Abstract
Platelet binding by Streptococcus gordonii strain M99 is mediated predominantly by the cell surface glycoprotein GspB. This adhesin consists of a putative N-terminal signal peptide, two serine-rich regions (SRR1 and SRR2), a basic region between SRR1 and SRR2, and a C-terminal cell wall anchoring domain. The glycosylation of GspB is mediated at least in part by Gly and Nss, which are encoded in the secY2A2 locus immediately downstream of gspB. This region also encodes two proteins (Gtf and Orf4) that are required for the expression of GspB but whose functions have not been delineated. In this study, we further characterized the roles of Gly, Nss, Gtf, and Orf4 by investigating the expression and glycosylation of a series of glutathione S-transferase-GspB fusion proteins in M99 and in gly, nss, gtf, and orf4 mutants. Compared with fusion proteins expressed in the wild-type background, fusion proteins expressed in the mutant strain backgrounds showed altered electrophoretic mobility. In addition, the fusion proteins formed insoluble aggregates in protoplasts of the gtf and orf4 mutants. Glycan detection and lectin blot analysis revealed that SRR1 and SRR2 were glycosylated but that the basic region was unmodified. When the fusion protein was expressed in Escherichia coli, glycosylation of this protein was observed only in the presence of both gtf and orf4. These results demonstrate that Gly, Nss, Gtf, and Orf4 are all involved in the intracellular glycosylation of SRRs. Moreover, Gtf and Orf4 are essential for glycosylation, which in turn is important for the solubility of GspB.
The interaction of platelets and microorganisms is thought to play a central role in the pathogenesis of infective endocarditis (6, 7, 8, 9, 11, 19, 20), and studies have indicated that platelet binding by Streptococcus gordonii strain M99 is mediated predominantly by the very large cell surface protein GspB (2). This protein consists of an N-terminal region predicted to contain an atypically long signal peptide, a serine-rich region (SRR1), a region rich in basic amino acid residues (basic region), a second serine-rich region (SRR2), and a carboxy-terminal cell wall sorting signal that includes in LPXTG motif (LPRTG; Fig. 1). GspB has been shown to be heavily glycosylated primarily with N-acetylglucosamine and glucose (1). The total carbohydrate accounts for ∼10% (wt/wt) of the GspB mass (1), although it is still not known which domains of GspB are glycosylated.
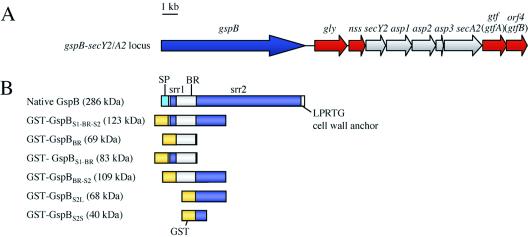
FIG. 1.
gspB-secY2A2 operon and GST-GspB fusion proteins. (A) Genetic map of the gspB-secY2A2 chromosomal region in M99. (B) Diagram of native GspB and the GST-GspB fusion proteins. The predicted molecular mass of each fusion protein is indicated in parentheses. SP, putative signal peptide; srr1, first serine-rich region; BR, basic region (the predicted pI of this region is approximately 9.4, compared with 4.0 for the entire GspB polypeptide); srr2, second serine-rich region.
Previously, it was demonstrated that a 14-kb chromosomal region (the secY2A2 locus) just downstream of the gspB structural gene is required for the expression of GspB (Fig. 1) (2). This region encodes SecY2 and SecA2, which are homologues of two highly conserved components of the general protein secretion (Sec) system. Disruption of either secY2 or secA2 results in a loss of GspB export and the accumulation of GspB in the bacterial cytoplasm, but it has no apparent effect on the transport of other proteins. These findings indicate that SecY2 and SecA2 selectively mediate the export of GspB (2). The same phenotype is also observed when any of three genes (asp1 to asp3) located between secY2 and secA2 (Fig. 1) are disrupted, indicating that Asp1, Asp2, and Asp3 are also essential components of the accessory Sec system required for GspB export (24).
In addition to these accessory Sec proteins, the secY2A2 locus also encodes four proteins (Gly, Nss, Gtf, and Orf4) that are either known or proposed to affect the glycosylation of GspB (Fig. 1A). Gly and Nss appear to be directly involved in the glycosylation of GspB. Compared with GspB purified from the parent strain, GspB isolated from gly and nss mutant strains has an altered carbohydrate composition; there is a reduction in the glucose content, and there are increases in the amounts of galactose, rhamnose, and mannose (24). In contrast, the precise roles of Gtf and Orf4 have been less clear. Mutagenesis of either gtf or orf4 resulted in an inability to detect the glycoprotein but had no effect on the transcription of gspB (24). This suggested that Gtf and Orf4 might be required either for the translation of GspB or for the stability of GspB. Because of some similarities of Gtf and Orf4 to proteins that are likely to be involved in protein glycosylation or carbohydrate metabolism, it was postulated that these proteins may contribute to the stability of GspB via glycosylation of the protein (24). However, there was no direct evidence that Gtf and Orf4 glycosylated GspB and that glycosylation contributed to the stability of the protein.
Since Gly, Nss, Gtf, and Orf4 do not have typical amino-terminal signal peptides, these proteins are predicted to function intracellularly (2, 24). Moreover, because GspB is glycosylated in protoplasts of secA2 or secY2 mutant strains, which do not export the protein, GspB appears to be glycosylated independent of export (1). However, it has not been determined whether GspB is glycosylated intracellularly, and whether Gly, Nss, Gtf, and Orf4 function intracellularly, when the accessory Sec system is fully functional.
To address these questions, we constructed a series of glutathione S-transferase (GST)-GspB fusion proteins and investigated the expression and glycosylation of the fusion proteins in strains derived from M99 that carried mutations in either gly, nss, gtf, or orf4. Our results show that Gly, Nss, Gtf, and Orf4 function intracellularly to mediate the glycosylation of the SRRs of GspB. In particular, Gtf and Orf4 are essential for glycosylation and can mediate the glycosylation of GspB in a heterologous host (Escherichia coli). Furthermore, glycosylation of GspB appears to be necessary for maintaining the protein in a soluble form.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and reagents.
