Abstract
The three-dimensional (3D) architecture of the cell nucleus plays an important role in protein dynamics and in regulating gene expression. However, protein dynamics within the 3D nucleus are poorly understood. Here, we present, to our knowledge, a novel combination of 1) single-objective based light-sheet microscopy, 2) photoconvertible proteins, and 3) fluorescence correlation microscopy, to quantitatively measure 3D protein dynamics in the nucleus. We are able to acquire >3400 autocorrelation functions at multiple spatial positions within a nucleus, without significant photobleaching, allowing us to make reliable estimates of diffusion dynamics. Using this tool, we demonstrate spatial heterogeneity in Polymerase II dynamics in live U2OS cells. Further, we provide detailed measurements of human-Yes-associated protein diffusion dynamics in a human gastric cancer epithelial cell line.
Introduction
The cell nucleus is composed of a dense three-dimensional (3D) architecture, with specialized regions essential for gene expression and regulation, and transcriptional regulators are in continual dynamic exchange between different compartments (1). Many key processes depend on the physical dimensions and spatial organization of the nucleus (2), therefore, quantifying the spatio-temporal protein dynamics in the full 3D nucleus could provide useful information on macromolecule behavior and nuclear organization (3). At present, microscopy methods for quantitative measurement of protein dynamics have restricted sensitivity due to high background and/or potential damage to the live cell from high photon loads. These limitations inhibit an understanding of dynamics in the inherently heterogeneous 3D live cell.
Fluorescence correlation spectroscopy (FCS) is a quantitative method for measuring protein dynamics and binding in live single cells and in vivo (4, 5). Recently, there have been two important advances in FCS technology: first, it has been extended from single point to multiplex imaging-FCS via use of light-sheet microscopy (SPIM) (6, 7, 8, 9), allowing simultaneous, spatially extended dynamic measurements; and second, the adoption of photoactivatable fluorophores has optimized the relative number of particles in the observation volume, significantly improving the quality of the autocorrelation functions (ACFs), which are central to analysis of FCS data ((5, 10), and see the Supporting Material). Furthermore, camera improvements (in both speed and sensitivity) have enabled faster and longer image acquisition times (8, 9), and these longer intensity traces can be autocorrelated temporally and spatially to create contiguous maps of diffusivity, concentration, and binding in two dimensions (8, 11, 12, 13, 14, 15). By being able to record an extended region of space, we can overcome the limitation of point-FCS for which results are highly sensitive to the specific regions imaged.
Extending multiplex imaging FCS to three dimensions requires either simultaneous multiplane illumination and acquisition, or fast sequential multiplane imaging. Simultaneous multiplane imaging systems require multi-light-sheet generation, controlled distance between the imaging planes, and simultaneous focusing of the excited planes on a camera. Multiple light-sheet microscopy (16) enables simultaneous excitation of multiple planes but with a ∼5–7 μm thick light-sheet and a fixed distance between planes (10–15 μm). In addition, detection of the multiple excited planes requires moving the sample or objective, leading to a sequential acquisition in practice. Simultaneous multiplane acquisition on a single camera can be achieved using a diffractive multifocus grating (17, 18); however, the use of wide field excitation schemes currently prevents their use for live multiplane FCS imaging. Sequential single-plane FCS imaging requires moving the sample through the light-sheet plane or adjusting the detection path according to the light-sheet depth into the sample. However, photobleaching typically drastically limits the effective number of planes where imaging FCS can be performed. Thus, multiplane FCS requires specific illumination and detection schemes, as well as labeling strategies limiting photobleaching effects.
Here, we present, to our knowledge, a novel combination of a single-objective based light-sheet microscope (soSPIM) (19) with photoconvertible fluorophores to perform sequential imaging FCS on up to eight planes (<1 μm apart). This enables quantification of protein diffusion effectively across the entire cell nucleus in a multiplexed-multiplane fashion. First, we tested and calibrated the performance of this soSPIM-FCS combination with organic dyes in a buffer solution, which showed similar sensitivity and calibration parameters as previously reported using SPIM setups (8, 20). We then tested the sensitivity of soSPIM-FCS by imaging enhanced green fluorescent proteins (eGFP) in live NIH3T3 cells. To explore 3D protein dynamics, we used photoconvertible Dendra2-tagged RNA Polymerase II (Dendra2-Pol II) protein to create multiplexed-multiplane protein diffusion maps that traverse the 3D cell nucleus. We acquired roughly an order of magnitude more correlation functions from a single cell nucleus than previously reported. Importantly, the improvement in the quality of correlation functions and the number of data points per cell was only possible due to the continuous photoconversion of a sparse subset of proteins in the acquisition channel leading to a high FCS signal-to-noise ratio (SNR) and significantly reduced photobleaching effects. Because the total data acquisition time (<5 min) is still less than the typical eukaryotic transcriptional time window ∼30 min (21, 22), soSPIM-FCS can access 3D dynamics within a biologically relevant timescale. Finally, we investigated the dynamics of Yes-associated protein (YAP) (23), a downstream effector of the Hippo signaling pathway. YAP is a transcriptional coactivator, whose nuclear localization is regulated by chemical and mechanical inputs (24). By imaging Dendra2-tagged YAP, our approach enabled us to estimate nuclear YAP diffusion kinetics for what we believe is the first time and demonstrate the dynamic heterogeneity of YAP in the nucleus. By measuring YAP dynamics throughout a large area of the cell nucleus, we expect our results to be unbiased due to localized variations in YAP dynamics (e.g., due to association with DNA via TEAD-binding).
Materials and Methods
Detailed description on imaging FCS, data fitting, microfabrication, and the protocol for cell culture can be found in the Supporting Material.
Experimental setup and data collection
The soSPIM system is composed of a high numerical aperture (NA) objective (CFI Plan Apochromat VC 60× WI 1.27 NA; Nikon Instruments, Melville, NY), a beam steering unit, and dedicated microfabricated devices containing mirrors angled at 45° alongside microwells (Fig. 1; Fig. S1 in the Supporting Material). The soSPIM components are mounted on a conventional inverted microscope (model No. Ti-E, with 1.5× lens of the microscope body and a 0.45× lens to ensure a pixel size of 160 nm in the image plane for sCMOS camera and 266 nm pixel size for EMCCD camera; Nikon Instruments). The microfabricated imaging chamber (Fig. S2 (19)) is placed on an axial translation piezo stage within a controlled environment chamber for live cell imaging. Fluorescence emission is collected through the same objective used for excitation and is captured in streaming mode on a sCMOS camera. The whole acquisition process is steered using MetaMorph software (Molecular Devices, Sunnyvale, CA).
