonication. Cell lysates with released C. burnetii were used to infect fresh confluent Vero cells in individual wells of a 24-well tissue culture plate. After a 5-day incubation, infected cells were fixed with cold 100% methanol, and FFU were stained by indirect immunofluorescence employing polyclonal rabbit antiserum generated against formalin-killed C. burnetii and Alexa Fluor 448-conjugated goat anti-rabbit immunoglobulin G serum (Molecular Probes, Eugene, Oreg.). FFUs were enumerated by fluorescence microscopy at ×320 magnification with a Ziess Axiovert 25 inverted microscope.
A TaqMan QPCR to quantify C. burnetii genomes was conducted by using DNA extracted from infected cells as template. Infected cells were harvested by scraping, and DNA was isolated with an UltraClean microbial DNA isolation kit (MoBio Laboratories Inc., Carlsbad, Calif.) as recommended by the supplier, with an additional incubation at 70°C for 10 min before physical disruption of the samples. The primer and probe sets used here and for transcriptional analysis (see below) were designed with PrimerExpress software (Applied Biosystems, Foster City, Calif.) and are listed in Table 1. The efficiencies of amplification were comparable for all primer and probe sets used in this study. Purified C. burnetii (Nine Mile strain in phase II) DNA in the range of 10 to 106 genome copies was used as template to generate standard curves as described previously (4, 34). QPCR and reverse transcriptase PCR (RT-PCR) (see below) were performed using TaqMan Universal PCR Master Mix and a Prism 7000 sequence detection system (Applied Biosystems).
TABLE 1.
Oligonucleotide PCR primers and TaqMan internal probes for C. burnetii genes
| Gene | Designation | Gene primer sequences (5′-3′) | Gene probe sequence (5′-3′) |
|---|---|---|---|
| rrs | Q16s-F | CCATGAAGTTGGAATCGCTAG | CGGTGAATACGTTCTCGGGCCTTGTAC |
| Q16s-R | ACTCCCATGGTGTGACGG | ||
| gltA | QgltA-F | CCGGTTCTACGGGAGCAAA | CCGTTTGCTTGTATTTCGGCGGG |
| QgltA-R | GGGCCCCAGAGAGCACTAAT | ||
| CBU0311 | Qp1-F | CGGCGATTGGCGTTTC | AACTGTTCAAAATCCGAAACGAGTCGCA |
| Qp1-R | GGTTGCGGTAATGCCGTTAA | ||
| rpoS | QrpoS-F | CGCGTTCGTCAAATCCAAATA | ACGCTCTGCAGCAATTACGCCA |
| QrpoS-R | GACGCCTTCCATTTCCAAAA | ||
| dotA | QdotA-F | GCGCAATACGCTCAATCACA | CCGGAGATACCGGCGGTGGG |
| QdotA-R | CCATGGCCCCAATTCTCTT | ||
| enhC | QenhC-F | TTTGATCTAACCGTCC CTGCTT | TCCTTTTGAAATGCCGCCTGGAGCT |
| QenhC-R | CTCGGGAGAGGGATTGAAAAG | ||
| hcbA | QhcbA-F | AGCTAAAGCGAAGAAAGATGCAA | CCGTAAACTTGCTAAACTTCGAAAAGAGGCC |
| QhcbA-R | TGGCAGCAGCTACTTTTCGA | ||
| scvA | QscvA-F | TGGAAAGACAAAATGTCCAACAA | ACGTGGAAAAGACCAACG |
| QscvA-R | GGTTAGAAGCACCCGGTCGT |
Quantitative RT-PCR.
Total RNA was purified from infected Vero cells cultivated in 25-cm2 tissue culture flasks. Adherent monolayers were directly lysed in 2.5 ml of RNA Wiz (Ambion, Inc., Austin, Tex.). Lysates were treated with RNase-free DNase, and total RNA was purified with an RNeasy Mini kit (QIAGEN, Valencia, Calif.). The concentration and purity of extracted RNA were determined by measuring the A260 and A280. One microgram of total RNA was converted to cDNA with a High Capacity cDNA Archive kit (Applied Biosystems) and subjected to PCR amplification. To control for DNA contamination of RNA samples, a PCR was also conducted on RNA that had not been reverse transcribed. Amplified cDNA was normalized to C. burnetii genomic equivalents (quantified as described above) and plotted as relative expression.
TEM.
Purified SCV and C. burnetti-infected Vero cells grown on 13-mm-diameter Thermanox coverslips (Nunc, Naperville, Ill.) were fixed overnight at 4°C with 2.5% glutaraldehyde-4% paraformaldehyde in 100 mM sodium cacodylate buffer (pH 7.2). Cells were postfixed with 0.5% osmium tetroxide-0.8% potassium ferricyanide in 100 mM sodium cacodylate buffer followed by 1% tannic acid in distilled water. Samples were stained overnight with 1% uranyl acetate, washed with distilled water, dehydrated with a graded ethanol series, and embedded in Spurr's resin. Thin sections were cut with an RMC MT-7000 ultramicrotome (Ventana, Tucson, Ariz.) and stained with 1% uranyl acetate and Reynold's lead citrate.
