Abstract
Interactions between the polyamine spermine and nucleic acids drive important cellular processes. Spermine condenses DNA and some RNAs, such as poly(rA):poly(rU). A large fraction of the spermine present in cells is bound to RNA but apparently does not condense it. Here, we study the effect of spermine binding to short duplex RNA and DNA, and compare our findings with predictions of molecular-dynamics simulations. When small numbers of spermine are introduced, RNA with a designed sequence containing a mixture of 14 GC pairs and 11 AU pairs resists condensation relative to DNA of an equivalent sequence or to 25 bp poly(rA):poly(rU) RNA. A comparison of wide-angle x-ray scattering profiles with simulation results suggests that spermine is sequestered deep within the major groove of mixed-sequence RNA. This prevents condensation by limiting opportunities to bridge to other molecules and stabilizes the RNA by locking it into a particular conformation. In contrast, for DNA, simulations suggest that spermine binds externally to the duplex, offering opportunities for intermolecular interaction. The goal of this study is to explain how RNA can remain soluble and available for interaction with other molecules in the cell despite the presence of spermine at concentrations high enough to precipitate DNA.
Introduction
Spermine is a tetravalent polyamine that is found in high concentrations in cells (1, 2, 3). It interacts with nucleic acids in diverse and important ways, functioning as a protective agent (4, 5, 6, 7, 8) and a modulator of DNA secondary structure (9). In cells, spermine is largely bound to RNA, with between one and four bound spermine molecules per 100 RNA phosphates (2, 10) (between 65% and 85% of the total spermine (9)). Crystal structures of spermine complexed with tRNA show spermine lying deep within the major groove (11).
With so many spermine molecules associating with RNA, it is important to consider the possibility of cation-induced condensation, in which small amounts of multivalent cations cause highly charged, ordinarily self-repelling nucleic acids to adopt a compact state occupying only a fraction of the volume available to them (12, 13, 14). Previous studies of spermine-induced RNA condensation had a limited scope but showed that spermine can condense RNAs with nonphysiological sequences or under nonphysiological experimental conditions, e.g., long duplex poly(rA):poly(rU) (15), tRNA at nonphysiological pHs (16), and viral RNA in the complete absence of other salts (17). More extensive studies of DNA condensation by spermine showed that even small numbers of spermines condense both long and short, natural and synthetic DNA duplexes (18, 19, 20). Since spermine is such an effective condensing agent, it seems plausible that it could also condense RNA in cells. Such condensation would lead to aggregation instead of ordered folding and could potentially interfere with important RNA functions. However, given that spermine plays roles in processes that could be impeded by condensation, such as mRNA translation (2), tRNA aminoacylation, and the formation of correct tRNA-ribosome interactions (21), it appears that spermine-induced RNA condensation may not occur in vivo. A correct and thorough understanding of spermine-RNA interactions could provide insight into whether spermine causes RNA condensation and how it facilitates the above key processes.
Previously, we studied the condensation of duplex DNA and RNA by the trivalent cation cobalt(III) hexammine (CoHex) and arrived at the surprising conclusion that RNA resists condensation under the same conditions that cause DNA of an equivalent sequence to condense readily (22, 23). The RNA duplexes used in that work have a well-defined composition and sequence (see Materials and Methods), containing a mixture of both GC and AU basepairs to imitate physiological RNA. This sequence favors duplex formation over stem loops in the individual strands and has a high melting temperature (61°C as calculated with OligoCalc (24)). We compared the experimental results with all-atom molecular-dynamics (MD) simulations to develop a model in which the distribution of bound CoHex ions relates to the condensation propensity: nucleic acids with more CoHex bound in the external binding shell (12–16 Å from the helical axis) condense most readily, whereas those with more ions bound to the internal shell (7–12 Å from the helical axis) resist condensation (23, 25). Although these studies produced interesting experimental findings, they were not biological. However, because of the straightforward representation of CoHex ions as spheres, both the distinction between different binding modes and their relevance to attraction were clearly delineated. In this work, we focused on spermine to explore the potential physiological relevance of the different binding modes of multivalent ions to nucleic acids. In particular, we asked whether the location of the spermine ions affects the forces between duplexes. We applied the above model in combination with experiments to examine RNA’s resistance to condensation by spermine, to probe spermine-nucleic acid interactions and spermine-induced structural changes, and to gain insight into how this biologically important polyamine affects the function of these molecules.
Materials and Methods
Materials
Single-stranded nucleic acid oligos were purchased from Integrated DNA Technologies (Coralville, IA) and annealed into double-stranded constructs by incubation in STE buffer (50 mM NaCl, 10 mM TRIS, 1 mM ethylenediaminetetraacetic acid) for 2 min at 94°C. Some of these double-stranded constructs (25 bp DNA and RNA with the sequence GCA TCT GGG CTA TAA AAG GGC GTC G, with the RNA strands containing U instead of T, were also examined in (23)). Additionally, duplex lyophilized 25 bp poly(rA):poly(rU) RNA was purchased from Integrated DNA Technologies and resuspended by vortexing in STE buffer. The annealed duplexes were dialyzed into pH 7 Na-MOPS buffer containing NaCl. Spermine tetrachloride powder (Sigma-Aldrich, St. Louis, MO) was dissolved in water.
UV absorption measurements of spermine-induced condensation
Each duplex was dialyzed into 1 mM Na-MOPS buffer at pH 7 with 20 mM NaCl and then separated into 90 μL aliquots at 44.4 μM concentration. Each aliquot was spiked with 10 μL of concentrated spermine and incubated at 4°C for 2 h. After incubation, the aliquots were centrifuged at 10,000 rpm for 10 min. The supernatant was removed and its absorption spectrum was recorded from 220 to 400 nm by a Cary 50 spectrophotometer. To calculate the amount of nucleic acid remaining in solution, the absorbance value at 260 nm was normalized by the absorbance at that wavelength of a sample containing no spermine. Error bars were determined using baseline differences at the longest wavelengths probed.
Circular dichroism
Circular dichroism (CD) was used to determine the helical geometry of each nucleic acid construct and to probe spermine-induced geometrical changes. Nucleic acids were dialyzed into 1 mM Na-MOPS with 100 mM NaCl. Mixed-sequence RNA and DNA were each separated into two 20 μL, 500 μM aliquots: one without spermine and one containing 3 mM spermine. Due to limited sample quantities, the poly(rA):poly(rU) RNA was separated into two 20 μL, 20 μM aliquots: one without spermine and one containing 0.04 mM spermine. The spermine/phosphate ratio used here was lower than that used for the mixed-sequence molecules due to condensation of the poly(rA):poly(rU) RNA at the higher ratio. The high NaCl concentration of these samples relative to those used in UV absorption measurements was chosen to prevent condensation of the CD samples, as in (23). Samples were loaded into a 0.01 mm demountable cuvette (0.1 mm path length in the case of poly(rA):poly(rU)). CD measurements were obtained on a MOS-450 spectrometer (BioLogic, Seyssinet-Pariset, France). Each spectrum is the average of five scans from 200 to 300 nm with 1 s/nm steps.