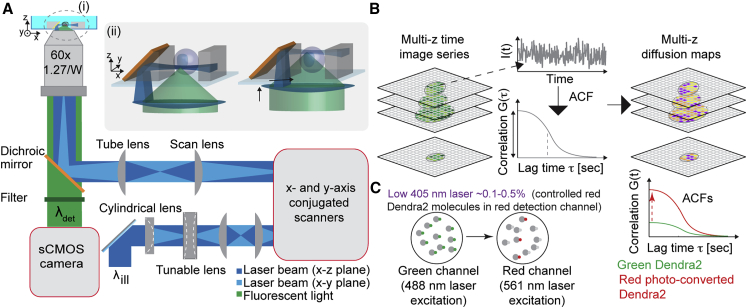
Figure 1.
Experimental setup of single-objective based multiplexed-multiplane FCS (soSPIM-FCS) measurements. (A) (i) Experimental setup of the soSPIM system. (ii) Closeup of the 45° micromirror angled to reflect the light-sheet beam orthogonal to the detection axis for imaging at two different depths. (B) Principle of multiplane FCS measurements. In multiplane imaging FCS, a sequential time image series is acquired at different z position in a cell and the time autocorrelation performed at every pixel of the plane to achieve multiplane diffusion and concentration maps. (C) Principle of using photoconvertible protein for FCS. (Left) The number of the photoconverted red molecule can be tuned in the observation volume by 405 nm laser excitation. (Right) Dendra2 protein in red form is expected to show increased ACF amplitude. To see this figure in color, go online.
The tunable lens and the galvanometric mirrors (Fig. 1 A) were precisely aligned and conjugated to the objective back focal plane to ensure the light sheet to be perpendicular to the microscope objective optical axis after reflection onto the 45° mirror, regardless of its reflection position. Drifting of the light sheet along the x axis when performing 3D imaging is compensated by calibrating the light-sheet position at two different depths. Further details of soSPIM setup are described in Galland et al. (19). A cylindrical lens (f = 150 mm; Thorlabs, Newton, MA) may be inserted thanks to a switchable mount into the optical path to focalize the laser beam in one direction at the objective back focal plane. It enables us to create a static light sheet in the sample for FCS acquisition. The cylindrical lens is mounted on a rotational mount to align its focalization direction with the 45° mirror’s long axis.
Imaging FCS characterization and calibration for soSPIM
The thickness of the excitation light-sheet was characterized by imaging 100 nm tetraspeck beads embedded in 1% agarose gel filling the microwells (1:100 ratio; Fig. S3). The light-sheet characteristics can subsequently be extracted from , where ω0 is the beam waist, representing half the minimum thickness e0 of the light-sheet; ZR is the Rayleigh length, corresponding to half the size of the field of view of the light-sheet; and x0 is the position of the beam focalization along the propagation direction. For a 3 mm diameter excitation beam at the cylindrical lens position, we measured e0 = 1.8 ± 0.2 μm and ZR = 13 ± 0.5 μm at 561 nm wavelength (Fig. S3).
The width of the static light sheet created by inserting a cylindrical lens into the beam steering optical path (f = 150 mm) was characterized by imaging the light-sheet through a fluorescent polymer after reflection on the mirror (Fig. S3 B). Fitting the width of the light-sheet by a Gaussian function, we estimate the width of the light-sheet to be 26 ± 1 μm (full width half-maximum).
The laser power density was then calculated as the power spread over an area of 18 × 26 μm2, which corresponds to the XY area of the light-sheet.
Imaging FCS data acquisition
For imaging, the growth media was replaced by 2 mL of imaging media composed of FluoroBrite media (Gibco/Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS and 10 mM of HEPES buffer in a 35 mm dish, to ensure minimal fluorescence background.
For FCS imaging, a region of interest (ROI) of between 32 × 32 and 100 × 60 pixels was chosen, centered on the sample inside the camera field of view. Time series of 10–20 k frames per plane were acquired at 2.4–4.3 ms integration time for live cell imaging, and time series in the range 50–100 k frames per planes were acquired at 155 μs integration times for beads and single fluorophore imaging. The objective axial position stability was ensured by the Perfect Focus System of the microscope body (Nikon Instruments). For live cell FCS imaging, up to eight time-series at different z positions (dz = 0.8 μm) were acquired. For FCS imaging of U2OS Dendra2-Pol II cells and MKN28 Dendra2-YAP cells, acquisitions were performed by excitation with the 561 nm laser (laser power in between 0.5 and 1.5 kW/cm2). The 405 laser power (0.05–0.2 kW/cm2) was adjusted to continuously photoconvert a sparse subset of Dendra2 proteins, ensuring sufficient SNR for FCS analysis while renewing photobleached fluorophores to allow up to eight planes to be acquired on a single cell. Tables S1 and S2 summarize the different camera settings, data acquisition, and postprocessing parameters used in the article.
All imaging FCS data were corrected for fluorescence bleaching, with detailed description of the data fitting, typical defects, and photobleaching correction available here in Krieger et al. (8), and the analysis was performed using QFit3.0 and/or ImageJ imaging FCS plugin (25, 26).