The bacterial strains and plasmids used in this study are listed in Table 1. S. gordonii strain M99 was isolated from the blood of a patient with infective endocarditis (21). M99 and strains derived from it were grown in Todd-Hewitt broth (Difco Laboratories) or on sheep blood agar (Hardy Diagnostics) at 37°C in a 5% CO2 environment. If required, antibiotics were added to the media at the following concentrations: 60 μg/ml for erythromycin and 100 μg/ml for spectinomycin. E. coli strains were grown in Luria-Bertani broth or agar containing 100 μg of ampicillin per ml or 200 μg of erythromycin per ml when appropriate. Primers used in this study are listed in Table 2. Mutanolysin, streptavidin-conjugated horseradish peroxidase, nisin, isopropyl-β-d-thiogalactopyranoside (IPTG), and Dulbecco's phosphate-buffered saline (DPBS) were obtained from Sigma. Biotinylated lectins were purchased from Vector Laboratories (Burlingame, Calif.).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| S. gordonii strains | ||
| M99 | Parent strain | 21 |
| PS624 | M99 Δgly::spc; Spcr | 24 |
| PS649 | M99 Δnss::spc; Spcr | 24 |
| PS666 | M99 Δgtf::spc; Spcr | 24 |
| PS668 | M99 Δorf4::spc; Spcr | 24 |
| PS768 | M99(pGST-GspBS1-BR) | This study |
| PS769 | M99(pGST-GspBBR) | This study |
| PS771 | PS624(pGST-GspBS1-BR) | This study |
| PS772 | PS649(pGST-GspBS1-BR) | This study |
| PS773 | PS666(pGST-GspBS1-BR) | This study |
| PS774 | PS668(pGST-GspBS1-BR) | This study |
| PS780 | PS624(pGST-GspBBR) | This study |
| PS781 | PS649(pGST-GspBBR) | This study |
| PS782 | PS666(pGST-GspBBR) | This study |
| PS783 | PS668(pGST-GspBBR) | This study |
| PS806 | M99(pGST-GspBS1-BR-S2) | This study |
| PS807 | PS624(pGST-GspBS1-BR-S2) | This study |
| PS808 | PS649(pGST-GspBS1-BR-S2) | This study |
| PS809 | PS666(pGST-GspBS1-BR-S2) | This study |
| PS810 | PS668(pGST-GspBS1-BR-S2) | This study |
| PS811 | M99(pGST-GspBBR-S2) | This study |
| PS812 | PS624(pGST-GspBBR-S2) | This study |
| PS813 | PS649(pGST-GspBBR-S2) | This study |
| PS814 | PS666(pGST-GspBBR-S2) | This study |
| PS815 | PS668(pGST-GspBBR-S2) | This study |
| PS826 | M99(pGST-GspBS2S) | This study |
| PS827 | PS624(pGST-GspBS2S) | This study |
| PS828 | PS649(pGST-GspBS2S) | This study |
| PS829 | PS666(pGST-GspBS2S) | This study |
| PS830 | PS668(pGST-GspBS2S) | This study |
| PS831 | M99(pGST-GspBS2L) | This study |
| PS832 | PS624(pGST-GspBS2L) | This study |
| PS833 | PS649(pGST-GspBS2L) | This study |
| PS834 | PS666(pGST-GspBS2L) | This study |
| PS835 | PS668(pGST-GspBS2L) | This study |
| PS868 | M99(pMSP3545) | This study |
| E. coli strains | ||
| DH5α | Host for cloning vectors | 15 |
| BL21 | Host for pGEX-3X derivatives | Novagen |
| PS873 | BL21(pGEX-3X) | This study |
| PS874 | BL21(pGEX-GspBS1-BR) | This study |
| PS875 | BL21(pGEX-GspBS1-BR-G-O4) | This study |
| PS876 | BL21(pGEX-GspBS1-BR-ΔG-O4) | This study |
| PS877 | BL21(pGEX-GspBS1-BR-G-ΔO4) | This study |
| Plasmids | ||
| pGEX-3X | pBR322 replication origin, gst laclq Ptac, MCS; Ampr | Amersham Biosciences |
| pUC19 | ColE1 replication origin, lacZ′, MCS; Ampr | 26 |
| pMSP3545 | pAMβ1 and ColE1 replication origins, nisRK PnisA, MCS; Ermr | 5 |
| pGST-GspBS1-BR-S2 | pMSP3545 carrying gst and a fragment of gspB spanning SRR1, the basic region, and part of SRR2 | This study |
| pGST-GspBBR | pMSP3545 carrying gst and a fragment of gspB which spans the basic region | This study |
| pGST-GspBS1-BR | pMSP3545 carrying gst and a fragment of gspB which spans SRR1 and the basic region | This study |
| pGST-GspBBR-S2 | pMSP3545 carrying gst and a fragment of gspB which spans the basic region and part of SRR2 | This study |
| pGST-GspBS2L | pMSP3545 carrying gst and a fragment of gspB which spans part of SRR2 | This study |
| pGST-GspBS2S | pMSP3545 carrying gst and a fragment of gspB which spans part of SRR2 | This study |
| pGEX-GspBBR | pGEX-3X carrying a fragment of gspB which spans the basic region | This study |
| pGEX-GspBS1-BR | pGEX-3X carrying a fragment of gspB which spans SRR1 and the basic region | This study |
| pGEX-GspBS1-BR-G-O4 | pGEX-GspBS1-BR carrying both gtf and orf4 | This study |
| pGEX-GspBS1-BR-ΔG-O4 | pGEX-GspBS1-BR-G-O4 with an in-frame deletion of the gtf coding region | This study |
| pGEX-GspBS1-BR-G-ΔO4 | pGEX-GspBS1-BR-G-O4 with an in-frame deletion of the orf4 coding region | This study |
Spcr, spectinomycin resistant; Ampr, ampicillin resistant; Ermr, erythromycin resistant; MCS, multiple cloning site; SRR1, first serine-rich region; SRR2, second serine-rich region.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′)a | Restriction enzymeb |
|---|---|---|
| RGspBgst1 | TCTCGGATCCCTGAAGAGGAACAAGCACAT | BamHI |
| RGspBgst2 | CAACGGATCCCAGAAGCTTCTAGTCAAACA | BamHI |
| RGspBgst3 | CGAAGAATTCACACTGATAGAATTGGAAGT | EcoRI |
| RGspBgst5 | ATACGAATTCACTAGTTGACGCGGACGTGC | EcoRI |
| RGspBgst8 | ACAAGGATCCGAAGTATATCTAAATCTCAG | BamHI |
| GGB1 | CTATCCATGGCCCCTATACTAGGTTATTGG | NcoI |
| GGB2 | GGCTTCTAGACAAGCTGTGACCGTCTC | XbaI |
| GGB3 | AAAGCTGCAGGAAACAGTATTCATGTCCCC | PstI |
| GGB4 | AGGCTTTCTAGAGTCACGACGTTGTAAAAC | XbaI |
| gtfC5 | CACTGAATTCCTAAGGAGAGGTCGTAATGT | EcoRI |
| orf4C4 | ATTTGAATTCTTTAGCCATGCTGACCTCCT | EcoRI |
The underlined sequences are the restriction sites. The bold-face sequence functions as a stop codon for GST-GspBS1-BR in pGEX-GspBS1-BR-G-O4, pGEX-GspBS1-BR-ΔG-O4, and pGEX-GspBS1-BR-G-ΔO4.
Restriction enzymes that can digest the underlined sequences.
Construction of GST-GspB fusion protein expression vectors.
pMSP3545, which contains origins of replication for both gram-positive and gram-negative bacteria and a nisin-inducible promoter, was used for the expression of GST-GspB fusion proteins. Plasmids designed for the expression of GST-GspBS1-BR and GST-GspBBR were constructed in two stages. First, gspBS1-BR and gspBBR fragments were amplified from M99 chromosomal DNA by PCR by using the RGspBgst1-RGspBgst3 and RGspBgst2-RGspBgst3 primer pairs, respectively. The amplified fragments were digested with BamHI and EcoRI and ligated into the multiple cloning site of pGEX-3X, which is located just downstream of the gst coding region. The resultant gst-gspB fragments were then amplified from the pGEX-3X derivatives (pGEX-GspBS1-BR and pGEX-GspBBR) with primers GGB1 and GGB2, digested with NcoI and XbaI, and cloned into NcoI- and XbaI-digested pMSP3545, which placed the cloned gst-gspB fusion protein genes under the control of a nisin-inducible promoter.
For the expression of GST-GspBS1-BR-S2 and GST-GspBBR-S2, gspB fragments were amplified with the RGspBgst1-RGspBgst5 and RGspBgst2-RGspBgst5 primer pairs, respectively. The amplified fragments were digested with BamHI and EcoRI and ligated into the multiple cloning site of pUC19. The gst fragment was then amplified from pGEX-3X with primers GGB2 and GGB3, digested with PstI and BamHI (a BamHI restriction site is located just downstream of the gst coding region), and cloned into pUC19 just upstream of the gspB fragments. The resultant gst-gspB fragments were amplified with primers GGB1 and GGB4 from the pUC19 derivatives and cloned into pMSP3545.
