Abstract
Coronaviruses encode the largest replicase polyprotein of any known positive-strand RNA virus. Replicase protein precursors and mature products are thought to mediate the formation and function of viral replication complexes on the surfaces of intracellular double-membrane vesicles. However, the functions of only a few of these proteins are known. For the coronavirus mouse hepatitis virus (MHV), the first proteolytic processing event of the replicase polyprotein liberates an amino-terminal 28-kDa product (p28). While previous biochemical studies have suggested that p28 is associated with viral replication complexes, the intracellular localization and interactions of p28 with other proteins during the course of MHV replication have not been defined. We used immunofluorescence confocal microscopy to show that p28 localizes to viral replication complexes in the cytoplasm during early times postinfection. However, at late times postinfection, p28 localizes to sites of M accumulation distinct from the replication complex. Furthermore, by yeast two-hybrid and coimmunoprecipitation analyses, we demonstrate that p28 specifically binds to p10 and p15, two coronavirus replicase proteins of unknown function. Deletion mutagenesis experiments determined that the carboxy terminus of p28 is not required for its interactions with p10 and p15. These results suggest that p28 may play a part at the replication complex by interacting with p10 and p15. Moreover, our findings highlight a potential role for p28 at virion assembly sites.
Coronaviruses are enveloped, positive-strand RNA viruses that cause a wide spectrum of disease in both humans and animals. The identification of a newly discovered human coronavirus as the causative agent of severe acute respiratory syndrome (SARS) underscores the pathogenic potential of this diverse virus family. However, the mechanisms by which coronaviruses replicate are not fully understood. Based on genome organization and phylogenetic analysis of gene 1 (replicase gene), coronaviruses are categorized into three distinct groups (1, 2, and 3), with the SARS coronavirus (SARS-CoV) being most closely related to group 2 (25, 34). Mouse hepatitis virus (MHV) is a prototype group 2 coronavirus and a model for studies of viral replication and pathogenesis. MHV and SARS-CoV share orthologous protein domains encoded by the 5′ end of the replicase gene that are absent in coronaviruses belonging to groups 1 and 3 (25, 34). Although the functions of these amino-terminal replicase proteins are not known, the similarity between MHV and SARS-CoV suggests that the proteins may have important roles during group 2-like coronavirus replication. Thus, determination of the intracellular localization and interactions of these unique replicase proteins during MHV replication may provide insight into processes by which the SARS-CoV replicates and causes disease.
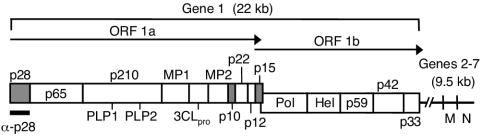
Following viral attachment and cell entry, the first event in the MHV life cycle is translation of the 22-kb replicase gene to yield two large polyproteins from two overlapping open reading frames (ORF1a and ORF1b). During an infectious cycle, ORF1a may be translated into a predicted 495-kDa polyprotein or ORF1a-ORF1b may be translated into an 803-kDa fusion polyprotein as a consequence of a −1 ribosomal frameshift (3, 6, 7, 24, 27). These polyproteins are cleaved both co- and posttranslationally by two papain-like proteinases (PLP1 and PLP2) and a picornavirus 3C-like proteinase (3CLpro) to generate >15 protein products. Mature MHV replicase proteins and precursors are presumed to mediate viral RNA synthesis at replication complexes on the surfaces of intracellular double-membrane vesicles (18, 28, 31, 39). However, aside from the viral proteinases, an RNA-dependent RNA polymerase (Pol), an RNA helicase (Hel), and putative RNA-processing enzymes, there are no known or predicted functions for many of the replicase proteins during MHV infection (17, 24, 35). Once genome replication is completed, MHV particle assembly occurs in the endoplasmic reticulum-Golgi intermediate compartment (ERGIC), distinct from sites of RNA synthesis (4, 5, 22, 23). These observations suggest that MHV uses a mechanism for delivering newly synthesized genome molecules to be packaged into virions. It is not known how MHV regulates the processes of RNA synthesis and virion assembly during an infectious cycle, and viral and cellular factors required for these events have not been identified.
The processing strategy of the replicase polyprotein is conserved among the studied coronaviruses, with the exception of proteins expressed from the amino terminus. Specifically, the group 2 coronaviruses cleave unique 28- and 65-kDa amino-terminal proteins (p28 and p65, respectively) (10, 12). The functions of p28 and p65 are not known, and neither protein is expressed during the infectious cycle of the group 1 and 3 coronaviruses. Like the group 2 coronaviruses, SARS-CoV is predicted to cleave analogous 20- and 70-kDa proteins (nsp1 and nsp2, respectively) (25, 34, 35). For MHV, the liberation of p28 by PLP1 is the first cleavage event in the maturation of the replicase polyprotein (10, 13, 16). p28 is likely cleaved immediately after translation of the PLP1 domain within p210, and p28 appears to be stable in infected cells for several hours after cleavage (15, 16). Determinants of p28 cleavage have been defined in vitro (16, 20, 21) and recently confirmed by direct mutagenesis of the cleavage site using a reverse genetic approach (11, 40). Interestingly, MHV mutants that are incapable of liberating p28 from the nascent polyprotein exhibit delayed growth, diminished peak titers, small plaques, and reduced RNA synthesis in comparison to wild-type controls (11). While it has not been proven that the defects in these mutant viruses are strictly related to p28 function, these results emphasize the importance of p28 cleavage for optimal viral RNA synthesis and suggest that p28 might play an important role in the function of MHV replication complexes.
MHV p28 is associated with intracellular membranes, but it does not have characteristics of an integral membrane protein (8, 18, 33). After homogenization and differential centrifugation of membranes from MHV-infected cells, p28 is detected in a membrane-containing fraction, along with several MHV proteins that are known components of MHV replication complexes. These proteins include the viral nucleocapsid (N), Hel, Pol, and at least two replicase proteins of unknown function (p65 and p22) (8, 33). Density gradient analysis of intracellular membranes from MHV-infected cells shows that p28 is distinct from Pol, p65, and p22 and instead cofractionates with Hel, N, and viral RNA in the denser, more soluble fractions (8, 33). Immunofluorescence confocal-microscopy experiments show that Hel and N are retained at viral replication complexes during early times of infection (4 to 6 h postinfection [p.i.]). However, at later times (6 to 12 h p.i.), these proteins colocalize with the virion structural protein M and markers for the ERGIC (5, 8, 23, 28). It is not known whether p28 localizes to viral replication complexes or whether it is a component of the Hel-N-RNA complex that may translocate to virion assembly sites. Because p28 lacks predicted enzymatic domains, it is possible that this protein functions by interacting with other proteins or viral RNA. Thus far, p28 has not been shown to bind viral RNA, and there has been no description of interactions between p28 and any viral or cellular proteins.
To better understand the roles of p28 during MHV replication, we defined the intracellular localization of p28 during an infectious cycle and identified viral proteins that interact with p28. Using immunofluorescence confocal microscopy, we found that p28 colocalizes with p65, p10, p22, p15, Pol, Hel, and N at MHV replication complexes at 6 h p.i. However, by 9 h p.i., p28 is distinct from MHV replication complexes and instead localizes to sites of M accumulation and virion assembly. Additionally, specific interactions between p28 and the replicase proteins p10 and p15 were identified using yeast two-hybrid and coimmunoprecipitation experiments. Deletion mutagenesis analysis demonstrated that residues within the carboxy-terminal 81-amino-acid region of p28 are not required for p10 and p15 binding. Together, these results suggest that p28 may play a role at the MHV replication complex by interacting with p10 and p15. Moreover, the localization of p28 to replication complexes at early times of MHV infection and to viral assembly sites at late times is consistent with the involvement of this protein in the delivery of genomic RNA for packaging into progeny virions.