Results
soSPIM-FCS characterization and validation
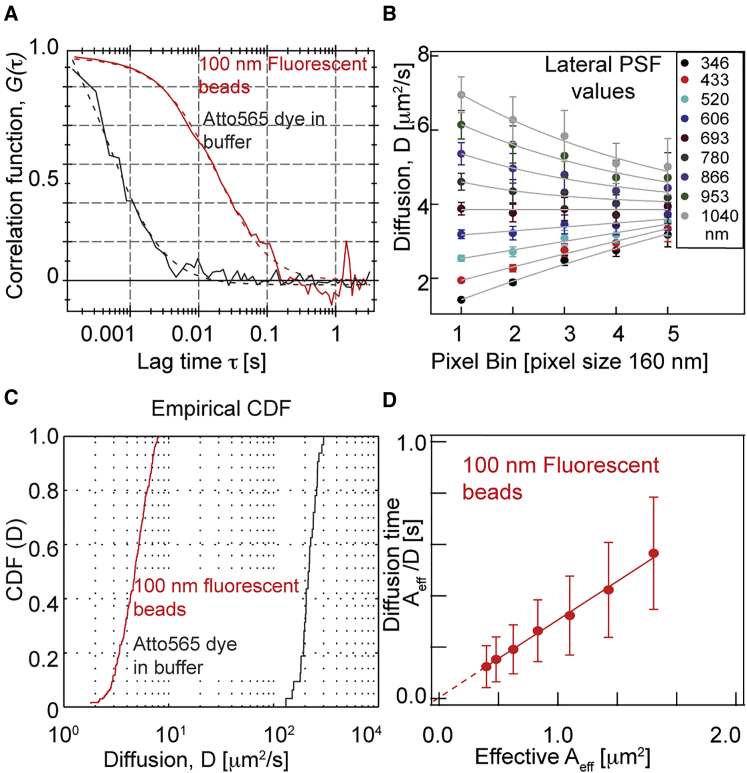
We calculated ACFs using fluorescent beads and organic dyes. To minimize variation due to self-aggregation of beads, buffer pH was kept at ∼10 (use M/10 NaOH solution to adjust pH) and the beads were sonicated before use, while Atto 565 dye was first dissolved in DMSO and further diluted in 1 × PBS buffer at pH7.4. Fig. 2 A shows typical ACFs for 100 nm fluorescent beads (red) and Atto 565 organic dyes (gray) in buffer solution, with dashed lines representing fits to Eq. S2 (see the Supporting Material; see also Fig. S4). By varying the effective camera pixel size (by postprocessing) we estimated the lateral extension of the point spread function (PSF)—the value at which the predicted diffusion coefficient is independent of pixel size—to be 0.7 μm (Fig. 2 B). The bead and Atto565 dye average diffusion coefficients were D22°C = 4.8 ± 1.8 μm2/s and 220 ± 40 μm2/s, respectively, at room temperature, although note this likely underestimates diffusivity for organic dyes due to slow camera exposure (27). The bead diffusion data were in good agreement with single point confocal measurements (see Fig. S4 and Table S3; setup as described in Maharana et al. (28)). Furthermore, the corresponding cumulative distribution function (CDF) for the fitted diffusion coefficients at room temperature (Fig. 2 C) were in a similar range to previously reported SPIM-FCS measurements (8, 20). Finally, in principle, a freely diffusing particle should have a linear relationship between the effective area (determined here by the camera pixel size and binning) and the diffusion time, with zero intercept (29, 30). By plotting the change of diffusion time (y axis) with the change in effective area (x axis, postprocess binning; Fig. 2 D), we see that the relationship is close to linear, with D22°C ∼ 3.3 ± 1.3 μm2/s at room temperature, in good agreement with the fitting to the ACFs from Eq. S2 (see the Supporting Material). These calibration and FCS sensitivity tests (8) show that the presented soSPIM-FCS setup can be reliably used for FCS measurements (see comparison with other methods in Table S3).
Figure 2.
Imaging FCS calibration and sensitivity of soSPIM-FCS. (A) Typical single pixel autocorrelation function for 100 nm tetra-spec fluorescent beads (red) and organic Atto565 dye (gray) in 1 × PBS buffer. Data were acquired with a sCMOS camera using a 32 × 32 (32 × 16 organic dyes) pixel ROI at 155 μs camera integration time. (Dashed lines) Fits of Eq. S1; see the Supporting Material. (B) Dependence of diffusion coefficient, D, at different camera pixel binning and lateral PSF (analysis was performed with varying binning values from 1 × 1 to 5 × 5 pixels). (C) The diffusion coefficient cumulative distribution function for 100 nm fluorescent beads and Atto 565 dyes in 1 × PBS. (D) Dependence of the diffusion time on the effective area (postacquisition camera binning). Inverse slope estimates the diffusion coefficient (D22°C = 3.3 ± 1.3 μm2/s for 100 nm fluorescent beads). Experiments were performed on a sCMOS camera with a pixel size of 160 nm. ACF analysis were performed with varying binning values from 1 × 1 to 5 × 5 pixels. To see this figure in color, go online.
To further test the FCS capabilities of soSPIM, we next performed sequential multiplane soSPIM-FCS to live NIH3T3 cells expressing stable eGFP-tagged H2B histone. Cells were prepared as described in the Supporting Material. The reflected light-sheet was positioned on the cell nucleus and multiple z planes were sequentially illuminated. For each section a time image series of 20 k frames was collected at 2.4 ms camera exposure on an EMCCD camera (using a cropped field of view). The time image series were autocorrelated (see Eq. S1) at each pixel and fitted with a one-diffusive component model, Eq. S2 (see also the Supporting Material) (8). Fig. S5, A and B (for two different cells), shows the diffusion coefficient maps and distribution at two different planes, with an average diffusion value 0.28 ± 0.08 μm2/s (average 2300 pixels). To test sensitivity, we measured the effect of DNA-binding drug Hoechst 33342 (which induces cell death) on histone diffusion in the same cell. After treatment, the average fluorescence signal showed higher intensity and the diffusion coefficient decreased to 0.16 ± 0.06 μm2/s (Fig. S5 C). The decreased diffusion coefficient may be due to shrinkage of the whole chromosome structure as the nucleus shrinks during apoptosis (31). The diffusion CDF was markedly changed after Hoechst drug treatment (Fig. S5 D), showing that soSPIM-FCS is sensitive to dynamic changes inside the nucleus.
The important result of this analysis is that the slow histone protein diffusion, particularly after Hoechst 33342 treatment, showed fast fluorescence bleaching. High-quality FCS measurements could thus not be achieved for more than three consecutive planes. Due to the high expression and low mobility of many nuclear localized proteins it is technically challenging to obtain good estimates of diffusion coefficients without significant photobleaching in live cells. To tackle these challenges, photoconvertible fluorophores are needed to control the number of active molecules in the focal volume to achieve high-quality FCS measurements across the whole nucleus.
Improved multiplex FCS using photoconvertible fluorophore Dendra2
FCS in living cells requires an optimal number of particles in the observation volume: too few and the signal is significantly affected by the detector background; too many and the ACFs are too flat and noisy. Controlling the protein expression of fluorescently tagged proteins is often difficult in practice. To overcome this challenge, prebleaching before FCS was previously used to reduce the effective number of fluorescent protein molecules in live cells (32, 33); however, this may induce severe photodamage and precludes multiplane imaging due to rapid loss of total fluorescence signal in a living sample. An alternative way to overcome protein overexpression without bleaching is to use photoactivatable fluorescent proteins (5, 10). The density of fluorescent proteins can be easily tuned by adjusting the intensity of the 405 nm laser, allowing a high SNR ratio for FCS, as demonstrated recently in the early mammalian embryo (5, 10).