To immunolabel ScvA, C. burnetii-infected Vero cells grown in 25-cm3 tissue culture flasks were trypsinized, washed, and pelleted in a microfuge tube. These pellets, and pellets of purified SCV, were fixed overnight with 0.25% glutaraldehyde-2% paraformaldehyde in 100 mM sodium cacodylate buffer. Samples were postfixed as described above, with the following modifications. Samples were dehydrated with a graded ethanol series, embedded in LR white resin and cured overnight at 55°C. Sections were collected on nickel grids and etched for 20 min with 4% sodium-m-periodate, followed by blocking for 30 min with 3% bovine serum albumin-100 mM Tris buffer. Sections were then incubated for 1 h with monospecific rabbit anti-ScvA serum (20) diluted 1:50 with 3% bovine serum albumin-100 mM Tris buffer, washed, and incubated for 1 h in a 1:25 dilution of goat anti-rabbit immunoglobulin G conjugated to 10-nm colloidal gold (BB International, Cardiff, United Kingdom). Then sections were cut as described above, washed with distilled H2O, and stained with 1% uranyl acetate. Sections were viewed at 80 kV on a Philips CM-10 transmission electron microscope (FEI, Hillsboro, Oreg.). Digital images were acquired with a digital camera (AMT, Chazy, N.Y.) and processed with Adobe Photoshop (version 7.0; Adobe Systems, Mountain View, Calif.). To enumerate the relative ScvA content of individual C. burnetii cells, the number of gold particles per organism on representative micrographs (at least 100 organisms per time point) was counted, and organisms were scored as having more or fewer than 10 gold particles.
RESULTS
Purification of infectious SCV.
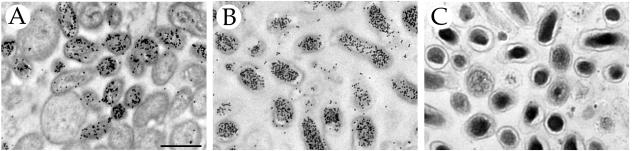
A temporal evaluation of C. burnetii development has been thwarted by an inability to synchronously infect host cells with a homogenous population of C. burnetii cell forms. Conventional methods of C. burnetii cultivation and purification yield a mixture of morphological forms (30). SCV and LCV can be purified to near homogeneity from a mixed population by cesium chloride density gradient centrifugation (20, 40); however, in our hands these preparations show little to no infectivity of cultured cells (19). We had previously observed by TEM that the percentage of SCV within the host PV increases with extended incubation times (unpublished observations). Therefore, to promote production of this cell form we cultured infected Vero cells for 4 weeks without replenishing the medium (see Materials and Methods). The SCV content of C. burnetii purified from these cultures was assessed as previously described (17, 20) by immunogold TEM with antibody directed against ScvA, an SCV-specific DNA binding protein, and by a TEM ultrastructural analysis. The specificity of anti-ScvA serum was confirmed by showing specific labeling of SCV among a mixed C. burnetii population (Fig. 1A). Organisms purified from cells infected for 4 weeks showed uniform heavy labeling by anti-ScvA serum (Fig. 1B). Moreover, the dominant cell morphology and ultrastructure were those of prototypic SCV, i.e., 0.2 to 0.5 μm in length with an electron-dense compacted chromatin (Fig. 1C; the fixation of C. burnetii depicted in Fig. 1A and B was optimized for retention of ScvA antigenicity, and consequently, the condensed chromatin of the SCV is not obvious in those panels). Collectively, these results indicate that homogeneous SCV can be purified from aged infected cell cultures.
FIG. 1.
Transmission electron micrograph showing SCV purified from aged Vero cell cultures. SCV were purified from infected cells cultured for 4 weeks as described in Materials and Methods. (A) Immunogold labeling of a mix of C. burnetii morphological forms showing specific labeling of SCV by serum against the SCV-specific protein, ScvA. (B) Immunogold labeling of C. burnetii purified from aged Vero cell cultures showing uniform heavy labeling with anti-ScvA serum. (C) C. burnetii purified from aged Vero cell cultures showing morphological and ultrastructural characteristics typical of SCV, e.g., size of 0.2 to 0.5 μm and electron-dense compacted chromatin. (The fixation of C. burnetii depicted in panels A and B was optimized for retention of ScvA antigenicity, and consequently, the condensed chromatin of the SCV is not obvious in these panels. The fixation of C. burnetii depicted in panel C was optimized for preservation of ultrastructure.) Bar, 0.5 μm.
Kinetics of Coxiella morphological differentiation.
