Abstract
Escherichia coli prefers growth in neutral pH environments but can withstand extremely acidic conditions (pH 2) for long periods. Of the four E. coli systems that contribute to acid resistance, one, the glutamate-dependent system, is remarkable in its efficacy and regulatory complexity. The resistance mechanism involves the intracellular consumption of protons by the glutamate decarboxylase isozymes GadA and GadB. The antiporter GadC then exports the product, γ-aminobutyric acid, in exchange for fresh glutamate. A microarray study using overexpressed regulators uncovered evgAS and ydeO as potential regulators of gadE, now known to encode the essential activator of the gadA and gadBC genes. Examination of evgA and ydeO under normal expression conditions revealed that their products do activate gadE expression but only under specific conditions. They were important during exponential growth in acidified minimal medium containing glucose but were unnecessary for gadE expression in stationary-phase cells grown in complex medium. The response regulator EvgA activates gadE directly and indirectly via induction of the AraC-like regulator ydeO. Evidence obtained using gadE-lacZ operon fusions also revealed that GadE was autoinduced. Electrophoretic mobility shift assays indicated that EvgA, YdeO, and GadE bind to different regions upstream of gadE, indicating they all act directly at the gadE promoter. Since GadE controls the expression of numerous genes besides gadA and gadBC, the relevance of these regulatory circuits extends beyond acid resistance.
Gastric acid is a formidable barrier for gastrointestinal pathogens. Penetrating this barrier requires either a massive assault by large numbers of organisms or a powerful acid resistance (AR) mechanism that allows small numbers of bacteria to survive until the stomach empties its contents into the more alkaline intestine. One remarkable microbe, the stomach pathogen Helicobacter pylori, is well regarded for its ability to survive gastric acidity. What is not widely recognized is that Escherichia coli is nearly equal to H. pylori in this respect, able to survive pH 2 for hours (12, 27).
E. coli possesses four phenotypically distinct systems of AR. AR system 1 is repressed by glucose, is evident in stationary-phase cells, and protects cells in minimal medium (pH 2.5). The other systems are not repressed by glucose and require the addition of glutamic acid (AR system 2), arginine (AR system 3), or lysine (AR system 4) in the pH 2.5 acid challenge medium (4, 11, 12). These amino acid-dependent systems utilize matched decarboxylases and antiporters to protect the cell. The decarboxylases involved are the glutamic acid decarboxylase isozymes GadA and GadB for system 2, the arginine decarboxylase AdiA for system 3, and presumably, the inducible lysine decarboxylase CadA for system 4 (4, 6, 9, 11). These enzymes all contain pyridoxyl phosphate and work by replacing the α-carboxyl groups of their amino acid substrates with a proton recruited from the cytoplasm. The end products are CO2 and γ-amino butyric acid, agmatine, and cadaverine, the end products of glutamate decarboxylase, arginine decarboxylase, and lysine decarboxylase, respectively. The cognate antiporters, GadC for glutamate, AdiC for arginine, and CadC for lysine, expel the decarboxylation product in exchange for new amino acid substrate. AR systems 2 and 3 increase intracellular pH and create a positive electrical potential inside the cell (22). AR system 4, a weaker system, has not been examined.
The most effective of these systems is the glutamate-dependent system. For a seemingly simple mechanism, glutamate-dependent AR is subject to extraordinary control. There are at least 10 regulatory proteins known to control the core gadA and gadBC loci. The gadA and gadBC genes are induced by growth under acidic conditions or by entry into stationary phase. GadE (formerly yhiE), a LuxR-family activator, is the central activator of gadA and gadBC expression (10, 13). GadE binds to a 20-bp sequence called the GAD box centered −63 bp from the transcriptional start sites of gadA and gadBC (3, 13). The other regulators form iterative control circuits designed to activate gadE expression under different growth conditions. The requirements and roles of these other regulators change with growth phase, aeration, and medium.
One circuit involves cyclic AMP receptor protein (CRP), RpoS, and two AraC-like regulators, GadX and GadW. This circuit plays a prominent role in cells grown in complex medium but also influences expression during growth in minimal medium. The two AraC-like regulators, GadX and GadW, reside downstream of gadA but are transcribed, for the most part, by independent promoters (15). GadX and GadW initially appeared to activate and repress the core gad genes, depending on the situation (13, 25). However, we now have evidence that these regulators activate gadE, and thus gadA and gadBC, during growth in complex medium but can also directly repress gadA and gadBC (S. Gong, Z. Ma, A. Sayed, and J. Foster, submitted for publication). GadX, GadW, and RpoS, the stress response alternative sigma factor, also form a regulatory loop that is influenced by cyclic AMP and CRP (14, 15, 27, 28).
Members of the second circuit were initially discovered when Masuda and Church found that overexpressing the EvgA response regulator at pH 7 resulted in highly acid-resistant cells (16). A series of gene array studies using strains that overexpressed these regulators suggested a regulatory circuit in which the EvgSA two-component regulatory system activates expression of the AraC-like regulator ydeO. YdeO then activates AR. We predicted that this activation occurs by inducing expression of the essential activator gadE and tested the proposed linear EvgA-YdeO-GadE regulatory scheme under physiological conditions. The data revealed that an EvgA regulatory circuit does activate gadE, but the circuit is used only in exponential-phase cultures growing at low pH (pH 5.5) in a minimal medium containing salts and glucose. It is not needed to activate gadE in stationary phase or during growth in a rich medium, such as Luria-Bertani medium (LB). Chromosomal knockout mutations indicate the system is a branched control circuit (diagrammatically summarized in the Discussion) where EvgA activates gadE expression directly and indirectly through a ydeO feed-forward loop and where gadE autoregulates its own expression. Consequently, the EvgA regulatory pathway is more complex than originally thought. The significance of this control extends beyond AR, since GadE, as well as EvgA and YdeO, affect multiple aspects of cell physiology (10, 17).
MATERIALS AND METHODS
Bacterial strains and growth media.