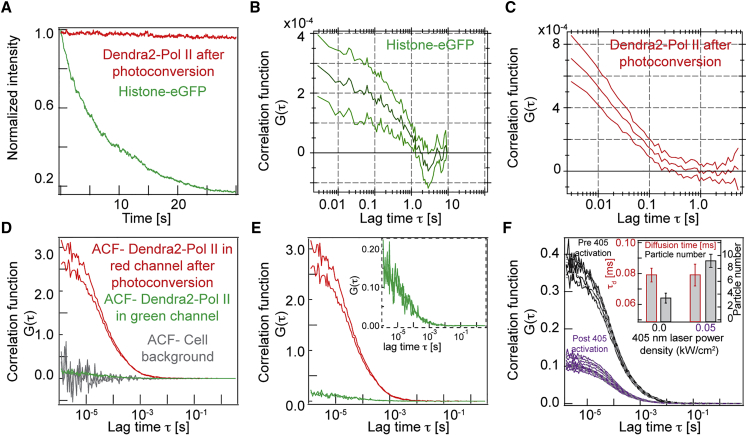
To test the sensitivity of photoconvertible tagged proteins for camera-based imaging FCS in live cells, we used stably expressing Dendra2-Pol II U2OS cells (34). Dendra2 was converted from green to red fluorescence using a 405 nm excitation laser (34, 35, 36), with the number of photoconverted molecules controlled by varying the 405 nm laser excitation power. Fig. 3 shows the characterization of eGFP- and Dendra2-tagged proteins for FCS in live cells. The fluorescence signal from the photoconverted red form of Dendra2 showed long-term stability (>3 min) compared to eGFP-tagged proteins (Fig. 3 A). Most importantly, the ACFs were significantly improved with Dendra2 compared to eGFP (Fig. 3, B and C), with lower noise and subsequently improved data fitting.
Figure 3.
Characterization of Dendra2 fluorescent protein for FCS imaging. (A) Typical intensity profiles for eGFP and red photoconverted Dendra2 protein in imaging FCS. (B and C) Average autocorrelation functions of eGFP (B) and Dendra2 (C). Upper and lower lines represent maximum extent of ACFs (n = 36). (D) ACFs, including cell background (gray), for Dendra2 protein (green), and photoconverted in the red channel with point FCS on a confocal FCS setup. (E) As for (D), but without background and the inset shows a magnified view of the ACF in the green channel. (F) Change in ACF amplitude for Dendra2-3xNLS under different 405-nm laser activation. (Inset) There is no change in diffusion time, but the particle number increases after photoconversion of Dendra2 green to red fluorophore in the red channel detection. Histone-eGFP experiments were performed on an EMCCD camera with effective pixel size = 266 nm and Dendra2-Pol II on a sCMOS sensor with effective pixel 160 nm. To see this figure in color, go online.
We compared the ACFs of the different Dendra2 emissions on confocal-based single-point FCS. The ACF amplitude (inversely proportional to concentration, Eq. S2; and see the Supporting Material) was significantly increased in photoconverted (red) Dendra2 expressing cells compared with the green channel (Fig. 3, D and E). Further, we confirmed using Dendra2-NLS (to avoid binding kinetics) that varying the activation of Dendra2 alters the observed particle number, but not the measured diffusion time, Figs. 3 F and S6. Hence, photoconversion allows precise control of the number of active fluorescent molecules for FCS analysis and this significantly improves the quality of fitting to the measured ACFs.
Sequential, multiplane imaging FCS
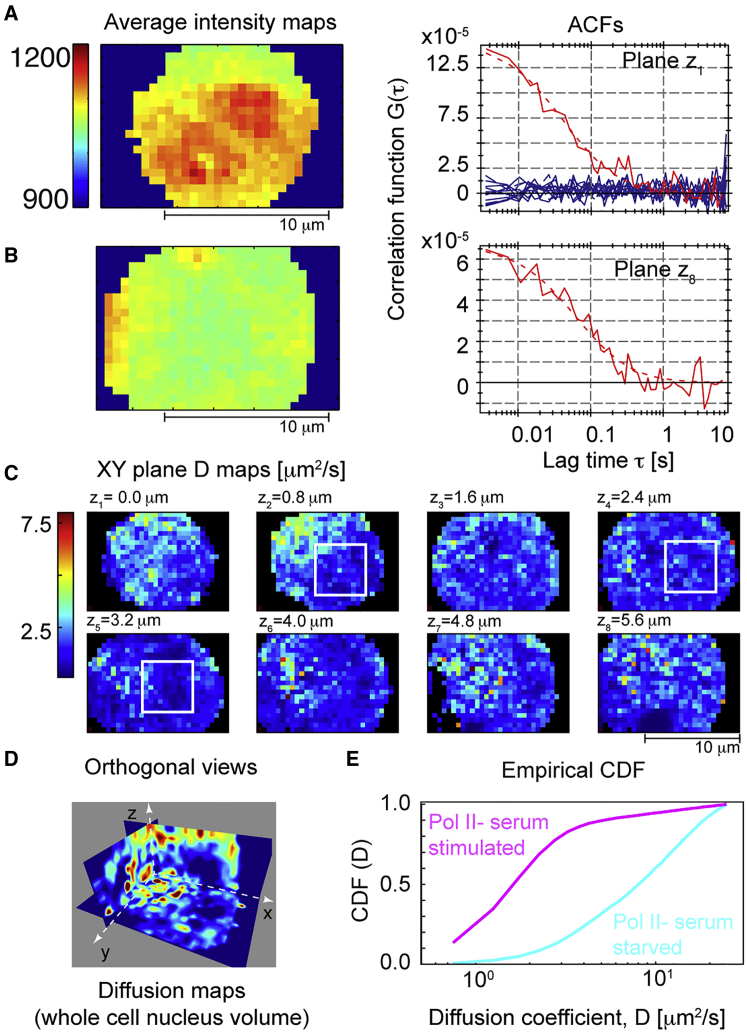
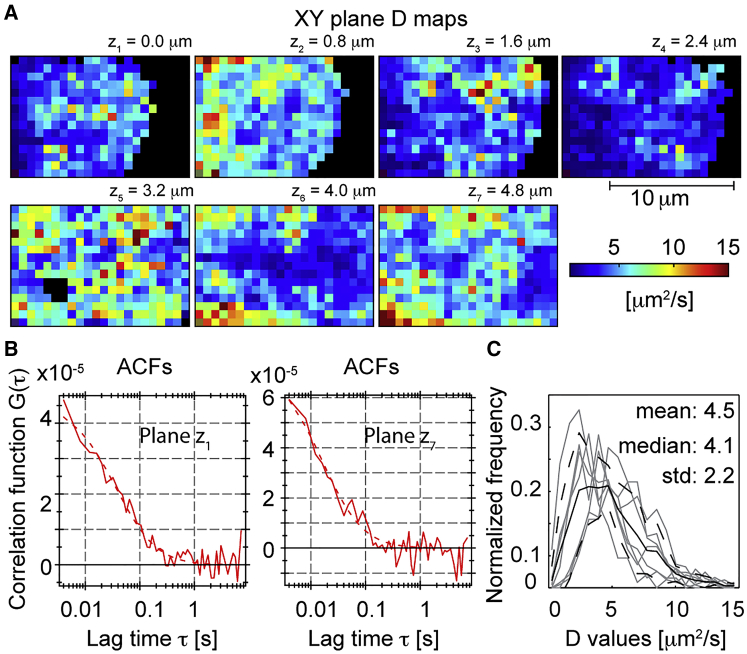
We now combined soSPIM-FCS with Dendra2 to obtain sequential multiplane, multiplexed FCS. We imaged stable Dendra2-Pol II U2OS cells using soSPIM-FCS, with a total of eight planes per cell with z spacing = 0.8 μm. A stable 561 nm laser power of ≈0.8 kW/cm2 was used with a stable low 405 nm activation dose (∼0.05 kW/cm2). The bottom and top planes of a serum-stimulated U2OS cell nucleus are shown in Fig. 4, A and B, respectively, along with their (unnormalized) ACFs (right side). Although there is a decrease in quality at the top plane (primarily due to photobleaching), the ACF was still of sufficient quality for reliable model fitting. The diffusion maps for all eight planes of a serum-stimulated cell are shown in Fig. 4 C (see Fig. S7 for further examples). The diffusion coefficient of RNA Pol II typically ranged from 1 to 5 μm2/s, which is similar to other transcription factors in the cell nucleus (3, 37). Fig. 4 D shows orthogonal views of the diffusion maps along x, y, and z axes. Structures are apparent in the diffusion maps; these are consistent with different chromosomal packing at different nuclear positions (2) (see also Movies S1 and S2).
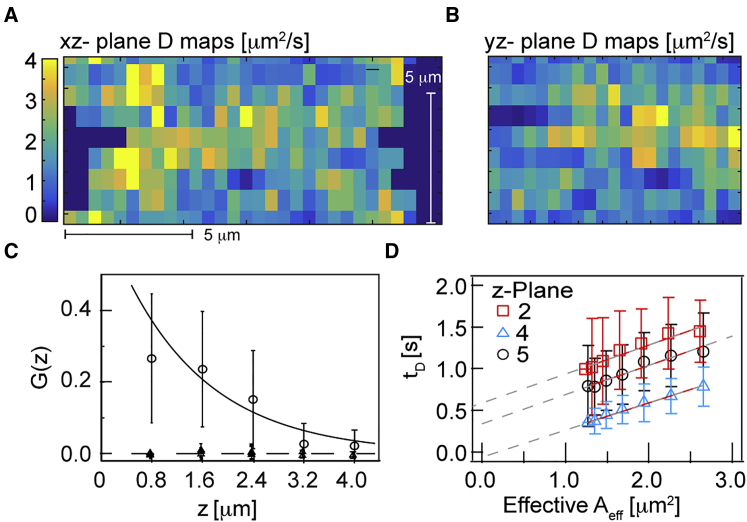
Figure 4.