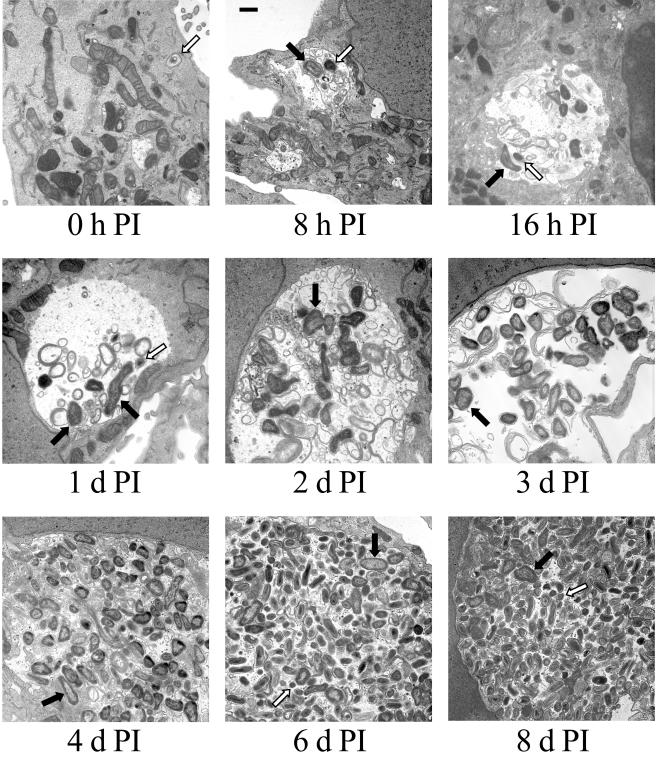
The purification of viable SCV allowed us to synchronously infect Vero cells with this cell form and to monitor morphological changes in the bacterial population over time. Vero cells were infected with SCV and processed for TEM at 0, 8, and 16 h and 1, 2, 3, 4, 6, and 8 days p.i. (Fig. 2). At 0 h p.i., SCV were observed to be tightly bound by the PV membrane. At 8 h p.i., PVs that contained both SCV and LCV were observed, indicating the initiation of SCV-to-LCV morphological differentiation. At this time point, the PV usually contained several organisms as the result of homotypic fusion of multiple C. burnetii-containing vacuoles (24). At 16 h and 1 day p.i., PVs harbored almost exclusively LCV, with occasional SCV observed. Obvious replication of LCV was evident at 2 days p.i., which coincided with the appearance of a large and spacious PV that was easily visible by light microscopy. Accumulation of cellular debris within the PV lumen was also clearly evident at this time point and presumably reflects PV fusion with autophagic vesicles (3). Numbers of LCV increased substantially from 2 to 6 days p.i., eventually tightly packing the PV lumen, which at 6 days p.i. encompassed the majority of the cell volume. At this point the PV became difficult to visualize by light microscopy. SCV reappeared at 6 days p.i., and at 8 days p.i. this cell form was estimated to comprise greater than 50% of the C. burnetii population of a typical PV.
FIG. 2.
Temporal analysis of C. burnetii morphological development in Vero cells. Vero cell monolayers were incubated with purified SCV for 1 h to allow for adherence and internalization. Extracellular organisms were then washed from cell monolayers, and fresh medium was added. This time was designated as 0 h p.i. Infected cells were fixed and processed for TEM at 0, 8, and 16 h and 1, 2, 3, 4, 6, and 8 days p.i. Prototypic SCV and LCV are designated in selected panels with white and black arrows, respectively. Bar, 0.5 μm.
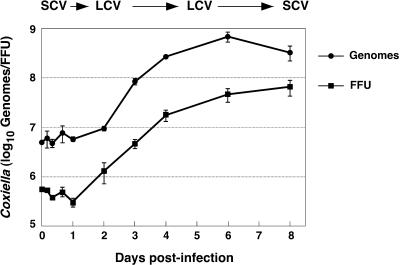
To correlate C. burnetii morphological differentiations with growth cycle phase, one-step growth curves were generated by using a replating FFU assay, which quantifies recoverable infectious organisms, and a QPCR of genome equivalents. Data from both assays resulted in similar growth cycle profiles (Fig. 3). The C. burnetii lag phase lasted approximately 2 days. Exponential growth was observed over the next 4 days, with stationary phase beginning at approximately 6 days p.i. Calculated generation times during exponential phase were 10.2 h (QPCR assay) and 11.7 h (replating assay).
FIG. 3.
One-step growth curves of C. burnetii. Vero cell monolayers were incubated with purified SCV for 1 h to allow for adherence and internalization. Extracellular organisms were then washed from cell monolayers, and fresh medium was added. This time was designated as 0 h p.i. Replating FFU and genome equivalent assays were conducted to quantify C. burnetii replication as described in Materials and Methods. The approximate times p.i. of C. burnetii morphological changes and LCV replication are indicated above the graph. The results are expressed as the mean from three experiments, with error bars representing the standard error of the mean.
The replicative properties of SCV and LCV can be established when growth cycle kinetics (Fig. 3) are considered together with the time course of morphological differentiation (Fig. 2). SCV do not appear to be replicative forms, as the lag phase was comprised primarily of SCV-to-LCV morphogenesis, with SCV reappearing in significant numbers only with the onset of stationary phase. Conversely, LCV appear to be responsible for C. burnetii replication, as they were observed almost exclusively during the exponential phase of growth. The cessation of robust replication occurs concurrently with the emergence of SCV that apparently result from LCV-to-SCV condensation.