In order to construct fusion proteins containing GST and portions of SRR2 (GST-GspBS2L and GST-GspBS2S), SRR2 fragments were amplified from the M99 chromosome with the RGspBgst5-RGspBgst8 primer pair. Primer RGspBgst5 can anneal at two sites in the SRR2 region (the original targeted site and another site 972 bp upstream of the original target site, which differs by three nucleotides), which results in two PCR products (approximately 1.4 and 0.4 kb). The 1.4- and 0.4-kb fragments were purified separately and used to construct plasmids for the expression of GST-GspBS2L and GST-GspBS2S, respectively, by using the strategy used for construction of plasmids for GST-GspBS1-BR-S2 and GST-GspBBR-S2.
Transformation of S. gordonii strains.
Introduction of pMSP3545 derivatives into S. gordonii strains was accomplished by natural transformation, as described previously (2).
Cloning of gtf and orf4 into pGEX-GspBS1-BR.
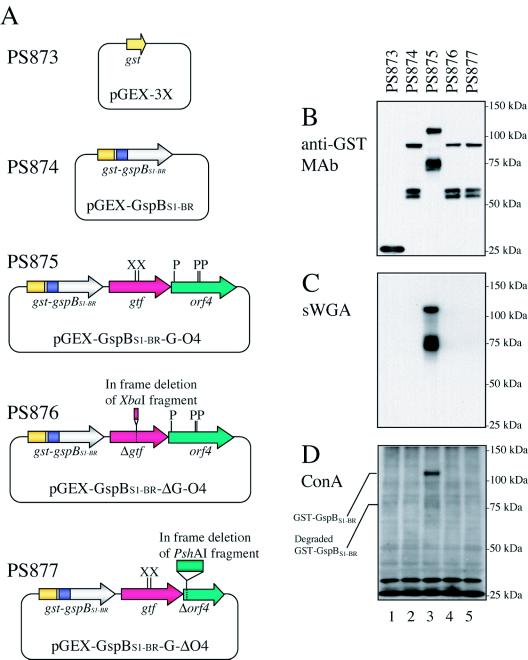
The gtf-orf4 coding region and native ribosome binding sites were amplified by PCR with primers gtfC5 and orf4C4. Primer gtfC5 includes a stop codon (Table 2) which, upon insertion of the fragment just downstream of the gst-gspBS1-BR fusion protein gene, functions as a stop signal for the translation of GST-GspBS1-BR. The amplified fragment was then digested with EcoRI and cloned into the EcoRI site of pGEX-GspBS1-BR. The direction of the cloned gene fragment was confirmed by digesting the resultant plasmid with BamHI and ClaI and by sequencing the cloned fragment. The resultant plasmid, in which the gtf-orf4 coding region was transcribed in the same direction as the gst-gspBS1-BR fusion protein gene, was designated pGEX-GspBS1-BR-G-O4 (Table 1; see Fig. 6A). In-frame deletions of gtf and orf4 were generated by removing a 66-bp XbaI fragment and a 651-bp PshAI fragment from pGEX-GspBS1-BR-G-O4, respectively. The translational frame of the deletions was confirmed by sequencing. The resultant plasmids with in-frame deletions of gtf and orf4 were designated pGEX-GspBS1-BR-ΔG-O4 and pGEX-GspBS1-BR-G-ΔO4, respectively (Table 1; see Fig. 6A).
FIG. 6.
GST-GspBS1-BR production and glycosylation in E. coli BL21. (A) Diagrams of expression vectors carried by E. coli strains. X, XbaI site; P, PshAI site. (B to D) Detection of GST-GspBS1-BR and carbohydrate linked to the fusion proteins expressed by E. coli strains. Whole-cell proteins of E. coli strains were separated by electrophoresis through 3 to 8% polyacrylamide gradient gels and then analyzed by Western blotting with the anti-GST monoclonal antibody (MAb) (A) or by lectin blotting analysis with sWGA (B) or ConA (C). Lane 1, PS873 [= BL21(pGEX-3X)] (negative control); lane 2, PS874 [= BL21(pGEX-GspBS1-BR)]; lane 3, PS875 [= BL21(pGEX-GspBS1-BR-G-O4)]; lane 4, PS876 [= BL21(pGEX-GspBS1-BR-ΔG-O4)]; lane 5, PS877 [= BL21(pGEX-GspBS1-BR-G-ΔO4)].
Analysis of fusion protein expression.
S. gordonii M99 derivatives carrying expression vectors for GST-GspB fusion proteins were grown at 37°C for 16 h and then diluted 10-fold with Todd-Hewitt broth containing erythromycin and 250 ng of nisin per ml. After incubation at 37°C for 6 to 8 h, protoplasts were prepared as described previously (24) and lysed by suspension in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and boiling for 10 min. In some experiments, protoplasts were first sonicated on ice with a model 550 Sonic Dismembrator (Fisher) as described previously (24) and then treated with DPBS or DPBS containing 6 M urea and 100 mM dithiothreitol at 4°C for 12 h before suspension in SDS-PAGE sample buffer. For analysis of protein expression in E. coli, BL21 derivatives were grown at 37°C for 16 h. The cultures were diluted 10-fold with Luria-Bertani broth containing ampicillin and incubated at 37°C for 90 min. After induction of protein expression with 0.4 mM IPTG for 1 h at 37°C, bacteria were lysed by suspension in SDS-PAGE sample buffer and boiling for 10 min. Proteins were separated by SDS-PAGE by using 3 to 8% Tris-acetate gels (Invitrogen) under reducing conditions and transferred to BioTrace NT nitrocellulose membranes (Pall Corporation) by using an XCell SureLock transfer apparatus (Invitrogen). For Western blot analysis, membranes were incubated for 16 h at 4°C in a suspension of 1× blocking reagent (Roche) in DPBS. The membranes were then incubated for 1 h with anti-GST serum (Molecular Probes), anti-recombinant GspB (rGspB) serum (1), or anti-GST monoclonal antibody (Covance) at room temperature. Antibody binding was detected by incubating the membranes for 1 h with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (anti-GST and anti-rGspB sera) (Sigma) or anti-mouse immunoglobulin G (anti-GST monoclonal antibody) (Sigma) at room temperature, followed by development with the SuperSignal West Pico chemiluminescent detection system (Pierce). Carbohydrate on the blotted proteins was detected by oxidation of the glycoproteins with periodate, followed by reaction of any resultant aldehyde groups with digoxigenin-labeled hydrazide by using a DIG-glycan detection kit as recommended by the manufacturer (Roche). The digoxigenin-labeled hydrazide was detected by using a peroxidase-conjugated anti-digoxigenin antibody (Roche).
Lectin blot analysis.
Succinylated wheat germ agglutinin (sWGA) and concanavalin A (ConA) were used for lectin blot analysis in this study. The protoplast-associated proteins of S. gordonii strains or the whole-cell proteins of E. coli strains were subjected to SDS-PAGE under reducing conditions, and the separated proteins were transferred to BioTrace NT nitrocellulose membranes. The membranes were treated with blocking buffer (10 mM Tris-HCl [pH 7.5], 0.15 M NaCl, 0.05% [vol/vol] Tween 20) for 16 h at 4°C and then incubated for 1 h at room temperature in blocking buffer containing either 0.4 μg of biotinylated sWGA per ml or 2 μg of ConA per ml. The treated membranes were washed three times with blocking buffer and probed with 0.1 μg of streptavidin-conjugated horseradish peroxidase per ml for 1 h at room temperature. The blots were then developed with the SuperSignal West Pico chemiluminescent substrate.