Multiplane, multiplexed diffusion maps of Dendra2-Pol II expressed in U2OS cells. (A and B) Average intensity maps of Dentra2-Pol II time image series (left) and corresponding ACFs (right). 10 k images were acquired at different z positions (A, z+ = 0.0 μm; B, z8 = 5.6 μm) with a spacing of ∼0.8 μm and all eight planes were sequentially imaged with a total time of 5 min. The regions outside the cell nucleus showed zero correlation (flat ACFs in A shown in blue). (C) Representative diffusion maps for Dendra2-Pol II in the cell nucleus at different z positions (see Movie S2). (D) 3D orthogonal view of diffusion maps along x, y, and z axes. (E) Cumulative probability distribution of diffusion coefficient in serum-starved (cyan) and stimulated (magenta) environments (average from four cells; >13,400 data points for serum-stimulated and >4800 data points for serum-starved conditions). All imaging performed on a sCMOS camera with effective pixel size = 160 nm. ACF analysis was performed with 3 × 3 pixel binning. To see this figure in color, go online.
To validate our analyses, we repeated the experiments on serum-starved cells (cells were starved at least 18 h before imaging), and diffusion values were compared between serum-stimulated and -starved conditions. In serum-starved cells, smaller Pol II clusters are predicted due to a lower transcriptional activity, hence an increase in the diffusion coefficient is expected. This was confirmed by comparison of the diffusion coefficient CDF for the two conditions (Fig. 4 E). Taken together, these results suggest that our approach is sensitive to spatial variations in Pol II dynamics. Although previous work has performed single molecule tracking of Pol II localization, this is inherently limited in number and spatial range. Here, we can describe the global (on the scale of the nucleus) Pol II dynamics in an unbiased manner. Qualitatively, we observe that diffusion appears slowest toward the cell center (Figs. 4 C and S7)—where chromosomes are more densely packed (38, 39)—although a more comprehensive analysis is needed to confirm this. Therefore, our approach here provides an effective tool for exploring nuclear protein dynamics.
Recent observations in live cells suggest that Pol II clusters are dynamic and transient in nature during serum stimulation (34, 40, 41). Consistent with this, we observe distinct regions of slow dynamics in the diffusivity maps (Fig. 4 D). To explore this further, we performed a more careful analysis of the interplane variation in the diffusion maps. Fig. 5, A and B, shows representative cross-sectional diffusion map views along the xz- and yz planes. The diffusion coefficient maps measured for each plane (∼36 s acquisition per plane) demonstrate spatial correlation in the z direction over a distance of 1.7 μm (∼3 planes along z direction) (Fig. 5 C). This diffusion correlation length is unlikely an artifact of the microscope setup, as planes are taken sequentially and hence correlations between planes in the diffusivity are most likely due to conserved Pol II dynamical structures, and not due to overlap of the PSF—we see correlations in diffusivity between planes imaged over a minute apart. Also, this naïve vertical diffusion correlation is likely an underestimate of structure extent, as most structures will not lie vertically in the z axis of the microscope. To test this further, we applied the FCS diffusion laws (as in Fig. 2 D (29, 30, 42)). Serum-stimulated cells in different z planes showed positive and varying y axis intercepts (Fig. 5 D; Fig. S8). A positive y intercept suggests possible spatially confined domains. Similar organization of Pol II and other nuclear proteins has been previously observed from single particle imaging and tracking (34, 36). Alternatively, spatially varying diffusivity might be from local changes in effective viscosity due to chromosome organization, and these regions can effect transcriptional activity (43). The dynamic heterogeneities we measure may arise from local spatial heterogeneities in transcription activity, evolving on timescales >1 min.