Wide-angle x-ray scattering
Wide-angle x-ray scattering (WAXS) was used as a complementary technique to CD to probe structural changes due to conformational shifts in the molecule or to tight association of spermine. The mixed-sequence nucleic acid constructs were dialyzed into 1 mM Na-MOPS with 100 mM NaCl and divided into two 30 μL, 500 μM aliquots containing 0 and 3 mM spermine. WAXS measurements were carried out at station G1 of the Cornell High Energy Synchrotron Source with a 0.4 m flight tube and 10.52 keV x-ray energy. Scattering images were taken with the 1024 × 1024 pixel FLICAM CCD detector and analyzed using MATLAB (The MathWorks, Natick, MA) code written in-house.
To directly compare our MD simulations with experimental scattering profiles, we used CRYSOL to calculate WAXS profiles from the Protein Data Bank (PDB) files of the mixed-sequence DNA and RNA models (26). We ran 200 PDB snapshots from simulations of each nucleic acid with and without spermine through CRYSOL using default parameters, and averaged the resulting scattering profiles. We did not allow CRYSOL to fit the experimental data, so the comparison between the simulated profiles and experiment is a worst-case agreement.
All-atom MD simulations
Mixed-sequence 25-bp DNA and RNA duplexes were simulated using the AMBER14 suite of programs (27, 28) with the parm10 force field, which includes parmbsc0 and parmbsc0_chiOL3 refinements (29, 30) for DNA and RNA, respectively. Spermine molecules were simulated using the general Amber force field (GAFF version 1.7). Nucleic Acid Builder (31) was used to construct initial DNA and RNA structures in the canonical B- and A-forms, respectively. To evaluate the distribution of spermine ions around nucleic acids, simulations were carried out in the absence of monovalent salt. This protocol avoids uncertainties associated with sodium ion force-field parameters (32) and is justified by the low sodium concentrations in the condensation experiments, since it is known that sodium concentrations below 40 mM do not affect CoHex bound to DNA (33) and therefore likely do not affect spermine. The duplexes were solvated with 16,880 TIP3P water molecules and a neutralizing amount of spermine (12 molecules). This amount is relevant to compare with experimental measurements of duplex condensation, which begins when ∼90% of the phosphate charge is neutralized by the counterions (13). An initial 2000-step water minimization was followed by a 0.5 ns equilibration time in a canonical NVT ensemble and another 0.5 ns in an isothermal-isobaric (NPT) ensemble using 1 fs time steps. The system achieved 1 atm pressure and 300 K temperature, which was maintained using Langevin dynamics with a collision frequency of 1 ps−1. Periodic boundary conditions and the particle mesh Ewald method were employed. All nucleic acid atoms were harmonically restrained with a 100 kcal/mol/Å2 force constant during the minimization and equilibration stages. After these stages, 600 ns production trajectories were generated for each system using an NVT ensemble and a 2 fs time step. The DNA atoms were restrained with a reduced force constant of 50 kcal/mol/Å2 in the production stage, whereas RNA atoms were unrestrained, as discussed previously (23). Spermine charge distributions were calculated as the average numbers of spermine nitrogen atoms (where the centers of the spermines’ positive charges are located) in 0.25-Å-thick, consecutive cylindrical layers around the duplexes. The first 100 ns of each trajectory were disregarded in these calculations. The robustness of the spermine distributions to the choice of water model was confirmed using the OPC water model (34).
To compare the MD results with WAXS experiments performed in 100 mM NaCl, we ran new simulations with the addition of 24 NaCl in each simulation box using monovalent ion force field parameters from (35). PDB snapshots from these simulations were run through CRYSOL as described above.
Results
Measurement of duplex condensation
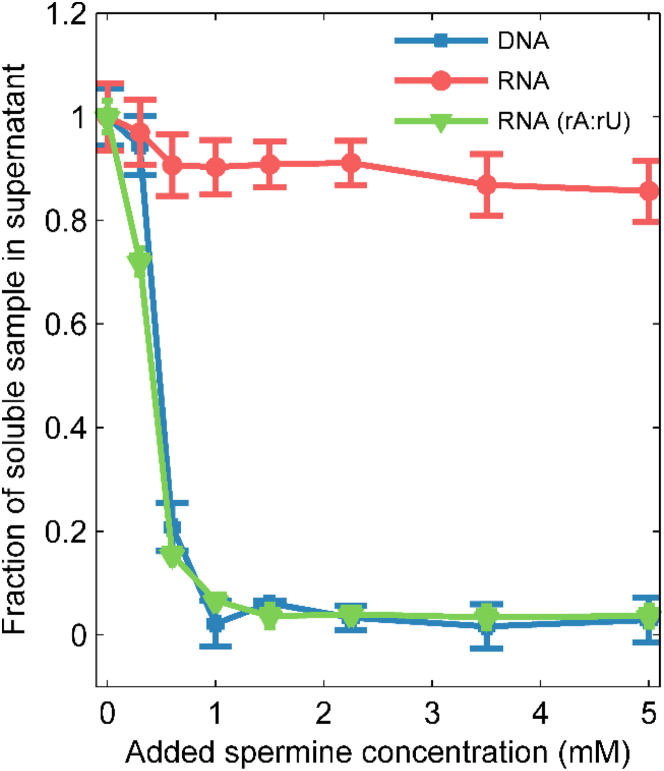
We used UV spectroscopy to probe the condensation propensities of our nucleic acid constructs. As previously described (22), we measured the concentration of nucleic acid remaining in the supernatant after the addition of spermine. Fig. 1 shows the differences in condensation among the three constructs, with a notable discrepancy in the behavior of the mixed-sequence RNA and that of the equivalently sequenced DNA construct. The mixed-sequence DNA precipitates from solution when small amounts of spermine are present, whereas the RNA remains soluble. This behavior agrees qualitatively with observations described for the same nucleic acid constructs in the presence of CoHex (23); however, the trends are exaggerated when spermine is used as the condensing agent. The mixed-sequence RNA is more resistant to condensation by spermine than by CoHex, whereas the DNA condenses more readily when spermine is present. The DNA duplexes precipitate completely when the spermine concentration is ∼1 mM, roughly half the concentration of phosphate in the duplexes. This agrees with previous work showing that a significant amount of spermine is required to precipitate DNA (13). Prior experiments using CoHex suggest that an even higher concentration is required to completely draw the DNA out of solution (19). It is interesting that the poly(rA):poly(rU) RNA condenses out of solution in the presence of small amounts of spermine, consistent with previous literature reports (15) and in contrast to our findings on the mixed-sequence duplex.
Figure 1.
Fraction of 25 bp nucleic acid duplexes remaining in solution, measured by UV absorption, as a function of the added spermine concentration. Individual spermine molecules are shown in addition to the nucleic acid structures. To see this figure in color, go online.