The bacterial strains and plasmids used in this study are listed in Table 1. The media used included minimal E medium containing 0.4% glucose (EG) (32), complex LB buffered with either 100 mM morpholinepropanesulfonic acid (MOPS) (pH 8) or 100 mM morpholinethanesulfonic acid (MES) (pH 5.5), LB containing 0.4% glucose (LBG), and brain heart infusion broth (BHI) containing 0.4% glucose (BHIG). When required, the following antibiotics at the following concentrations were used: carbenicillin, 100 μg/ml; ampicillin, 100 μg/ml; kanamycin, 25 μg/ml; tetracycline, 20 μg/ml; and chloramphenicol, 30 μg/ml. Where indicated, tryptophan was added at 0.3 mM, and isopropyl-β-d-thiogalactopyranoside (IPTG) was used at 0.3 or 1.5 mM, as indicated. All strains were grown at 37°C with aeration. The growth rates of all strains used were approximately equal when the strains were tested in similar growth media. The average growth rates or doubling times for these isogenic strains were 60 min (±3 min) in minimal EG at pH 7.7, 67 min (±4 min) in EG at pH 5.5, 30 min (±3 min) in BHIG, and 30 min (±3 min) in LBG. Inducing plasmid-containing strains with IPTG did not significantly affect their growth rate relative to those of non-IPTG controls.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source, reference, or construction |
|---|---|---|
| Strains | ||
| EK298 | (TE2680) λ− F− IN(rrnD-rrnE) Δ(lac)X74 rpsL galK2 recD1903::Tn10dTc trpDC::putPA1303 | 8 |
| EK432 | Δ(araD-araB)567 Δ(lacA-lacZ)515(::cat) lacIp-4000(lacIq) Δ(rhaD-araB)568 hsdR514 | 5 |
| EK551 | Δ(gadE) yhiE::Km | 29 |
| EK584 | TB1ara Δ(lac-proAB)rpsL(φ80lacZΔM15)hndR | New England Biolabs |
| EK592 | MG1655 (wild-type K-12) | 17 |
| EK593 | ΔevgA | 17 |
| EK594 | ΔevgAS | 17 |
| EK595 | ΔydeO | 17 |
| EK596 | ΔevgAS ΔydeO | 17 |
| EK616 | ΔgadE(yhiE) | 17 |
| EK619 | ΔyhiUV | 17 |
| EF1108 | K-12/pQEyhiE (gadE) | This study |
| EF1148 | K-12/pQE80L | This study |
| EF1149 | K-12/pQEevgA | This study |
| EF1151 | ΔevgA/pQEevgA | This study |
| EF1155 | ΔgadE(yhiE)::Km | EK592 × EK551 |
| EF1156 | ΔydeO/pQEevgA | EK595 × pQEevgA |
| EF1157 | ΔgadE/pQEevgA | EK1155 × pQEevgA |
| EF1164 | K-12/pQEydeO | EK592 × pQEydeO |
| EF1165 | ΔevgA/pQEydeO | EK593 × pQEydeO |
| EF1166 | ΔgadE::Km/pQEydeO | EF1155 × pQEydeO |
| EF1206 | EK298 trpDC::putPA1303-Km-gadE−804 to +28ATG-lacZ(Oc)a | EK298 × pMF537 |
| EF1207 | TB1ara Δ(lac-proAB)rpsL(φ80lacZΔM15)hndR | EK584 × pMF539 |
| EF1223 | EK298 trpDC::putPA1303-Km-evgA-lacZ(Oc) | EK298 × pMF546 |
| EF1238 | ΔevgAS ΔydeO ΔgadE::Km | EK596 × EK551 |
| EF1239 | ΔevgAS ΔydeO ΔgadE | EF1238 × pCP20 |
| EF1240 | K-12 Δlac::cat | EK592 × EK432 |
| EF1241 | Δlac::cat ΔevgAS | EK594 × EK432 |
| EF1242 | Δlac::cat ΔydeO | EK595 × EK432 |
| EF1243 | Δlac::cat ΔgadE | EK616 × EK432 |
| EF1244 | trpDC::putPA1303-Km-gadE−804 to +28ATG-lacZ(Oc) Δlac::cat | EF1240 × EF1206 |
| EF1245 | trpDC::putPA1303-Km-gadE−804 to +28ATG-lacZ(Oc) Δlac::cat ΔevgAS | EF1241 × EF1206 |
| EF1246 | trpDC::putPA1303-Km-gadE−804 to +28ATG-lacZ(Oc) Δlac::cat ΔydeO | EF1242 × EF1206 |
| EF1247 | trpDC::putPA1303-Km-gadE−804 to +28ATG-lacZ(Oc) Δlac::cat ΔgadE | EF1243 × EF1206 |
| EF1248 | Δlac::cat ΔevgAS ΔydeO ΔgadE | EF1239 × EK432 |
| EF1256 | trpDC::putPA1303-Km-gadE−804 to +28ATG-lacZ(Oc) Δlac::cat ΔevgAS ΔydeO ΔgadE | EF1248 × EF1206 |
| EF1257 | K-12 ΔevgAS ΔydeO | EK596 × EK432 |
| EF1258 | trpDC::putPA1303-Km-gadE−804 to +28ATG-lacZ(Oc) Δlac::cat ΔevgAS ΔydeO | EF1257 × EF1206 |
| EF1285 | EK298 trpDC::putPA1303-Km-gadE−804 to +331ATG-lacZ(Oc) | EK298 × pMF555 |
| EF1286 | EK298 trpDC::putPA1303-Km-gadE−360 to +331ATG-lacZ(Oc) | EK298 × pMF556 |
| EF1288 | K298 trpDC::putPA1303-Km-gadE−195 to +331ATG-lacZ(Oc) | EK298 × pMF557 |
| EF1294 | trpDC::putPA1303-Km-evgA-lacZ(Oc) Δlac::cat | EF1240 × EF1223 |
| EF1295 | trpDC::putPA1303-Km-evgA-lacZ(Oc) Δlac::cat ΔevgAS | EF1241 × EF1223 |
| EF1296 | trpDC::putPA1303-Km-evgA-lacZ(Oc) Δlac::cat ΔydeO | EF1242 × EF1223 |
| EF1297 | trpDC::putPA1303-Km-evgA-lacZ(Oc) Δlac::cat ΔgadE | EF1243 × EF1223 |
| EF1303 | trpDC::putPA1303-Km-gadE−804 to +331ATG-lacZ(Oc) | EF1240 × EF1285 |
| EF1304 | trpDC::putPA1303-Km-gadE−360 to +331ATG-lacZ(Oc) | EF1240 × EF1286 |
| EF1305 | trpDC::putPA1303-Km-gadE−195 to +331ATG-lacZ(Oc) | EF1240 × EF1288 |
| EF1348 | ΔevgAS ΔydeO/pQEyhiE (gadE) | EK595 × pQEyhiE(gadE) |
| Plasmids | ||
| pCP20 | FLP recombinase | 5 |
| pQE80L | His tag cloning vector | Qiagen |
| pRS551 | Transcriptional lacZ fusion vector | 26 |
| pQEyhiE | gadE(yhiE) ORF cloned into pQE80L, lac promoter | 17 |
| pQEevgA | evgA ORF cloned into pQE80L, lac promoter | 17 |
| pQEydeO | ydeO ORF cloned into pQE80L, lac promoter | 17 |
| pMALc2E | MBP fusion vector | New England Biolabs |
| pMF533 | pMAL2cEgadE | 13 |
| pMF539 | pMAL2cEydeO | This study |
| pMF537 | gadE-lacZ fusion−804 to +28ATG in pRS551 | This study |
| pMF546 | evgA-lacZ fusion−420 to +150ATG in pRS551 | This study |
| pMF555 | gadE-lacZ fusion−804 to +331ATG in pRS551 | This study |
| pMF556 | gadE-lacZ fusion−360 to +331ATG in pRS551 | This study |
| pMF557 | gadE-lacZ fusion−195 to +331ATG in pRS551 | This study |
| pUCevgA | evgA with native promoter in pUC19 | 17 |
Numbers reflect the extent of upstream sequence fused to lacZ.
Molecular biology techniques.
Phage P1 transduction, transformation with CaCl2, and electroporation were performed by standard methods (18). General DNA manipulations, kanamycin cassette insertions, and deletions were all performed as described earlier (5, 23). Oligonucleotide primers are listed in Table 2.
TABLE 2.
Oligonucleotide primers used in this study
| Oligo-nucleo-tide | Sequence |
|---|---|
| 377 | 5′-GGAGTTCGAAATGGACCAGAAG-3′ |
| 378 | 5′-AGTTTCGGGTGATCGCTGAG-3′ |
| 540 | 5′-CAAGTTATGATTTTTCTCATGACGAA-3′ |
| 541 | 5′-CTAAAAATAAGATGTGATACCCAG-3′ |
| 544 | 5′-CGGGATCCATGTCGCTCGTTTGTTCTGTT-3′ |
| 545 | 5′-CGCAAGCTTAAATAATCAAATAGCTAAAGC-3′ |
| 586 | 5′-GTTAAATGTTTATATTATAAAAAGTCGTTT-3′ |
| 587 | 5′-AAACGACTTTTTATAATATAAACATTTAAC-3′ |
| 598 | 5′-CTAGTGATTTCAACCTACT-3′ |
| 599 | 5′-AAGAATCTTTCGTCATGA-3′ |
| 600 | 5′-TTCATTATTTACATCCTTGTC-3′ |
| 601 | 5′-TGGCAATTGGATTGCCAGCTT-3′ |
| 609 | 5′-GCCGAATTCCAATAATTACCCCGGTTGTCAC-3′ |
| 610 | 5′-GCGGATCCAAGAATCTTTCGTCATGAGAA-3′ |
| 624 | 5′-GCCAAAAGCCCTGTAAAAGAAAAGAATC-3′ |
| 625 | 5′-CCTTGTCCGAATCGTTGTTCAATATAG-3′ |
| 626 | 5′-GTCTGGAGACACGGATATTTATGCAATG-3′ |
| 627 | 5′-CGCGAATTCAGGAATCTTACTTAGGATCAATAT-3′ |
| 633 | 5′-CGCGAATTCATTAATCTGTTCCACTATTATC-3′ |
| 634 | 5′-CCGGATCCGATGACGATATCAGGCTTAAGT-3′ |
| 661 | 5′-TTGCCAGCTTAAGTCGAAACAAGG-3′ |
| 662 | 5′-ATTCCTGGTTGTTATCAGCTTGTA-3′ |
| 663 | 5′-TGACTACGGAAAATATCAGCCAT-3′ |
| 700 | 5′-TTCCTTGCCGAATTCCAATAATTACCCCGGTTGTCAC-3′ |
| 701 | 5′-CGCACGCATGAATTCAGGAATCTTACTTAGGATCAATAT-3′ |
| 702 | 5′-TTCCTTGCCGAATTCTTGCCAGCTTAAGTCGAAACA-3′ |
| 704 | 5′-CGCCGGATTGGATCCTCTTATGGGGCAAGTGTTTAC-3′ |
| 724 | 5′-TCGGACAAGGATGTAAATAATGAAAGGATGAC-3′ |
| 725 | 5′-GTCATCCTTTCATTATTTACATCCTTGTCCGA-3′ |
| 726 | 5′-TTCTTATAGGCGTTTACTATATTGAACAACGA-3′ |
| 727 | 5′-TCGTTGTTCAATATAGTAAACGCCTATAAGAA-3′ |
| 728 | 5′-CAAACGTTAACTTTTTGTTTGCTATTTACAAGCTGA-3′ |
| 729 | 5′-TCAGCTTGTAAATAGCAAACAAAAAGTTAACGTTTG-3′ |
Western blot analysis.