Figure 5.
Spatial analysis of Dendra2-Pol II sequential multiplane, multiplexed diffusion maps. (A and B) Cross-section diffusion map views along xz- and yz planes (pixel size: 0.48 × 0.8 μm). (C) Spatial Pearson correlation coefficient along z direction (n = 4 cells), where the line is fit to se−z/λ: s = 0.75 and λ = 1.7 (with 95% confidence interval [0.5, 3.0]). (D) Representative diffusion law plots for Dendra2-Pol II in serum-stimulated condition shows varying y axis intercepts (ROI for diffusion law analysis is shown by white rectangle for planes 2, 4, and 5 in Fig. 4C). Experimental FCS details as in Fig. 4. To see this figure in color, go online.
Dendra2-YAP dynamics
We next extended our analysis to investigate the dynamics of YAP, a transcriptional coactivator. It is known that shuttling of YAP between the cytoplasm and nucleus is important for YAPs function in regulating the Hippo pathway. Thus we anticipate that YAP will be present in both the cytoplasm and nucleus, and we expect that YAP will exhibit reduced and heterogeneous diffusion dynamics in the nucleus as it can associate with DNA-binding transcription factors to regulate numerous downstream target genes.
MKN28 cells stably expressing Dendra2-YAP were generated (see the Supporting Material) and imaged with the soSPIM-FCS setup using a stable 561 nm laser power (≈0.5–0.8 kW/cm2), and a low 405 nm activation dose to continuously activate a sparse subset of the Dendra2 protein. Time-series of 10 k frames were acquired at 4.3 ms integration time on 90 × 60 pixels ROI every 0.8 μm on up to seven planes in a single cell, which covers a significant portion of the nuclear volume.
Fig. 6 A shows the diffusion maps (akin to Fig. 4 C) for Dendra2-YAP, with corresponding intensity maps shown in Fig. S9 A (see Fig. S9 for other representative cells as well). Heterogeneity of YAP diffusion is apparent, with some spatial structure preserved between planes. This structure may indicate possible binding events between YAP and its targets within spatially conserved regions. The measured ACFs were well fitted by diffusion-dominated dynamics (Fig. S7 B); however, due to the temporal resolution of the cameras, we are principally identifying the dynamics of the slow (bound) fraction. To estimate the effective mean YAP diffusivity, we averaged all intranuclear ACFs and found DYAP ∼ 4.5 ± 2.2 μm2 s−1 (compared with DYAP ∼4.2 ± 2.0 μm2 s−1 using eGFP-YAP in the cell nucleus on confocal FCS; Fig. S10). Despite having a large standard deviation, the diffusion probability functions for different planes are comparable (Fig. 6 C), suggesting that this is a reliable estimate of YAP diffusivity. Because nuclear-localized YAP associates with DNA-binding proteins, YAP diffusion in the nucleus was slower compared to cytoplasmic diffusion (note, to capture the faster cytoplasmic dynamics—DYAP,CYT ∼80 μm2 s−1—we used confocal FCS; Fig. S10). Because we measure the diffusivity across nearly the entire nucleus, our results represent a good estimate of the overall effective YAP nucleus diffusion. To our knowledge, the data presented here is the first quantitative measurement of YAP dynamics within the cell nucleus. With further improvements in detector sensitivity, we anticipate being able to dissect spatial variation in nuclear YAP dynamics in the future as we can access faster timescales in a spatially extended manner. An interesting future direction would be to explore the fraction of freely available YAP simultaneously in the cytoplasm and nucleus (including transport into the nucleus), and how this responds to external stimuli such as mechanical stress (44, 45). This may give important insight into the dynamics of YAP as a transcriptional coactivator and how the cell responds rapidly to external stress.
Figure 6.
Multiplane diffusion maps of Dendra2-YAP protein in cell nucleus. (A) Dendra2-YAP diffusion maps (similar to Fig. 4C) at different planes in the cell nucleus (z spacing 0.8 μm). See Fig. S9 for mean intensity images. (B) Representative ACF of Dendra2-YAP in the cell nucleus (fit in dashed line). (C) Diffusion coefficient probability distribution (solid black line) of Dendra2-YAP for all seven planes (dashed lines represent ±1 SD). (Gray) Distributions for individual planes. All images were performed on a sCMOS camera with effective pixel size = 160 nm. ACF analysis was performed with 4 × 4 pixel binning. To see this figure in color, go online.
Discussion
Heterogeneity of protein dynamics are often the signature of local changes in protein activity. Developing techniques that enable mapping of such spatial heterogeneity could provide key inputs into our understanding of protein function. soSPIM-FCS provides a 3D mapping of the dynamics of cellular processes. The unique soSPIM-FCS architecture reported here provides distinct improvements on previous multiplex FCS methods: 1) high sectioning capability at the single cell level; 2) high mechanical stability as compared to the standard two-objective SPIM setup; 3) the possibility to use high numerical aperture objectives for efficient signal collection ensuring sensitivity at the single molecule level (19); and 4) the ability to combine localized activation (and other quantitative techniques such as fluorescence recovery after photobleaching, FRAP) using the inverted microscope with the soSPIM sensitivity and low photobleaching and phototoxicity. The power to combine photoconversion of fluorophores with FCS in a spatially extended manner is perhaps the strongest element of this approach. Further, all the functionality of a standard inverted microscope (differential interference contrast, phase, epifluorescence, etc.) can be readily used to position and select the sample. This provides clear practical advantages over the classical SPIM approaches.
Data presented here demonstrates that the soSPIM configuration is stable and sensitive enough to produce high quality ACFs. Coupling the SPIM configuration with photoconvertible fluorophores allowed us to sufficiently reduce protein photobleaching so that we can measure up to eight diffusion maps spaced by 0.8 μm. This approach provided a more precise 3D reconstruction of the nuclear dynamic environment. We validated the efficacy of our setup with two quite different proteins in the nucleus. We established that Dendra2-Pol II has a nonhomogeneous distribution of diffusivity at different spatial positions within the nucleus, consistent with single molecule tracking approaches (34). However, our approach encompasses orders-of-magnitude more measurements per cell. We demonstrated the power of our approach by providing an estimate of the effective nuclear diffusion of the transcriptional coactivator YAP. Such an estimate is not significantly affected by local structures in the nucleus due to the large number of data points collected for each cell. Of course, camera-based FCS still has limitations, particularly in terms of the relatively slow frame rates (milliseconds here versus microseconds on confocal-based FCS) and bleaching effects for longer imaging time windows. We have carefully assessed that photophysics effects from using a photoconvertible fluorophore do not distort our conclusions. The results presented here represent a significant improvement over most previous camera-based FCS studies and we have validated results using confocal FCS.