MD results
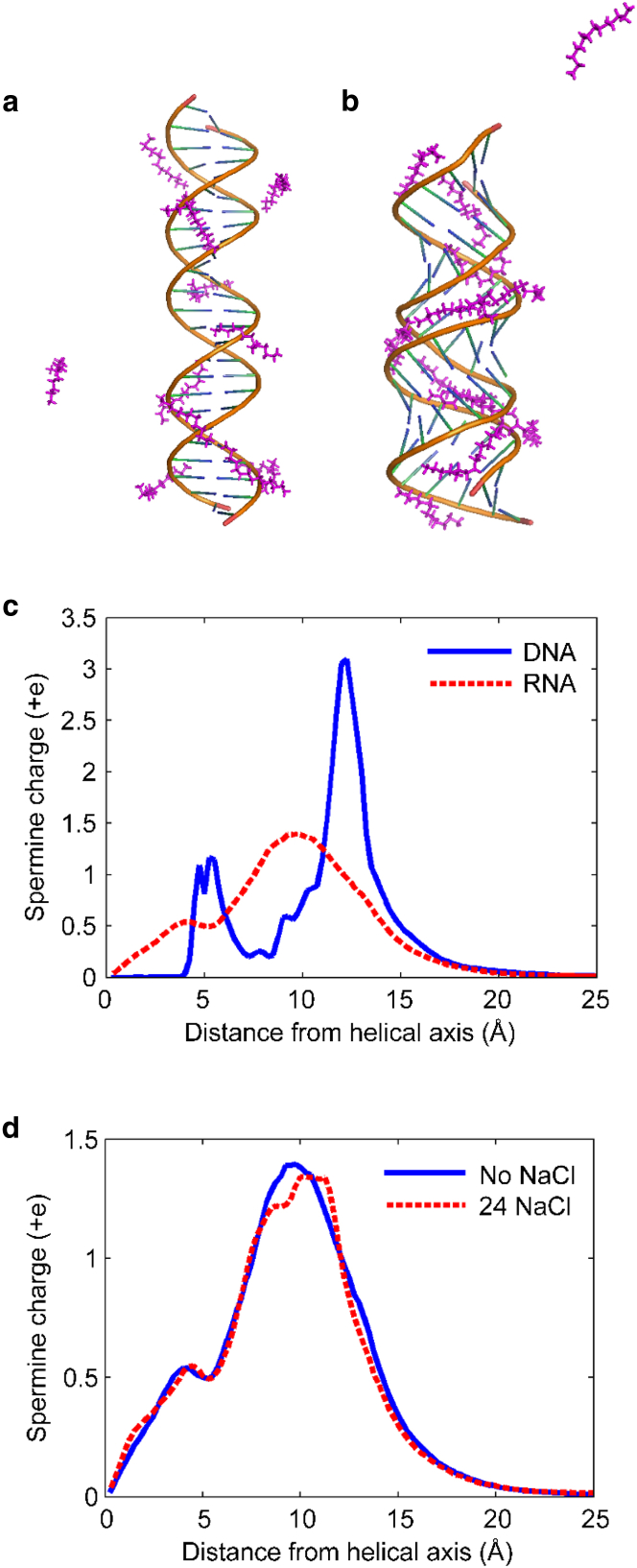
All-atom MD simulations give insight into the connection between how spermine binds to nucleic acids and its ability to condense them. Fig. 2 shows the distribution of spermine charge around simulated mixed-sequence DNA and RNA duplexes. In both systems, the spermine nitrogen atoms bind preferentially to the phosphate groups centered ∼10 Å from the helical axis of the duplex. However, the spermine binding modes vary between the DNA and RNA, likely due to structural differences such as the width of the major groove and the relative orientation of the phosphate group oxygens, and to the more negative electrostatic potential in the major groove of A-form RNA compared with B-form DNA (23, 36). The electrostatic potential of each nucleic acid is shown in Fig. 4 of Ref. (23). Although the quantitative picture of the electrostatic potential difference may be more complex due to dielectric saturation and other effects (e.g., as discussed in Ref. (37)), the qualitative picture obtained from Poisson-Boltzmann estimations is in good agreement with results obtained for CoHex.
Figure 2.
(a and b) Representative snapshots of mixed-sequence (a) DNA and (b) RNA duplexes simulated with spermine. Spermine molecules are shown in magenta. On average, ∼11 of the 12 spermine molecules are bound to the nucleic acids in each snapshot. These frames were taken from Movies S1 and S2 of the simulations. (c) Distribution of the charge of bound spermine counterions around 25 bp mixed-sequence DNA and RNA duplexes derived from 500 ns of MD trajectories. Interestingly, this plot qualitatively resembles the distributions computed for CoHex (Fig. 3 of Ref. (23)). (d) Spermine charge distributions around RNA simulated with no NaCl (for comparison with condensation data) and with 24 NaCl per simulation box (80 mM, for comparison with WAXS). The distributions are qualitatively unchanged by the addition of NaCl. To see this figure in color, go online.
Most of the spermine charge is bound to the DNA phosphate backbone outside the major groove, with the distribution peak at ∼12–13 Å from the helical axis. This type of counterion binding was characterized in (23) as the external binding mode, which promotes counterion-mediated attraction between duplexes. A small fraction of spermine charge is found deeply buried inside the DNA major groove, where spermine molecules are bound to the oxygen atoms of guanine bases.
Simulations of RNA show most of the bound spermine ions buried within the major groove, with the distribution peak 9.5 Å from the helical axis. This type of counterion binding, previously defined as the internal binding mode (23), does not appear to facilitate a counterion-induced attraction between duplexes. As seen in the DNA simulations, a small fraction of spermine charge is bound to guanine bases deep inside the RNA major groove.
MD trajectories of spermine around mixed-sequence DNA and RNA are provided in the Supporting Materials and Methods in the Supporting Material.