Strains were grown at 37°C in media containing the required antibiotics as indicated. Genes encoding EvgA, YdeO, or GadE were cloned into the pQE80L vector and conditionally expressed by adding 1.5 mM IPTG to EG containing 100 μg of carbenicillin. At an optical density at 600 nm (OD600) of 0.4 (log phase) or 3.8 (late stationary phase), cells were collected by centrifugation and then resuspended in 0.01% sodium dodecyl sulfate (SDS) solution. Protein concentrations were determined using Bio-Rad protein assay reagent. Samples (5 μg of protein) were prepared and subjected to Western blot analysis as described earlier (13). Membranes were probed with rat anti-GAD, followed by the anti-rat secondary monoclonal antibodies coupled to peroxidase (Sigma) (diluted 1:10,000) (4). Antibody-tagged protein bands on the probed membranes were detected using an ECL Western blot detection kit (Amersham). Results were analyzed by densitometry using Scion Image software.
Northern blot analysis.
Total RNA was extracted by using the RNeasy Mini kit (Qiagen) from log-phase cell cultures (OD600 of 0.4) grown under both alkaline and acidic conditions in minimal medium containing glucose. RNA (5 μg) was separated through a denaturing formaldehyde-agarose gel (1.2% formaldehyde-agarose) and subjected to Northern blot analysis as described previously (13, 23). Membranes were probed with a 0.534-kb gadE or 0.762-kb ydeO probe generated by PCR using oligonucleotide 540 or 541 and oligonucleotide 544 or 545, respectively. Probes were labeled with [α-32P]dCTP (ICN) using DECA prime II random-priming DNA labeling kit (Ambion). Both ydeO and gadE probes correspond to the entire open reading frames (ORFs) of ydeO and gadE, respectively. For a control, the membranes were also hybridized with a 23S rRNA probe (oligonucleotide 379) end labeled with [γ-32P]ATP. Northern blot quantitations were determined by densitometry using Scion Image software.
Primer extension.
To determine the gadE transcriptional start site, six primers (oligonucleotides 599, 624, 625, 662, 626, and 663) that span the hdeD-gadE intergenic region were used for primer extension analysis. One picomole each of the primers was 5′ end labeled with [γ-32P]ATP T4 using polynucleotide kinase (Promega). Reverse transcription of gadE mRNA, isolated from pH 5.5 log-phase cells (EG medium), was performed using the Promega primer extension system. RNA (5 μg) and end-labeled primers were annealed for 20 min at 58°C, and then 10 U of avian myeloblastosis virus reverse transcriptase was added to the 20-μl reaction mixture and incubated at 42°C for 1 h. The reaction was halted by adding 20 μl of kit loading dye. Sequencing reactions were performed using the thermal Sequenase cycle sequencing kit (U.S. Biochemicals) and run in parallel with the cDNA primer extension transcripts to map the 5′ end of gadE mRNA. A PCR product produced by oligonucleotide 700 or 704, which contains the entire hdeD-gadE intergenic region and part of the gadE ORF were used as templates in the sequencing reactions.
Construction of gadE-lacZ and evgA-lacZ transcriptional fusions.
Chromosomal gadE-lacZ operon fusions were constructed by the method of Elliott (8). An 804-bp fragment containing the hdeD-gadE intergenic region plus part of the gadE ORF (positions −804 to +28 relative to the gadE start codon) was amplified using oligonucleotides 609 and 610 engineered to include EcoRI and BamHI restriction sites, respectively. The fragments were then digested and cloned into EcoRI/BamHI-digested pRS551 (Kmr Ampr), which is a lacZ operon fusion vector (26). The resulting plasmid, pMF537, was linearized by XhoI digestion and transformed into the E. coli recD strain EK298 (Cmr Kms) containing a Kms-lacZ cassette inserted into a putPA operon that was itself inserted into the trp locus of E. coli. Recombination between the plasmid and chromosome produced an Amps Cms Kmr merodiploid strain containing an intact gadE gene and a gadE-lacZ transcriptional fusion located at the putPA operon. Correct insertion of the gadE-lacZ fusion was confirmed by PCR with oligonucleotide 609 to the gadE promoter and oligonucleotide 194 for lacZ. The gadE-lacZ fusion was transduced to wild-type E. coli strains by P1 phage transduction. Additional fusions between lacZ and various fragments of the hdeD-gadE intergenic region were prepared using oligonucleotide 700 or 704 (positions −804 to +331 relative to start codon ATG), oligonucleotide 702 or 704 (positions −360 to +331), and oligonucleotide 701 or 704 (positions −195 to +331). The PCR fragments from these oligonucleotides were cloned into pRS551, resulting in plasmids pMF555, pMF556, and pMF567, respectively, which were linearized and transformed into strain EK298 as described above.
An evgA-lacZ operon fusion was also constructed. A 570-bp DNA fragment containing the evgA promoter region was generated by PCR using oligonucleotide 633 or 634 containing engineered EcoRI and BamHI restriction sites, respectively. This fragment was cloned into pRS551, creating plasmid pMF546. A chromosomal merodiploid evgA-lacZ transcriptional fusion strain was constructed as described above. β-Galactosidase activities were measured as described previously (18).
Expression and purification of MBP-GadE, MBP-YdeO, and His6-EvgA fusion proteins.
Maltose binding protein (MBP)-GadE fusion protein was purified previously (13). An MBP-YdeO fusion protein was constructed by excising ydeO from pQEydeO (17) using BamHI and HindIII and religating the fragment downstream of MalE in plasmid pMALc2E (New England Biolabs). The resulting plasmid, pMF539, was transformed into strain TB1 (New England Biolabs). MBP-YdeO fusion protein was expressed and purified as described previously for MBP-GadE (13). The protein was purified to homogeneity as determined by Coomassie blue-stained SDS-polyacrylamide gel electrophoresis (PAGE) (data not shown).
To purify EvgA, a His6-EvgA fusion vector, pQEevgA, was constructed and transformed into an evgAS mutant background as described previously (17). A culture grown overnight was diluted (1:10) into 500 ml of fresh LB medium with 100 μg of carbenicillin per ml and incubated at 37°C with shaking until reaching an OD600 of 0.5. The fusion was induced with 1.5 mM IPTG and incubated for an additional 3 h. The culture was centrifuged for 15 min at 10,000 × g, resuspended in 30 ml of lysis buffer (50 mM NaH2PO4, 300 mM NaCl,10 mM imidazole [pH 8.0]), and passed three times through a French pressure cell at 20,000 lb/in2. Cell lysates were cleared by centrifugation as described above and passed through a 0.45-μm-pore-size filter. The cleared extract was mixed with 4.5 ml of Ni-nitrilotriacetic acid agarose (Invitrogen) and gently shaken at 4°C for 1 h. The supernatant-agarose mixture was loaded into a column, washed with 65 ml of wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole [pH 8.0]). His6-EvgA protein was eluted with 15 ml of elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole [pH 8.0]) and collected in 1.0-ml fractions. Samples (about 3 ml) of the three highest protein content fractions were combined and desalted through a PD-10 Sephadex column (Amersham Pharmacia). The purity of the purified His6-EvgA was checked by 10% HCl SDS-PAGE (data not shown).
EMSA.