Gene expression during C. burnetii development.
We next conducted an evaluation of gene expression during the C. burnetii developmental cycle using TaqMan QPCR. Genes selected for this analysis included those that encode ScvA (scvA) and Hq1 (hcbA), basic proteins preferentially expressed by SCV and likely involved in chromatin condensation (18, 20), and the stationary-phase sigma factor RpoS (rpoS) and the porin P1 (CBU0311), proteins preferentially expressed by LCV (35, 39). Expression of the presumed housekeeping genes encoding 16S rRNA (rrs) and citrate synthase (gltA) was also assessed, in addition to that of genes encoding DotA (dotA) and EnhC (enhC), a structural protein of the C. burnetii type IV secretory apparatus and a possible type IV effector protein, respectively (9, 34). Transcriptional activity was expressed as relative expression with transcript copy number normalized to the number of C. burnetii genomes present in each sample.
Carryover transcript has been described for obligate intracellular Chlamydia trachomatis and is proposed to represent residual transcript of genes expressed late in the organism's life cycle (2, 37). Accordingly, the relative expression of C. burnetii genes observed at 0 h p.i. probably reflects carryover transcript, as the 1-h incubation of the organism with Vero cells prior to extraction of message is unlikely to result in significant acid activation of C. burnetii metabolism (15) and de novo production of transcript (Fig. 4). However, de novo synthesis of message is clearly indicated by increased expression relative to the 0-h time point. This increase occurred between 0 and 8 h p.i. for gltA and dotA; 8 h and 1 day p.i. for rrs, rpoS, CBU0311, and hcbA; 1 and 3 days p.i. for enhC; and 3 and 6 days p.i. for scvA. No carryover transcript was observed for enhC. With the exception of scvA, the highest relative expression of all genes occurred at 3 days p.i., concurrent with mid-exponential phase and robust LCV replication. At this time point, the highest relative expressions were observed for rrs and CBU0311, while the lowest relative expressions were observed for enhC, gltA, and rpoS. scvA was the only gene to show maximal activity at 8 days p.i., which was coincident with the appearance of significant numbers of SCV (Fig. 1). Moreover, the stability and abundance of scvA transcript during the growth cycle approached that of rrs. Interestingly, hcbA did not show the same expression pattern as scvA, with peak expression observed at 3 days p.i.
FIG. 4.
Relative expression levels of selected C. burnetii genes during morphological differentiation as detected by quantitative RT-PCR. Assays were performed using TaqMan primers and probes specific for each gene. Vero cell monolayers were incubated with purified SCV for 1 h to allow for adherence and internalization. Extracellular organisms were then washed from cell monolayers, and fresh medium was added. This time was designated as 0 h p.i. Total RNA was extracted at the indicated times. Transcriptional activity is expressed as relative expression, with transcript copy number normalized to the number of C. burnetii genomes present in each sample. The results are expressed as the mean from three experiments, with error bars representing the standard error of the mean.
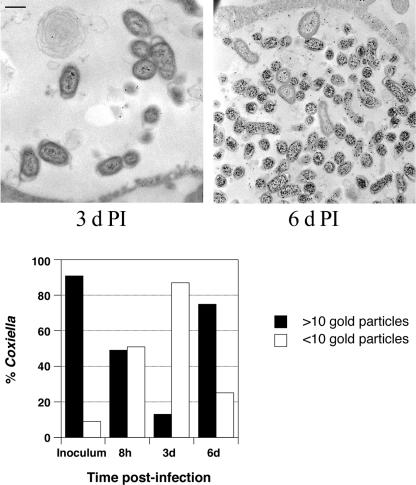
To compare temporal scvA transcription with translation, we used quantitative immunogold TEM to quantify the amount of ScvA present in cell forms during the developmental cycle. As expected, the SCV inoculum purified from aged infected cell cultures was heavily labeled for ScvA, with 91% of C. burnetii having >10 gold particles (Fig. 5). At 8 h and 3 days p.i., the percentages of C. burnetii with >10 gold particles decreased to 49 and 13%, respectively. Thus, during the transition of SCV to LCV, and the onset of LCV exponential replication, there is a coordinate degradation of ScvA without a similar decrease in the amount of scvA message. At 6 days p.i., the percentage of C. burnetii with >10 gold particles rebounded to 75%, which correlated with an increase in scvA transcription and the onset of stationary phase.
FIG. 5.
Quantification of immunogold labeling of ScvA. Vero cells were infected with SCV and processed for immunogold TEM at 8 h, 3 days, and 6 days p.i. by using rabbit polyclonal anti-ScvA serum. The purified SCV inoculum was also labeled. The relative ScvA content of individual C. burnetii organisms (at least 100 organisms per time point) was determined by counting the number of gold particles per organism on representative micrographs. Organisms were scored as having more or fewer than 10 gold particles. Representative micrographs of immunogold labeling of C. burnetii at 3 and 6 days p.i. are depicted above the graph. Bar, 0.5 μm.