Being able to measure dynamics in three dimensions is an important step to link the existence of local dynamical heterogeneities to biological structures. Indeed, local fluctuation of diffusion coefficients may transiently arise in the cell or may be due to intrinsic noise of the detection system. However, the existence of correlated regions of diffusion coefficient heterogeneities at different z positions for Pol II strongly suggests that the observed dynamical variations arise from differential local interactions of proteins. In future, we anticipate that the combination of soSPIM-FCS and photoconvertible proteins may become a powerful quantitative bioimaging tool to create differential diffusion and interaction maps of transcription factors and other proteins in vivo. Such information may be important, for example, in deciphering the role of transcription factor binding to determine cell fate (5), or understanding the role of spatio-temporal changes in the effective viscosity within the nucleus due to chromosomal reorganization and its effect on the rate of transcriptional kinetics (3). Single-particle tracking can also be achieved with this technique (46). Finally, the laser driving unit to implement the soSPIM is essentially a modified FRAP head. Our approach could thus readily combine FCS, single-particle tracking, and FRAP on the same system to probe a spectrum of protein dynamics ranging from millisecond to minutes.
Author Contributions
A.P.S., and R.G. contributed equally to this work; A.P.S., R.G., V.V., and T.E.S. designed the project; A.P.S. and R.G. performed experiments and collected the raw data and analyzed the FCS data; M.L.F.-E. cloned the 3xNLS-Dendra2, Dendra2-YAP, and eGFP-YAP constructs, and generated the stably expressing cell lines; G.G. designed and implemented the microwells; R.G., J.-B.S., V.S., and V.V. designed and built soSPIM and adapted it for performing FCS; A.P.S., R.G., V.V., and T.E.S. wrote the article; and all authors participated in discussion and editing of the article.
Acknowledgments
Authors thank Prof. Shivashankar for stable H2B-eGFP NIH3T3 cells, Prof. Xavier Darzacq for stable Dendra2- Pol II U2OS cells, and the Electronic Imaging Center of the Bordeaux Imaging Center for the metallization of the devices. The authors acknowledge the Michael W. Davidson group, The Florida State University, Tallahassee, FL for provision of all Dendra2 and eGFP cDNAs.
A.P.S. was supported under a National Research Foundation (NRF) Fellowship awarded to T.E.S. (grant No. 2012NRF-NRFF001-094). This work was also supported by the Ministère de l'Enseignement Supérieur et de la Recherche (ANR soSPIM, Labex BRAIN and ANR-10-INBS-04 France-BioImaging), the Centre National de la Recherche Scientifique and the Conseil Régional d'Aquitaine.
Editor: Paul Wiseman.
Footnotes
Anand P. Singh and Remi Galland contributed equally to this work.
Supporting Materials and Methods, ten figures, four tables, and two movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)34265-5.
Contributor Information
Virgile Viasnoff, Email: virgile.viasnoff@espci.fr.
Timothy E. Saunders, Email: dbsste@nus.edu.sg.
Supporting Material
References
- 1.Woringer M., Darzacq X., Izeddin I. Geometry of the nucleus: a perspective on gene expression regulation. Curr. Opin. Chem. Biol. 2014;20:112–119. doi: 10.1016/j.cbpa.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Makhija E., Jokhun D.S., Shivashankar G.V. Nuclear deformability and telomere dynamics are regulated by cell geometric constraints. Proc. Natl. Acad. Sci. USA. 2016;113:E32–E40. doi: 10.1073/pnas.1513189113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hager G.L., McNally J.G., Misteli T. Transcription dynamics. Mol. Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macháň R., Wohland T. Recent applications of fluorescence correlation spectroscopy in live systems. FEBS Lett. 2014;588:3571–3584. doi: 10.1016/j.febslet.2014.03.056. [DOI] [PubMed] [Google Scholar]
- 5.White M.D., Angiolini J.F., Plachta N. Long-lived binding of Sox2 to DNA predicts cell fate in the four-cell mouse embryo. Cell. 2016;165:75–87. doi: 10.1016/j.cell.2016.02.032. [DOI] [PubMed] [Google Scholar]
- 6.Capoulade J., Wachsmuth M., Knop M. Quantitative fluorescence imaging of protein diffusion and interaction in living cells. Nat. Biotechnol. 2011;29:835–839. doi: 10.1038/nbt.1928. [DOI] [PubMed] [Google Scholar]
- 7.Singh A.P., Wohland T. Applications of imaging fluorescence correlation spectroscopy. Curr. Opin. Chem. Biol. 2014;20:29–35. doi: 10.1016/j.cbpa.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Krieger J.W., Singh A.P., Wohland T. Imaging fluorescence (cross-) correlation spectroscopy in live cells and organisms. Nat. Protoc. 2015;10:1948–1974. doi: 10.1038/nprot.2015.100. [DOI] [PubMed] [Google Scholar]
- 9.Struntz P., Weiss M. Multiplexed measurement of protein diffusion in Caenorhabditis elegans embryos with SPIM-FCS. J. Phys. D Appl. Phys. 2016;49:044002. [Google Scholar]
- 10.Kaur G., Costa M.W., Plachta N. Probing transcription factor diffusion dynamics in the living mammalian embryo with photoactivatable fluorescence correlation spectroscopy. Nat. Commun. 2013;4:1637. doi: 10.1038/ncomms2657. [DOI] [PubMed] [Google Scholar]
- 11.Hebert B., Costantino S., Wiseman P.W. Spatiotemporal image correlation spectroscopy (STICS) theory, verification, and application to protein velocity mapping in living CHO cells. Biophys. J. 2005;88:3601–3614. doi: 10.1529/biophysj.104.054874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiseman P.W. Chapter Ten—Image correlation spectroscopy: mapping correlations in space, time, and reciprocal space. In: Sergey Y.T., editor. Methods in Enzymology. Academic Press; New York: 2013. pp. 245–267. [DOI] [PubMed] [Google Scholar]
- 13.Hedde P.N., Stakic M., Gratton E. Rapid measurement of molecular transport and interaction inside living cells using single plane illumination. Sci. Rep. 2014;4:7048. doi: 10.1038/srep07048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krieger J.W., Singh A.P., Langowski J. Dual-color fluorescence cross-correlation spectroscopy on a single plane illumination microscope (SPIM-FCCS) Opt. Express. 2014;22:2358–2375. doi: 10.1364/OE.22.002358. [DOI] [PubMed] [Google Scholar]