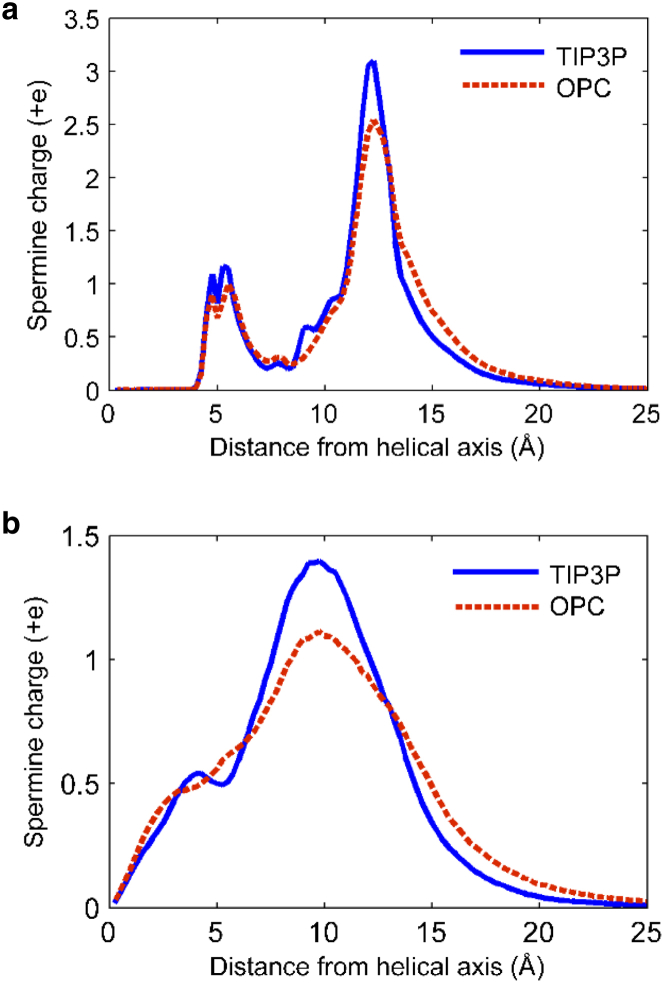
Robustness of simulation results
We tested spermine charge distribution convergence by doubling the production trajectory lengths to 1 μs. The difference between these distributions and those obtained from the 600 ns MD simulation is negligible. We have also verified that the spermine charge distributions are not sensitive to the water model used. The difference between the spermine distributions obtained with the four-point OPC water model (33) and three-point TIP3P model is negligible, as shown in Fig. 3. These simulations also indicate that the distributions resulting from 600 ns trajectories are insensitive to the initial conditions.
Figure 3.
(a and b) Distribution of spermine charge around (a) mixed-sequence DNA and (b) mixed-sequence RNA with the four-point OPC water model (34) and three-point TIP3P model. To see this figure in color, go online.
CD
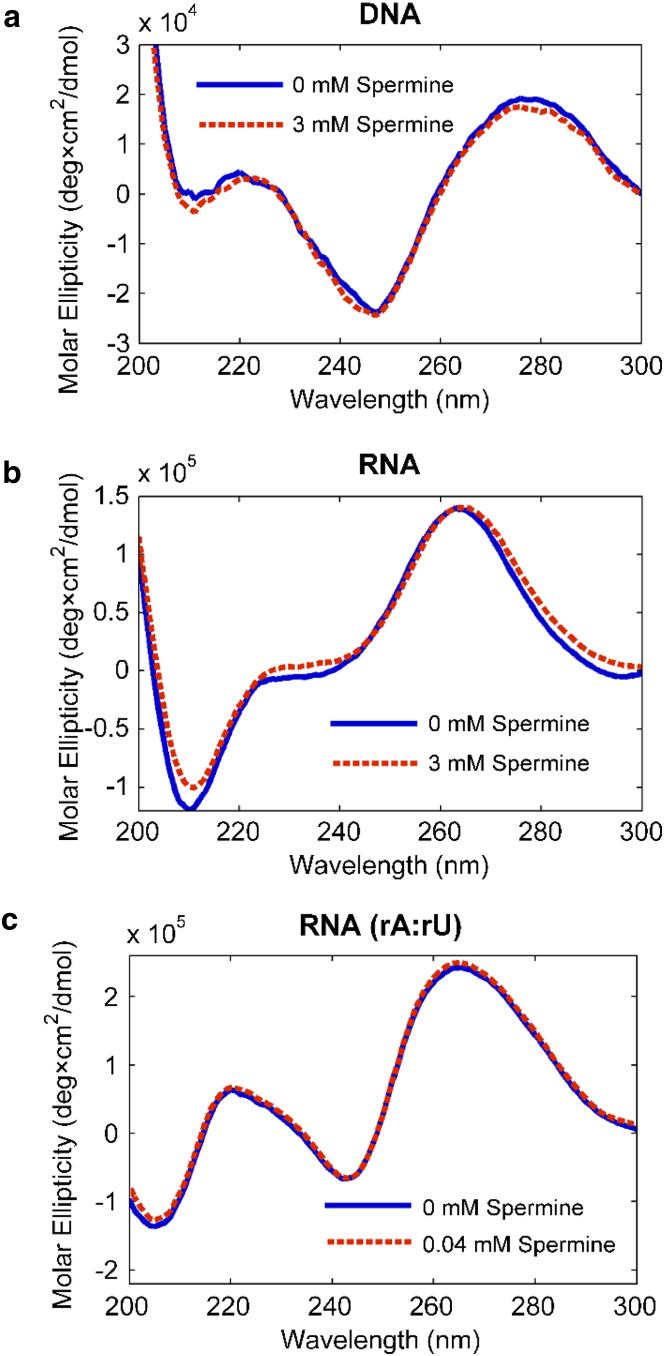
To explore the effect of helical geometry on condensation propensity and to determine whether spermine binding results in conformational changes of our nucleic acids, we measured CD spectra for all three constructs with and without spermine (Fig. 4). CD differentiates between nucleic acids with A- and B-form geometries (38), with B-DNA typically displaying local maxima at 220 and 275 nm and a local minimum at 245 nm, and A-RNA showing a maximum at 260 nm and a minimum at 210 nm.
Figure 4.
(a–c) CD spectra for (a) mixed-sequence DNA, (b) mixed-sequence RNA, and (c) poly(rA):poly(rU) RNA with and without added spermine. The wavelengths of the peaks and valleys shift slightly when spermine is added, suggesting a geometrical change. Panels (b) and (c) highlight the differences between the structures of the mixed-sequence duplex (specified in Materials and Methods) and the poly(rA):poly(rU) duplex. To see this figure in color, go online.
Our DNA construct displays a canonical B-form spectrum. The CD spectra of the RNA constructs are A-like, but are distinct for the two different sequences probed (the single mixed sequence and the poly(rA):poly(rU) complex). A subtle change is observed in the CD spectra of mixed-sequence RNA when spermine is added. The relative magnitudes of the extrema change and their wavelengths vary slightly. Because these changes are small, we employed WAXS to monitor subtle structural shifts.