DISCUSSION
In this study we describe a model to examine C. burnetii development that employs synchronous infection of Vero cells with homogeneous SCV harvested from aged infected cell cultures. The SCV morphological form is highly resistant and environmentally stable; consequently, it is likely to be responsible for most natural infections (29, 41). This infection model allows a systematic analysis of C. burnetii morphological differentiation, growth kinetics, and temporal transcription and translation.
C. burnetii exhibited a growth cycle typical of a closed bacterial system with defined lag, exponential, and stationary phases. Lag phase extended to approximately 2 days p.i. and was composed primarily of SCV-to-LCV morphogenesis. Exponential phase occurred over the next 4 days, with PVs harboring replicating LCV almost exclusively. The generation times calculated during this phase with the replating FFU assay (11.7 h) and QPCR assay (10.3 h) are in agreement with results from a previous study where a generation time of 12.4 h was estimated for C. burnetii in Vero cells by using fluorescence microscopy to quantify the content of 4′,6′-diamidino-2-phenylindole (DAPI)-stained organisms within individual PVs (43). Stationary phase began at approximately 6 days p.i., concomitantly with the appearance of SCV. C. burnetii growth cycle kinetics will presumably vary depending on the host cell and culture conditions. The QPCR method developed in this study to characterize the growth cycle is technically straightforward and should prove useful in defining developmental transitions of C. burnetii cultivated under different experimental conditions.
The replicative properties of LCV and SCV can be established when C. burnetii morphogenesis is viewed together with growth cycle phase. The notion that SCV are not replicative forms is supported by their paucity during exponential phase and their appearance in significant numbers during stationary phase. SCV apparently arise in stationary phase via condensation of the LCV through a continuum of intermediate forms and increase in number as the phase progresses without further C. burnetii replication. Infection with SCV results in reasonably synchronous morphogenesis of LCV during lag phase. The idea of LCV as the replicative form of C. burnetii is supported by their predominance in PVs during the exponential phase of growth.
There are similarities between the morphological development of C. burnetii and that of Chlamydia spp. and Legionella pneumophila. SCV and the chlamydial elementary body (EB) are both adapted for extracellular survival and have a characteristic histone-induced condensed chromatin (16, 17). Both differentiate into large, more metabolically active cell types, namely the LCV of C. burnetii and the reticulate body (RB) of Chlamydia, that are responsible for exponential growth. LCV-to-SCV condensation during stationary phase mimics chlamydial RB-to-EB differentiation. However, unlike the situation with C. burnetii, a one-step growth cycle of Chlamydia based on genome equivalents lacks a defined stationary phase because development becomes asynchronous at about 18 h p.i., with some RBs differentiating into EBs while others continue to divide by binary fission until host cell lysis (37). Also, as reported here and elsewhere (16, 17), LCV differ from RBs in being infectious. L. pneumophila alternates in HeLa cells between a replicative form and a cyst-like form termed a mature intracellular form (MIF) (12, 13). MIFs are observed only during infection and exhibit traits similar to those of broth-grown stationary-phase organisms that enhance environmental transmission, including motility and resistance to osmotic shock (5, 12, 13). Thus, to a first approximation, MIFs and replicative forms would appear to represent the functional equivalents of SCV and LCV, respectively. MIFs are also structurally similar to SCV in having laminations of intracytoplasmic membranes, but they lack a histone-containing condensed chromatin (12, 13).
The synchronous infection model described in this study allowed an initial analysis of developmentally regulated gene expression in C. burnetii. All genes tested, with the exception of scvA, demonstrated their highest expression levels during mid-exponential phase (3 days p.i.). De novo expression of scvA was evident at 3 days p.i., with expression levels increasing throughout stationary phase. Chlamydial genes expressed at high levels late in the infectious cycle demonstrate the highest level of carryover of transcript (2). This pattern is also the case with scvA, which encodes an abundant transcript that is stable through the purification of C. burnetii and into lag and early exponential phase. Unlike scvA transcript, the abundance of SCV protein directly correlates with the abundance of SCV forms. Thus, SCV-to-LCV morphogenesis is associated with degradation of ScvA, with the reciprocal process observed during LCV-to-SCV morphogenesis. The abundance of scvA transcript in the absence of significant translation suggests that this gene may be posttranscriptionally regulated. Surprisingly, hcbA, which encodes the SCV-specific histone homolog Hq1, shows an expression pattern very different from that of scvA, with peak expression occurring at 3 days p.i. Perhaps LCV harbor a supply of nontranslated hcbA transcript that, upon sensing an environmental signal for production of SCV, allows rapid production of the abundant histone-like protein and consequent condensation of chromatin.