Electrophoretic mobility shift assays (EMSA) were used to test whether His-tagged EvgA, MBP-YdeO, and MBP-GadE proteins bound to different regions within the hdeD-gadE intergenic sequence. The entire intergenic region and three subfragments were amplified with oligonucleotide 597 or 599 (spanning nucleotides [nt] −804 to +28 relative to the translational start codon), oligonucleotide 627 or 599 (designated F1, spanning nt −195 to +28), oligonucleotide 661 or 662 (designated fragment F2, spanning nt −360 to −190), and oligonucleotide 598 or 601 (designated fragment F3, spanning nt −682 to −355). These fragments were used in binding reactions with purified regulator fusion proteins. All DNA fragments were end labeled with [γ-32P]ATP by T4 polynucleotide kinase. Radiolabeled DNA probes (5 ng) were incubated with different concentrations of His6-EvgA, MBP-YdeO, and MBP-GadE fusion proteins at room temperature for 30 min in 20 μl of binding buffer [20 mM HEPES (pH 8.0), 5 mM MgCl2, 50 mM potassium glutamate, 0.01 mM EDTA, 1 mM NaH2PO4, 20 mM NaCl, 1 mM dithiothreitol, 30 μg of bovine serum albumin per ml, 50 μg of poly(dI-dC) per ml]. Where indicated, an excess (100 ng) of specific, unlabeled DNA was added for competitive binding. Samples were loaded onto a 5% Tris-borate-EDTA (TBE) nondenaturing ready gel (Bio-Rad) and electrophoresed at room temperature in 0.5× TBE buffer with 1.2% glycerol. The gels were dried and exposed to X-Omat Kodak film at −70°C for 3 h. Each EMSA experiment was repeated in triplicate.
AR assays.
AR was tested using stationary-phase cultures (OD600 of >2.0) and exponential-phase cultures (OD600 of 0.4). Stationary-phase cells were prepared for AR system 1 by growth in LB with MES (pH 5.5) and LB with MOPS (pH 8.0) for 22 h. EG minimal medium was used to prepare cells to test AR system 2 (glutamate dependent), while cells grown in BHIG were used to test AR system 3 (arginine dependent). Extracellular glutamate is not needed to induce the glutamate-dependent system. Stationary-phase cultures were diluted 1:1,000 into prewarmed EG medium at pH 2.5 to test AR (final concentration of 2 × 106 cells/ml). Dilutions were made in unsupplemented EG medium (pH 2.5) for AR system 1, EG medium (pH 2.0) supplemented with 1.5 mM glutamate for AR system 2, and EG medium (pH 2.5) containing 1.0 mM arginine for AR system 3.
To test AR in log-phase cells, cultures (108 CFU/ml) were diluted 1:10 into prewarmed EG medium (pH 2.0), yielding a final pH of 2.5 and a final cell density of 107 CFU/ml. Viable counts were determined at time zero, 1, 2, and 4 h after acid challenge. A previous report suggesting that log-phase cells grown in minimal medium were sensitive to acid did not use a sufficiently high cell density to detect resistance (4). A more recent report has shown that log-phase cells grown at pH 5.5 can survive pH 2.5 acid challenge (2). Results are presented as the averages ± standard errors of the means for three experiments.
RESULTS
Overexpression of EvgA, YdeO, and GadE affects only glutamate-dependent AR.
A previous study found that overexpressing EvgA or YdeO in log-phase cells grown in LB will induce AR to a level equal to that of LB (pH 2.5) (17). However, E. coli contains four distinct systems of AR. The microarray data suggested that two of those systems, the glutamate decarboxylase-dependent (AR system 2) and arginine decarboxylase-dependent (AR system 3) systems, might be affected by EvgA but did not directly test this prediction (17). Consequently, we asked which of the AR systems (AR system 1, 2, or 3) was activated by overexpressing these regulators. The lysine-dependent system (AR system 4) was not examined.
Strains containing the overexpressing plasmids pQEevgA, pQEydeO, or pQEgadE(yhiE) were grown to log phase in minimal medium (pH 7.7) containing glucose, rich LB, or BHIG medium and then tested for the three AR systems. Table 3 presents the results from cells grown on minimal medium containing glucose. In each case, overexpression of the regulatory gene induced only the glutamate-dependent AR system. Similar results were found using BHIG, a medium containing cofactors needed to optimally induce arginine-dependent AR (data not shown). Thus, EvgA, YdeO, and GadE appear to affect only induction of glutamate-dependent AR.
TABLE 3.
Effects of EvgA, YdeO, and GadE overexpression on AR mechanisms
| Strain | Plasmid | % Survivala
|
||
|---|---|---|---|---|
| No addition | Glutamate | Arginine | ||
| EF1148 | pQE80L | <0.06 | <0.06 | <0.06 |
| EF1149 | pQEevgA | <0.03 | 100 ± 10 | <0.03 |
| EF1164 | pQEydeO | <0.07 | 85 ± 12 | <0.07 |
| EF1108 | pQEgadE | <0.06 | 90 ± 10 | <0.06 |
Wild-type cells containing control plasmid pQE80L or plasmids with inserts were grown overnight in EG medium at pH 7.7 with 1.5 mM IPTG and tested for AR. Cells were diluted into fresh EG medium and grown to an OD600 of 0.4. At that point the culture was diluted 1/10 into medium at pH 2.5 (final pH) with no supplement (no addition) or containing 1.5 mM glutamic acid or 1.0 mM arginine. The percent survival in EG medium at pH 2.5 4 h after the addition of glutamate or arginine is shown.
Overexpression of EvgA or YdeO increases glutamate decarboxylase production at pH 7.7 in a GadE-dependent fashion.
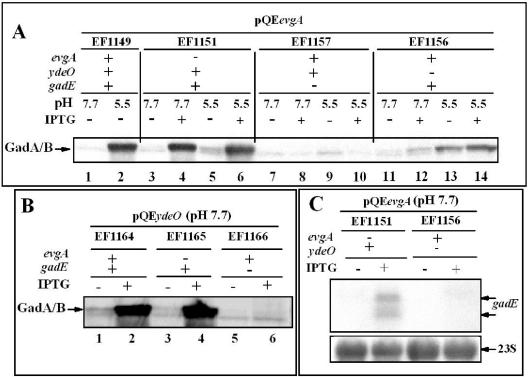
The published overexpression study found that the increase in AR caused by overexpressing EvgA was dependent on YdeO and GadE (17). Whether increased AR was the result of concomitant overexpression of glutamate decarboxylase was not tested. Glutamate decarboxylase is essential for high-level AR (4). The results of Western blot analysis presented in Fig. 1 indicate that elevating EvgA levels will increase GAD production in log-phase cells grown in minimal medium (Fig. 1A, compare strains EF1149 [lane 1] and EF1151 [lanes 3 and 4]). The results in Fig. 1A, lanes 4 and 6 versus lanes 8 and 10, illustrate that when EvgA was overexpressed, GAD production increased in a GadE-dependent fashion but was only partially YdeO-dependent (Fig. 1A, lanes 12 and 14). The latter result is the first indication that EvgA may also affect GAD induction through a YdeO-independent route (see below).
FIG. 1.
Induction of GadA or GadB by overexpressing EvgA and YdeO is dependent on GadE. Cells were grown to log phase in minimal EG medium with 1.5 mM IPTG at the pH values indicated. IPTG was added (+) to induce expression of cloned genes evgA (A) and ydeO (B). Five micrograms of protein from each extract was separated on an SDS-10% polyacrylamide gel and probed with anti-Gad antibody. (C) RNA was extracted from log-phase cells grown in minimal medium at pH 7.7. RNA (5 μg) was loaded onto 1.2% agarose-formaldehyde denaturing gels and probed for gadE mRNA. The smaller, cross-reacting band seen in these figures is not related to either GadA or GadB. The band is still observed in a gadA gadB double mutant. 23S rRNA was used as the loading standard in the lower blot.
The linear EvgA-dependent pathway, as originally proposed, proceeded from EvgA to YdeO to GadE (17). Consistent with this idea, overexpressing YdeO also increased GAD production and relied on GadE to do so (Fig. 1B, compare lanes 2 and 6). The effect was not dependent on EvgA (Fig. 1B, compare lanes 2 and 4). The Northern blot shown in Fig. 1C indicates that EvgA overexpression does induce GadE and that this induction is heavily dependent on YdeO. These results are consistent with the AR phenotypes reported earlier and argue that overexpression-dependent AR is due, at least in part, to increases in GadE, and thus, GadA, GadB, and GadC (Table 3) (16).
Under natural inducing conditions, evgA and ydeO contribute to glutamate-dependent AR additively.