- 15.Szaloki N., Krieger J.W., Vamosi G. Evidence for homodimerization of the c-Fos transcription factor in live cells revealed by fluorescence microscopy and computer modeling. Mol. Cell. Biol. 2015;35:3785–3798. doi: 10.1128/MCB.00346-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohan K., Purnapatra S.B., Mondal P.P. Three dimensional fluorescence imaging using multiple light-sheet microscopy. PLoS One. 2014;9:e96551. doi: 10.1371/journal.pone.0096551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abrahamsson S., Chen J., Gustafsson M.G.L. Fast multicolor 3D imaging using aberration-corrected multifocus microscopy. Nat. Methods. 2013;10:60–63. doi: 10.1038/nmeth.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prevedel R., Yoon Y.-G., Vaziri A. Simultaneous whole-animal 3D imaging of neuronal activity using light-field microscopy. Nat. Methods. 2014;11:727–730. doi: 10.1038/nmeth.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galland R., Grenci G., Sibarita J.B. 3D high- and super-resolution imaging using single-objective SPIM. Nat. Methods. 2015;12:641–644. doi: 10.1038/nmeth.3402. [DOI] [PubMed] [Google Scholar]
- 20.Singh A.P., Krieger J.W., Wohland T. The performance of 2D array detectors for light sheet based fluorescence correlation spectroscopy. Opt. Express. 2013;21:8652–8668. doi: 10.1364/OE.21.008652. [DOI] [PubMed] [Google Scholar]
- 21.Ben-Ari Y., Brody Y., Shav-Tal Y. The life of an mRNA in space and time. J. Cell Sci. 2010;123:1761–1774. doi: 10.1242/jcs.062638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molina N., Suter D.M., Naef F. Stimulus-induced modulation of transcriptional bursting in a single mammalian gene. Proc. Natl. Acad. Sci. USA. 2013;110:20563–20568. doi: 10.1073/pnas.1312310110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sudol M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene. 1994;9:2145–2152. [PubMed] [Google Scholar]
- 24.Yu F.X., Zhao B., Guan K.L. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wohland T. National University of Singapore; 2015. imFCS Plugin for ImageJ.http://www.dbs.nus.edu.sg/lab/BFL/imfcs_image_j_plugin.html [Google Scholar]
- 26.Krieger, J. W., and J. Langowski. 2015. QuickFit 3.0 (GPL 3.0). https://github.com/jkriege2/QuickFit3.
- 27.Sankaran J., Bag N., Wohland T. Accuracy and precision in camera-based fluorescence correlation spectroscopy measurements. Anal. Chem. 2013;85:3948–3954. doi: 10.1021/ac303485t. [DOI] [PubMed] [Google Scholar]
- 28.Maharana S., Sharma D., Shivashankar G.V. Dynamic organization of transcription compartments is dependent on functional nuclear architecture. Biophys. J. 2012;103:851–859. doi: 10.1016/j.bpj.2012.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bag N., Huang S., Wohland T. Plasma membrane organization of epidermal growth factor receptor in resting and ligand-bound states. Biophys. J. 2015;109:1925–1936. doi: 10.1016/j.bpj.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wawrezinieck L., Rigneault H., Lenne P.-F. Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys. J. 2005;89:4029–4042. doi: 10.1529/biophysj.105.067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu S.-W., Wang H., Dawson V.L. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 32.Kim S.A., Heinze K.G., Schwille P. Fluorescence correlation spectroscopy in living cells. Nat. Methods. 2007;4:963–973. doi: 10.1038/nmeth1104. [DOI] [PubMed] [Google Scholar]
- 33.Altan-Bonnet N., Altan-Bonnet G. Fluorescence correlation spectroscopy in living cells: a practical approach. Curr. Protoc. Cell Biol. 2009;4:24. doi: 10.1002/0471143030.cb0424s45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cisse I.I., Izeddin I., Darzacq X. Real-time dynamics of RNA polymerase II clustering in live human cells. Science. 2013;341:664–667. doi: 10.1126/science.1239053. [DOI] [PubMed] [Google Scholar]
- 35.Buchholz J., Krieger J.W., Langowski J. FPGA implementation of a 32x32 autocorrelator array for analysis of fast image series. Opt. Express. 2012;20:17767–17782. doi: 10.1364/OE.20.017767. [DOI] [PubMed] [Google Scholar]
- 36.Izeddin I., Récamier V., Darzacq X. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. eLife. 2014;3:e02230. doi: 10.7554/eLife.02230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorski S.A., Dundr M., Misteli T. The road much traveled: trafficking in the cell nucleus. Curr. Opin. Cell Biol. 2006;18:284–290. doi: 10.1016/j.ceb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Cremer T., Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 39.Horn P.J., Peterson C.L. Molecular biology. Chromatin higher order folding—wrapping up transcription. Science. 2002;297:1824–1827. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- 40.Zhao Z.W., Roy R., Xie X.S. Spatial organization of RNA polymerase II inside a mammalian cell nucleus revealed by reflected light-sheet superresolution microscopy. Proc. Natl. Acad. Sci. USA. 2014;111:681–686. doi: 10.1073/pnas.1318496111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X., Wei M., Sun Y. Study of RNA Polymerase II clustering inside live-cell nuclei using Bayesian nanoscopy. ACS Nano. 2016;10:2447–2454. doi: 10.1021/acsnano.5b07257. [DOI] [PubMed] [Google Scholar]
- 42.Ng X.W., Teh C., Wohland T. The secreted signaling protein Wnt3 is associated with membrane domains in vivo: a SPIM-FCS study. Biophys. J. 2016;111:418–429. doi: 10.1016/j.bpj.2016.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grünwald D., Martin R.M., Cardoso M.C. Probing intranuclear environments at the single-molecule level. Biophys. J. 2008;94:2847–2858. doi: 10.1529/biophysj.107.115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halder G., Dupont S., Piccolo S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- 45.Santinon G., Pocaterra A., Dupont S. Control of YAP/TAZ activity by metabolic and nutrient-sensing pathways. Trends Cell Biol. 2016;26:289–299. doi: 10.1016/j.tcb.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 46.Chen J., Zhang Z., Liu Z. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.