MATERIALS AND METHODS
Virus, cells, and antisera.
Delayed brain tumor (DBT) cell monolayers (19) were infected with MHV-A59 at a multiplicity of infection of 10 PFU per cell in Dulbecco modified Eagle medium (DMEM) that was supplemented to contain 10% fetal calf serum (FCS) for all experiments.
Polyclonal antisera used for biochemical experiments have been previously described. These include α-p65 (33), B1 (α-hel) (14), VU145 (α-pol) (8), α-p22, α-p12, α-p10, and α-p15 (4). Two monoclonal antibodies specific for the structural proteins nucleocapsid (α-N; J.3.3) and M (α-M; J.1.3) were generously provided by J. Fleming (University of Wisconsin, Madison).
Guinea pig polyclonal α-p28 antiserum (GP3) and rabbit polyclonal α-p28 antiserum (VU221) were generated by Cocalico, Inc., using a recombinant p28 protein as an antigen. All p28 nucleotide and amino acid numbers correspond to the MHV-A59 sequence reported by Bonilla et al. (3). Nucleotides (nt) 210 to 950 (gene 1 amino acids [aa] M1 to G247) were amplified by reverse transcription-PCR using purified MHV-A59 genomic RNA as a template. Primer-generated restriction sites (5′ EcoRI and 3′ XhoI) were used to subclone the PCR fragment into the pET23a bacterial expression vector (Novagen). A 28-kDa histidine-tagged protein was expressed in Escherichia coli BL21 cells, isolated using nickel resin chromatography according to the manufacturer's protocol, and further purified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electroelution (Bio-Rad) as previously described (4).
Radiolabeling MHV proteins and immunoprecipitation of cell lysates.
Confluent monolayers of DBT cells in 60-mm-diameter cell culture dishes (∼3 × 106 cells) were either infected with MHV or mock infected with 10% FCS DMEM. At 2.5 h p.i., the medium was replaced with fresh 5% FCS DMEM lacking methionine and cysteine and supplemented with actinomycin D (5 μg/ml). The cells were incubated from 5 to 8 h p.i. with 100 μCi of [35S]methionine-cysteine (Translabel; ICN)/ml. The cells in each 60-mm-diameter dish were washed using 500 μl of 1 M Tris and then lysed in 300 μl of buffer A (150 mM NaCl, 1% NP-40, 0.5% deoxycholate [DOC], 50 mM Tris, pH 8.0). The lysates were subjected to centrifugation at 3,500 × g to remove cell nuclei. Immunoprecipitations were performed in a final volume of 1 ml, using protein A-Sepharose beads (Sigma), 100 μl of radiolabeled lysate (derived from ∼106 cells), and 2 to 10 μl of polyclonal antiserum in either buffer A (150 mM NaCl, 1% NP-40, 0.5% DOC, 50 mM Tris, pH 8.0) or buffer B (150 mM NaCl, 1% NP-40, 1% DOC, 0.1% SDS, 50 mM Tris, pH 7.4) or, after the lysate was boiled for 5 min in 1% SDS, in buffer C (150 mM NaCl, 1% NP-40, 1% DOC, 1% SDS, 10 mM Tris, pH 7.4). Protein-bead conjugates were washed three times in the same buffer used for immunoprecipitations, and the proteins were eluted from the beads, followed by boiling for 5 min in 2× protein loading buffer (200 mM dithiothreitol, 100 mM Tris, pH 6.8, 0.04% bromophenol blue, 20% glycerol). The proteins were resolved by SDS-PAGE in 5 to 18% polyacrylamide gradient gels and analyzed by fluorography. The [14C] high-molecular-weight standard (Gibco) and full-range rainbow marker (Invitrogen) were used as molecular weight standards. Estimated molecular mass determinations of coprecipitating proteins were calculated from regression curves of log molecular weight versus relative mobility established using these standards.
Immunofluorescence assays.
DBT cells grown on glass coverslips were either mock infected or infected with MHV and then rocked gently at 25°C for 30 min. Following virus adsorption, the infected medium was replaced with fresh prewarmed 10% FCS DMEM, and the cells were incubated at 37°C. At specific times p.i., the cells were fixed and permeabilized with −20°C 100% methanol. Indirect immunofluorescence assays were performed as previously described (5). Guinea pig polyclonal α-p28 antiserum (GP3) was used at 1:2,000 dilution, and all rabbit polyclonal antisera were used at 1:100 dilution, with the exception of α-pol VU145, which was used at 1:500. Mouse monoclonal antibodies were used at 1:1,000 dilution. Secondary antibodies conjugated to fluorophores were used at 1:1,000 dilution and included α-guinea pig-Alexa 546, α-rabbit-Alexa 488, and α-mouse-Alexa 633. Immunofluorescence was detected using a Zeiss LSM 510 laser scanning confocal microscope with a 40× oil immersion objective. Image analysis and merging was performed using Adobe Photoshop version 7.0.
Quantitation of percent colocalization between two viral proteins was performed using Metamorph Imaging software (Universal Imaging). Background fluorescence was determined empirically by staining mock-infected cells with immune sera. The lower-limit threshold was set to exclude pixels below background. The upper-limit level was set to exclude saturated pixels. For colocalization measurements of each protein pair at each time point, three independent images (∼5 cells per image) were acquired and processed.
Cloning MHV protein-coding domains into yeast expression vectors.
Protein-coding regions corresponding to p28, p65, p10, p15, Hel, Pol, and N were cloned into the Matchmaker System 3 yeast expression vectors (Clontech). pGBKT7 [GAL4 (amino acids 1 through 147) DNA binding domain (BD) TRP1 Kanr c-Myc epitope tag] was used for bait constructs, and pGADT7 [GAL4 (amino acids 768 through 881) DNA activation domain (AD) LEU2 Ampr influenza virus hemagglutinin (HA) epitope tag] was used for prey constructs. PCR was used to amplify the region of interest, and primer-generated restriction sites allowed the cloning of PCR products into pGBKT7 and pGADT7. All nucleotide and amino acid sequence numbers correspond to the MHV-A59 sequence reported by Bonilla et al. and Bredenbeek et al. (3, 6). Plasmids used for constructing the MHV infectious clone (fragments A, D, E, F, and G) were used as PCR templates (40). The protein-coding regions of all constructs were sequenced to verify the fidelity of PCR amplification and cloning.
Plasmids pGBK-p28 and pGAD-p28 were generated by using PCR to amplify nt 210 to 950 (aa M1 to G247) from fragment A and subcloned using 5′ NdeI and 3′ EcoRI. Plasmids pGBK-p65 and pGAD-p65 were generated by amplification of nt 951 to 2705 (aa V248 to A832) from fragment A and subcloned using 5′ SfiI and 3′ BamHI. Plasmids pGBK-p10 and pGAD-p10 (nt 11973 to 12248; aa S2180 to Q2275) and plasmids pGBK-p15 and pGAD-p15 (nt 13161 to 13571; aa A4320 to Q4456) were amplified from fragment D and fragment E, respectively, and subcloned using 5′ SfiI and 3′ EcoRI. Plasmids pGBK-pol and pGAD-pol were generated by transferring the Pol-coding domain from pEGFP-C1-pol (8) into the yeast expression vectors using 5′ EcoRI and 3′ BamHI. Plasmids pGBK-hel and pGAD-hel were generated by amplifying nt 16355 to 18154 (aa S5385 to Q5984) from fragment F and subcloned using 5′ EcoRI to 3′ BamHI. For plasmids pGBK-N and pGAD-N, the N-coding domain was amplified from fragment G (pMH54) and subcloned using 5′ NdeI to 3′ BamHI.
Deletion mutagenesis of p28.
