Abstract
To investigate the role of the hepatitis C virus internal ribosome entry site (HCV IRES) domain IV in translation initiation and regulation, two chimeric IRES elements were constructed to contain the reciprocal domain IV in the otherwise HCV and classical swine fever virus IRES elements. This permitted an examination of the role of domain IV in the control of HCV translation. A specific inhibitor of the HCV IRES, vitamin B12, was shown to inhibit translation directed by all IRES elements which contained domain IV from the HCV and the GB virus B IRES elements, whereas the HCV core protein could only suppress translation from the wild-type HCV IRES. Thus, the mechanisms of translation inhibition by vitamin B12 and the core protein differ, and they target different regions of the IRES.
Hepatitis C virus (HCV) is classified within the genus Hepacivirus of the Flaviviridae family. All members of the family Flaviviridae are small, icosahedral, enveloped viruses that contain a positive-sense RNA genome (2, 5). The genomes share a common organization, in which a single large open reading frame is flanked by 5′ and 3′ untranslated regions (UTR). The open reading frame encodes a large polyprotein which is processed into individual structural and nonstructural proteins. The family Flaviviridae consists of three genera: Flavivirus, Pestivirus, and Hepacivirus. It was suggested that three new proposed members of the Flaviviridae, the GB agents (GB virus [GBV] A, B, and C) should be classified in a separate genera within the Flaviviridae or in a subgenera of Hepacivirus (20).
The genomes of the pestiviruses (viz., bovine viral diarrhea virus and classical swine fever virus [CSFV]), HCV, and the GB agents have highly structured 5′ UTR of similar lengths (341 to 445 nucleotides [nt]) that contain multiple AUG triplets preceding the authentic polyprotein initiation codon. These structured RNA segments function as an internal ribosome entry site (IRES) to initiate polyprotein translation (6, 16, 31). Among these IRES elements, the HCV and the GBV-B IRESs are very similar in terms of proposed structure and function and differ from the IRES elements of the picornaviruses (16). Although there are slight differences in the proposed structures, the IRES elements of HCV, GBV-B, and pestiviruses were classified as type 2 IRES elements (6). The central domain of these IRES elements (domain III) is highly conserved and their predicted secondary structures, first proposed by Brown et al. (3) and modified at a later date (11), are superimposable. The structural integrity of domain III is essential for IRES activity (25, 32). However, a major difference is that the HCV and GBV-B IRES elements contain a stem-loop within domain IV whereas this feature is missing in the pestivirus IRES elements (11). The function of the stem-loop which comprises domain IV of the HCV IRES element is unclear.
There is still some uncertainty over the requirement for the inclusion of the core protein coding sequence for optimal HCV IRES function (for a review, see reference 26), and this point may be refined to question if stem-loop IV is necessary, since the core coding sequence forms part of stem-loop IV. Early studies demonstrated that the inclusion of nt 1 to 32 of the core protein coding sequences was essential for efficient IRES activity (19, 23). A recent study suggested that IRES activity was retained in the absence of stem-loop IV and suggested that it is necessary to avoid the formation of a stable RNA structure downstream of the initiator AUG for optimal HCV IRES activity (24).
A previous study demonstrated that the stability of the stem-loop in domain IV of the HCV IRES was closely related to IRES activity (11). Mutations which led to increased stability of stem-loop IV resulted in a sharp reduction of IRES efficiency. It was suggested that a self-regulatory mechanism may influence HCV translation if a gene product can bind to stem-loop IV and increase its stability (11). However, no evidence has been found to support this hypothesis, although recent studies suggested that the HCV core, NS4, and NS5 proteins can regulate HCV IRES-directed translation (17, 28, 33).