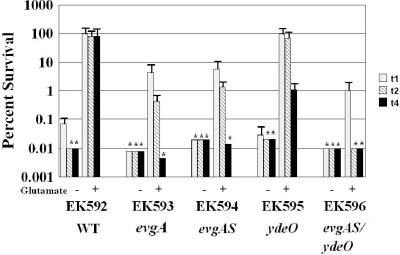
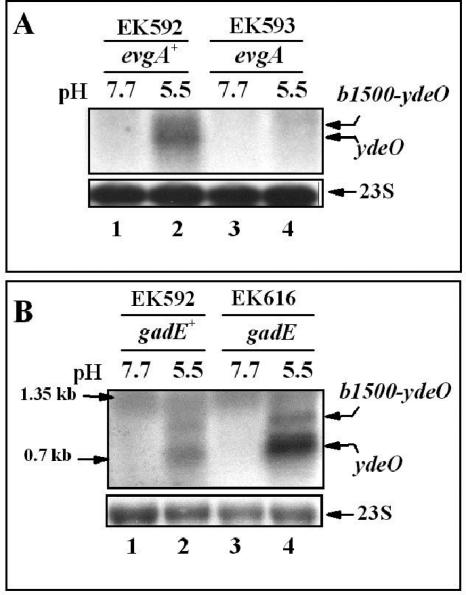
The overexpression results suggested a linear control circuit proceeding from EvgA to YdeO to GadE, which would then activate the gadA or gadBC genes (13, 17). However, as powerful as overexpression strategies are at revealing regulatory circuits, abnormally high levels of a regulatory protein could lead to inadvertent, and perhaps physiologically irrelevant, cross talk between regulatory systems (31, 33). Consequently, we tested the relevance of the proposed EvgA-YdeO-GadE circuit under conditions that do not involve overexpression. Mutants defective in these regulators were grown under log-phase and stationary-phase growth conditions that naturally lead to AR. Neither single nor double mutations in evgA and ydeO affected stationary-phase-induced glutamate-dependent AR (data not shown), but when cells were grown to log phase in minimal medium, evgA and ydeO mutants exhibited reduced AR (Fig. 2). However, in contrast to what one would predict from a linear regulatory pathway, the evgA mutation had a greater effect on AR than the ydeO mutation, suggesting that EvgA and YdeO have separable effects on AR (Fig. 2, compare EK592 to EK593 to EK595).
FIG. 2.
Effects of evgA, ydeO, and gadE mutations on glutamate-dependent AR. The effects of the individual mutations on AR are shown. Cells were grown to exponential phase in minimal EG medium at pH 5.5 and tested for AR at pH 2.5 as described in Table 3, footnote a. All cultures grown at pH 7.7 were acid sensitive (data not shown). Cells were diluted 1:10 as described in Materials and Methods. The final pH was pH 2.5. The medium either contained no additives (−) or contained glutamate as indicated (+). Asterisks indicate that the result was below the limit of detection indicated by the bar. Viable counts were determined 1, 2, and 4 h after acid challenge (t1, t2, and t4, respectively). WT, wild type.
EvgA and YdeO activate GAD expression in exponential-phase cells grown in minimal medium.
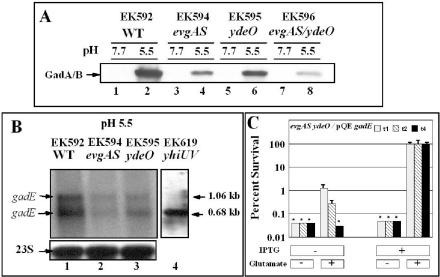
The previous result established that EvgA and YdeO activate glutamate-dependent AR under physiological conditions. We next examined whether the AR effects of evgA and ydeO mutations correlated with decreases in glutamate decarboxylase expression. The gadA and gadBC genes are normally induced at pH 5.5 in exponential-phase cultures growing in minimal medium containing glucose. Densitometric analysis of GAD Western blots revealed that GadA and GadB expression in an evgA mutant grown at pH 5.5 was four- to fivefold lower than that in the wild-type control (Fig. 3A, EK594). The ydeO mutation lowered expression only approximately twofold (Fig. 3A, EK595). Thus, EvgA has a greater effect than YdeO on gadA or gadBC expression.
FIG. 3.
Effects of evgA and ydeO on gadA and gadB expression (A), gadE expression (B), and complementation of the evgA ydeO AR phenotype by GadE (C). Cells were grown to exponential phase (OD600 of 0.4) in minimal EG medium and analyzed for GadA and GadB protein levels by Western blotting (A) as noted in the legend to Fig. 2 and for gadE mRNA by Northern blotting (B). Northern blots were performed on 5 μg of total RNA that was electrophoresed through 1.2% agarose-formaldehyde denaturing gels and probed with a 0.534-kb gadE probe. C. Complementation of the evgA ydeO acid-sensitive phenotype by GadE expression. Cells (EF1348) were grown as in panel A but with (+) or without (−) IPTG to induce the pQEgadE plasmid. The slight increase in resistance seen without glutamate is thought to be due to the effects of GadE overexpression on target genes other than gadABC that aid AR. Viable counts were determined 1, 2, and 4 h after acid challenge (t1, t2, and t4, respectively). WT, wild type.
The effects of EvgA and YdeO on GAD production also appear additive (Fig. 3A, EK596). If regulation occurred only via the linear EvgA-YdeO-GadE circuit, one would predict that GadA or GadB expression in an evgA ydeO double mutant would be no different than in an evgA single mutant. These results paralleled the AR data shown above and suggest that under physiological conditions much of the EvgA effect on GAD production occurs by a route other than through EvgA control over ydeO.
EvgA and YdeO affect transcription of gadE.
Next we tested whether EvgA and YdeO affected gadE expression under physiological conditions. The results of Northern blot analysis shown in Fig. 3B revealed that EvgA (compare lanes 1 and 2) had a greater effect on gadE transcription than YdeO (compare lanes 1 and 3). This finding was consistent with the decarboxylase Western blot (Fig. 3A) and AR (Fig. 2) results. These results also confirm that EvgA affects gadE expression in YdeO-dependent and -independent pathways.
We have, to this point, been assuming that the acid-sensitive phenotypes of evgA and ydeO mutants were due to the loss of glutamate decarboxylase. However, evgA and/or ydeO mutants might be acid sensitive because of decreased expression of other genes under their control. Since GadE is the essential activator of gadA and gadBC, we tested whether overexpression of GadE could suppress the acid-sensitive phenotype of an evgA ydeO mutant. Figure 3C illustrates that the acid-sensitive phenotype of an evgA ydeO mutant was completely reversed by overexpressing GadE. This would not happen if YdeO or EvgA activated other genes necessary for AR. Thus, the acid-sensitive phenotype of evgA and ydeO mutants appears to be directly and solely due to the loss of gadE expression and the subsequent loss of glutamate decarboxylase.
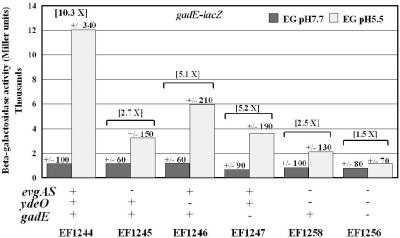
GadE is autoregulated.
A variety of gadE-lacZ operon fusion strains were constructed to confirm the Northern blot results and begin detailed studies of the gadE control region (Fig. 4). The control strain, EF1244, clearly showed that gadE expression is induced by acid (10-fold). When gadE-lacZ expression was tested in the presence of various mutations, evgA had a greater effect on expression than did ydeO (Fig. 4, compare EF1245 and EF1246). These results correlate with those of the gadE Northern and GAD Western blots shown above and indicate the fusion acts appropriately.
FIG. 4.
Effects of evgAS, ydeO, and gadE on gadE-lacZ expression. Strains containing a gadE-lacZ operon fusion (positions −804 to +28 relative to the gadE translational start) inserted at the trp operon were grown to exponential phase (OD600 of 0.4) in minimal EG medium (containing 0.3 mM tryptophan) at different pH values and assayed for β-galactosidase activity. Values are means ± standard errors of the means (error bars) for four experiments.
Data from Fig. 3 suggested that EvgA and YdeO might not be the only activators of gadE, since an evgA ydeO double mutant still exhibited acid-induced expression of gadA or gadB (Fig. 3A, lanes 7 and 8). Since the gadE promoter region contained three potential GAD boxes (see below) and because GadE binds to the known GAD box, we questioned whether gadE itself might be that other regulator. To answer this question, we used the chromosomal gadE-lacZ operon fusion inserted at the trp locus to create a gadE+/gadE-lacZ merodiploid situation. The expression of gadE-lacZ in gadE+ and gadE mutant backgrounds was then compared. The data presented in Fig. 4 indicate that GadE does help activate its own expression (Fig. 4, compare EF1244 and EF1247). LacZ production in the gadE mutant exhibited a decrease of approximately threefold from that of the wild type under acid conditions but could still be induced by acid (showing a fivefold increase). A strain in which all three regulators were mutated failed to induce gadE-lacZ at all (Fig. 4, EF1256). The accumulated data suggest that EvgA, YdeO, and GadE all contribute to gadE induction in response to acid. The results also reveal that the gadE and evgA ydeO mutations lowered pH 7.7 expression, further indication that these genes have a general role in gadE expression.