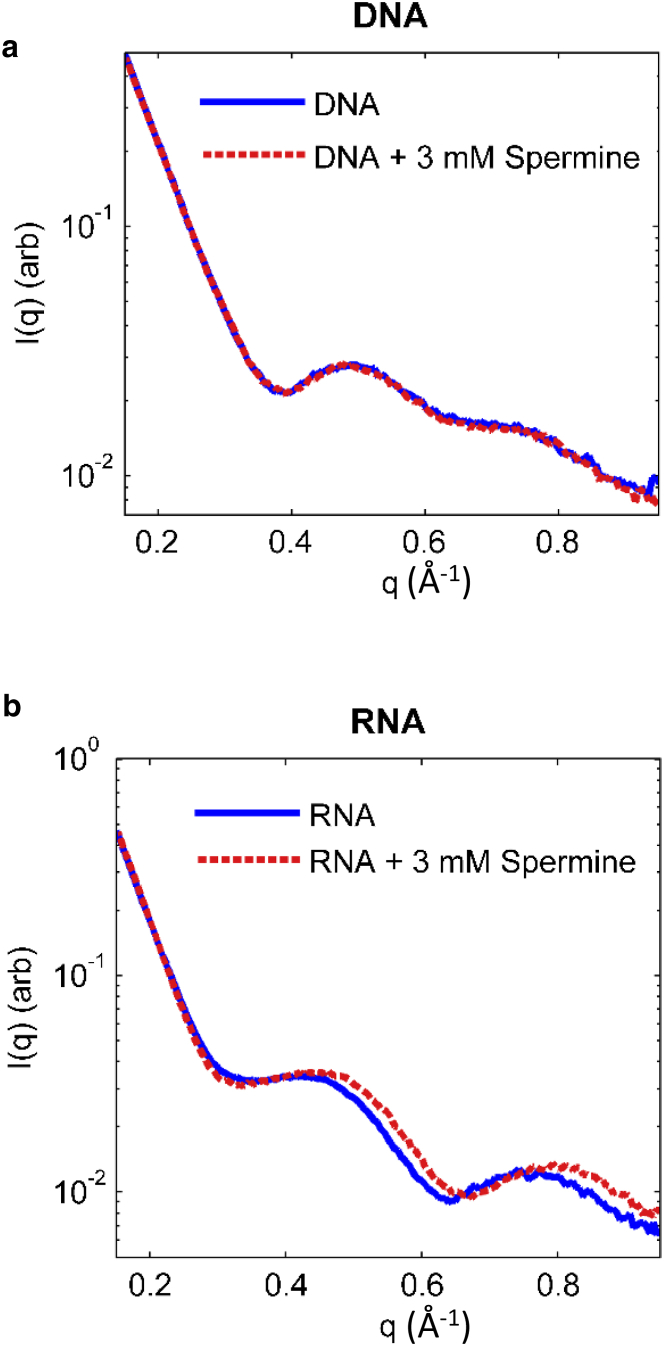
WAXS
WAXS is a sensitive probe of small conformational changes (39). The length scales probed in our WAXS experiments begin at ∼7 Å. This range includes features on the order of the helical radius. Our CD results suggest that the nucleic acid geometry is potentially changed by spermine, and therefore the observed changes in the WAXS profiles may reflect either these changes or tight spermine binding to the nucleic acid. WAXS profiles for mixed-sequence DNA and RNA, with and without spermine, are shown in Fig. 5. The profile of the DNA changes only slightly with the addition of spermine. It is known from previous small-angle x-ray scattering studies of DNA (40) that spermine associates with short duplexes. The profile of the mixed-sequence RNA shows larger shifts.
Figure 5.
(a and b) WAXS profiles for (a) mixed-sequence DNA and (b) mixed-sequence RNA with and without spermine. Larger changes are observed in the profile of the RNA. To see this figure in color, go online.
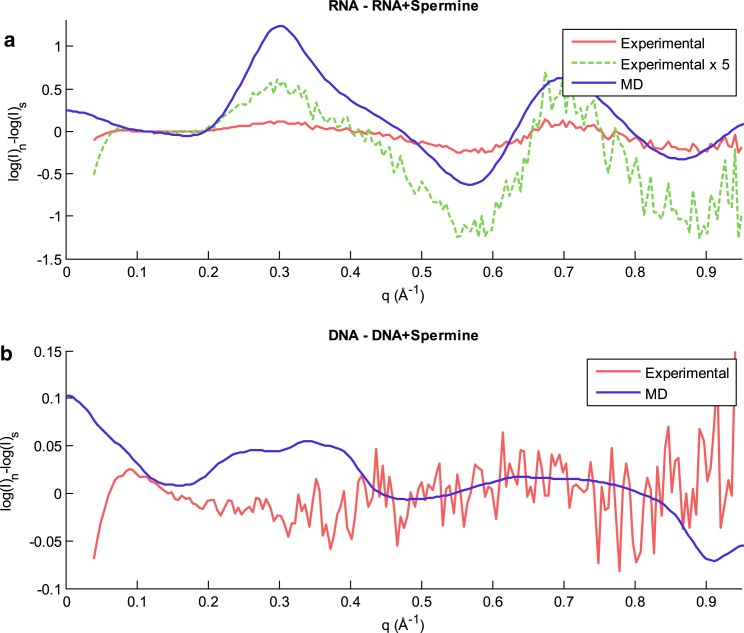
To determine whether our MD simulations could explain the changes in these scattering profiles, we used CRYSOL to calculate WAXS profiles from the PDB files of the mixed-sequence DNA and RNA models obtained from simulation snapshots. Although CRYSOL can be inaccurate at the large scattering angles used for WAXS, an examination of the difference between two WAXS profiles often yields good agreement between experimental data and scattering profiles calculated from accurate models (39). This WAXS difference signal reflects changes in the structure, as opposed to hard-to-model details such as the hydration shell. Here, we compare the differences in profiles for each nucleic acid with and without spermine. All four scattering profiles (experimental and simulated) were scaled to match at low q-values before subtraction. As shown in Fig. 6, the differences in the calculated profiles for mixed-sequence RNA with and without spermine agree qualitatively with the differences in experimental profiles for q > 0.08 Å−1, based on the locations of extrema, validating the MD models. The differences in the calculated profiles for DNA with and without spermine do not agree qualitatively with the differences in the experimental profiles, as extrema that exist in one of the difference curves do not appear in the other curve.
Figure 6.
(a and b) Differences in the scattering profiles for (a) mixed-sequence RNA and (b) DNA with and without spermine. Differences were taken from the logs of the intensities of the original profiles to allow easier examination of differences at higher q-values. The noisier (red) profiles are experimental data, while the smoother (blue) profiles show the average of profiles generated with CRYSOL from 200 different MD snapshots. The RNA plot also shows the experimental curve multiplied by a factor of 5 to emphasize the locations of extrema. To see this figure in color, go online.
Discussion
Mechanism of mixed-sequence RNA’s resistance to condensation by spermine
Spermine assists in a wide variety of RNA functions, protects RNA from damage, and complexes with RNA in cells. With so much interaction between spermine and RNA, it seems surprising that condensation does not occur. Such precipitation could be catastrophic, causing aggregation instead of ordered folding and reducing functionality. Our condensation experiments and MD modeling provide some insight into the mechanism of mixed-sequence RNA’s resistance to condensation by spermine, which has not been previously reported.
The nucleic acid constructs examined in this work show the same trends with spermine as were observed with CoHex-induced condensation in (23). However, the differences between constructs are enhanced in the presence of spermine. Mixed-sequence DNA condenses at lower multivalent cation concentrations when spermine is present rather than CoHex, and the RNA is even more resistant to condensation.
This variation could result from the different geometries of the two condensing agents: spermine is elongated, whereas CoHex is spherical. The simulations show spermine lying down inside the deep major grooves of RNA, but spanning the grooves on the outside of DNA. Due to its smaller diameter (4.4 Å (41)), spermine may be able to bury itself more completely in the RNA’s grooves than CoHex (diameter 6 Å (42)), resulting in fewer opportunities to bridge nucleic acid molecules or to form an electrostatic zipper (43). On the other hand, these extended molecules located on the outside of DNA could provide better bridging by allowing more ways for two nucleic acid molecules to contact each other.