Several small molecules and proteins which can regulate HCV IRES activity have been identified. Polypyrimidine tract binding protein (8), La protein (1), and heterogeneous nuclear ribonucleoprotein L (9) stimulate HCV IRES-directed translation. Two DNA ribonucleases, Dz2 and Dz4, were found to inhibit HCV IRES activity (21). Cyanocobalamin (vitamin B12) was recently demonstrated to selectively inhibit HCV IRES-directed translation (18, 30). Inhibition by vitamin B12 and HCV core and NS5A proteins were demonstrated to be specific for HCV IRES-directed translation and had no effect on translation from CSFV and encephalomyocarditis virus IRES elements (10, 17, 18). Since the stem-loop in domain IV only exists in the HCV and GBV-B IRES elements, this may indicate that stem-loop IV is responsible for any effect which is specific to these IRES elements.
The aim of this study was to examine the role of the stem-loop in the HCV IRES domain IV in the regulation of IRES function. Although the pestivirus IRES elements do not have a recognized domain IV, for convenience we have used the term domain IV(A) (assumed) when referring to the analogous region in the CSFV IRES element. We used two chimeric IRES elements, in which domain IV/IV(A) of the HCV and CSFV IRES elements was exchanged (Fig. 1). To avoid any possible influence on other IRES domains, the splice sites were chosen downstream of a common GUA motif, located downstream of the pseudoknot structures in both IRES elements. As HCV IRES activity can be influenced by long-range RNA-RNA interactions (13, 15), all IRES elements were used as monocistronic reporter molecules. We examined the effect of two known regulators of the HCV IRES, vitamin B12 and the HCV core protein, to determine the effect on translation from these two chimeric IRES elements.
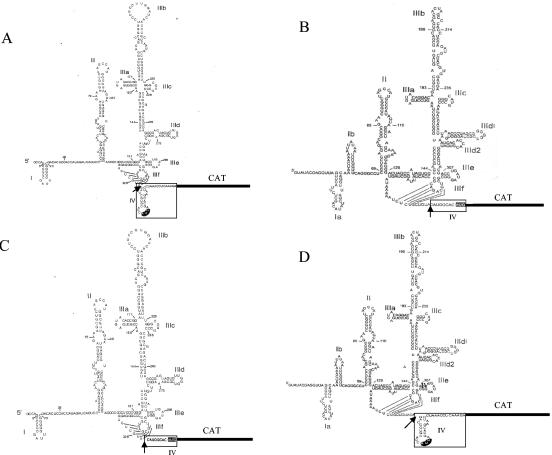
FIG. 1.
The construction of two chimeric IRES elements with the reciprocal domain IV/IV(A) of the HCV and CSFV IRES elements. pHCV-IRES-CAT (A) and pCSFV-IRES-CAT (B) represent wild-type HCV and CSFV IRES elements ligated in frame upstream of a CAT gene (18). The shaded AUG in domain IV/IV(A) represents the authentic polyprotein initiation codon. The chimeric IRES element pHCV1-3/CSFV4 (C) contains the HCV IRES domains I to III and the CSFV IRES domain IV(A), and pCSFV1-3/HCV4 (D) contains the CSFV IRES domains I to III and the HCV IRES domain IV. The splice site used to switch the domain IV/IV(A) regions is shown by an arrow, and the respective domain IV regions are outlined.