The peak expression levels in exponential-phase C. burnetii organisms of genes encoding the porin protein P1 (CBU0311) and the alternative sigma factor RpoS (rpoS) are in agreement with published results showing differential synthesis of these proteins by LCV (35, 39). Upregulation of P1 during exponential phase is presumably an adaptation by LCV to acquire nutrients from the lysosomal milieu (39). Conversely, the peak expression level of rpoS by exponential-phase organisms presents a conundrum, as this alternative sigma factor is typically involved in the regulation of genes required for survival in stationary phase (22). It has been suggested that C. burnetii rpoS functions as a positive regulator of genes that protect the LCV from toxic lysosomal products (35). RpoS is known to upregulate catalase genes of enteric bacteria during exponential phase (22). The function of RpoS in some bacteria is complex. For example, the peak transcription level of L. pneumophila rpoS is observed during exponential phase, while protein levels are highest in post-exponential-phase organisms (1). Bachman and Swanson (1) concluded that L. pneumophila RpoS has an unusual bifunctional property, where a low level of RpoS in exponential-phase organisms downregulates expression of transmission traits (i.e., motility, osmotic resistance, infectivity for macrophages, and evasion of lysosomal targeting), while a high level of RpoS in post-exponential (stationary)-phase organisms, induced by the stringent response, upregulates expression of transmission traits. The C. burnetii genome contains genes of the stringent response pathway, including relA, spoT, gacA/S, and csrA (6, 34). C. burnetii biphasic development is likely to be regulated by a sophisticated interplay of this pathway and rpoS-regulated functions.
The C. burnetii genome contains a nearly complete copy of the L. pneumophila dot/icm genes (36), and some of these paralogs rescue the corresponding L. pneumophila type IV secretion mutant (42, 44). By analogy to the vacuolar biology of L. pneumophila and other intracellular bacteria (31), it is logical to assume that secreted C. burnetii type IV effectors mediate PV maturation. In this study we sought to associate type IV function with C. burnetii growth phase by monitoring the expression of genes encoding a structural component of the secretion apparatus (DotA) and a putative secreted effector protein (EnhC). enhC was originally described as encoding a function that enhances L. pneumophila entry into epithelial cells and monocytes (7) and subsequently shown to be translocated by type IV secretion (9). De novo expression of C. burnetii dotA occurs between 0 and 8 h p.i., with peak expression occurring at mid-exponential phase. A previous study demonstrated detectable expression of C. burnetii icmQ, icmS, icmW, and dotB at approximately 1 day p.i. by using a less sensitive agarose gel visualization of RT-PCR products (42). Interestingly, in this study de novo and peak transcription of enhC occurs during mid-exponential phase, making enhC a clear example of an LCV-specific developmentally regulated gene. Initiation of C. burnetii replication during early exponential phase coincides with the appearance of a large and spacious PV visible by light microscopy. Moreover, the spacious PV collapses and loses fusogenicity with other endocytic vacuoles upon cessation of C. burnetii protein synthesis (24). Collectively, these data suggest that C. burnetii is actively modifying its PV via type IV secretion relatively late after infection and concurrently with LCV replication. This situation is unlike that for L. pneumophila, where type IV secretion is required only coincident with and immediately following infection for establishment of a replicative PV (8, 32).
The relative expression levels of rrs and gltA, encoding 16S rRNA and citrate synthase, respectively, are approximately 10-fold higher during mid-exponential phase than during lag or stationary phases. Gene expression is often normalized against presumed “housekeeping” genes, such as rrs (2). This normalization would be inappropriate in the case of C. burnetii, as expression levels of both genes are not consistent on a per genome basis throughout the growth cycle. Nonetheless, expression levels of these genes can be viewed as a general indicator of the metabolic status of C. burnetii and further indicate that SCV are less metabolically active than LCV (23, 27, 29).
The environmental conditions that drive C. burnetii development are unknown. C. burnetii metabolism is dependent on the moderately low pH (∼5) of its PV (15). Because the pH of the PV is stable within this range over many weeks in persistently infected cells (25), it is unlikely that this condition regulates morphogenesis. Rather, development is likely to be a response to a decline in the nutritional status of the host, due in part to the parasitic burden imposed by C. burnetii growth. Heavily infected host cells that are degenerating may reduce trafficking of nutrient-laden vesicles of the endocytic and/or autophagic pathway(s) to the C. burnetii-containing vacuole (3, 21). Depletion of critical metabolites such as amino acids is known to regulate prokaryotic development such as bacterial sporulation (11) and may similarly drive development of C. burnetii to a population dominated by the SCV.
The functional relevance of C. burnetii LCV and SCV can be proposed based on their known biological properties. Evidence presented here and elsewhere (29) indicates that LCV are more metabolically and replicatively active than SCV. As such, they might play a more important role than SCV in cell-to-cell spread during acute infection, a process that may be facilitated by the display of unique LCV antigens, such as P1 (39). Perhaps the LCV are also the “secretion-competent” form of C. burnetii and thus able to translocate molecules (possibly via type IV secretion) that promote PV fusion with nutrient- and lipid-rich vesicles of the endocytic and/or autophagic pathway(s) (3, 21). Indeed, a dramatic expansion of the PV occurs concomitantly with the appearance of replicating LCV. The resistance properties of the SCV strongly implicate this form as responsible for long-term extracellular survival and aerosol transmission of C. burnetii. The organism lacks an obvious mechanism for active egress from host cells and is frequently transmitted by an aerosol of infected tissues that have slowly desiccated in the soil. In vivo, the metabolically quiescent SCV may persist in chronic Q fever infections, such as endocarditis, and contribute to the refractory nature of these infections to antibiotic therapy (26). A similar scenario has been proposed for dormant mycobacteria in latent tuberculosis (38).