This effect could also be driven by the higher charge of spermine (4+) relative to CoHex (3+). This higher charge may allow spermine to bury itself deeper in the electronegative major groove of RNA than CoHex, resulting in fewer bridging opportunities. For DNA, the higher spermine charge located on the outside of the molecule could enhance attraction between duplexes and improve opportunities for condensation. The counterion’s charge density may also be a critical parameter (44).
Mechanism of poly(rA):poly(rU) condensation
The two RNA constructs examined in this work exhibit a striking difference in condensation behavior: the mixed-sequence RNA resists condensation even at high spermine concentrations, whereas the poly(rA):poly(rU) RNA condenses readily when even small amounts of spermine are added. One possible source of this discrepancy is the ability of two poly(rA):poly(rU) duplexes to convert back and forth to triplex poly(rA):poly(rU):poly(rU) and a poly(rA) strand, as reported in previous studies (45, 46, 47). We have no direct measure of such a duplex-to-triplex conversion; this speculation is based on those previous reports. Divalent ions such as Ni(II) and Mg(II) can bias the triplex or tetra-aggregate forms of this construct even at room temperature (47), and spermine is known to stabilize the triplex form of poly(dA):poly(dT) DNA (48, 49). Therefore, it is possible that the addition of spermine entices poly(rA):poly(rU) to transition to either a triplex or tetra-aggregate form. Such a transition has been observed to cause a small change in the CD signal (a broadening of the positive band (50)), which was not seen here. However, it is possible that molecules that have converted to the triplex form condense out of solution, leaving only duplexes and causing no detectable CD change.
The triplex geometry may lead to enhanced opportunities for condensation. With a third strand of negative charge wound inside the major groove of the helix, it may no longer be favorable or even possible for the spermine to remain deep within the groove of this RNA, and different spermine locations could facilitate condensation. To explore this possibility, we used all-atom MD to model the spermine distribution around a short poly(rA):poly(rU):poly(rU) triplex taken from PDB: 3P22 RNA structure (51). We found that although half of the bound spermine was located in the remnants of the major groove, the rest was bound to the phosphate groups at the external surface of the RNA triplex. Perhaps this externally bound spermine, located at the external part of the RNA molecule, provides opportunities for bridging between triplexes and promotes condensation. Another possibility is that aggregation occurs due to interconnected spermine-induced duplexes and triplexes, as has been discussed for poly(dA):poly(dT) DNA (52).
Validity of MD models
The model constructed in (23), in which the distribution of bound counterions determines the condensation behavior of the nucleic acids, does accurately predict the relative condensation behaviors of the mixed-sequence nucleic acid constructs in the presence of spermine. Differences in the WAXS profiles calculated from our MD simulations for mixed-sequence RNA with and without spermine reproduce the experimental trends. Confidence in the MD models is further enhanced by comparison with crystal structures of spermine complexed with tRNA and with A-form DNA, which also show spermine lying deep within the major groove (11, 53). Therefore, the models for RNA seem accurate and allow insight into conformational changes and spermine binding modes.
The differences in the calculated WAXS profiles for mixed-sequence DNA with and without spermine do not agree with the experimental differences. This is in agreement with previous work that compared MD simulations of nucleic acids with WAXS data (54, 55). Our model of spermine association to DNA is consistent with NMR studies, which suggest either that the spermine diffuses rapidly along the DNA molecule or that there are not discrete binding sites (56), and with the conclusions of Raman spectroscopy studies that also support non-specific electrostatic binding (57). It could also be consistent with a recent comparison of MD simulations and Förster resonance energy transfer data for a spermine-DNA system, which indicated that the methyl groups of thymine can sterically prevent spermine from binding to the major grooves of AT-rich DNA (58). Therefore, the simulations appear globally correct, but gaining more specific insights into the binding of polyamine to DNA and the mechanism of condensation will require improved force fields (59, 60).
Spermine bound deep in the major grooves stabilizes mixed-sequence RNA
One of spermine’s many functions is to stabilize the structure of RNA (2, 6, 21), and this stabilization has many important repercussions. For example, spermine-induced stabilization of tRNA prevents incorrect basepairing and increases the fidelity of translation (21), and the resulting binding and stabilization also create binding sites for divalent cations (61). Given that spermine-RNA interactions are critical for these types of specific binding events, it is important to gain insight into how spermine stabilizes RNA.
Fig. 5 shows that the addition of spermine does not significantly change the WAXS profile of DNA, suggesting that structural features on the length scales probed (∼10 Å) are unmodified by the addition of the multivalent partner. In contrast, and as demonstrated more strikingly in Ref. (55), the measured features in the WAXS curves for RNA become sharper when counterions are added. Importantly, the spermine molecules themselves contribute negligibly to the scattering. No differences are detected in the computed scattering profiles of RNA plus spermine when the spermines are present or (computationally) deleted, as shown in the Supporting Material. Fig. 5 b shows a (slightly) deeper first minimum and a larger trough-to-peak variation in the range of 0.3 Å−1 < q < 0.5 Å−1. The sharpening of features in the SAXS/WAXS curves reflects an increased definition of structure. In other words, the appearance of better-defined features can be explained by decreased molecular flexibility and more consistently determined molecular dimensions. Taken together, this interpretation of the WAXS data and the results of the MD predictions support a decrease in RNA flexibility when spermine is present. We therefore propose that the elongated spermine, deeply sequestered in the grooves of the RNA, fills the space and leaves less room for the molecule’s conformation to fluctuate, locking in its structure and stabilizing the molecule. This switch to the stabilized conformation could be reflected in the changes measured by WAXS and CD. Therefore, even though the RNA is intrinsically flexible, the spermine fixes it into a particular conformation. This stabilization reduces RNA conformational entropy but may increase selectivity, affecting the way in which an RNA binds to other molecules.
Conclusion
In this work, we combined experimental and simulation studies to elucidate the interactions between spermine and nucleic acids. Mixed-sequence duplex RNA, which imitates the basepaired regions of physiological RNA, resists condensation relative to DNA of an equivalent sequence. Poly(rA):poly(rU) RNA condenses out readily when small amounts of spermine are added, likely due to the formation of triplexes or higher aggregates. Models developed in previous work accurately predict the measured condensation propensities. These models suggest that spermines associate within the major grooves of the mixed-sequence RNA but loosely span the grooves of DNA. These different binding modes can explain the observed resistance of mixed-sequence RNA to condensation. These data, along with previous reports that spermine stabilizes RNA, suggest that the long spermine strands infiltrate the groove and lock the RNA into a rigid conformation that selectively enhances interactions with other molecules in the cell. Future refinement of the model of spermine distribution presented here will provide a more accurate and atomically detailed picture of spermine binding to both RNA and DNA, and may lend insight into the many important biological processes that require polyamine-nucleic acid interactions.