To generate carboxy-terminal p28 truncations, PCR was used to amplify the p28 coding sequence with progressive deletions of the 3′ end. The sequence of the left primer for both mutants was 5′ GGC CAT GGA GGC CAT GGC AAA GAT GGG CAA A 3′. The sequences of the right primers were 5′ GAA TTC ACG GCC ATT ACC CAG GCA 3′ (p28ΔC1) and 5′ GAA TTC ATC TAC AAA AAT ATC ACG 3′ (p28ΔC2). PCR products corresponding to p28ΔC1 (aa M1 to R166) and p28ΔC2 (aa M1 to D83) were cloned into pGEM-T-Easy (Promega) and sequenced by using primers corresponding to the T7 and SP6 promoter sequences. Primer-generated restriction sites (5′ SfiI and 3′ EcoRI) were used to subclone p28ΔC1 and p28ΔC2 sequences into pGADT7 and pGBKT7 (Clontech).
Yeast two-hybrid assay.
Saccharomyces cerevisiae strain AH109 (Clontech) with the HIS3, ADE2, and MEL1 reporter genes downstream of a GAL4-responsive promoter element, was cotransformed with pairwise combinations of bait and prey vectors with lithium acetate (Matchmaker System 3; Clontech), as described in the manufacturer's protocol. Yeast cells were plated onto synthetic dropout medium lacking tryptophan, histidine, leucine, and adenine in the presence of 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (X-α-Gal; Sigma) to select for yeast containing interacting proteins.
In vitro translation and coimmunoprecipitations of epitope-tagged proteins.
Epitope-tagged proteins were expressed from the bait and prey vectors using rabbit reticulocyte lysate (TNT T7 Quick-coupled Transcription/Translation kit; Promega) in either the presence or absence of 100 μCi of [35S]methionine (Translabel; ICN)/ml. Equal amounts of radiolabeled and nonradiolabeled reticulocyte lysates were mixed together at room temperature for 90 min and then subjected to immunoprecipitation with either mouse α-c-Myc monoclonal antibodies or rabbit polyclonal α-HA antibodies conjugated to protein G-Sepharose beads (Sigma) in a buffer containing 300 mM NaCl and 1% Triton X-100. The bead-protein conjugates were washed six times with immunoprecipitation buffer. Proteins were eluted from the beads and resolved by SDS-PAGE in 4 to 20% polyacrylamide gradient gels (Ready Gel; Bio-Rad) and analyzed by fluorography.
RESULTS
Detection and intracellular localization of p28 in MHV-infected cells.
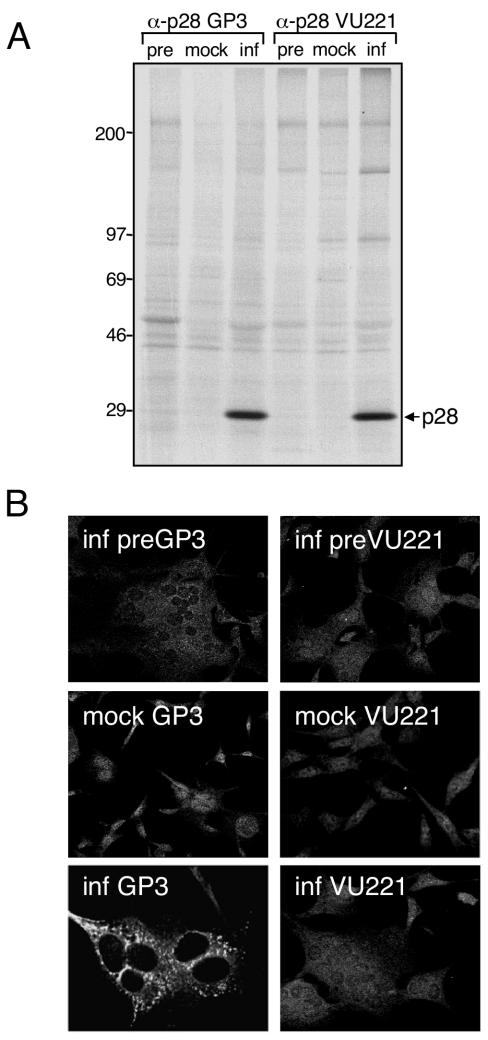
To define the intracellular localization and protein interactions of p28 during MHV infection, we generated polyclonal α-p28 antibodies that would recognize p28 in both biochemical and immunofluorescence assays. Guinea pig (GP3) and rabbit polyclonal α-p28 antisera (VU221) were raised against a recombinant p28 protein (Fig. 1) and used to probe mock-infected and MHV-infected DBT cells by immunoprecipitation (Fig. 2A) and indirect immunofluorescence (Fig. 2B). When lysates from radiolabeled MHV-infected cells were immunoprecipitated using either GP3 or VU221, a 28-kDa protein band consistent with the size of p28 following SDS-PAGE was detected (Fig. 2A). No 28-kDa protein was detected after immunoprecipitation of lysate from mock-infected cells with either GP3 or VU221 or after immunoprecipitation of lysate from infected cells with preimmune serum from the same animal (Fig. 2A). These results demonstrate that both α-p28 antisera (GP3 and VU221) specifically recognize p28 in MHV-infected cells.
FIG. 1.
MHV genome organization. The diagram shows the organization of the 22-kb MHV-A59 gene 1 (replicase gene) and the locations of genes 6 and 7, encoding the structural proteins M and N, respectively. Gene 1 is composed of two open reading frames (ORF1a and ORF1b). The ORF1a-ORF1b fusion replicase polyprotein is shown, with mature replicase proteins represented as boxes. Proteins with confirmed or predicted functions include two papain-like proteinases (PLP1 and PLP2), the 3C-like proteinase (3CLpro), two transmembrane proteins (MP1 and MP2), the RNA-dependent RNA polymerase (Pol), and the RNA helicase (Hel). All other proteins are labeled based on their molecular masses in kilodaltons. The gray boxes represent replicase proteins of interest in the present study: the amino-terminal cleavage product, p28, and two carboxy-terminal ORF1a proteins, p10 and p15. The region of the gene 1 polyprotein (amino acids M1 to G247) used to generate α-p28 guinea pig (GP3) and rabbit (VU221) polyclonal antisera is indicated.
FIG. 2.
Detection of p28 by immunoprecipitation and immunofluorescence. (A) Immunoprecipitation of p28. Lysates from either mock-infected (mock) or MHV-infected (inf) radiolabeled DBT cells were immunoprecipitated using antiserum raised against p28 (GP3 or VU221) in buffer C as described in Materials and Methods. Lysate from infected cells was also immunoprecipitated using preimmune sera from the same animal (pre). Bands corresponding to p28 are indicated on the right of the gel, and molecular mass standards (in kilodaltons) are shown on the left. (B) Immunofluorescence analysis using p28 antisera. DBT cells grown on glass coverslips were either mock infected or infected with MHV for 6 h, fixed and permeabilized with 100% methanol, and incubated with GP3, VU221, or preimmune sera using an indirect immunofluorescence assay. The cells were imaged using a Zeiss LSM 510 confocal microscope at 546 nm. The images are single confocal slices obtained using a 40× objective and are representative of the cell population. Multinucleated cells are a cytopathic effect of MHV replication.
To determine whether GP3 or VU221 recognizes p28 during immunofluorescence assays, DBT cells grown on glass coverslips were either mock infected or infected with MHV, fixed and permeablized with methanol, and incubated with preimmune or immune sera in an indirect immunofluorescence assay. Protein staining was detected using a laser scanning confocal microscope (Fig. 2B). Although the α-p28 rabbit antiserum (VU221) was specific for p28 during immunoprecipitation experiments, VU221 did not detect p28 in immunofluorescence assays. Cells stained with VU221 displayed a diffuse granular pattern of fluorescence in both mock-infected and MHV-infected cells, similar to that in infected cells incubated with preimmune sera (Fig. 2B). In contrast, MHV-infected cells incubated with α-p28 guinea pig antiserum (GP3) demonstrated bright, punctate foci of specific fluorescence strictly limited to the cell cytoplasm (Fig. 2B). This pattern of p28 staining resembles that of other MHV proteins that are known to localize to viral replication complexes on intracellular membranes (2, 4, 8, 14, 33, 39). Diffuse staining of both the cytoplasm and the nucleus was detected in mock-infected cells probed with GP3 and infected cells probed with preimmune sera (Fig. 2B), suggesting that the staining observed at these sites was not specific for p28.
p28 localizes to MHV replication complexes at 6 h p.i. and is associated with sites of M accumulation by 9 h p.i.