The data in Fig. 4 also indicate that the three regulators have independent effects on gadE-lacZ expression. For example, the ydeO, ydeO evgA, and ydeO evgA gadE mutants demonstrate a stepwise decrease in gadE-lacZ expression relative to the wild type. The decreases were 2-, 5-, and 12-fold, respectively (12,000 ± 300 Miller units for the wild type, 5,960 ± 150 Miller units for the ydeO mutant, 2,100 ± 190 Miller units for the ydeO evgA mutant, and 1,100 ± 70 Miller units for the ydeO evgA gadE mutant). The accumulated results indicate that each regulator affects gadE expression individually and in an additive manner.
Location of the gadE promoter.
Primer extension analysis was then used to identify the gadE transcriptional start site. The results indicate that the start site is an A positioned 21 bp upstream of the translational start codon (Fig. 5). Figure 1C and 3B revealed two gadE transcripts, one approximately 0.680 kb and the other around 1.06 kb in size. Attempts to find a second, upstream promoter failed. Six other oligonucleotides spanning the 777-bp region upstream of the identified start site failed to hybridize to either transcript. We have also demonstrated that the second transcript does not represent cross hybridization, since both transcripts disappeared in a gadE deletion mutant (13). The second, smaller transcript seen in Fig. 1C and 3B is likely the result of alternative termination or processing sites downstream of gadE (see below).
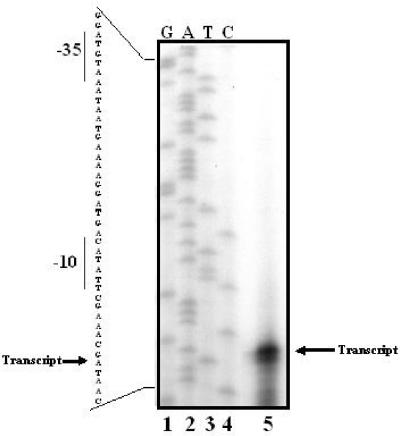
FIG. 5.
Primer extension analysis of gadE transcription. RNA was extracted from log-phase cells grown in minimal medium at pH 5.5 (OD600 of 0.4). The RNA was subjected to primer extension analysis as described in Materials and Methods. Dideoxy sequencing ladder of the fragment used for primer extension is shown in lanes 1 to 4. Primer extensions were performed on RNA extracts using labeled oligonucleotide 624. The DNA sequence using the same oligonucleotide is shown to the left. However, the sequence autoradiograph was reversed to show the sequence of the coding strand.
Hommais et al. reported two promoters for gadE located 91 and 125 bp upstream of the start codon (10). We performed primer extensions using oligonucleotides 624 and 599, which bind to gadE mRNA +18 and +5 bp from the ATG start codon, respectively. These oligonucleotides should have easily revealed any significant transcripts within at least 200 bp. Only the transcript starting at −21 bp from the presumed ATG was identified. We cannot offer an explanation for the absence of the other transcripts other than that the earlier study used an hns mutant and a plasmid to express gadE.
If the two gadE transcripts shown in Fig. 3B initiate from the same promoter but reflect different termination sites downstream of gadE, then mutations in evgA or ydeO should reduce both transcripts similarly. Figure 3B illustrates that mutations in either regulator caused parallel decreases of both gadE transcripts, although evgA had a greater effect on these transcripts than ydeO did. We also demonstrated that the larger gadE transcript disappears when the yhiUV genes, located 338 bp downstream of gadE, were deleted (EK619 [Fig. 3B]). The gadE ORF itself is 528 bp long. This suggests that termination of the larger transcript occurs within the yhiU ORF region.
EvgA, GadE, and YdeO directly bind the gadE promoter.
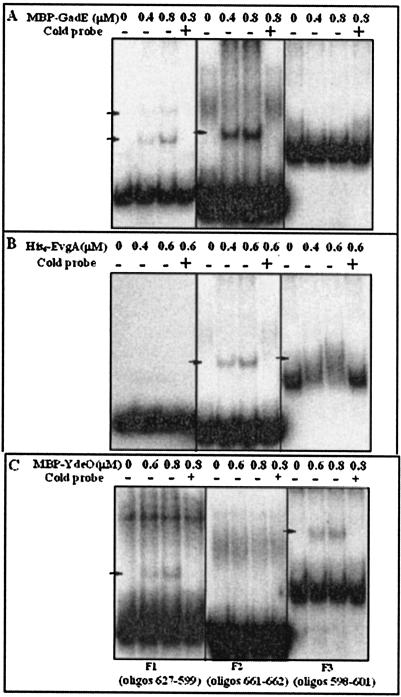
Initial EMSA experiments revealed that all three regulators bound to a 748-bp fragment representing most of the 798-bp intergenic region between hdeD and gadE (data not shown). We then divided this large fragment into three smaller fragments of 223 bp (F1), 171 bp (F2), and 373 bp (F3) as illustrated in Fig. 6. Figure 7 indicates that GadE bound to fragments F1 and F2 (Fig. 7A), EvgA bound to F2 and F3 (Fig. 7B), while YdeO bound to F1 and F3 but not to F2 (Fig. 7C). The results are consistent with all three regulators binding directly at the gadE promoter region.
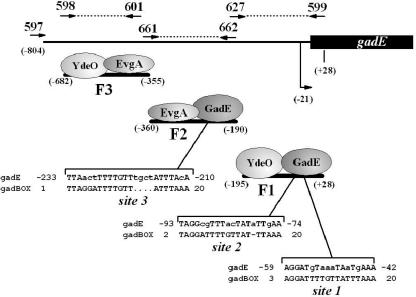
FIG. 6.
hdeD-gadE intergenic region. The primers used to generate fragments F1, F2, and F3 for EMSA are numbered. Nucleotide distances relative to the gadE translational start are noted in parentheses. The location of the gadE transcriptional start site is indicated by the bent arrow. Sequences in the gadE upstream region that exhibit homology to GAD boxes are identified in fragments 1 and 2 and are shown aligned with the 20-bp GAD box sequence; nonaligned bases are indicated by lowercase letters. EMSA results presented in Fig. 7 are summarized using ovals to represent the different purified regulators.
FIG. 7.
Electrophoretic mobility shifts of gadE promoter region fragments by EvgA, YdeO, and GadE. Fragments corresponding to those described in the legend to Fig. 6 were radiolabeled as described in Materials and Methods, and various concentrations of purified protein were added prior to electrophoresis through a 5% polyacrylamide gel. Some lanes also contained (+) unlabeled, specific competitor DNA (cold probe) as indicated. A. MBP-GadE. B. His6-EvgA. C. MBP-YdeO. Bands in the middle of lanes 5 to 8 in panels A and C were artifacts of PCR. oligos, oligonucleotides.
A 20-bp, totally conserved DNA sequence called the GAD box was previously identified upstream of the gadA and gadBC genes (3). Each is centered at −63 bp from the transcriptional start sites. The GAD boxes in both of those control regions are essential for expression of the downstream gene and bind purified GadE protein in EMSA experiments (3, 13). Until now, gadA and gadB were the only two functional GAD boxes known, although others have been predicted (30). Figure 6 illustrates that three potential GAD box sequences were found in fragments 1 and 2, but none were noted in fragment 3. This is consistent with the GadE binding results shown in Fig. 7. Fragment 1 contains two potential GAD box sequences, one site contains 14 of 20 bases (70% identity), while the other site contains 12 of the 20 GAD box bases (60% identity). Fragment 2 possesses one GAD box sequence exhibiting 80% identity to the published GAD box, but the sequence in fragment 2 is interrupted by an imperfect 4-bp repeat TGCT. Site 2 in fragment 1 is centered −63 bp from the gadE transcriptional start site, a distance equivalent to the distance of the GAD box sequences of gadA and gadB from their transcriptional start sites. Of the two other potential GAD boxes, site 1 is only 20 bp from the transcriptional start (overlapping the putative −35 site), while site 3 lies 189 bp from the transcriptional start site.