Two monocistronic IRES plasmids, which reflect the physiological use of viral IRES elements, were used to construct the chimeric IRES elements. pHCV-IRES-CAT directs the expression of chloramphenicol acetyltransferase (CAT) from the HCV IRES (18) and includes the first 27 nt of the core coding sequence to retain the structure of domain IV (Fig. 1A). pCSFV-IRES-CAT contains 375 nt of the CSFV 5′ UTR to control expression of the CAT gene (Fig. 1B). To construct the chimeric IRES elements pCSFV1-3/HCV4, which contains the CSFV IRES domain I-III and the HCV IRES domain IV, and pHCV1-3/CSFV4, which contains the HCV IRES domain I-III and the CSFV IRES domain IV(A), the reciprocal domains were substituted downstream of the pseudoknot loop (Fig. 1). This feature, which is highly conserved in the native IRES elements, was retained in pHCV-IRES-CAT and pCSFV-IRES-CAT and the two chimeric IRES elements (Fig. 1C and 1D). The CAT gene was ligated in frame downstream of each IRES element. Compared with wild-type pHCV-IRES-CAT, pHCV1-3/CSFV4 has no stem-loop IV, whereas pCSFV1-3/HCV4 has a stem-loop which contains the authentic initiator AUG and is similar to the HCV IRES domain IV (Fig. 1C and 1D). No other mutations were made that might influence translation efficiency. No other change was noted in the structures of the two chimeric IRES elements as determined by M-fold analysis (data not shown) using the conditions and constraints outlined in the initial publication (3).
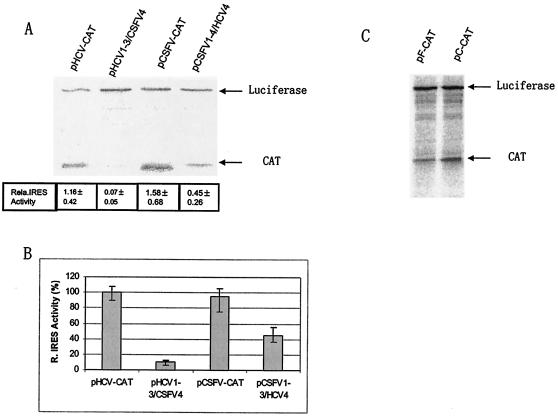
Initially, to determine the translational activity of the two chimeric IRES elements, an in vitro translation reaction was performed in a rabbit reticulocyte lysate (RRL) loaded with 80 ng of the respective RNAs. Each reaction contained 50 ng of capped-luciferase RNA as an internal control (18). The translation reactions were carried out for 90 min at 30°C in a reaction volume of 25 μl. The [35S]methionine-labeled products were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and quantitated by PhosphorImager analysis. The activity of each IRES element was calculated by comparing the level of CAT produced relative to the expression of luciferase. Compared with the wild-type HCV IRES, the relative activity of pHCV1-3/CSFV4 was reduced to ∼5% (Fig. 2A), consistent with virtual loss of IRES function. The translational efficiency of pCSFV1-3/HCV4 was also decreased relative to CSFV IRES, but the IRES was clearly functional (Fig. 2A) and showed an efficiency of approximately 30 to 40% relative to the CSFV and HCV IRES elements, respectively. The experiment was repeated with various amounts of template RNA and the results were similar.
FIG. 2.
Analysis of the translation efficiency of each IRES element either in monocistronic (A and B) or bicistronic forms (C). In vitro translation was performed in a RRL reaction. The products from the translation reactions were resolved by SDS-PAGE (A) and quantified by PhosphorImager analysis. The IRES efficiency was defined as the ratio of CAT product to luciferase (relative IRES efficiency = CAT/LUC). The translation efficiency of each IRES element was also determined in HepG2 cells. Capped-luciferase RNA was cotransfected with each IRES RNA to adjust the transfection efficiency. The relative level of CAT in the pHCV-IRES-CAT-transfected cells was normalized to 100% and the relative efficiency of each IRES was calculated (B). The efficiency of pF-CAT-directed translation was also assessed in a bicistronic form and compared against pcCAT (see reference 17).
The translational activity of the chimeric IRES elements was then tested in cells. The respective IRES elements and capped-luciferase RNA were cotransfected into HepG2 cells. The cells were harvested 5 h posttransfection and the expression of CAT and luciferase were analyzed as described previously (17). The results of this experiment showed that the wild-type HCV and CSFV IRES elements were translationally efficient as expected (Fig. 2B). The activity of pHCV1-3/CSFV4 was reduced to ∼10% of pHCV-IRES-CAT, while the translation efficiency of pCSFV1-3/HCV4 was approximately 40% of that of the wild-type pCSFV-IRES-CAT. To discount the possibility that decreased RNA stability accounted for the reduction in the efficiency of the chimeric IRES elements, the stability of all the RNA species was examined by incubating radiolabeled RNA in rabbit reticulocyte lysates for up to 90 min followed by acid precipitation and analysis of the recovered counts, but no differences were found (data not shown).