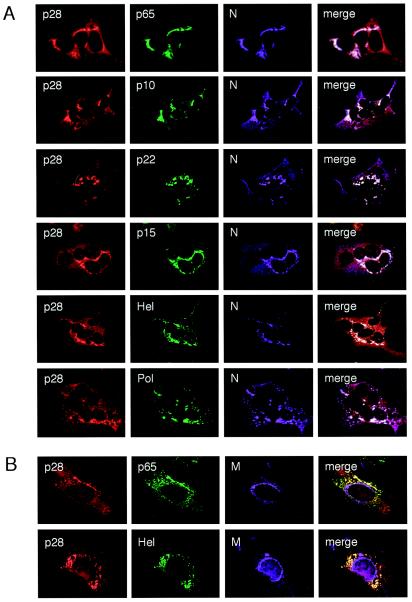
To determine whether the α-p28 GP3 staining in infected cells was positive for markers of viral replication complexes, triple-label immunofluorescence experiments were performed (Fig. 3). DBT cells grown on glass coverslips were infected with MHV for 6 h, fixed and permeabilized with methanol, and incubated with antibodies specific for various MHV replicase and structural proteins. In all cases, p28 colocalized predominantly with proteins that are components of the viral replication complex, including Pol, Hel, N, and four replicase proteins of unknown function (p65, p10, p22, and p15) (Fig. 3A). In contrast, p28 did not colocalize with the viral M protein, which accumulates at sites of virion assembly in the ERGIC (22, 23) (Fig. 3B). These findings demonstrate that at 6 h p.i., p28 localizes to viral replication complexes distinct from sites of M accumulation.
FIG. 3.
Triple-label immunofluorescent images of MHV-infected DBT cells at 6 h p.i. (A) p28 colocalizes with MHV replication complexes. DBT cells grown on glass coverslips were infected with MHV for 6 h, fixed and permeabilized with 100% methanol, and incubated with antibodies against p28 (GP3) (red), various MHV replicase proteins (green), and N (purple). Colocalization of all three colors is shown as white pixels. The cells were imaged using a Zeiss LSM 510 confocal microscope at 488, 546, and 633 nm. The images are single confocal slices obtained using a 40× objective and are representative of the cell population. (B) p28 is distinct from sites of M accumulation. Cells were incubated with antibodies against p28 (GP3) (red), replicase protein p65 or Hel (green), and the viral M protein (purple) as a marker of virion assembly sites. Colocalization of red and green is shown as yellow pixels.
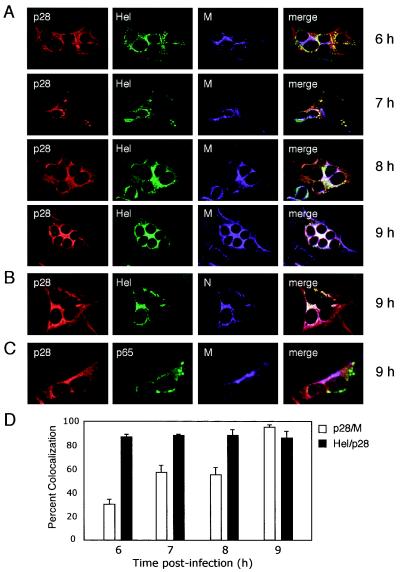
Previous studies have shown that Hel and N are detected at replication complexes at 6 h p.i. but then are observed at sites of MHV assembly at late times p.i (5, 8, 28). To determine whether p28 colocalizes with Hel and N at virion assembly sites or remains associated with replication complexes, DBT cells grown on glass coverslips were infected, fixed at 6, 7, 8, and 9 h p.i., and incubated with antibodies against p28, p65, Hel, N, and M (Fig. 4). At 6 and 7 h p.i., p28 colocalized with Hel in punctate cytoplasmic structures that did not predominantly overlap with M staining (Fig. 4A). In contrast, at 8 and 9 h p.i., the pattern of p28 staining changed from punctate cytoplasmic foci to a contiguous perinuclear pattern that resembled ERGIC staining (5) (Fig. 4A). p28 remained colocalized with Hel and N over the entire time course analyzed and significantly overlapped with M staining by 9 h p.i. (Fig. 4A and B). In contrast, p65 remained in cytoplasmic foci distinct from both p28 and M (Fig. 4C). The percent colocalization of p28 with Hel, and of p28 with M, at various times p.i. was calculated after quantitation of immunofluorescence images using Metamorph computer imaging software (Fig. 4D). The percentage of p28 colocalized with M changed from 25% at 6 h p.i. to nearly 95% by 9 h p.i., while the percent colocalization of p28 with Hel remained ∼90% over the time course. These results suggest that at late times of infection, p28 is separate from replication complexes and instead colocalizes with Hel and N at sites of M accumulation and virion assembly.
FIG. 4.
Time course of p28 localization during MHV infection. MHV-infected DBT cells grown on glass coverslips were fixed at the times p.i. shown on the right of the images and stained with antibodies against viral replicase and structural proteins. (A) p28 colocalizes with Hel over the course of MHV infection. Individual coverslips were stained with antibodies against p28 (GP3) (red), Hel (green), and M (purple), a marker for sites of virion assembly. (B) p28 is associated with Hel and N at 9 h p.i. Coverslips from the 9-h time point were stained with antibodies against p28 (GP3) (red), Hel (green), and N (purple). Colocalization of all three colors is shown as white pixels. (C) p28 is distinct from replication complexes at 9 h p.i. Coverslips from the 9-h time point were incubated with antibodies against p28 (GP3) (red), M (purple), and the replicase protein p65 (green) as a marker for replication complex staining at late times p.i. Colocalization of red and purple is shown as pink pixels. (D) Quantitation of percent colocalization of p28, Hel, and M. Percent colocalization was determined using Metamorph Imaging software. The background fluorescence was determined empirically by staining mock-infected cells with immune sera. The lower-limit threshold was set to exclude pixels below background. The upper-limit level was set to exclude saturated pixels. For colocalization measurements of each protein pair at each time point, three independent images (∼5 cells per image) were acquired and processed. The error bars indicate standard deviations.
Replicase proteins p28, p10, and p15 interact in yeast two-hybrid and coimmunoprecipitation assays.