RESULTS
Gly and Nss function intracellularly.
Previous studies demonstrated that gly and nss contribute to the glycosylation of GspB (24). It has also been shown that GspB could be glycosylated intracellularly in secA2 and secY2 mutant strains (1). However, whether the glycosylation of GspB by Gly and Nss occurs intracellularly when the accessory Sec system is fully functional was still uncertain. To address this issue, we constructed vectors that allowed high-level expression of GST fused N terminally to various domains of GspB. The fusion proteins lacked an amino-terminal export signal and were thus anticipated to remain in the protoplasts of M99 and its derivatives. In addition, we reasoned that if Gly and Nss function intracellularly, glycosylation of the fusion proteins should be altered in protoplasts of the gly and nss mutant strains compared with protoplasts of wild-type strain M99.
We first examined whether the GST-GspBS1-BR-S2 fusion protein, which contained SRR1, the basic region, and a portion of SRR2 (Fig. 1B), could be glycosylated in a wild-type background. On a Western blot with either the anti-GST serum or the anti-rGspB serum, nothing was detected in the protoplasts of negative control strain PS868 (M99 carrying only the pMSP3545 expression vector) (Fig. 2A and B, lane 1). In contrast, the antisera reacted with a single protein in the protoplasts of PS806 (M99 carrying pGST-GspBS1-BR-S2). However, the apparent molecular mass of the fusion protein (more than 250 kDa) was far greater than the predicted mass (123 kDa) (Fig. 2A and B, lane 2), suggesting that the GST-GspBS1-BR-S2 fusion protein produced by PS806 was heavily glycosylated. To verify that GST-GspBS1-BR-S2 underwent glycosylation in PS806, we looked for the presence of carbohydrate on the fusion protein by using lectins that recognize β-N-acetylglucosamine (sWGA) and α-mannose-α-glucose (ConA), in addition to a periodate-based glycan detection assay. As shown in Fig. 3A and B (lane 2), both sWGA and ConA bound a protein among the PS806 protoplast proteins which was absent from the protoplasts of negative control strain PS868 (Fig. 3A and B, lane 1). Similarly, a single glycoprotein was detected in PS806 by the glycan detection assay (Fig. 3C, lane 2). The glycoprotein detected in PS806 either by the lectins or by the glycan detection assay had the same electrophoretic mobility as GST-GspBS1-BR-S2, as determined by Western blotting (Fig. 2). These results indicate that GST-GspBS1-BR-S2 is indeed glycosylated in the cytoplasm of PS806 and that the high apparent molecular mass of the fusion protein is likely due to linked carbohydrates. In addition, because native GspB is primarily glycosylated with N-acetylglucosamine and glucose (1), the lectin blotting results also suggest that the glycosylation of GST-GspBS1-BR-S2 is similar or identical to that of full-length GspB.
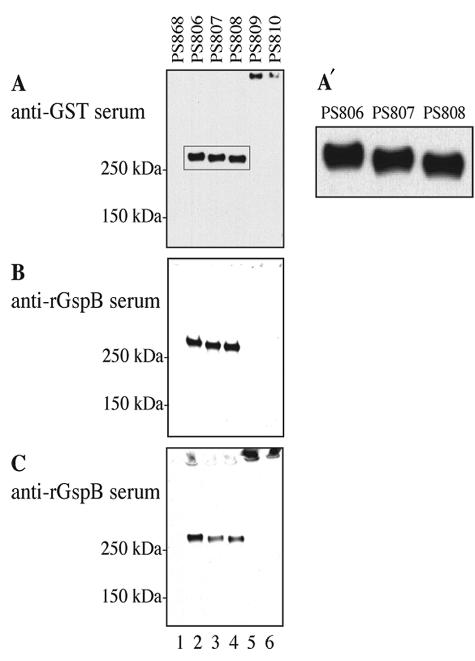
FIG. 2.
GST-GspBS1-BR-S2 production by the wild type and the gly, nss, gtf, and orf4 mutant strains. Protoplast-associated proteins were subjected to electrophoresis through 3 to 8% polyacrylamide gradient gels and Western blot analysis with anti-GST serum (A) or anti-rGspB serum (B and C). Samples in panel C were sonicated prior to SDS-PAGE. The rectangle in panel A indicates the area enlarged in panel A′. Lane 1, negative control strain PS868 (M99 with pMSP3545); lane 2, fusion protein expressed in the M99 background; lanes 3 to 6, fusion proteins expressed in the gly, nss, gtf, and orf4 mutant backgrounds, respectively. The relatively minor amounts of native GspB that may have been present in the samples in lanes 1 to 4 are not evident, since the anti-rGspB serum reacts only weakly with native, fully glycosylated GspB (1).
FIG. 3.
Detection of carbohydrate linked to GST-GspBS1-BR-S2 by lectin blot analysis with sWGA (A) or ConA (B) and by the glycan detection assay (C). Lane 1, negative control strain PS868 (M99 with pMSP3545); lane 2, fusion protein expressed in the M99 background; lanes 3 to 6, fusion proteins expressed in the gly, nss, gtf, and orf4 mutant backgrounds, respectively.
In order to determine whether Gly and Nss contribute to the intracellular glycosylation of GST-GspBS1-BR-S2, the expression of the fusion protein in gly and nss mutant strains (PS807 and PS808, respectively) was investigated. In each of these strains, expression of GST-GspBS1-BR-S2 was also detected by both anti-GST and anti-rGspB sera (Fig. 2A and B, lanes 3 and 4). However, the apparent molecular masses of the fusion proteins in the mutant strains were slightly less than that of GST-GspBS1-BR-S2 in PS806 (Fig. 2A′), suggesting that the glycosylation of GST-GspBS1-BR-S2 was altered. In order to establish that the differences in migration were not a result of any mutations that might have occurred to the expression vector, pGST-GspBS1-BR-S2 was extracted from PS807 and PS808 and reintroduced into M99. The expression of GST-GspBS1-BR-S2 was then examined. The molecular masses of GST-GspBS1-BR-S2 expressed by PS806 and rederived strains were indistinguishable (data not shown), indicating that there were no major changes in pGST-GspBS1-BR-S2 that could account for the differences in the apparent molecular masses. Therefore, the altered migration of GST-GspBS1-BR-S2 during SDS-PAGE for the gly and nss mutants appeared to be due to changes in the modification of the fusion protein in these strains.
To further examine whether the glycosylation of GST-GspBS1-BR-S2 in PS807 and PS808 was different from that in PS806, the presence of carbohydrate on the fusion proteins was examined by using the lectin blot and glycan detection assays described above. When binding of sWGA and ConA was assessed, there were no obvious differences between the GST-GspBS1-BR-S2 proteins expressed by the wild type and the mutant strains (Fig. 3A and B, lanes 2 to 4). However, the GST-GspBS1-BR-S2 produced by the mutant strains differed considerably from that produced by the parent strain when the products were examined by the glycan detection assay. As shown in Fig. 3C (lanes 3 and 4), the signal intensities of the fusion proteins produced by PS807 and PS808 were notably weaker than those of GST-GspBS1-BR-S2 produced by PS806 (Fig. 3C, lane 2). This was not due to differences in the amount of protein loaded in each lane, since comparable levels of antibody binding were seen on Western blots (Fig. 2A and B, lanes 2 to 4). These observations, in conjunction with differences in the apparent molecular masses of GST-GspBS1-BR-S2, indicate that disruption of either gly or nss affects the intracellular glycosylation of GST-GspBS1-BR-S2. It is noteworthy that the effects of Gly and Nss on the fusion protein are consistent with those reported for native GspB (24). It appears, therefore, that the GST-GspB fusion protein expression system is suitable for characterizing proteins that are involved in GspB production or modification.