As the chimeric pHCV1-3/CSFV4 IRES only showed low activity, the remainder of the study was restricted to the CSFV1-3/HCV4 chimeric IRES. Although uncapped RNA is not translationally competent in our hands, it was necessary to demonstrate that the chimeric pCSFV1-3/HCV4 IRES functioned as an IRES in a bicistronic vector. Consequently, the HCV IRES in a bicistronic vector, pcCAT (17), was replaced with the chimeric IRES, CSFV1-3/HCV4, in a novel bicistronic vector, pF-CAT, so that the expression of firefly luciferase (LUC) was controlled by a cap-dependent mechanism and the expression of CAT was controlled by the intergenic chimeric IRES, essentially as described previously (17). In vitro-transcribed RNA was added to a rabbit reticulocyte lysate as described above, and the products were analyzed by SDS-PAGE. The results show that the downstream CAT gene was expressed with an efficiency of approximately 40% relative to the HCV IRES and confirmed that the chimera represented a functional IRES (Fig. 2C).
The position of the AUG codon relative to other structural elements of the IRES is likely to be critical since the 40S ribosomal subunit binds directly to these flavivirus elements in such a way that the ribosomal P site is placed in the immediate vicinity of the initiation codon (12). To confirm that the HCV stem-loop in the CSFV1-3/HCV4 chimera is still folded correctly, we performed a toeprinting analysis on the RNA in the presence of ribosomes, as described previously (30). This assay, which results in a strong stop in the product of reverse transcription of the RNA, highlights the 3′ position of the RNA that is protected by the ribosome and has been described previously (22, 30) as 15 nucleotides downstream of the AUG codon. The results of this experiment showed that the toeprint of the wild-type HCV IRES and the CSFV1-3/HCV4 chimeric IRES were identical (Fig. 3) and identified a strong stop at U356, 15 nucleotides downstream of the AUG initiation codon. Thus, we can conclude that the HCV IRES domain IV is folded correctly in this chimera and the AUG initiator is in the same context as it is in the HCV IRES.
FIG. 3.
Toeprint analysis of the HCV IRES and the CSFV1-3/HCV4 IRES after formation of the initiation complex prior to and after incubation in rabbit reticulocyte lysates. The arrows indicate the positions of the start codon for the polyprotein and the ribosomal toeprint at U356. The reference lanes marked T, G, C, and A are dideoxy sequencing reactions of HCV IRES cDNA. Note that the pattern in the HCV and CSFV1-3/HCV4 IRES lanes in the presence of the rabbit reticulocyte lysate is identical in the lower part of the figure, and the pattern only diverges upstream of the HCV domain IV.
A previous report demonstrated that vitamin B12 selectively inhibited HCV IRES function, but not CSFV IRES-directed translation, in an in vitro translation system (18). We now wished to determine if an interaction between vitamin B12 and the HCV IRES domain IV is responsible for this translation inhibition. RNA transcribed from pCSFV1-3/HCV4, which contains the HCV IRES domain IV, was incubated with vitamin B12 for 10 min at room temperature and then added to a RRL to initiate the translation reaction. The concentration of B12, 2.5 mM, was previously shown to be optimal, as described previously (18). Consistent with the previous report, the HCV IRES activity was reduced by 60% by prior incubation with vitamin B12, whereas CSFV IRES-directed translation was not affected (Fig. 4A). The effect of vitamin B12 on pCSFV1-3/HCV4 was similar to that of HCV IRES, i.e., translational activity of pCSFV1-3/HCV4 IRES was reduced by approximately 50 to 60%. This level of inhibition is similar to that reported in previous studies (18, 30). These results imply that the HCV IRES domain IV is responsible for translational inhibition by vitamin B12.