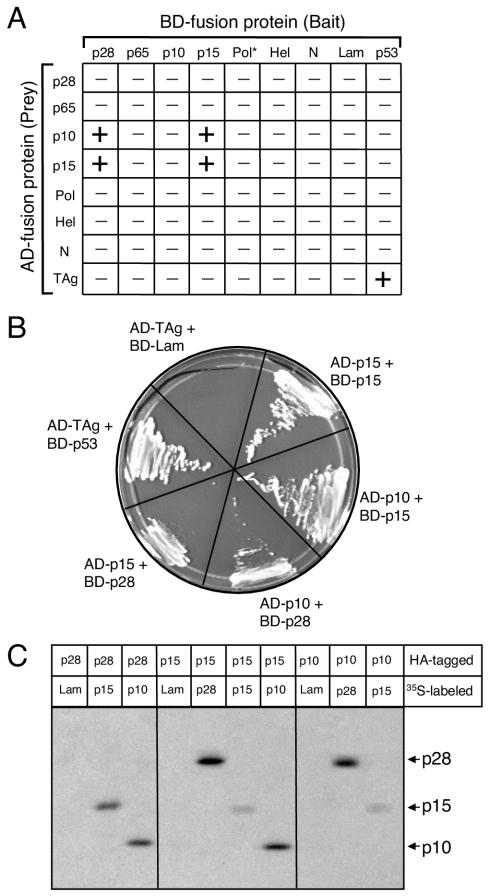
Having demonstrated that p28 associates with the MHV replication complex at early times of infection and colocalizes with Hel and M at late times of infection, we next sought to determine whether p28 specifically interacts with other viral proteins. Sequences encoding p28, p65, p10, p15, Hel, Pol, and N were cloned either as bait, fused to the carboxy terminus of the GAL4 DNA BD, or as prey, fused to the carboxy terminus of the GAL4 AD, for use in yeast two-hybrid experiments. The bait and prey fusion proteins were expressed in pairwise combinations in the budding yeast strain AH109 (Fig. 5A). Interactions of bait and prey proteins cause the activation of the reporter genes ADE2, HIS3, and MEL1, allowing the survival of yeast on minimal media and blue-white screening of yeast in the presence of X-α-Gal. The MHV proteins expressed alone did not induce reporter gene activation, with the exception of the Pol bait fusion protein (BD-Pol), which resulted in autonomous activation of MEL1 and expression of α-Gal in the yeast.
FIG. 5.
Interactions between MHV proteins using yeast two-hybrid and coimmunoprecipitation assays. (A) Pairwise cotransformations. Various MHV replicase proteins and N were expressed as either bait (fused to the GAL4 BD) or prey (fused to the GAL4 AD) in pairwise combinations using the yeast two-hybrid system. Yeast expressing both bait and prey fusion proteins was scored for the capacity to grow on medium lacking Trp, Leu, Ade, and His and by blue-white screening in the presence of X-α-Gal. In the matrix shown, − indicates lack of growth and + indicates both growth and blue color. The interaction between TAg and p53 served as a positive control for the experiment; TAg and lamin C (Lam) served as a negative control. The asterisk indicates that BD-Pol activates the reporter gene MEL1. (B) Interactions among p28, p10, and p15. Single yeast colonies containing the indicated bait and prey plasmids were streaked onto medium lacking Trp, Leu, Ade, or His to confirm growth. (C) Coimmunoprecipitation of in vitro-expressed proteins. Proteins were translated, using reticulocyte lysate, as fusions to c-Myc epitope tags in the presence of [35S]methionine or as fusions to HA epitope tags in the absence of radiolabel. Equal amounts of radiolabeled and nonradiolabeled lysates were combined, and the proteins were immunoprecipitated using rabbit polyclonal α-HA antiserum. Interacting 35S-labeled proteins were resolved in an SDS-12% polyacrylamide gel and analyzed by fluorography. The identities of coprecipitating proteins are shown on the right of the gel.
Following the pairwise cotransformations, p28 bait (BD-p28) interacted with prey proteins p10 (AD-p10) and p15 (AD-p15). Furthermore, p15 bait (BD-p15) interacted with prey p10 (AD-p10) and prey p15 (AD-p15) (Fig. 5A and 5B). Reciprocal interactions between BD-p10 or BD-p15 and AD-p28 were not detected, nor was an interaction between BD-p10 and AD-p15. Whether the lack of reciprocal interactions was due to differences in bait and prey protein folding or structure remains to be determined. The well-established interaction between the simian virus 40 large T antigen (AD-TAg) and the cellular protein p53 was used as a positive control, while AD-TAg and the 27-kDa lamin C protein (BD-Lam) was used as a negative control (Fig. 5A and B). The MHV bait and prey fusion proteins did not interact with AD-TAg, BD-Lam, or empty vector bait and prey controls, suggesting that p28, p10, and p15 interactions in yeast are specific (Fig. 5A). No additional interactions between any other MHV proteins were detected.
We next conducted experiments to determine whether p28, p10, and p15 interact when coexpressed in cultured cells. However, we found that exogenous expression of p28 using either a retroviral vector or a cytomegalovirus immediate-early promoter-driven mammalian expression vector induced growth arrest and death in multiple cell types (data not shown). Therefore, to provide further evidence that p28, p10, and p15 physically interact in the absence of other viral proteins, we expressed c-Myc and HA epitope-tagged fusion proteins in coupled transcription-translation reactions for use in coimmunoprecipitation experiments (Fig. 5C). To verify protein expression, the epitope-tagged proteins were translated in vitro using reticulocyte lysates in the presence of [35S]methionine and immunoprecipitated using either α-HA rabbit polyclonal antiserum or a mouse monoclonal α-c-Myc antibody and resolved by SDS-PAGE. As anticipated, all constructs were expressed as epitope-tagged fusion proteins of the appropriate size (data not shown). For coimmunoprecipitation studies, c-Myc-tagged proteins were translated in the presence of [35S]methionine, while HA-tagged proteins were translated in the absence of radiolabel. After equal amounts of radiolabeled and nonradiolabeled lysates were mixed in different combinations, the proteins were immunoprecipitated using α-HA rabbit polyclonal antiserum. Radiolabeled c-Myc-tagged interacting proteins were detected following SDS-PAGE and fluorography (Fig. 5C).
Lysates expressing unlabeled HA-p28 coimmunoprecipitated radiolabeled 10- and 15-kDa proteins but did not coimmunoprecipitate the control 27-kDa protein lamin C (Fig. 5C) or the replicase protein p65 (data not shown). Likewise, HA-p10 or HA-p15 coimmunoprecipitated a radiolabeled 28-kDa protein but not lamin C, suggesting that p28 interacts directly and reciprocally with both p10 and p15 (Fig. 5C). Furthermore, HA-p15 coprecipitated radiolabeled 10- and 15-kDa proteins, and HA-p10 coprecipitated a radiolabeled 15-kDa protein (Fig. 5C). Thus, these coimmunoprecipitation experiments confirm the yeast two-hybrid assay results and suggest that interactions occur between p28 and p10, p28 and p15, p15 and p10, and p15 and itself.
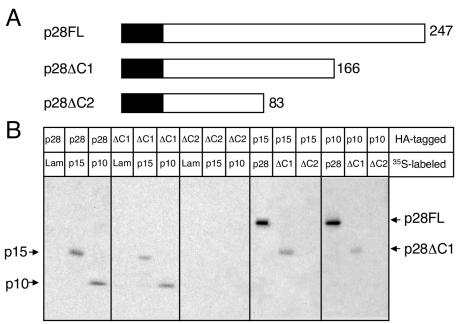
The carboxy terminus of p28 is not required for binding to p10 and p15.
To define a region of p28 required for interactions with p10 and p15, we generated epitope-tagged carboxy-terminal p28 truncation mutants (p28ΔC1 and p28ΔC2) (Fig. 6A). The p28 truncation mutants were translated in vitro using reticulocyte lysates and tested for the capacity to bind p10 and p15 using coimmunoprecipitation assays (Fig. 6B). Each of the constructs was expressed as an epitope-tagged fusion protein of the appropriate mass: p28FL (28 kDa), p28ΔC1 (18 kDa), and p28ΔC2 (9 kDa) (data not shown). Lysates expressing either HA-p28FL or HA-p28ΔC1 were capable of specific coimmunoprecipitation of radiolabeled p10 and p15. In contrast, HA-p28ΔC2 was incapable of coprecipitating p15 or p10. Likewise, HA-p10 and HA-p15 coprecipitated p28FL and p28ΔC1 but not p28ΔC2 (9 kDa). The control protein lamin C was not coimmunoprecipitated by p28FL, p28ΔC1, p28ΔC2, p10, or p15 (Fig. 6B). These results suggest that the carboxy-terminal residues F167 to G247 are not required for p28 to interact with p10 and p15 (Fig. 6B). This result further suggests that residues within the region E84 to R166 may be required for p10 and p15 binding.