The experimental model for studying C. burnetii development described in this report will allow a global characterization of the proteome and transcriptome of SCV and LCV. Characterization of the programmed gene expression associated with C. burnetii morphogenesis may reveal novel cell-form-specific proteins that account for the unique pathogenesis of this organism.
Acknowledgments
We thank the Heinzen lab, Harlan Caldwell, Ted Hackstadt, and Shelly Robertson for review of the manuscript, Amanda Bestor for technical assistance, and Gary Hettrick for graphics.
REFERENCES
- 1.Bachman, M. A., and M. S. Swanson. 2004. Genetic evidence that Legionella pneumophila RpoS modulates expression of the transmission phenotype in both the exponential phase and the stationary phase. Infect. Immun. 72:2468-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belland, R. J., G. Zhong, D. D. Crane, D. Hogan, D. Sturdevant, J. Sharma, W. L. Beatty, and H. D. Caldwell. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. USA 100:8478-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beron, W., M. G. Gutierrez, M. Rabinovitch, and M. I. Colombo. 2002. Coxiella burnetii localizes in a Rab7-labeled compartment with autophagic characteristics. Infect. Immun. 70:5816-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan, R. E., and J. E. Samuel. 2003. Evaluation of Coxiella burnetii antibiotic susceptibilities by real-time PCR assay. J. Clin. Microbiol. 41:1869-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byrne, B., and M. S. Swanson. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66:3029-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 7.Cirillo, S. L., J. Lum, and J. D. Cirillo. 2000. Identification of novel loci involved in entry by Legionella pneumophila. Microbiology 146:1345-1359. [DOI] [PubMed] [Google Scholar]
- 8.Coers, J., J. J. Monahan, and C. R. Roy. 1999. Modulation of phagosome biogenesis by Legionella pneumophila creates an organelle permissive for intracellular growth. Nat. Cell Biol. 1:183-188. [DOI] [PubMed] [Google Scholar]
- 9.Conover, G. M., I. Derre, J. P. Vogel, and R. R. Isberg. 2003. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 48:305-321. [DOI] [PubMed] [Google Scholar]
- 10.Davis, G. E., and H. R. Cox. 1938. A filter-passing infectious agent isolated from ticks. I. Isolation from Dermacentor andersonii, reactions in animals, and filtration. Public Health Rep. 53:2259-2282. [Google Scholar]
- 11.Errington, J. 1993. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol. Rev. 57:1-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faulkner, G., and R. A. Garduno. 2002. Ultrastructural analysis of differentiation in Legionella pneumophila. J. Bacteriol. 184:7025-7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garduno, R. A., E. Garduno, M. Hiltz, and P. S. Hoffman. 2002. Intracellular growth of Legionella pneumophila gives rise to a differentiated form dissimilar to stationary-phase forms. Infect. Immun. 70:6273-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hackstadt, T., R. Messer, W. Cieplak, and M. G. Peacock. 1992. Proteolytic cleavage of the 120-kilodalton outer membrane protein of rickettsiae: identification of an avirulent mutant deficient in processing. Infect. Immun. 60:159-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hackstadt, T., and J. C. Williams. 1981. Biochemical stratagem for obligate parasitism of eukaryotic cells by Coxiella burnetii. Proc. Natl. Acad. Sci. USA 78:3240-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatch, T. P. 1999. Developmental biology, p. 29-67. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. American Society for Microbiology, Washington, D.C.
- 17.Heinzen, R. A. 1997. Intracellular development of Coxiella burnetii, p. 99-129. In B. Anderson, M. Bendinelli, and H. Friedman (ed.), Rickettsial infection and immunity. Plenum Publishing Corp., New York, N.Y.
- 18.Heinzen, R. A., and T. Hackstadt. 1996. A developmental stage-specific histone H1 homolog of Coxiella burnetii. J. Bacteriol. 178:5049-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinzen, R. A., T. Hackstadt, and J. E. Samuel. 1999. Developmental biology of Coxiella burnetii. Trends Microbiol. 7:149-154. [DOI] [PubMed] [Google Scholar]
- 20.Heinzen, R. A., D. Howe, L. P. Mallavia, D. D. Rockey, and T. Hackstadt. 1996. Developmentally regulated synthesis of an unusually small, basic peptide by Coxiella burnetii. Mol. Microbiol. 22:9-19. [DOI] [PubMed] [Google Scholar]
- 21.Heinzen, R. A., M. A. Scidmore, D. D. Rockey, and T. Hackstadt. 1996. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect. Immun. 64:796-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hengge-Aronis, R. 1996. Regulation of gene expression during entry into stationary phase, p. 1497-1512. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 23.Howe, D., and L. P. Mallavia. 1999. Coxiella burnetii infection increases transferrin receptors on J774A.1 cells. Infect. Immun. 67:3236-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howe, D., J. Melnicakova, I. Barak, and R. A. Heinzen. 2003. Maturation of the Coxiella burnetii parasitophorous vacuole requires bacterial protein synthesis but not replication. Cell. Microbiol. 5:469-480. [DOI] [PubMed] [Google Scholar]
- 25.Maurin, M., A. M. Benoliel, P. Bongrand, and D. Raoult. 1992. Phagolysosomes of Coxiella burnetii-infected cell lines maintain an acidic pH during persistent infection. Infect. Immun. 60:5013-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maurin, M., and D. Raoult. 1999. Q fever. Clin. Microbiol. Rev. 12:518-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCaul, T. F. 1991. The developmental cycle of Coxiella burnetii, p. 223-258. In J. C. Williams and H. A. Thompson (ed.), Q fever: the biology of Coxiella burnetii. CRC Press, Boca Raton, Fla.