Author Contributions
L.P., N.B., and A.V.O. designed the research. I.S.T. carried out simulations. A.M.K. and S.A.P. performed the experiments and analyzed the data. All authors contributed to writing the article.
Acknowledgments
We thank members of L.P.’s group for helpful discussions.
This work was funded by National Institutes of Health grant R01 GM099450 and in part by National Science Foundation (NSF) grant CNS-0960081, and the HokieSpeed supercomputer at Virginia Tech. This work is based on research conducted at the Cornell High Energy Synchrotron Source, which is supported by the NSF and the National Institute of General Medical Sciences, National Institutes of Health, under NSF award DMR-1332208. A.M.K. received an NSF Graduate Research Fellowship under grant DGE-1144153.
Editor: Tamar Schlick.
Footnotes
Supporting Materials and Methods, two figures, and two movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)31047-5.
Supporting Material
References
- 1.Tabor H., Tabor C.W. Spermidine, spermine, and related amines. Pharmacol. Rev. 1964;16:245–300. [PubMed] [Google Scholar]
- 2.Lightfoot H.L., Hall J. Endogenous polyamine function—the RNA perspective. Nucleic Acids Res. 2014;42:11275–11290. doi: 10.1093/nar/gku837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tabor C.W., Tabor H. Polyamines. Annu. Rev. Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 4.Tabor H. The protective effect of spermine and other polyamines against heat denaturation of deoxyribonucleic acid. Biochemistry. 1962;1:496–501. doi: 10.1021/bi00909a021. [DOI] [PubMed] [Google Scholar]
- 5.Mandel M. The interaction of spermine and native deoxyribonucleic acid. J. Mol. Biol. 1962;5:435–441. [Google Scholar]
- 6.Goldstein J. Resistance of RNA to thermal denaturation in the presence of polyamines. Biochim. Biophys. Acta. 1966;123:620–623. doi: 10.1016/0005-2787(66)90231-0. [DOI] [PubMed] [Google Scholar]
- 7.Kaiser D., Tabor H., Tabor C.W. Spermine protection of coliphage λ DNA against breakage by hydrodynamic shear. J. Mol. Biol. 1963;6:141–147. doi: 10.1016/s0022-2836(63)80130-8. [DOI] [PubMed] [Google Scholar]
- 8.Pegg A.E. Toxicity of polyamines and their metabolic products. Chem. Res. Toxicol. 2013;26:1782–1800. doi: 10.1021/tx400316s. [DOI] [PubMed] [Google Scholar]
- 9.Igarashi K., Kashiwagi K. Modulation of cellular function by polyamines. Int. J. Biochem. Cell Biol. 2010;42:39–51. doi: 10.1016/j.biocel.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe S., Kusama-Eguchi K., Igarashi K. Estimation of polyamine binding to macromolecules and ATP in bovine lymphocytes and rat liver. J. Biol. Chem. 1991;266:20803–20809. [PubMed] [Google Scholar]
- 11.Quigley G.J., Teeter M.M., Rich A. Structural analysis of spermine and magnesium ion binding to yeast phenylalanine transfer RNA. Proc. Natl. Acad. Sci. USA. 1978;75:64–68. doi: 10.1073/pnas.75.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teif V.B., Bohinc K. Condensed DNA: condensing the concepts. Prog. Biophys. Mol. Biol. 2011;105:208–222. doi: 10.1016/j.pbiomolbio.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Bloomfield V.A. DNA condensation by multivalent cations. Biopolymers. 1997;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 14.He S., Arscott P.G., Bloomfield V.A. Condensation of DNA by multivalent cations: experimental studies of condensation kinetics. Biopolymers. 2000;53:329–341. doi: 10.1002/(SICI)1097-0282(20000405)53:4<329::AID-BIP5>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 15.Huang S.L., Felsenfeld G. Solubility of complexes of polynucleotides with spermine. Nature. 1960;188:301–302. doi: 10.1038/188301a0. [DOI] [PubMed] [Google Scholar]
- 16.Cantoni G.L. Action of spermine on soluble ribonucleic acid and some polynucleotides. Nature. 1960;188:300–301. doi: 10.1038/188300a0. [DOI] [PubMed] [Google Scholar]
- 17.Mitra S., Kaesberg P. Interaction of polyamines with turnip yellow mosaic virus RNA. Biochem. Biophys. Res. Commun. 1963;11:146–151. [Google Scholar]
- 18.Gosule L.C., Schellman J.A. DNA condensation with polyamines I. Spectroscopic studies. J. Mol. Biol. 1978;121:311–326. doi: 10.1016/0022-2836(78)90366-2. [DOI] [PubMed] [Google Scholar]
- 19.Pelta J., Livolant F., Sikorav J.-L. DNA aggregation induced by polyamines and cobalthexamine. J. Biol. Chem. 1996;271:5656–5662. doi: 10.1074/jbc.271.10.5656. [DOI] [PubMed] [Google Scholar]
- 20.Raspaud E., Durand D., Livolant F. Interhelical spacing in liquid crystalline spermine and spermidine-DNA precipitates. Biophys. J. 2005;88:392–403. doi: 10.1529/biophysj.104.040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kućan Z., Naranda T., Weygand-Durasević I. Effect of spermine on transfer RNA and transfer RNA-ribosome interactions. Adv. Exp. Med. Biol. 1988;250:525–533. doi: 10.1007/978-1-4684-5637-0_47. [DOI] [PubMed] [Google Scholar]
- 22.Li L., Pabit S.A., Pollack L. Double-stranded RNA resists condensation. Phys. Rev. Lett. 2011;106:108101. doi: 10.1103/PhysRevLett.106.108101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tolokh I.S., Pabit S.A., Onufriev A.V. Why double-stranded RNA resists condensation. Nucleic Acids Res. 2014;42:10823–10831. doi: 10.1093/nar/gku756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kibbe W.A. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 2007;35:W43–W46. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tolokh I.S., Drozdetski A.V., Onufriev A.V. Multi-shell model of ion-induced nucleic acid condensation. J. Chem. Phys. 2016;144:155101. doi: 10.1063/1.4945382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petoukhov M.V., Konarev P.V., Svergun D.I. ATSAS 2.1—towards automated and web-supported small-angle scattering data analysis. J. Appl. Cryst. 2007;40:s223–s228. [Google Scholar]
- 27.Case D.A., Cheatham T.E., 3rd, Woods R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salomon-Ferrer R., Case D.A., Walker R.C. An overview of the Amber biomolecular simulation package. WIREs Comput. Mol. Sci. 2013;3:198–210. [Google Scholar]
- 29.Pérez A., Marchán I., Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheatham T.E., 3rd, Cieplak P., Kollman P.A. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 31.Macke T., Case D. Modeling unusual nucleic acid structures. In: Leontis N.B., SantaLucia J. Jr., editors. Molecular Modeling of Nucleic Acids. American Chemical Society; Washington, DC: 1997. [Google Scholar]