FIG. 6.
The carboxy terminus of p28 is not required for interactions with p10 and p15. (A) p28 deletion mutagenesis. A schematic of full-length 247-amino-acid p28 (p28FL) is shown with the two carboxy-terminal truncation mutants, p28ΔC1 and p28ΔC2, below p28FL. The number of p28 amino acid residues remaining is indicated to the right of each protein. The c-Myc or HA epitope tag is represented as a black box fused to the amino terminus of each p28 protein and is not drawn to scale. (B) Coimmunoprecipitation of in vitro-expressed proteins. The proteins were translated, using reticulocyte lysates, as fusions to c-Myc epitope tags in the presence of [35S]methionine or as fusions to HA epitope tags in the absence of radiolabel. Equal amounts of radiolabeled and nonradiolabeled lysates were combined, and the proteins were immunoprecipitated using rabbit polyclonal α-HA antiserum. Interacting 35S-labeled proteins were resolved in an SDS-12% polyacrylamide gel and analyzed by fluorography. The identities of coprecipitating proteins are shown on the right of the gel.
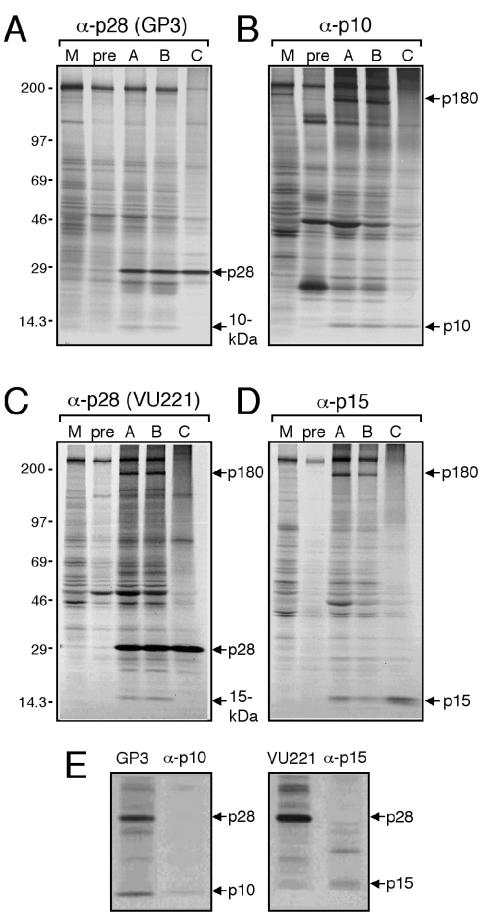
p28 coimmunoprecipitates with 10- and 15-kDa proteins in MHV-infected cell lysates.
To determine whether p28 interacts with p10 and p15 in the context of MHV infection, DBT cells were either mock infected or infected with MHV and radiolabeled with [35S]methionine-cysteine. Cytoplasmic extracts were subjected to immunoprecipitation using antisera against p28, p10, and p15 and buffer conditions of various stringencies (Fig. 7). Using both α-p28 antisera (GP3 and VU221), a 28-kDa protein (p28) was immunoprecipitated specifically from MHV-infected lysates using each of the buffer conditions tested (Fig. 7A and C). Using the guinea pig α-p28 antiserum GP3, a 10-kDa band coimmunoprecipitated with p28 (Fig. 7A). This 10-kDa p28-interacting protein migrated with the same electrophoretic mobility as p10 immunoprecipitated with α-p10 antiserum (Fig. 7B and E). Interestingly, the rabbit α-p28 antiserum VU221 coimmunoprecipitated a 15-kDa band, along with p28 (Fig. 7C). This 15-kDa protein migrated with the same electrophoretic mobility as p15 immunoprecipitated with α-p15 antiserum (Fig. 7D and E). Interactions between p28 and the 10- and 15-kDa proteins were detected only in infected cell lysates using 1% NP-40 (buffer A; low-stringency) and 0.1% SDS (buffer B; medium-stringency) buffer conditions. Neither the 10-kDa protein nor the 15-kDa protein was detected in mock-infected cell lysates or in infected cell lysates incubated with preimmune sera (Fig. 7A and C). Boiling the lysate prior to immunoprecipitation using 1% SDS buffer conditions (buffer C; high stringency) abolished the coprecipitations, suggesting that the 10- and 15-kDa proteins are likely interacting with p28 rather than being precipitated nonspecifically by α-p28 antiserum (Fig. 7A and C). Furthermore, in vitro-expressed epitope-tagged p10 and p15 fusion proteins were not detected following immunoprecipitation using either GP3 or VU221 (data not shown), suggesting that α-p28 antibodies do not cross-react with p10 and p15. These coimmunoprecipitation results are consistent with the hypothesis that p28 interacts with both p10 and p15 during MHV replication.
FIG. 7.
Coimmunoprecipitation of proteins from MHV-infected cell lysates. Mock-infected or MHV-infected cells were radiolabeled for 3 h at 5 h p.i., and cytoplasmic extracts were immunoprecipitated using buffer A (1% NP-40 buffer), buffer B (0.1% SDS buffer), or buffer C (1% SDS plus preboiling lysate) and antiserum against replicase protein p28, p10, or p15. Buffer A was used for control immunoprecipitations, including those with mock-infected lysate immunoprecipitated with immune sera (M) and infected lysate immunoprecipitated with preimmune sera (pre). Proteins were analyzed following SDS-PAGE in 5 to 18% polyacrylamide gradient gels and by fluorography. Molecular mass markers (in kilodaltons) are shown on the left, and the proteins of interest are indicated on the right of the gels. (A) α-p28 (GP3). (B) α-p10. (C) α-p28 (VU221). (D) α-p15. (E) p28 coprecipitating proteins have the same mobility as p10 and p15. Radiolabeled infected cell lysate was immunoprecipitated with antiserum specific for p28 (GP3 or VU221), p10 (α-p10), or p15 (α-p15) using buffer A conditions. The proteins were analyzed following SDS-PAGE in a 12% polyacrylamide gradient gel and fluorography. The identities of precipitated proteins are indicated to the right of the gels.
In addition to the interactions between p28 and p10 and p15, a protein band with the calculated molecular mass of ∼180 kDa (p180) also coprecipitated specifically in MHV-infected lysates using VU221 and either buffer A or buffer B conditions (Fig. 7C). Similarly, using α-p10 and α-p15 antisera, p180 was also detected specifically in MHV-infected cell lysates (Fig. 7B and D). Although the identity of this protein is not known, p180 might represent the precursor product that spans the carboxy terminus of ORF1a and includes MP1, 3CLpro, MP2, p10, p22, p12, and p15, as has been described for MHV-JHM (30).
DISCUSSION
In the present study, we defined the intracellular localization of p28 over the course of MHV infection and identified protein interactions between p28 and the carboxy-terminal ORF1a replicase proteins p10 and p15. We demonstrated that p28 localizes to MHV replication complexes in the cytoplasm at early times p.i., where it likely interacts with p10 and p15. At late times p.i., p28 is separate from replication complexes and is instead associated with Hel and N at sites of M accumulation. These results are in contrast to a previous study reporting that p28 is diffusely distributed in the cytosol, as well as in granular foci (2). While it is possible that some p28 is diffusely localized in infected cells, especially if the localization of p28 changes with time, our results show that the majority of detectable p28 is associated with MHV replication complexes at 6 h p.i. and with M at 9 h p.i. One reason for the discrepancy between the previous study and the present report may be due to differences in α-p28 antibody specificity or epitope recognition during immunofluorescence assays. Specifically, it is possible that the antibody used in the present study is not capable of recognizing cytosolic p28. The analysis of p28 intracellular localization and protein interactions does not allow us to draw conclusions regarding the specific function of p28 during MHV replication. Nonetheless, the results of this study corroborate previous biochemical data and provide new ideas about the involvement of p28 in viral RNA synthesis at the replication complex, as well as a possible role for p28 in the delivery of genomic RNA to sites of virion assembly.