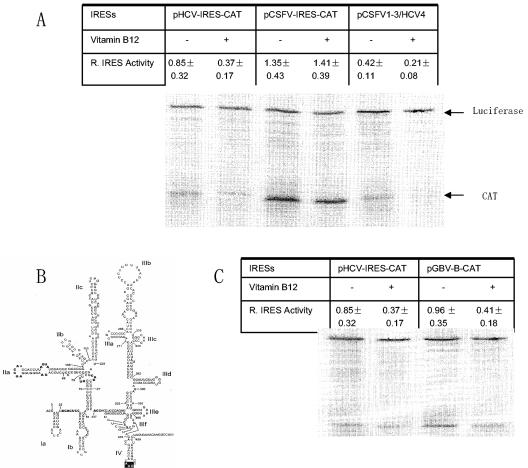
FIG. 4.
Vitamin B12 inhibits pCSFV1-3/HCV4- and GBV-B IRES-directed translation. The effect of vitamin B12 on translation directed by these IRES elements was examined by incubating vitamin B12 with the RNA for 10 min at room temperature prior to translation in a RRL. RNA with the same volume of water represented the control. pHCV-IRES-CAT and pCSFV-IRES-CAT translations in the presence and absence of vitamin B12 represented positive and negative controls for inhibition by vitamin B12 (A). The predicted structure of the GBV-B IRES is similar to that of the HCV IRES, including domain IV (B). (C) The effect of vitamin B12 on GBV-B IRES-directed translation.
To provide additional evidence that vitamin B12 interacts with IRES domain IV, we then examined the effect of vitamin B12 on GBV-B IRES-directed translation. The proposed structure of the HCV and GBV-B IRES elements is similar and each contains a stem-loop which encompasses the polyprotein initiation codon in domain IV (Fig. 1A and 4B). To facilitate this, pGBV-B-CAT, which contains the GBV-B IRES ligated in frame to the CAT gene sequence, was constructed to include the first 27 nt of the GBV-B core gene. Prior incubation with vitamin B12 resulted in a ca. 60% reduction in the level of CAT translation from the GBV-B IRES (Fig. 4C). This result suggests that vitamin B12 does not specifically inhibit HCV IRES activity but also selectively inhibits translation from the IRES of a closely related virus.
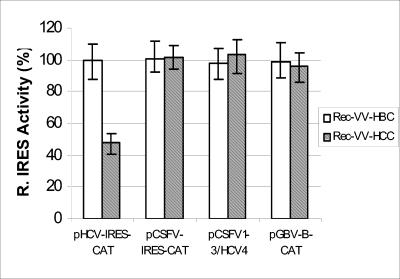
It has previously been shown (17, 28, 33) that the HCV core protein can inhibit translation from the HCV IRES. Furthermore, it was shown that the HCV core protein had no effect on CSFV and encephalomyocarditis virus IRES-directed translation in HepG2 cells (17). To determine if the HCV IRES domain IV is also the target for inhibition of translation by the HCV core protein, a recombinant vaccinia virus (RecVV-HCC) was used to express the HCV core protein in HepG2 cells. The cells were infected with RecVV-HCC or a RecVV-HBC to express the hepatitis B virus (HBV) core protein as control and then transfected with the RNA as described previously (17). Cotransfected capped-luciferase RNA was used to adjust transfection efficiency. The cells were harvested 5 h posttransfection, and the levels of CAT and luciferase were analyzed. The IRES activity in RecVV-HBC-infected cells was normalized to 100% (Fig. 5). The HCV IRES activity in the RecVV-HCC-infected cells was reduced by 51%. The absolute values for CAT and LUC differ from those in Fig. 2, because the transfection efficiency of RecVV-infected cells is higher than that of uninfected cells. However, expression of the HCV core protein had no effect on CSFV IRES-, pCSFV1-3/HCV4-, and GBV-B IRES-directed translations (Fig. 5). Thus, although the primary sequence and structure of the GBV-B IRES is very similar to that of the HCV IRES, this result shows that the HCV core protein regulates HCV IRES-directed translation in a highly specific manner, and furthermore, the target of the core protein is unlikely to be—or is not restricted to—domain IV.