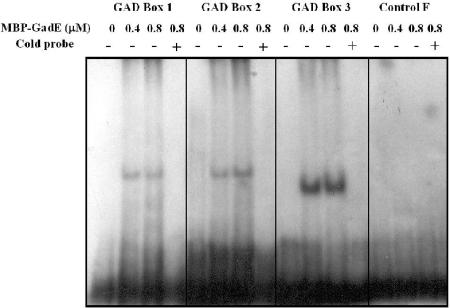
We then examined whether purified GadE would bind to any of these putative GAD box sequences in vitro. Sequences of 32 to 36 bp containing the different GAD box sites were synthesized and used in EMSA experiments with purified GadE. The results, shown in Fig. 8, reveal that GadE will bind each of the GAD box sites, but not to a fragment lacking a GAD box (fragment F).
FIG. 8.
Binding of MBP-GadE to potential GAD box sequences in the gadE control region. Sequences (32 bp) containing GAD box site 1 (oligonucleotide 724 annealed to oligonucleotide 725) and GAD box site 2 (oligonucleotide 726 annealed to oligonucleotide 727) and a 36-bp sequence containing GAD box site 3 (oligonucleotide 728 annealed to oligonucleotide 729) were made and used in EMSA experiments as described in Materials and Methods and in the legend to Fig. 7. The negative-control fragment was a sequence from the gadA promoter region made using oligonucleotides 586 and 587 and does not contain a GAD box.
To confirm a role for fragments 2 and 3 in regulating gadE, we constructed gadE-lacZ fusion strains in which one or both of these fragments was missing through deletion. The results presented in Table 4 illustrate that loss of fragment 3 (EF1304) reduced pH 5.5 expression approximately 2.5-fold (from 3,520 Miller units in strain EF1303 to 1,350 Miller units in strain EF1304). Loss of fragments 3 and 2 (strain EF1305) reduced gadE expression almost sixfold, indicating that both fragments contribute to overall GadE production. The deletions also lowered pH 7.7 expression. Interestingly, removing these upstream sequences did not eliminate acid induction of gadE, suggesting that either all three regulators contribute to pH control (Fig. 4) or another, undetermined factor regulates acid induction of gadE. Full-length fusions were expressed differently in the strains in Table 4 (EF1303) than in the strains in Fig. 4 (EF1244) because of the larger amount of gadE ORF included in the Table 4 strains.
TABLE 4.
Contribution of upstream DNA sequences to gadE-lacZ operon fusions
| Operon fusion | β-Galactosidase activitya
|
Fold induction | |
|---|---|---|---|
| EG at pH 7.7b | EG at pH 5.5 | ||
| EF1303 (−804 to +331)c | 729 ± 60 | 3,520 ± 90 | 5× |
| EF1304 (−360 to +331) | 180 ± 30 | 1,350 ± 25 | 7.5× |
| EF1305 (−195 to +331) | 210 ± 50 | 720 ± 70 | 3.5× |
β-Galactosidase activity is expressed in Miller units. Values are the means ± standard errors of the means for three experiments. For ease of construction, the fusions were made to nt +331 rather than +28, as shown in Fig. 5.
Cells were grown to exponential phase (approximately 2 × 108 cells/ml).
Numbers reflect the extent of upstream sequence fused to lacZ.
EvgA-YdeO-GadE form a bifurcated regulatory circuit with partial feedback control.
We presented evidence above that EvgA and YdeO directly activate gadE. Figure 9A illustrates, using a Northern blot, that ydeO is also acid induced and that EvgA is needed to activate ydeO expression. When the effect of GadE on ydeO expression was examined, we found that GadE exhibited partial negative control over ydeO expression (Fig. 9B, compare lanes 2 and 4). An evgA-lacZ fusion gene was then constructed to examine whether this operon was also under acid control and whether YdeO or GadE affected expression. The results indicated that evgAS is not acid induced and is not subject to autoinduction and that neither YdeO nor GadE had any major effect on evgA expression (data not shown). The fact that EvgA did not autoregulate its expression seems to contradict previous in vitro gel shift results, indicating that purified EvgA caused a shift in an upstream evgA sequence that contained a putative EvgA box (17).
FIG. 9.
Northern blot analysis of the effects of EvgA (A) and GadE (B) on ydeO expression. Cells were grown to exponential phase, and total RNA was extracted and processed for Northern blot analysis as described in the legend to Fig. 3B. Five milligrams of RNA was run per lane. 23S rRNA was used as the loading control.
DISCUSSION
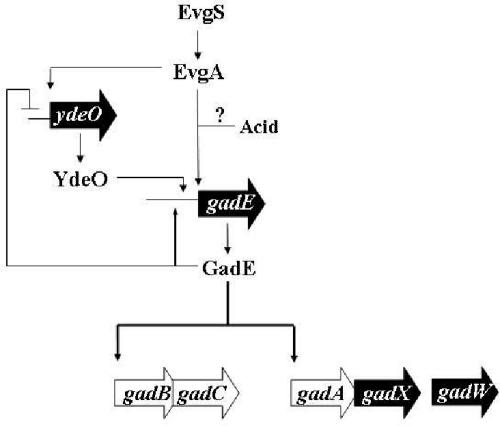
E. coli is a remarkably acid-resistant neutralophilic microorganism that prefers growth near neutral pH, but is able to withstand transient exposures to pH 2 environments for hours. This property appears designed to protect the organism as it moves through the acidic stomach toward the more hospitable pH environment of the intestine. Glutamate-dependent AR is the most efficient of the four systems that provide this protection (4, 11). The results of this and prior publications indicate that the system is regulated by multiple complex regulatory loops that allow the cell to anticipate future encounters with extreme acid stress. The Crp-RpoS-GadX-GadW loop (called the GadXW circuit) is most important in complex medium, while the EvgA-YdeO-GadE activation circuit, described here, is important in exponential-phase cells growing in minimal medium (Fig. 10).
FIG. 10.
EvgA-YdeO-GadE branched pathway regulating glutamate-dependent AR. See text for details.
The two-component regulators EvgS (sensor-kinase) and EvgA (response regulator) are highly homologous to the virulence-related BvgAS system of Bordetella pertussis. EvgAS is a two-component regulatory system with unknown function but has been the subject of several studies designed to determine its physiological role. The first hint of function was found in a spontaneous mutation that led to constitutively active EvgS and overexpression of the EmrKY multicomponent drug efflux pump (7). Subsequent microarray studies have exposed numerous other genes affected by this constitutively active mutant or by the overexpression of EvgA. Many of the genes uncovered were associated with drug resistance (7, 16, 17, 19, 21). Work by Masuda and Church first implicated EvgAS in control over AR (16, 17).
The EvgA-dependent GadE activation circuit functions in exponential-phase cells growing in minimal medium containing glucose. EvgA directly activates gadE without assistance from YdeO, and YdeO can activate gadE without EvgA. However, EvgA can also activate ydeO. The data further indicates that GadE activates itself and represses ydeO in a partial feedback loop. Thus, as GadE is produced, it will begin to shut down the YdeO activation pathway but stimulate its own synthesis. Our results verify an earlier gene array study suggesting that GadE might autoregulate expression (10).
EvgA, YdeO, and GadE proteins all demonstrated an ability to bind different fragments within the 798-bp intergenic region between hdeD and gadE. GadE binding sites were discovered in the upstream intergenic sequences shown to bind GadE (Fig. 6). However, no sites were found corresponding to a predicted EvgA binding consensus sequence, and nothing is known of potential YdeO binding sites. The EvgA consensus sequence was recently proposed on the basis of sequence comparisons between six EvgA-regulated genes (17). It is possible that the EvgA binding site is broader that expected. For example, another report found that EvgA could bind upstream of yhiU in a region that also lacks the proposed EvgA consensus site (20).
An interesting question posed by these studies is how the three regulators might collaborate to mediate control of gadE. The EMSA results indicate that each regulator is capable of binding to different pairs of three regions within the gadE promoter region. This pattern could indicate complex DNA looping arrangements, although protein-protein interactions between regulators have not yet been demonstrated. The promoter region for gadE (Fig. 6) looks like a classical positively regulated promoter with negligible −35 motif conservation and a weak −10 motif strengthened by TG immediately 5′ to the putative −10 motif (1). We predict that these three binding proteins function to facilitate RNA polymerase recruitment and/or binding to this promoter region.