Mutation of either gtf or orf4 results in the formation of GST-GspBS1-BR-S2 aggregates.
In a previous study, disruption of either gtf or orf4 was shown to abolish the expression of GspB. Since these mutations had no effect on gspB transcription, the results suggested that Gtf and Orf4 might be required either for the translation or for the stability of GspB (24). To examine these possibilities, we employed GST-GspBS1-BR-S2 to monitor the expression of the fusion protein in the gtf and orf4 mutant strains (PS809 and PS810, respectively) by Western blotting with the anti-GST and anti-rGspB sera.
In PS809 and PS810, no fusion protein was detected at either the predicted molecular mass or the apparent molecular mass of GST-GspBS1-BR-S2 produced by PS806 (Fig. 2, lanes 5 and 6). These results correlate well with the effect of gtf and orf4 mutations on native GspB expression (24). For both strains, however, a single band was detected at the top of the gel when it was probed with the anti-GST serum (Fig. 2A, lanes 5 and 6). The protein band was also detected by the anti-rGspB serum, but only after sonication of the protoplasts (Fig. 2B and C, lanes 5 and 6). Because these bands were not observed in the negative control samples (Fig. 2A to C, lane 1), the results suggest that the proteins detected at the top of the gel for PS809 and PS810 were GST-GspBS1-BR-S2 and that the fusion proteins in these strains apparently formed aggregates or nondissociable complexes. It is noteworthy that the aggregates did not dissociate after treatment with 6 M urea and 100 mM dithiothreitol (data not shown). The combined data indicate that the GST-GspBS1-BR-S2 fusion protein is expressed in the gtf and orf4 mutants. However, the loss of either Gtf or Orf4 results in the formation of GST-GspBS1-BR-S2 aggregates in the S. gordonii protoplasts.
Gtf and Orf4 are involved in the intracellular glycosylation of GspB.
It is known that glycosylation of proteins is often critical for proper protein conformation (13, 17, 25). Gtf is similar to the polyglycerol phosphate α-glucosyltransferase of Bacillus subtilis (2) and is predicted to be a glycosyltransferase. Orf4 has some similarity to Gly (24), suggesting that it may also be a glycosyltransferase. Therefore, it was possible that GST-GspBS1-BR-S2 was not glycosylated in the gtf and orf4 mutant strains, which resulted in aggregation of the fusion protein. In order to determine whether GST-GspBS1-BR-S2 underwent glycosylation in the protoplasts of these mutant strains, the presence of carbohydrate on GST-GspBS1-BR-S2 was examined by the lectin blot and glycan detection assays. As shown in Fig. 3 (lanes 5 and 6), no GST-GspBS1-BR-S2 was detected in PS809 and PS810 by either the lectin blot analysis with sWGA and ConA or the glycan detection assay. These results indicate that Gtf and Orf4 are both essential for the intracellular glycosylation of GspB, which is likely to maintain the protein in a soluble form.
Serine-rich repeat regions of GspB are targets for glycosylation by Gly, Nss, Gtf, and Orf4.
To determine the domains of GspB that become glycosylated, we examined the electrophoretic mobility and glycosylation of five different GST-GspB fusion proteins (GST-GspBBR, GST-GspBS1-BR, GST-GspBBR-S2, GST-GspBS2L, and GST-GspBS2S) produced by M99 and the mutant strains. These fusion proteins consist of an amino-terminal GST region and different combinations of GspB domains (Fig. 1B).
We first examined the expression of GST-GspBBR, in which GST was fused to the basic region of GspB (Fig. 1B), by Western blotting with the anti-GST serum. A single protein, which migrated near the predicted mass (69 kDa), was detected in the wild type (PS769) and each of the mutant backgrounds (PS780 [Δgly::spc], PS781 [Δnss::spc], PS782 [Δgtf::spc], and PS783 [Δorf4::spc]) (Fig. 4A, lanes 2 to 6). No signal was observed for the negative control sample (Fig. 4A, lane 1), indicating that the protein detected was indeed GST-GspBBR. The GST-GspBBR fusion protein produced by these strains was not detected by either the glycan detection assay or lection blot analysis with sWGA and ConA (data not shown). These results indicate that neither GST nor the basic region was glycosylated.
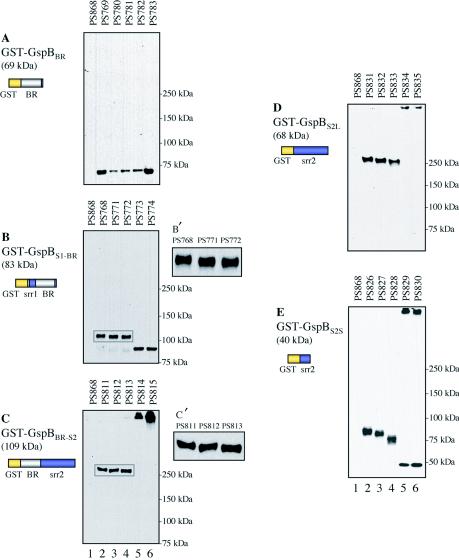
FIG. 4.
Production of GST-GspBBR (A), GST-GspBS1-BR (B), GST-GspBBR-S2 (C), GST-GspBS2L (D), and GST-GspBS2S (E) by the wild type and the gly, nss, gtf, and orf4 mutant strains. Protoplast-associated proteins were separated by electrophoresis through 3 to 8% polyacrylamide gradient gels and then subjected to Western blot analysis with the anti-GST serum. A schematic diagram of each fusion protein and its predicted molecular mass are to the left of each panel. The rectangles in panels B and C indicate areas enlarged in panels B′ and C′, respectively. Lane 1, negative control strain PS868 (M99 with pMSP3545); lane 2, fusion proteins expressed in the M99 background; lanes 3 to 6, fusion proteins expressed in the gly, nss, gtf, and orf4 mutant backgrounds, respectively. srr1, first serine-rich region; BR, basic region; srr2, second serine-rich region.
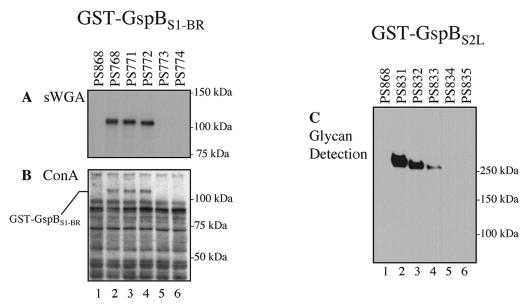
Expression of the GST-GspBS1-BR fusion protein, in which GST was fused to SRR1 and the basic region of GspB, was readily detected in each of the strains examined (Fig. 4B). However, the apparent molecular mass of GST-GspBS1-BR (∼110 kDa) in PS768 (wild-type background), PS771 (Δgly::spc), and PS772 (Δnss::spc) was greater than the predicted mass (83 kDa), and as observed with GST-GspBS1-BR-S2, the mobility of the protein differed slightly among the three strains (Fig. 4B′). In contrast, GST-GspBS1-BR expressed by PS773 (Δgtf::spc) and PS774 (Δorf4::spc) migrated closer to 83 kDa (Fig. 4B, lanes 5 and 6). It was noteworthy that minor amounts of GST-GspBS1-BR aggregates were also seen at the top of the gel only for PS773 and PS774, when the film was overexposed (data not shown). As shown above, GST and the basic regions do not undergo glycosylation in S. gordonii. Therefore, the observed differences in the mobility of GST-GspBS1-BR among the wild-type and mutant strains suggest that SRR1 is the domain targeted for glycosylation by Gly, Nss, Gtf, and Orf4. To confirm the glycosylation of SRR1 by the proteins, the presence of carbohydrate on GST-GspBS1-BR was examined by the glycan detection assay. However, no carbohydrate-containing proteins were detected in any of the strains expressing GST-GspBS1-BR (data not shown). Because it was possible that small amounts of carbohydrates associated with the fusion proteins could not be detected by this method, a lectin blot analysis was also performed with sWGA and ConA. sWGA bound GST-GspBS1-BR produced by PS768, PS771, and PS772 but not GST-GspBS1-BR produced by PS773 and PS774 (Fig. 5A). When the blot was probed with ConA, more than 10 protein bands were detected for each strain (Fig. 5B). However, a 110-kDa glycoprotein was present only in the protoplast proteins of PS768, PS771, and PS772. The combined results indicate that SRR1 is indeed glycosylated.