FIG. 5.
The HCV core specifically inhibits HCV IRES-directed translation in HepG2 cells. HepG2 cells were infected with RecVV-HBC or RecVV-HCC for 15 h to allow the expression of the HBV and HCV core proteins, respectively. The cells were then cotransfected with the respective IRES elements and capped-luciferase RNA. The levels of IRES-directed translation in HBV core protein-positive cells were normalized to 100% after adjusting for the transfection efficiency as determined by luciferase activity, and the relative IRES efficiencies from HCV core-positive cells were calculated.
The structure and function of domains II and III in the HCV and CSFV IRES elements are similar (14, 16). The heart of the IRES element is domain III, which is essential for IRES activity. Previous studies showed that domain III recruits the 40S ribosomal subunit and eIF3 (12, 14). However, the proposed structure in the vicinity of the start codon, known as domain IV, is quite different in the HCV and CSFV IRES elements. To examine the role of domain IV in the HCV IRES in translation regulation, the HCV domain IV and the CSFV IRES IV(A) were exchanged and two chimeric IRES elements were created. No significant change in the predicted structure of domains I, II, and III of the two chimeric IRES elements compared to the structure of the wild-type IRES elements was found by M-fold analysis. The analysis showed that a stem-loop containing the polyprotein start codon in the loop was introduced into the chimeric pCSFV1-3/HCV4 and removed from pHCV1-3/CSFV4.
Compared with the wild-type IRES elements, the pHCV1-3/CSFV4(A) IRES was barely functional while the other, pCSFV1-3/HCV4, showed reduced functional activity in vitro and in cells. This finding is similar to that in a recent report in which the substitution of downstream coding sequences of the HCV and CSFV IRES elements resulted in reduction of IRES activity (7).
We have previously identified two inhibitors of HCV IRES function, viz., vitamin B12 and the HCV core protein (17, 18). Vitamin B12 selectively inhibits HCV IRES-directed translation in RRL but has no effect on CSFV IRES-directed translation. We also showed that vitamin B12 traps 80S ribosomal complexes on the HCV IRES resulting in translational inhibition (30). However, the precise mechanism of this interaction is unclear, although vitamin B12 does not bind directly to the IRES but appears to stabilize the 80S/IRES interaction (30). In this present study, vitamin B12 was shown to inhibit translation directed by HCV, pCSFV1-3/HCV4, and GBV-B IRES elements with similar efficiencies. A common feature of these IRES elements is a stem-loop within domain IV, suggesting that this structure may be necessary for the interaction between vitamin B12, the ribosome, and the IRES. The proposed structure of the CSFV IRES is similar to that of the HCV IRES, and the inclusion of the HCV IRES stem-loop IV in pCSFV1-3/HCV4 that results in vitamin B12 sensitivity suggests that the general mechanisms of initiation are similar. The inhibition of GBV-B IRES-directed translation by vitamin B12 represents yet another feature of this virus that shows similarities with HCV (4, 27, 29). We think it very likely that vitamin B12 binds to an intermediate structure that is formed during the IRES/ribosomal complex interaction and prevents initiation by stabilizing this complex.