Role of p28 at viral replication complexes.
The results of yeast two-hybrid, coimmunoprecipitation, and colocalization studies suggest that p28 plays a part in viral replication complex formation or function by interacting with p10 and p15. Specifically, we found that p28 coimmunoprecipitated 10- and 15-kDa proteins from MHV-infected cell lysates. Because neither the α-p10 nor the α-p15 antiserum recognizes p10 or p15 in immunoblotting assays (data not shown), we cannot entirely exclude the possibility that the 10- and 15-kDa proteins are cellular proteins that bind to p28. However, antisera to all three proteins coprecipitate a 180-kDa protein (p180) only in infected cell lysates, suggesting that p180, p28, p15, and p10 are associated in a complex. The p28 deletion mutagenesis experiments further support the conclusion that the interactions with p10 and p15 are specific. We determined that the mutant p28 protein lacking the carboxy-terminal residues F167 to G247 (p28ΔC1) is capable of binding to both p10 and p15. In contrast, the p28 deletion mutant that lacks residues E84 to R166 (p28ΔC2) is incapable of interacting with either p10 or p15. These results demonstrate that the carboxy terminus of p28 is not necessary for its interactions with p10 and p15 using the in vitro system and suggest that the 83-aa region (E84 to R166) contains residues that may mediate p10 and p15 binding. It is not apparent from our studies whether this region is sufficient for p28 interactions with p10 and p15; therefore, the minimal interaction domain might extend amino-terminal to residue E84. It is also possible that large deletions of p28 resulted in protein misfolding, thus abolishing p10 and p15 interactions. Nonetheless, these results demonstrate the specificity of p28-protein interactions.
What is the functional significance of p28 interactions with p10 and p15? One possibility is that p10 and p15 tether p28 to membranes of the replication complex. Because p28 appears to be a peripheral membrane protein (18), it likely is bound to the replication complex by trans-acting protein-protein or protein-RNA interactions. Consistent with this idea, both p10 and p15 appear to be tightly associated with cellular membranes (data not shown). Alternatively, the precursor protein that contains MP1, 3CLpro, MP2, p10, p22, p12, and p15 may serve as a scaffolding component of the MHV replication complex, with which other mature proteins bind. Thus, it is possible that p28 binds to this precursor via the p10 or p15 subdomain, thereby retaining it in the replication complex. In support of this possibility, we detected a protein of ∼180 kDa (p180) in infected cell lysates, using antisera against p28, p10, and p15, which may represent this large precursor. However, it is also possible that p180 is a cellular protein that is intimately associated with the MHV replication complex. Additional experiments are required to unequivocally determine the relationship of this high-molecular-weight protein with p28, p10, and p15.
Besides securing p28 to membranes of the replication complex, it is possible that interactions among p28, p10, and p15 are required for viral RNA synthesis. Engineered viruses that are incapable of processing p28 from the polyprotein have diminished levels of viral RNA, supporting the hypothesis that p28 might function directly or indirectly in viral RNA synthesis (11). Neither p10 nor p15 has known functions; however, immunofluorescence studies show that both proteins localize to sites of RNA synthesis at viral replication complexes (4). The temperature-sensitive MHV-A59 mutant tsLA6 has a mutation in p15 and defects in negative-strand RNA synthesis, implying a possible function for the protein in genome replication and discontinuous transcription (32). In both yeast two-hybrid and coimmunoprecipitation experiments, p15 interacts with itself and p10. Interestingly, the coronavirus infectious bronchitis virus p15 homolog (p16) dimerizes via multiple cysteine residues (26). MHV p15 shows 47% primary sequence identity with infectious bronchitis virus p16, including complete conservation of cysteine residues, suggesting that these proteins have similar functions and interactions during viral replication. Ongoing studies are aimed at determining the functions of p28, p10, and p15 interactions in viral RNA synthesis using reverse genetic approaches.
Role of p28 at sites of M accumulation.
Several lines of evidence support the conclusion that sites of MHV virion assembly are distinct from sites of RNA synthesis (4, 5, 22, 23). Therefore, MHV must have a mechanism for delivering newly synthesized genomic RNA from replication complexes to the sites where it will be packaged into virion particles. It has been demonstrated using immunofluorescence confocal microscopy and immunoelectron microscopy that by 9 h p.i., the viral M protein accumulates at sites of MHV particle assembly in the ERGIC (4, 5, 22, 23). In the present study, we show that p28, Hel, and N colocalize throughout MHV infection, and by 9 h p.i. these proteins predominantly localize to sites of M accumulation. p28 and Hel have not been detected in mature virion particles, suggesting that these proteins are not directly involved in virion assembly (10, 14). Instead, we hypothesize that the Hel-N-p28 complex is involved in shuttling genomic RNA to the ERGIC (5). The capacity of N to bind viral RNA has been experimentally demonstrated, and based on a calculated isoelectric point of 9.5, p28 also might bind viral RNA (1, 9, 29). Moreover, N coimmunoprecipitates with Hel, suggesting that these proteins interact (14). Therefore, it is reasonable to propose that p28, Hel, and N bind to each other or to genomic RNA molecules, which are then transported to the ERGIC. In this study, the colocalization of p28, Hel, and N at sites of budding MHV particles was not verified using immunoelectron microscopy. Thus, we cannot exclude the possibility that p28, Hel, N, and M colocalize at late times of infection in an alternative subcellular compartment distinct from the ERGIC. Additional experiments are also required to determine the functions of p28 and Hel at sites of M accumulation and to elucidate the viral and cellular determinants regulating changes in their localization over time.
Dynamics of p28 interactions during MHV infection.
The separation of p28 from the replication complex at late times of infection leads to new questions about the nature of p28-protein interactions. Both p10 and p15 are tightly associated with membranes and remain localized in the replication complex throughout the course of MHV infection. If p28 interacts with membrane-bound p10 and p15, then how does it change its localization to sites of M accumulation in the ERGIC? One possible explanation is that the p28, p10, and p15 interactions are in dynamic flux and the recruitment of cellular proteins to the replication complex at late times of infection disrupts viral protein interactions and mediates p28 translocation. In support of the possibility that p28, p10, and p15 interactions are transient is the fact that the α-p28 antisera raised in rabbits and guinea pigs coimmunoprecipitate different proteins. Specifically, α-p28 rabbit antiserum (VU221) coimmunoprecipitates p15 but not p10, and α-p28 guinea pig antiserum (GP3) coimmunoprecipitates p10 but not p15. Thus, the α-p28 antibodies may compete for, or sterically hinder, protein-binding sites within p28. Furthermore, there is precedent for regulation of nidovirus replicase protein localization due to changes in the expression or localization of cellular proteins. For the arterivirus equine arteritis virus, the amino-terminal replicase protein (nsp1) predominantly localizes to the cell nucleus at early times p.i. and to cytoplasmic replication complexes later in infection (36, 37). Cellular proteins expressed during different stages of viral infection and cell cycle progression have been implicated in the regulation of equine arteritis virus nsp1 localization (37, 38). We think it is possible that, at late times p.i., the MHV Hel-N-p28 complex interacts with host cell proteins involved in intracellular transport, thus shuttling these proteins to viral assembly sites. Interestingly, during times of peak MHV RNA synthesis, Hel, N, and p28 are localized to replication complexes. If p28 and Hel are required for RNA synthesis, then the separation of these proteins away from the replication complex might function to regulate the transition between MHV RNA synthesis and particle assembly.