- 28.McCaul, T. F., N. Banerjee-Bhatnagar, and J. C. Williams. 1991. Antigenic differences between Coxiella burnetii cells revealed by postembedding immunoelectron microscopy and immunoblotting. Infect. Immun. 59:3243-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCaul, T. F., T. Hackstadt, and J. C. Williams. 1981. Ultrastructural and biological aspects of Coxiella burnetii under physical disruptions, p. 267-280. In W. Burgdorfer and R. L. Anacker (ed.), Rickettsiae and rickettsial diseases. Academic Press, New York, N.Y.
- 30.McCaul, T. F., and J. C. Williams. 1981. Developmental cycle of Coxiella burnetii: structure and morphogenesis of vegetative and sporogenic differentiations. J. Bacteriol. 147:1063-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagai, H., and C. R. Roy. 2003. Show me the substrates: modulation of host cell function by type IV secretion systems. Cell. Microbiol. 5:373-383. [DOI] [PubMed] [Google Scholar]
- 32.Roy, C. R., K. H. Berger, and R. R. Isberg. 1998. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol. Microbiol. 28:663-674. [DOI] [PubMed] [Google Scholar]
- 33.Seshadri, R., L. R. Hendrix, and J. E. Samuel. 1999. Differential expression of translational elements by life cycle variants of Coxiella burnetii. Infect. Immun. 67:6026-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seshadri, R., I. T. Paulsen, J. A. Eisen, T. D. Read, K. E. Nelson, W. C. Nelson, N. L. Ward, H. Tettelin, T. M. Davidsen, M. J. Beanan, R. T. Deboy, S. C. Daugherty, L. M. Brinkac, R. Madupu, R. J. Dodson, H. M. Khouri, K. H. Lee, H. A. Carty, D. Scanlan, R. A. Heinzen, H. A. Thompson, J. E. Samuel, C. M. Fraser, and J. F. Heidelberg. 2003. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl. Acad. Sci. USA 100:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seshadri, R., and J. E. Samuel. 2001. Characterization of a stress-induced alternate sigma factor, RpoS, of Coxiella burnetii and its expression during the development cycle. Infect. Immun. 69:4874-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sexton, J. A., and J. P. Vogel. 2002. Type IVB secretion by intracellular pathogens. Traffic 3:178-185. [DOI] [PubMed] [Google Scholar]
- 37.Shaw, E. I., C. A. Dooley, E. R. Fischer, M. A. Scidmore, K. A. Fields, and T. Hackstadt. 2000. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol. Microbiol. 37:913-925. [DOI] [PubMed] [Google Scholar]
- 38.Stewart, G. R., B. D. Robertson, and D. B. Young. 2003. Tuberculosis: a problem with persistence. Nat. Rev. Microbiol. 1:97-105. [DOI] [PubMed] [Google Scholar]
- 39.Varghees, S., K. Kiss, G. Frans, O. Braha, and J. E. Samuel. 2002. Cloning and porin activity of the major outer membrane protein P1 from Coxiella burnetii. Infect. Immun. 70:6741-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiebe, M. E., P. R. Burton, and D. M. Shankel. 1972. Isolation and characterization of two cell types of Coxiella burnetii phase I. J. Bacteriol. 110:368-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams, J. C. 1991. Infectivity, virulence, and pathogenicity of Coxiella burnetii for various hosts, p. 21-71. In J. C. Williams and H. A. Thompson (ed.), Q fever: the biology of Coxiella burnetii. CRC Press, Boca Raton, Fla.
- 42.Zamboni, D. S., S. McGrath, M. Rabinovitch, and C. R. Roy. 2003. Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 49:965-976. [DOI] [PubMed] [Google Scholar]
- 43.Zamboni, D. S., R. A. Mortara, and M. Rabinovitch. 2001. Infection of Vero cells with Coxiella burnetii phase II: relative intracellular bacterial load and distribution estimated by confocal laser scanning microscopy and morphometry. J. Microbiol. Methods 43:223-232. [DOI] [PubMed] [Google Scholar]
- 44.Zusman, T., G. Yerushalmi, and G. Segal. 2003. Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. Infect. Immun. 71:3714-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]