FIG. 5.
Detection of carbohydrate linked to GST-GspBS1-BR and GST-GspBS2L. (A and B) Protoplast-associated proteins of S. gordonii strains expressing GST-GspBS1-BR probed with sWGA (A) or ConA (B). (C) Protoplast-associated proteins of S. gordonii strains expressing GST-GspBS2L analyzed by the glycan detection assay. Lane 1, negative control strain PS868 (M99 with pMSP3545); lane 2, fusion proteins expressed in the M99 background; lanes 3 to 6, fusion proteins expressed in the gly, nss, gtf, and orf4 mutant backgrounds, respectively.
We then examined the expression and glycosylation of GST-GspBBR-S2 and GST-GspBS2L to determine whether Gly, Nss, Gtf, and Orf4 glycosylate SRR2. As shown in Fig. 1B, both of these fusion proteins lack SRR1 but contain approximately one-third of SRR2. When the fusion proteins were expressed in the wild-type or gly or nss mutant background, GST-GspBBR-S2 and GST-GspBS2L were found to migrate at an apparent molecular mass that was more than 150 kDa greater than the predicted value (Fig. 4C and D, lanes 2 to 4). Moreover, the apparent masses of the fusion proteins differed slightly among these three strains (Fig. 4C′ and D). In contrast, when produced in the gtf or orf4 mutant background, both of the fusion proteins migrated at the top of the gel (Fig. 4C and D, lanes 5 and 6), indicating that aggregates were formed.
It is noteworthy that when we analyzed the expression of GST-GspBS2S, which contains only a short stretch of SRR2 (Fig. 1B), the apparent molecular masses of the fusion proteins expressed by PS826 (wild-type background), PS827 (Δgly::spc), and PS828 (Δnss::spc) were greater than the predicted value (40 kDa). Again, the three variants migrated differently (Fig. 4E, lanes 2 to 4). In protoplast proteins of PS829 (Δgtf::spc) and PS830 (Δorf4::spc), two bands were detected for each mutant (Fig. 4E, lanes 5 and 6). One band migrated at the top of the gel, and the other migrated close to the predicted mass (40 kDa). These results suggest that even a very short subdomain of SRR2 can be glycosylated and that nonglycosylated SRRs promote aggregation of the fusion proteins.
To verify that SRR2 was glycosylated, we evaluated the GST-GspBS2L fusion protein by the glycan detection assay. As shown in Fig. 5C, GST-GspBS2L produced by PS831 (wild-type background), PS832 (Δgly::spc), and PS833 (Δnss::spc) showed a positive hydrazide reaction. In contrast, no reactivity was seen in PS834 (Δgtf::spc) or PS835 (Δorf4::spc). As observed with GST-GspBS1-BR-S2, the intensities of the signals produced by GST-GspBS2L differed for PS831, PS832, and PS833, although comparable amounts of protein were present, as indicated by Western blotting (Fig. 4D, lanes 2 to 4). The results confirm that Gly, Nss, Gtf, and Orf4 also contribute to the glycosylation of SRR2. The combined data indicate that SRR1 and SRR2, but not the basic region, are targets for glycosylation by Gly, Nss, Gtf, and Orf4 and that glycosylation of SRR2 by Gtf and Orf4 is necessary to prevent the aggregation of GspB in S. gordonii.
Gtf and Orf4 directly mediate the glycosylation of GspB.
Although Gtf and Orf4 are essential for the glycosylation of GspB in S. gordonii, it was conceivable that Gtf and Orf4 are not directly responsible for GspB glycosylation but rather regulate the expression of other proteins that modify GspB. To examine whether Gtf and Orf4 can directly glycosylate GspB, we expressed GST-GspBS1-BR in E. coli and assessed the glycosylation of the fusion protein in the absence and presence of Gtf and Orf4. We reasoned that since S. gordonii and E. coli are only distantly related, the latter species was unlikely to contain glycosyltransferases that could modify GspB. Thus, if expression of Gtf and Orf4 in E. coli resulted in the glycosylation of GspB, this would indicate that these proteins directly mediated the modification of GspB.
We first cloned both the gtf and orf4 coding regions just downstream of the gst-gspBS1-BR fusion protein gene of pGEX-GspBS1-BR, generating pGEX-GspBS1-BR-G-O4 (Table 1 and Fig. 6A). Whole-cell proteins of E. coli BL21 transformed with either plasmid were then analyzed by Western blotting by using an anti-GST monoclonal antibody. As a control, we also analyzed whole-cell proteins of BL21 transformed with pGEX-3X. As expected, GST was the only protein detected by the antibody in control strain PS873, which carried only pGEX-3X (Fig. 6B, lane 1). In the E. coli strain harboring pGEX-GspBS1-BR (PS874), the antibody reacted with three proteins (Fig. 6B, lane 2). One protein migrated near the predicted mass (83 kDa), and the others migrated with apparent molecular masses that were approximately 30 kDa smaller than the predicted values, suggesting that the largest protein was intact GST-GspBS1-BR and that the smaller proteins were degraded fusion proteins. The antibody also bound three proteins from PS875, which carried pGEX-GspBS1-BR-G-O4 (Fig. 6B, lane 3). The largest protein migrated at an apparent molecular mass that was approximately 30 kDa greater than those of the other two bands. However, the apparent molecular mass of each protein detected in PS875 was greater than that of the corresponding protein in PS874, suggesting that the full-length and degraded fusion proteins were all glycosylated in PS875.
To verify that GST-GspBS1-BR undergoes glycosylation in PS875, the presence of carbohydrate on GST-GspBS1-BR was examined by lection blot analysis. As shown in Fig. 6C (lanes 1 and 2), GST and GST-GspBS1-BR made in PS873 and PS874, respectively, were not bound by sWGA. In contrast, sWGA reacted with both intact and degraded GST-GspBS1-BR produced by PS875. When the blots were probed with ConA, more than 10 protein bands were detected for each strain (Fig. 6D, lanes 1 to 3). However, the fusion protein-specific bands were present only in the whole-cell proteins of PS875. These results indicate that Gtf and Orf4 mediate the direct glycosylation of GST-GspBS1-BR.