It is also not apparent how the GadE activation circuit shuts off. Efficient activation likely requires interactions between the various regulators and small signal molecules that accumulate under different environmental conditions. There is evidence that, in the absence of signal, GadE protein might naturally degrade and not be replaced. For instance, in spite of what appears to be ample gadE mRNA, it has been impossible to observe native intracellular GadE levels by Western blotting. The anti-GadE antibody used easily reveals MBP-GadE in whole-cell extracts, but not native nor His6-tagged GadE, suggesting that the smaller proteins may be subject to rapid turnover (data not shown). Rapid turnover of GadE coupled with changes in coeffector concentrations as cells leave inducing environments could shut down expression.
The data also do not fully explain the acid induction of gadE. Induction is not due to pH effects on the production of EvgA on the basis of evgA-lacZ results (data not shown). It is tempting to propose that acid pH alters the phosphorylation status of EvgA, which in turn would influence the acid induction of ydeO and gadE. However, induction of gadE cannot be due solely to a potential change in EvgA activity, since gadE was induced by acid in an evgA mutant, although to a reduced level (Fig. 4). In fact, no one regulator could be linked to acid induction, although ydeO was itself induced by acid (Fig. 9A). The results suggest that these regulators either all contribute to acid induction or there is some other regulatory feature that dictates pH control. Hommais et al. suggest that the promoter regions of gadE and other genes within the proposed acid fitness island encompassing gadE have a high propensity for helix disruption that might make them easily transcribed (10). They proposed the presence of an efficient locking mechanism involving H-NS that prevents inadvertent expression during normal growth. If this is correct, the decrease in internal pH from 7.8 to around 7.4 during growth at pH 5.5 might be enough to destabilize the locking mechanism. The regulatory proteins noted here could either assist in destabilization to unlock the promoter or perform tasks subsequent to unlocking, such as communicating with RNA polymerase.
The results of the microarray study also suggested that AR caused by YdeO overexpression depended on the slp-yhiF, hdeA, and hdeD genes (17). However, under conditions commonly used to naturally induce AR, none of these genes were required (data not shown). This suggests that another pathway of AR might exist, but the actual inducing conditions remain obscure. A recent report has suggested the existence of one such pathway employing the asr product (24).
In addition to demonstrating an EvgA-dependent pathway, the results here suggest the existence of an EvgA-independent pathway that activates gadE in stationary-phase cells grown in complex medium. Clearly, the regulatory web governing glutamate-dependent AR is vast, now encompassing 10 regulators.
Why does AR require all this regulation? It is important to note that the induction of AR probably does not occur in the stomach, at least not to a great extent. We predict that induction occurs some time before ingestion (e.g., during stationary-phase growth in the intestine). Thus, one possible reason for this extensive control is that the system may be rigged to induce under many different environmental conditions that could presage an encounter with extreme acid stress, such as gastric acidity. Each growth condition might trigger a different metabolic signal recognized by different regulators within the system. This complexity must reflect the importance of surviving extreme acid stress.
The EvgA-YdeO-GadE circuit also appears to affect cell physiology beyond AR. Gene array studies examining the regulatory reach of the GadX, GadE, and EvgA regulators indicate that each regulator controls numerous genes (7, 10, 17, 19, 21, 30). Some are clearly connected to AR, others display characteristics hinting at a possible connection, while several lack any obvious relationship to acid stress survival. The broad impact these regulators have on gene expression suggests a metabolic importance that remains unexplored.
Acknowledgments
We thank Patricia Couling for help in preparing the manuscript.
This work was supported by National Institutes of Health award R01-GM61147.
REFERENCES
- 1.Barne, K. A., J. A. Bown, S. J. Busby, and S. D. Minchin. 1997. Region 2.5 of the Escherichia coli RNA polymerase σ70 subunit is responsible for the recognition of the ′extended −10′ motif at promoters. EMBO J. 16:4034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhagwat, A. A. 2003. Regulation of the glutamate-dependent acid-resistance system of diarrheagenic Escherichia coli strains. FEMS Microbiol. Lett. 227:39-45. [DOI] [PubMed] [Google Scholar]
- 3.Castanie-Cornet, M. P., and J. W. Foster. 2001. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709-715. [DOI] [PubMed] [Google Scholar]
- 4.Castanie-Cornet, M. P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Biase, D., A. Tramonti, F. Bossa, and P. Visca. 1999. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 32:1198-1211. [DOI] [PubMed] [Google Scholar]
- 7.Eguchi, Y., T. Oshima, H. Mori, R. Aono, K. Yamamoto, A. Ishihama, and R. Utsumi. 2003. Transcriptional regulation of drug efflux genes by EvgAS, a two-component system in Escherichia coli. Microbiology 149:2819-2828. [DOI] [PubMed] [Google Scholar]
- 8.Elliott, T. 1992. A method for constructing single-copy lac fusions in Salmonella typhimurium and its application to the hemA-prfA operon. J. Bacteriol. 174:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hersh, B. M., F. T. Farooq, D. N. Barstad, D. L. Blankenshorn, and J. L. Slonczewski. 1996. A glutamate-dependent acid resistance gene in Escherichia coli. J. Bacteriol. 178:3978-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hommais, F., E. Krin, J. Y. Coppee, C. Lacroix, E. Yeramian, A. Danchin, and P. Bertin. 2004. GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiology 150:61-72. [DOI] [PubMed] [Google Scholar]
- 11.Iyer, R., C. Williams, and C. Miller. 2003. Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli. J. Bacteriol. 185:6556-6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin, J., I. S. Lee, J. Frey, J. L. Slonczewski, and J. W. Foster. 1995. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 177:4097-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma, Z., S. Gong, H. Richard, D. L. Tucker, T. Conway, and J. W. Foster. 2003. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol. Microbiol. 49:1309-1320. [DOI] [PubMed] [Google Scholar]
- 14.Ma, Z., H. Richard, and J. W. Foster. 2003. pH-dependent modulation of cyclic AMP levels and GadW-dependent repression of RpoS affect synthesis of the GadX regulator and Escherichia coli acid resistance. J. Bacteriol. 185:6852-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma, Z., H. Richard, D. L. Tucker, T. Conway, and J. W. Foster. 2002. Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW). J. Bacteriol. 184:7001-7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masuda, N., and G. M. Church. 2002. Escherichia coli gene expression responsive to levels of the response regulator EvgA. J. Bacteriol. 184:6225-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masuda, N., and G. M. Church. 2003. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48:699-712. [DOI] [PubMed] [Google Scholar]
- 18.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Nishino, K., Y. Inazumi, and A. Yamaguchi. 2003. Global analysis of genes regulated by EvgA of the two-component regulatory system in Escherichia coli. J. Bacteriol. 185:2667-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishino, K., and A. Yamaguchi. 2002. EvgA of the two-component signal transduction system modulates production of the YhiUV multidrug transporter in Escherichia coli. J. Bacteriol. 184:2319-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishino, K., and A. Yamaguchi. 2001. Overexpression of the response regulator evgA of the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters. J. Bacteriol. 183:1455-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richard, H., and J. W. Foster. 2004. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J. Bacteriol. 186:6032-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Seputiene, V., D. Motiejunas, K. Suziedelis, H. Tomenius, S. Normark, O. Melefors, and E. Suziedeliene. 2003. Molecular characterization of the acid-inducible asr gene of Escherichia coli and its role in acid stress response. J. Bacteriol. 185:2475-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin, S., M. P. Castanie-Cornet, J. W. Foster, J. A. Crawford, C. Brinkley, and J. B. Kaper. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 41:1133-1150. [DOI] [PubMed] [Google Scholar]
- 26.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 27.Small, P., D. Blankenhorn, D. Welty, E. Zinser, and J. L. Slonczewski. 1994. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 176:1729-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tramonti, A., P. Visca, M. De Canio, M. Falconi, and D. De Biase. 2002. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 184:2603-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tucker, D. L., N. Tucker, and T. Conway. 2002. Gene expression profiling of the pH response in Escherichia coli. J. Bacteriol. 184:6551-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tucker, D. L., N. Tucker, Z. Ma, J. W. Foster, R. L. Miranda, P. S. Cohen, and T. Conway. 2003. Genes of the GadX-GadW regulon in Escherichia coli. J. Bacteriol. 185:3190-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verhamme, D. T., J. C. Arents, P. W. Postma, W. Crielaard, and K. J. Hellingwerf. 2002. Investigation of in vivo cross-talk between key two-component systems of Escherichia coli. Microbiology 148:69-78. [DOI] [PubMed] [Google Scholar]
- 32.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 33.Wanner, B. L. 1992. Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? J. Bacteriol. 174:2053-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]