In contrast, the HCV core protein employs a different mechanism to suppress HCV IRES-directed translation. The results in this study showed that the HCV core protein specifically inhibited HCV IRES-directed translation in HepG2 cells but had no effect on translation directed by the IRES elements of GBV-B, CSFV, and CSFV1-3/HCV4. As the HCV1-3/CSFV4(A) chimeric IRES was not functional, we were unable to determine if removal of the stem-loop in domain IV from the HCV IRES resulted in elimination of the interaction with the core protein. Previous studies have shown that the HCV core protein binds specifically to the HCV IRES, suggesting that translation inhibition by the core protein results from direct binding (28, 33). This specific interaction signal is unlikely to involve the IRES domain IV.
As vitamin B12 is stored in hepatocytes, vitamin B12 and core protein suppression of HCV IRES-directed translation may be one mechanism by which the virus has evolved to reduce the level of replication and promote persistence.
Acknowledgments
This work was supported by grant number 191805 from the National Health and Medical Research Council of Australia. The HCV Laboratory of the Burnet Institute is a member of the Australian Centre for Hepatitis Virology, Inc.
REFERENCES
- 1.Ali, N., and A. Siddiqui. 1997. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc. Natl. Acad. Sci. USA 94:2249-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 3.Brown, E. A., H. Zhang, L. H. Ping, and S. M. Lemon. 1992. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 20:5041-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Tomassi, A., M. Pizzuti, R. Graziani, A. Sbardellati, S. Altamura, G. Paonessa, and C. Traboni. 2002. Cell clones selected from the Huh7 human hepatoma cell line support efficient replication of a subgenomic GB virus B replicon. J. Virol. 76:7736-7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fields, B. N., D. M. Knipe, and P. M. Howley. 1996. Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 6.Fletcher, S. P., and R. J. Jackson. 2002. Pestivirus internal ribosome entry site (IRES) structure and function: elements in the 5′ untranslated region important for IRES function. J. Virol. 76:5024-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fletcher, S. R., I. K. Ali, A. Kaminski, P. Digard, and R. J. Jackson. 2002. The influence of viral coding sequences on pestivirus IRES activity reveals further parallels with translation initiation in prokaryotes. RNA 8:1558-1571. [PMC free article] [PubMed] [Google Scholar]
- 8.Gosert, R., K. H. Chang, R. Rijnbrand, M. Yi, D. V. Sangar, and S. M. Lemon. 2000. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol. Cell. Biol. 20:1583-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahm, B., Y. K. Kim, J. H. Kim, T. Y. Kim, and S. K. Jang. 1998. Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J. Virol. 72:8782-8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He, Y., W. Yan, C. Coito, Y. Li, M. Gale, Jr., and M. G. Katze. 2003. The regulation of hepatitis C virus (HCV) internal ribosome-entry site-mediated translation by HCV replicons and nonstructural proteins. J. Gen. Virol. 84:535-543. [DOI] [PubMed] [Google Scholar]
- 11.Honda, M., E. A. Brown, and S. M. Lemon. 1996. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA 2:955-968. [PMC free article] [PubMed] [Google Scholar]
- 12.Kieft, J. S., K. Zhou, R. Jubin, and J. A. Doudna. 2001. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7:194-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, Y. K., S. H. Lee, C. S. Kim, S. K. Seol, and S. K. Jang. 2003. Long-range RNA-RNA interaction between the 5′ nontranslated region and the core-coding sequences of hepatitis C virus modulates the IRES-dependent translation. RNA 9:599-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolupaeva, V. G., T. V. Pestova, and C. U. Hellen. 2000. Ribosomal binding to the internal ribosomal entry site of classical swine fever virus. RNA 6:1791-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lafuente, E., R. Ramos, and E. Martinez-Salas. 2002. Long-range RNA-RNA interactions between distant regions of the hepatitis C virus internal ribosome entry site element. J. Gen. Virol. 83:1113-1121. [DOI] [PubMed] [Google Scholar]