To determine whether both gtf and orf4 are necessary for the glycosylation of GST-GspBS1-BR in E. coli, fusion protein expression vectors pGEX-GspBS1-BR-ΔG-O4 and pGEX-GspBS1-BR-G-ΔO4, which had in-frame deletions in the gtf and orf4 coding regions, respectively, were then generated from pGEX-GspBS1-BR-G-O4 (Fig. 6A), and the effects of the deletions on the glycosylation of GST-GspBS1-BR in E. coli were examined. As shown in Fig. 6B (lanes 4 and 5), the mobility of GST-GspBS1-BR expressed by PS876 and PS877, which carried pGEX-GspBS1-BR-ΔG-O4 and pGEX-GspBS1-BR-G-ΔO4, respectively, was the same as that of the protein made in PS874 (Fig. 6B, lane 2). When the blot was probed with sWGA and ConA, no fusion protein-specific bands were detected for PS876 and PS877 (Fig. 6C and D, lanes 4 and 5). These findings indicate that the in-frame deletion in either gtf or orf4 resulted in the loss of glycosylation of the fusion proteins in E. coli. The combined results demonstrate that both Gtf and Orf4 are necessary and sufficient for the glycosylation of GST-GspBS1-BR in E. coli and that these proteins directly mediate the glycosylation of GspB in the bacterial cytoplasm.
DISCUSSION
Although a number of glycoproteins have been identified in bacterial pathogens in recent years (reviewed in references 3, 14, 16, and 22), information about the mediators of glycosylation, as well as the biological role of the associated carbohydrates, is still limited. In this report, we show that at least four proteins (Gly, Nss, Gtf, and Orf4) encoded in the gspB-secY2A2 locus are involved in the intracellular glycosylation of the S. gordonii platelet-binding glycoprotein GspB. It is noteworthy that Gtf and Orf4 are essential for the glycosylation of GspB, which is necessary for maintaining the protein in a soluble form. In view of their glycosyltransferase activity, Gtf and Orf4 have been renamed GtfA and GtfB, respectively.
As for the domains of GspB that undergo glycosylation, our data indicate that the two SRRs, but not the basic region, are targets for modification. This finding is consistent with two recent reports on Hsa (23) and Fap1 (18), the GspB homologues in S. gordonii DL1 and Streptococcus parasanguis FW213, respectively. The serine-rich regions of these proteins were also found to be glycosylated. Thus, the SRRs may be common targets for glycosylation in this family of proteins.
In both eukaryotic and prokaryotic glycoproteins, carbohydrates are typically attached to the peptide backbone either via the amide nitrogen of an Asn residue (N glycosylation) or via the hydroxyl groups of Ser or Thr residues (O glycosylation) (12). Although there are several Asn residues in the SRRs, a previous study showed that the peptide:N-glycosidase F, which can remove most N-linked oligosaccharides from eukaryotic glycoproteins, had no effect on the electrophoretic mobility of GspB derivatives (1). This suggests that little or no N-linked glycan is associated with GspB. In contrast to the small number of Asn residues, there are more than 1,000 Ser and Thr residues in the SRRs (2). It is likely, therefore, that the carbohydrate moieties associated with GspB are attached to Ser or Thr residues via an O linkage.
The specific enzymatic functions of Gly, Nss, GtfA, and GtfB are still unknown. In eukaryotes, the biosynthesis of O-linked glycan chains bound to Ser or Thr residues appears to occur almost entirely by sequential addition of monosaccharides, where the product of one glycosyltransferase is utilized as an acceptor substrate for another glycosyltransferase (4). As demonstrated in this study, GtfA and GtfB are both essential for the glycosylation of GspB. These proteins thus may contribute to the first step of O-linked glycosylation of the protein (i.e., attachment of a proximal sugar residue to Ser or Thr residues in the SRRs of the GspB polypeptide). Since GtfA has a conserved domain of known glycosyltransferases in its C-terminal region (unpublished observations), this protein may directly transfer the proximal sugar residues to the target amino acid residues. GtfB does not have conserved domains with known functions, nor is it highly similar to well-characterized proteins. However, alignment of the deduced amino acid sequences of GtfB and Gly with predicted sequences of glycosyltransferases encoded in the secY2A2 locus of other gram-positive bacteria revealed short conserved sequences within the C-terminal regions (Fig. 7). Although we cannot currently predict the function of these conserved sequences, the alignment data imply that these regions might constitute a newly recognized functional domain that is important for O-linked glycosylation of proteins in gram-positive bacteria.
FIG. 7.
Alignment of the deduced amino acid sequences of GtfB and Gly of S. gordonii with putative glycosyltransferases encoded in the secY2A2 loci of S. pneumoniae TIGR4 and S. agalactiae 2603V/R. The conserved sequences identified by BLAST searches were aligned by using CLUSTAL W (http://www.ddbj.nig.ac.jp/search/clustalw-j.html). The amino acid residues that are the same as those of S. gordonii GtfB in the alignment are indicated by red type. Amino acid residues that are not present in GtfB but appear in at least 5 of the 10 aligned proteins are indicated by blue type. The numbers indicate the positions of the amino acid residues in the proteins. Dashes indicate gaps in the aligned sequences. The accession numbers for the sequences are as follows: S. gordonii GtfB, AAS86345; S. gordonii Gly, AAK16995; S. pneumoniae SP1765, AAK75840; S. pneumoniae SP1766, AAK75841; S. pneumoniae SP1767, AAK75842; S. pneumoniae SP1770, AAK75844; S. pneumoniae SP1771, AAK75845; S. agalactiae SAG1454, AAN00323; S. agalactiae SAG1459, AAN00327; and S. agalactiae SAG1460, AAN00328.
A previous report showed that disruption of either gly or nss results in a reduced glucose content and an increase in the amount of galactose, rhamnose, and mannose associated with GspB (24). Because the N-terminal domain of Gly resembles a conserved domain of several known glycosyltransferases (unpublished observations), this protein may contribute to the addition of terminal sugar residues from an activated donor substrate to the proximal sugar residue transferred by GtfA and GtfB. Nss is similar to the E. coli nucleotide sugar synthetase (2). Therefore, it is possible that Nss may be required for generating a pool of charged nucleotide sugars (e.g., UDP-glucose) that are utilized by Gly to decorate the proximal sugar residues added by GtfA and GtfB. Determination of the precise acceptor sequences for the GspB-linked glycan chain, along with the length and structure of the glycan chain, is needed to better define the precise functions of Gly, Nss, GtfA, and GtfB in GspB glycosylation.
For most bacterial glycoproteins, it is not known whether glycosylation occurs in the cytoplasm, in the periplasm (of gram-negative species), or extracellularly. However, the HMW1C protein of Haemophilus influenzae, which is required for glycosylation of the HMW1 adhesin, appears to function in the cytoplasm (10). Similarly, the E. coli adhesins AIDA-I and TibA may also undergo glycosylation in the cytoplasm by the autotransporter adhesin heptosyltransferase and TibC heptosyltransferases, respectively (3). In contrast, N-linked glycosylation of multiple proteins in Campylobacter is thought to take place in the periplasm (22). Thus, it appears that glycosylation may occur in different cellular compartments.
A previous study indicated that GspB was likely to undergo glycosylation prior to export (1). The results presented here confirm that Gly, Nss, GtfA, and GtfB mediate the intracellular glycosylation of GspB. Glycosylation of the GST-GspB fusion proteins appears to be highly efficient, since few or no nonglycosylated forms of GST-GspB fusion proteins were seen in the wild-type background. In addition to the finding that nonglycosylated forms of GspB aggregate, the results imply that the glycosylation of this large protein may occur cotranslationally.
At present, the role of protein glycosylation in bacteria is known in only a few cases. The proposed and defined functions of carbohydrates include altered antigenicity, maintenance of protein conformation, protection against proteolytic degradation, control of enzymatic activity, and direct mediation of adhesion (reviewed in references 3, 12, 16, and 25). As shown in this study, the GST-GspB fusion proteins form aggregates, if they are not glycosylated. Therefore, the carbohydrates on native GspB may also be crucial for maintaining the protein in a soluble form in S. gordonii. In addition to a role in protein solubility, previous data indicated that the glycosylation of GspB by Gly and Nss enhances the platelet-binding activity of the protein (24). The enhanced binding may be due to optimization and stabilization of the GspB conformation by the terminal sugar residues modified by Gly and Nss. Alternatively, it is also possible that the terminal sugar residues themselves directly mediate binding.