- 32.Yoo J., Aksimentiev A. Improved parametrization of Li+, Na+, K+, and Mg2+ ions for all-atom molecular dynamics simulations of nucleic acid systems. J. Phys. Chem. Lett. 2012;3:45–50. [Google Scholar]
- 33.Braunlin W.H., Anderson C.F., Record M.T., Jr. Competitive interactions of Co(NH3)6(3+) and Na+ with helical B-DNA probed by 59Co and 23Na NMR. Biochemistry. 1987;26:7724–7731. doi: 10.1021/bi00398a028. [DOI] [PubMed] [Google Scholar]
- 34.Izadi S., Anandakrishnan R., Onufriev A.V. Building water models: a different approach. J. Phys. Chem. Lett. 2014;5:3863–3871. doi: 10.1021/jz501780a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joung I.S., Cheatham T.E., 3rd Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chin K., Sharp K.A., Pyle A.M. Calculating the electrostatic properties of RNA provides new insights into molecular interactions and function. Nat. Struct. Biol. 1999;6:1055–1061. doi: 10.1038/14940. [DOI] [PubMed] [Google Scholar]
- 37.Wang R., Wang Z.G. Inhomogeneous screening near the dielectric interface. J. Chem. Phys. 2016;144:134902. doi: 10.1063/1.4945011. [DOI] [PubMed] [Google Scholar]
- 38.Kypr J., Kejnovská I., Vorlícková M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makowski L. Characterization of proteins with wide-angle X-ray solution scattering (WAXS) J. Struct. Funct. Genomics. 2010;11:9–19. doi: 10.1007/s10969-009-9075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu X., Andresen K., Pollack L. Abrupt transition from a free, repulsive to a condensed, attractive DNA phase, induced by multivalent polyamine cations. Phys. Rev. Lett. 2008;101:228101. doi: 10.1103/PhysRevLett.101.228101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Araneda R.C., Lan J.Y., Bennett M.V. Spermine and arcaine block and permeate N-methyl-D-aspartate receptor channels. Biophys. J. 1999;76:2899–2911. doi: 10.1016/S0006-3495(99)77445-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng H., Bloomfield V.A. Structural effects of cobalt-amine compounds on DNA condensation. Biophys. J. 1999;77:1556–1561. doi: 10.1016/S0006-3495(99)77003-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kornyshev A.A. Electrostatic zipper motif for DNA aggregation. Phys. Rev. Lett. 1999;82:4138–4141. [Google Scholar]
- 44.Koculi E., Thirumalai D., Woodson S.A. Counterion charge density determines the position and plasticity of RNA folding transition states. J. Mol. Biol. 2006;359:446–454. doi: 10.1016/j.jmb.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 45.Blagoi Y., Gladchenko G., He Y. Phase equilibrium in poly(rA).poly(rU) complexes with Cd2+ and Mg2+ ions, studied by ultraviolet, infrared, and vibrational circular dichroism spectroscopy. Biopolymers. 2005;78:275–286. doi: 10.1002/bip.20288. [DOI] [PubMed] [Google Scholar]
- 46.Biver T. Stabilisation of non-canonical structures of nucleic acids by metal ions and small molecules. Coord. Chem. Rev. 2013;257:2765–2783. [Google Scholar]
- 47.Biver T., Busto N., Venturini M. Mg(II) and Ni(II) induce aggregation of poly(rA)poly(rU) to either tetra-aggregate or triplex depending on the metal ion concentration. J. Inorg. Biochem. 2015;151:115–122. doi: 10.1016/j.jinorgbio.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 48.Hampel K.J., Crosson P., Lee J.S. Polyamines favor DNA triplex formation at neutral pH. Biochemistry. 1991;30:4455–4459. doi: 10.1021/bi00232a012. [DOI] [PubMed] [Google Scholar]
- 49.Thomas T., Thomas T.J. Selectivity of polyamines in triplex DNA stabilization. Biochemistry. 1993;32:14068–14074. doi: 10.1021/bi00213a041. [DOI] [PubMed] [Google Scholar]
- 50.Brahms J. Optical activity and the conformation of polynucleotide models of nucleic acids. J. Mol. Biol. 1965;11:785–801. doi: 10.1016/s0022-2836(65)80036-5. [DOI] [PubMed] [Google Scholar]
- 51.Mitton-Fry R.M., Degregorio S.J., Steitz J.A. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science. 2010;330:1244–1247. doi: 10.1126/science.1195858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zozulya V.N., Nesterov A.B., Blagoi Y.P. Conformational transitions and aggregation in poly(dA)-poly(dT) system induced by Na+ and Mg2+ ions. Int. J. Biol. Macromol. 2003;33:183–191. doi: 10.1016/j.ijbiomac.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 53.Jain S., Zon G., Sundaralingam M. Base only binding of spermine in the deep groove of the A-DNA octamer d(GTGTACAC) Biochemistry. 1989;28:2360–2364. doi: 10.1021/bi00432a002. [DOI] [PubMed] [Google Scholar]
- 54.Zuo X., Cui G., Tiede D.M. X-ray diffraction “fingerprinting” of DNA structure in solution for quantitative evaluation of molecular dynamics simulation. Proc. Natl. Acad. Sci. USA. 2006;103:3534–3539. doi: 10.1073/pnas.0600022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pabit S.A., Katz A.M., Pollack L. Understanding nucleic acid structural changes by comparing wide-angle x-ray scattering (WAXS) experiments to molecular dynamics simulations. J. Chem. Phys. 2016;144:205102. doi: 10.1063/1.4950814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wemmer D.E., Srivenugopal K.S., Morris D.R. Nuclear magnetic resonance studies of polyamine binding to a defined DNA sequence. J. Mol. Biol. 1985;185:457–459. doi: 10.1016/0022-2836(85)90418-8. [DOI] [PubMed] [Google Scholar]
- 57.Deng H., Bloomfield V.A., Thomas G.J., Jr. Structural basis of polyamine-DNA recognition: spermidine and spermine interactions with genomic B-DNAs of different GC content probed by Raman spectroscopy. Nucleic Acids Res. 2000;28:3379–3385. doi: 10.1093/nar/28.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoo J., Kim H., Ha T. Direct evidence for sequence-dependent attraction between double-stranded DNA controlled by methylation. Nat. Commun. 2016;7:11045. doi: 10.1038/ncomms11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoo J., Aksimentiev A. Improved parameterization of amine-carboxylate and amine-phosphate interactions for molecular dynamics simulations using the CHARMM and AMBER force fields. J. Chem. Theory Comput. 2016;12:430–443. doi: 10.1021/acs.jctc.5b00967. [DOI] [PubMed] [Google Scholar]
- 60.Yoo J., Aksimentiev A. The structure and intermolecular forces of DNA condensates. Nucleic Acids Res. 2016;44:2036–2046. doi: 10.1093/nar/gkw081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nöthig-Laslo V., Weygand-Durasević I., Kućan Z. Binding of spermine to tRNATyr stabilizes the conformation of the anticodon loop and creates strong binding sites for divalent cations. Eur. J. Biochem. 1981;117:263–267. doi: 10.1111/j.1432-1033.1981.tb06332.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.