- 16.Lemon, S. M., and M. Honda. 1997. Internal ribosome entry sites within the RNA genomes of hepatitis C virus and other flaviviruses. Semin. Virol. 8:274-288. [Google Scholar]
- 17.Li, D., S. T. Takyar, W. B. Lott, and E. J. Gowans. 2003. Amino acids 1-20 of the hepatitis C virus (HCV) core protein specifically inhibit HCV IRES-dependent translation in HepG2 cells, and inhibit both HCV IRES- and cap-dependent translation in HuH7 and CV-1 cells. J. Gen. Virol. 84:815-825. [DOI] [PubMed] [Google Scholar]
- 18.Lott, W. B., S. S. Takyar, J. Tuppen, D. H. Crawford, M. Harrison, T. P. Sloots, and E. J. Gowans. 2001. Vitamin B12 and hepatitis C: molecular biology and human pathology. Proc. Natl. Acad. Sci. USA 98:4916-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu, H. H., and E. Wimmer. 1996. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc. Natl. Acad. Sci. USA 93:1412-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muerhoff, A. S., T. P. Leary, J. N. Simons, T. J. Pilot-Matias, G. J. Dawson, J. C. Erker, M. L. Chalmers, G. G. Schlauder, S. M. Desai, and I. K. Mushahwar. 1995. Genomic organization of GB viruses A and B: two new members of the Flaviviridae associated with GB agent hepatitis. J. Virol. 69:5621-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oketani, M., Y. Asahina, C. H. Wu, and G. Y. Wu. 1999. Inhibition of hepatitis C virus-directed gene expression by a DNA ribonuclease. J. Hepatol. 31:628-634. [DOI] [PubMed] [Google Scholar]
- 22.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reynolds, J. E., A. Kaminski, H. J. Kettinen, K. Grace, B. E. Clarke, A. R. Carroll, D. J. Rowlands, and R. J. Jackson. 1995. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 14:6010-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rijnbrand, R., P. J. Bredenbeek, P. C. Haasnoot, J. S. Kieft, W. J. Spaan, and S. M. Lemon. 2001. The influence of downstream protein-coding sequence on internal ribosome entry on hepatitis C virus and other flavivirus RNAs. RNA 7:585-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rijnbrand, R., T. van der Straaten, P. A. van Rijn, W. J. Spaan, and P. J. Bredenbeek. 1997. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J. Virol. 71:451-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rijnbrand, R. C., and S. M. Lemon. 2000. Internal ribosome entry site-mediated translation in hepatitis C virus replication. Curr. Top. Microbiol. Immunol. 242:85-116. [DOI] [PubMed] [Google Scholar]
- 27.Scarselli, E., A. Urbani, A. Sbardellati, L. Tomei, R. De Francesco, and C. Traboni. 1997. GB virus B and hepatitis C virus NS3 serine proteases share substrate specificity. J. Virol. 71:4985-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimoike, T., S. Mimori, H. Tani, Y. Matsuura, and T. Miyamura. 1999. Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J. Virol. 73:9718-9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simons, J. N., T. J. Pilot-Matias, T. P. Leary, G. J. Dawson, S. M. Desai, G. G. Schlauder, A. S. Muerhoff, J. C. Erker, S. L. Buijk, M. L. Chalmers, et al. 1995. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc. Natl. Acad. Sci. USA 92:3401-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takyar, S. S., E. J. Gowans, and W. B. Lott. 2002. Vitamin B12 stalls the 80 S ribosomal complex on the hepatitis C internal ribosome entry site. J. Mol. Biol. 319:1-8. [DOI] [PubMed] [Google Scholar]
- 31.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, C., S. Y. Le, N. Ali, and A. Siddiqui. 1995. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA 1:526-537. [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, J., O. Yamada, H. Yoshida, T. Iwai, and H. Araki. 2002. Autogenous translational inhibition of core protein: implication for switch from translation to RNA replication in hepatitis C virus. Virology 293:141-150. [DOI] [PubMed] [Google Scholar]