Strains of several gram-positive bacteria, including Streptococcus pneumoniae, Streptococcus agalactiae, Staphylococcus aureus, and Staphylococcus epidermidis, also possess homologues of the gspB-secY2A2 locus, which encode a GspB-like protein and proteins predicted to be involved in the export and glycosylation of the GspB homologue (2, 24). In all of these loci, gtfA and gtfB homologues are well conserved. In contrast, the number of putative glycosyltransferase genes between the gspB-like gene and secY2 varies. No putative glycosyltransferase genes are present between the gspB-like gene and secY2 in the staphylococcal species, whereas S. pneumoniae TIGR4 and S. agalactiae 2603V/R each have one nss homologue and more than three putative glycosyltransferase genes in the locus. Although it is still not known whether the GspB-like proteins of these staphylococcal and streptococcal species are glycosylated, it is conceivable that the GtfA and GtfB homologues may also mediate the attachment of the proximal sugar residues to the nascent polypeptides of the GspB homologues. Moreover, the diversity of the putative glycosyltransferases encoded between the gspB-like gene and secY2 may contribute to the synthesis of different glycan structures through the addition of additional mono- or polysaccharides to the proximal sugar. The diverse glycan structures may produce differences in the binding properties or antigenicities of the glycoproteins among different gram-positive organisms. The heterologous protein expression and glycosylation system developed for the work presented here should be a valuable tool for assessing these possibilities in the future.
Acknowledgments
This work was supported by grants R01 AI041513 and R01 AI057433 from the National Institutes of Health and by the Department of Veterans Affairs.
The pMSP3545 shuttle vector was a gift from Gary M. Dunny. We thank Craig Rubens, Ian Siboo, and Julie Higashi for their helpful scientific and editorial suggestions.
REFERENCES
- 1.Bensing, B. A., B. W. Gibson, and P. M. Sullam. 2004. The Streptococcus gordonii platelet binding protein GspB undergoes glycosylation independently of export. J. Bacteriol. 186:638-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bensing, B. A., and P. M. Sullam. 2002. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol. Microbiol. 44:1081-1094. [DOI] [PubMed] [Google Scholar]
- 3.Benz, I., and M. A. Schmidt. 2002. Never say never again: protein glycosylation in pathogenic bacteria. Mol. Microbiol. 45:267-276. [DOI] [PubMed] [Google Scholar]
- 4.Berger, E. G., E. Buddecke, J. P. Kamerling, A. Kobata, J. C. Paulson, and J. F. Vliegenthart. 1982. Structure, biosynthesis and functions of glycoprotein glycans. Experientia 38:1129-1162. [DOI] [PubMed] [Google Scholar]
- 5.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183-190. [DOI] [PubMed] [Google Scholar]
- 6.Durack, D. T. 1975. Experimental bacterial endocarditis. IV. Structure and evolution of very early lesions. J. Pathol. 115:81-89. [DOI] [PubMed] [Google Scholar]
- 7.Durack, D. T., and P. B. Beeson. 1972. Experimental bacterial endocarditis. I. Colonization of a sterile vegetation. Br. J. Exp. Pathol. 53:44-49. [PMC free article] [PubMed] [Google Scholar]
- 8.Ferguson, D. J., A. A. McColm, D. M. Ryan, and P. Acred. 1986. Experimental staphylococcal endocarditis and aortitis. Morphology of the initial colonization. Virchows Arch. A Pathol. Anat. Histopathol. 410:43-48. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson, D. J., A. A. McColm, T. J. Savage, D. M. Ryan, and P. Acred. 1986. A morphological study of experimental rabbit staphylococcal endocarditis and aortitis. I. Formation and effect of infected and uninfected vegetations on the aorta. Br. J. Exp. Pathol. 67:667-678. [PMC free article] [PubMed] [Google Scholar]
- 10.Grass, S., A. Z. Buscher, W. E. Swords, M. A. Apicella, S. J. Barenkamp, N. Ozchlewski, and J. W. St Geme III. 2003. The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis. Mol. Microbiol. 48:737-751. [DOI] [PubMed] [Google Scholar]
- 11.McGowan, D. A., and R. Gillett. 1980. Scanning electron microscopic observations of the surface of the initial lesion in experimental streptococcal endocarditis in the rabbit. Br. J. Exp. Pathol. 61:164-171. [PMC free article] [PubMed] [Google Scholar]
- 12.Moens, S., and J. Vanderleyden. 1997. Glycoproteins in prokaryotes. Arch. Microbiol. 168:169-175. [DOI] [PubMed] [Google Scholar]
- 13.Olden, K., B. A. Bernard, M. J. Humphries, T.-K. Yeo, K.-T. Yeo, S. L. White, S. A. Newton, H. C. Bauer, and J. B. Parent. 1985. Function of glycoprotein glycans. Trends Biochem. Sci. 10:78-82. [Google Scholar]
- 14.Power, P. M., and M. P. Jennings. 2003. The genetics of glycosylation in Gram-negative bacteria. FEMS Microbiol. Lett. 218:211-222. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Schmidt, M. A., L. W. Riley, and I. Benz. 2003. Sweet new world: glycoproteins in bacterial pathogens. Trends Microbiol. 11:554-561. [DOI] [PubMed] [Google Scholar]
- 17.Sharon, N. 1984. Glycoproteins. Trends Biochem. Sci. 9:198-202. [Google Scholar]
- 18.Stephenson, A. E., H. Wu, J. Novak, M. Tomana, K. Mintz, and P. Fives-Taylor. 2002. The Fap1 fimbrial adhesin is a glycoprotein: antibodies specific for the glycan moiety block the adhesion of Streptococcus parasanguis in an in vitro tooth model. Mol. Microbiol. 43:147-157. [DOI] [PubMed] [Google Scholar]
- 19.Sullam, P. M. 1994. Host-pathogen interactions in the development of bacterial endocarditis. Curr. Opin. Infect. Dis. 7:304-309. [Google Scholar]
- 20.Sullam, P. M., U. Frank, M. R. Yeaman, M. G. Tauber, A. S. Bayer, and H. F. Chambers. 1993. Effect of thrombocytopenia on the early course of streptococcal endocarditis. J. Infect. Dis. 168:910-914. [DOI] [PubMed] [Google Scholar]
- 21.Sullam, P. M., F. H. Valone, and J. Mills. 1987. Mechanisms of platelet aggregation by viridans group streptococci. Infect. Immun. 55:1743-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szymanski, C. M., S. M. Logan, D. Linton, and B. W. Wren. 2003. Campylobactor—a tale of two protein glycosylation systems. Trends Microbiol. 11:233-238. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi, Y., A. Yajima, J. O. Cisar, and K. Konishi. 2004. Functional analysis of the Streptococcus gordonii DL1 sialic acid-binding adhesin and its essential role in bacterial binding to platelets. Infect. Immun. 72:3876-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takamatsu, D., B. A. Bensing, and P. M. Sullam. 2004. Genes in the accessory sec locus of Streptococcus gordonii have three functionally distinct effects on the expression of the platelet-binding protein GspB. Mol. Microbiol. 52:189-203. [DOI] [PubMed] [Google Scholar]
- 25.Upreti, R. K., M. Kumar, and V. Shankar. 2003. Bacterial glycoproteins: functions, biosynthesis and applications. Proteomics 3:363-379. [DOI] [PubMed] [Google Scholar]
- 26.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]