The results of the present study regarding the localization and interactions of MHV p28 may extend to other coronaviruses. Specifically, it is possible that the putative amino-terminal 20-kDa protein (nsp1) of SARS-CoV mediates functions similar to those mediated by MHV p28. Studies of the amino-terminal MHV proteins may lead to an enhanced understanding of the replication strategy of this diverse family of RNA viruses and identify novel targets for development of pharmacological inhibitors of coronavirus replication.
Acknowledgments
We express our appreciation to Valerie McMurray for technical assistance and to Rachel Graham, Paul McDonald, Erik Prentice, Jennifer Sparks, and Steven Sperry for critical advice and reviews of the manuscript.
This work was supported by Public Health Service award T32 HL07751 (S.M.B.) from the National Heart, Lung, and Blood Institute; the Howard Hughes Medical Institute (T.R.P.); Public Health Service awards AI32539 (T.S.D.) and AI50083 (M.R.D.) from the National Institute of Allergy and Infectious Diseases; and the Elizabeth B. Lamb Center for Pediatric Research. Additional support was provided by Public Health Service award CA68485 for the Vanderbilt DNA Sequencing Shared Resource and the Molecular Imaging Shared Resource of the Vanderbilt-Ingram Cancer Center.
REFERENCES
- 1.Baric, R. S., G. W. Nelson, J. O. Fleming, R. J. Deans, J. G. Keck, N. Casteel, and S. A. Stohlman. 1988. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J. Virol. 62:4280-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi, W., J. D. Pinon, S. Hughes, P. J. Bonilla, K. V. Holmes, S. R. Weiss, and J. L. Leibowitz. 1998. Localization of mouse hepatitis virus open reading frame 1A derived proteins. J. Neurovirol. 4:594-605. [DOI] [PubMed] [Google Scholar]
- 3.Bonilla, P. J., A. E. Gorbalenya, and S. R. Weiss. 1994. Mouse hepatitis virus strain A59 RNA polymerase gene ORF 1a: heterogeneity among MHV strains. Virology 198:736-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bost, A. G., R. H. Carnahan, X. T. Lu, and M. R. Denison. 2000. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 74:3379-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bost, A. G., E. Prentice, and M. R. Denison. 2001. Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection. Virology 285:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bredenbeek, P. J., C. J. Pachuk, A. F. Noten, J. Charite, W. Luytjes, S. R. Weiss, and W. J. Spaan. 1990. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 18:1825-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brierley, I., P. Digard, and S. C. Inglis. 1989. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell 57:537-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brockway, S. M., C. T. Clay, X. T. Lu, and M. R. Denison. 2003. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 77:10515-10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cologna, R., J. F. Spagnolo, and B. G. Hogue. 2000. Identification of nucleocapsid binding sites within coronavirus-defective genomes. Virology 277:235-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denison, M., and S. Perlman. 1987. Identification of putative polymerase gene product in cells infected with murine coronavirus A59. Virology 157:565-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denison, M., B. Yount, S. M. Brockway, R. L. Graham, A. C. Sims, and R. S. Baric. 2004. Cleavage between replicase proteins p28 and p65 of mouse hepatitis virus is not required for virus replication. J. Virol. 78:5957-5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denison, M. R., S. A. Hughes, and S. R. Weiss. 1995. Identification and characterization of a 65-kDa protein processed from the gene 1 polyprotein of the murine coronavirus MHV-A59. Virology 207:316-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denison, M. R., and S. Perlman. 1986. Translation and processing of mouse hepatitis virus virion RNA in a cell-free system. J. Virol. 60:12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denison, M. R., W. J. Spaan, Y. van der Meer, C. A. Gibson, A. C. Sims, E. Prentice, and X. T. Lu. 1999. The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis. J. Virol. 73:6862-6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denison, M. R., P. W. Zoltick, S. A. Hughes, B. Giangreco, A. L. Olson, S. Perlman, J. L. Leibowitz, and S. R. Weiss. 1992. Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology 189:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong, S., and S. C. Baker. 1994. Determinants of the p28 cleavage site recognized by the first papain-like cysteine proteinase of murine coronavirus. Virology 204:541-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 17:4847-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirano, N., K. Fujiwara, and M. Matumoto. 1976. Mouse hepatitis virus (MHV-2). Plaque assay and propagation in mouse cell line DBT cells. Jpn. J. Microbiol. 20:219-225. [PubMed] [Google Scholar]
- 20.Hughes, S. A., P. J. Bonilla, and S. R. Weiss. 1995. Identification and analysis of MHV-A59 P28 cleavage site. Adv. Exp. Med. Biol. 380:453-458. [DOI] [PubMed] [Google Scholar]
- 21.Hughes, S. A., P. J. Bonilla, and S. R. Weiss. 1995. Identification of the murine coronavirus p28 cleavage site. J. Virol. 69:809-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klumperman, J., J. K. Locker, A. Meijer, M. C. Horzinek, H. J. Geuze, and P. J. Rottier. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68:6523-6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krijnse-Locker, J., M. Ericsson, P. J. Rottier, and G. Griffiths. 1994. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 124:55-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, H. J., C. K. Shieh, A. E. Gorbalenya, E. V. Koonin, N. La Monica, J. Tuler, A. Bagdzhadzhyan, and M. M. Lai. 1991. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology 180:567-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 26.Ng, L. F., and D. X. Liu. 2002. Membrane association and dimerization of a cysteine-rich, 16-kilodalton polypeptide released from the C-terminal region of the coronavirus infectious bronchitis virus 1a polyprotein. J. Virol. 76:6257-6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pachuk, C. J., P. J. Bredenbeek, P. W. Zoltick, W. J. Spaan, and S. R. Weiss. 1989. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus, strain A59. Virology 171:141-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prentice, E., W. G. Jerome, T. Yoshimori, N. Mizushima, and M. R. Denison. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 279:10136-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robbins, S. G., M. F. Frana, J. J. McGowan, J. F. Boyle, and K. V. Holmes. 1986. RNA-binding proteins of coronavirus MHV: detection of monomeric and multimeric N protein with an RNA overlay-protein blot assay. Virology 150:402-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schiller, J. J., A. Kanjanahaluethai, and S. C. Baker. 1998. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology 242:288-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi, S. T., J. J. Schiller, A. Kanjanahaluethai, S. C. Baker, J. W. Oh, and M. M. Lai. 1999. Colocalization and membrane association of murine hepatitis virus gene 1 products and de novo-synthesized viral RNA in infected cells. J. Virol. 73:5957-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siddell, S., D. Sawicki, Y. Meyer, V. Thiel, and S. Sawicki. 2001. Identification of the mutations responsible for the phenotype of three MHV RNA-negative ts mutants. Adv. Exp. Med. Biol. 494:453-458. [DOI] [PubMed] [Google Scholar]
- 33.Sims, A. C., J. Ostermann, and M. R. Denison. 2000. Mouse hepatitis virus replicase proteins associate with two distinct populations of intracellular membranes. J. Virol. 74:5647-5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 36.Tijms, M. A., and E. J. Snijder. 2003. Equine arteritis virus non-structural protein 1, an essential factor for viral subgenomic mRNA synthesis, interacts with the cellular transcription co-factor p100. J. Gen. Virol. 84:2317-2322. [DOI] [PubMed] [Google Scholar]
- 37.Tijms, M. A., Y. van der Meer, and E. J. Snijder. 2002. Nuclear localization of non-structural protein 1 and nucleocapsid protein of equine arteritis virus. J. Gen. Virol. 83:795-800. [DOI] [PubMed] [Google Scholar]
- 38.Tijms, M. A., L. C. van Dinten, A. E. Gorbalenya, and E. J. Snijder. 2001. A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive-stranded RNA virus. Proc. Natl. Acad. Sci. USA 98:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. Spaan, and J. K. Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]