Abstract
During latency, Epstein-Barr virus (EBV) is stably maintained as a circular plasmid that is replicated once per cell cycle and partitioned at mitosis. Both these processes require a single viral protein, EBV nuclear antigen 1 (EBNA1), which binds two clusters of cognate binding sites within the latent viral origin, oriP. EBNA1 is known to associate with cellular metaphase chromosomes through chromosome-binding domains within its amino terminus, an association that we have determined to be required not only for the partitioning of oriP plasmids but also for their replication. One of the chromosome-binding domains of EBNA1 associates with a cellular nucleolar protein, EBP2, and it has been proposed that this interaction underlies that ability of EBNA1 to bind metaphase chromosomes. Here we demonstrate that EBNA1's chromosome-binding domains are AT hooks, a DNA-binding motif found in a family of proteins that bind the scaffold-associated regions on metaphase chromosomes. Further, we demonstrate that the ability of EBNA1 to stably replicate and partition oriP plasmids correlates with its AT hook activity and not its association with EBP2. Finally, we examine the contributions of EBP2 toward the ability of EBNA1 to associate with metaphase chromosomes in human cells, as well as support the replication and partitioning of oriP plasmids in human cells. Our results indicate that it is unlikely that EBP2 directly mediates these activities of EBNA1 in human cells.
The latent Epstein-Barr virus (EBV) genome is stably retained in cells extrachromosomally as a circular plasmid. Stable replication of the EBV genome relies on the latently expressed viral protein EBV nuclear antigen 1 (EBNA1) binding to both the family of repeats (FR) within oriP, as well as to cellular chromosomes. This tethering of viral plasmids to cellular chromosomes is hypothesized to be necessary for their replication during S phase and partitioning during mitosis.
Homodimers of EBNA1 are bound via a carboxy-terminal DNA-binding domain (DBD; amino acids [aa] 451 to 640) to two clusters of binding sites within oriP (60) called the FR and the dyad symmetry element (DS), whereas the amino terminus of EBNA1 (aa 1 to 450) is required for an association with cellular chromosomes (37, 48). During S phase, in concert with cellular chromosomes, oriP-plasmids are semiconservatively replicated (1, 2, 11, 47, 61), a process mediated by EBNA1's binding at the DS (7). However, the DBD of EBNA1 is by itself insufficient to recruit the licensed cellular replication apparatus to DS (28) but requires that the amino terminus of EBNA1 contain domain(s) that associate with mitotic chromosomes (48). We have recently demonstrated that the entire amino terminus of EBNA1 can be replaced by a cellular protein that specifically associates with mitotic chromosomes and that such a fusion supports the stable replication and partitioning of oriP plasmids similarly to wild-type EBNA1. In contrast, a fusion in which the amino terminus associates with interphase chromatin, but not mitotic chromosomes, is unable to support either the replication or the partitioning of oriP plasmids (48). This result strongly suggests that the ability of wild-type EBNA1 to support both of these processes is mediated by the ability of the amino terminus of EBNA1 to associate with mitotic chromosomes.
EBV genomes or oriP plasmids are thought to be partitioned via a “piggyback” mechanism in which viral episomes are tethered to sister chromatids as they are separated by the cellular mitotic apparatus (9). This hypothesis is supported by fluorescence in situ hybridization analysis which indicates that episomal EBV genomes are associated with sister chromatids (14) and by indirect immunofluorescence performed by others (37) and us (48) indicating that EBNA1 is associated in punctate dots with sister chromatids. Whatever the mechanism of partitioning, it is very efficient, with an estimated rate of loss of between 2 and 6% per cell generation (29, 53), a finding similar to that observed with centromere-containing plasmids in budding yeast (22, 30).
EBNA1 associates with mitotic chromosomes through two regions in its amino terminus that we term domains A (aa 33 to 89) and B (aa 328 to 378). When each of these regions is fused to green fluorescent protein (GFP), the fusion stains metaphase chromosomes as punctate dots (37). These regions share similarity in sequence, i.e., they both contain repeats of glycine and arginine (GR repeats), and at least one common biochemical property: DNA linking. When fused to a heterologous DBD, the A and B regions link the cognate binding site for the heterologous DBD into a large macromolecular complex (33, 35). A previous study indicates that EBNA1 derivatives with either domain A or domain B deleted are equivalent in function. Similarly, derivatives containing two copies of either domain, but none of the other, were also equivalent in function and are also approximately equivalent in function to wild-type EBNA1 that contains one copy of each domain (34).
In contrast to these studies, it was reported that EBNA1 with domain B deleted supports the replication of oriP plasmids up to 1 week posttransfection in C33A cells but not stably beyond that time (50). In contrast, derivatives that contained domain B supported transient and stable replication of oriP plasmids in the same cells (50). Furthermore, domain B, but not domain A, associated with a cellular nucleolar protein (EBP2/p40) (50), and the expression of EBP2 significantly enhanced the ability of wild-type EBNA1 to retain ARS/FR plasmids in budding yeast (27). The authors of that study have also found that a fusion between EBP2 and GFP associates with mitotic chromosomes in human cells (26). On the basis of these data, it has been proposed that the association of EBNA1 with mitotic chromosomes is indirect and mediated via EBP2.
In the present study, we have reexamined the mechanism by which EBNA1 associates with mitotic chromosomes for the following reasons. We and others have demonstrated that the cellular protein HMGA1a can functionally replace the amino terminus of EBNA1 (24, 48). The resulting fusion, HMGA1a-DBD, supports the replication of oriP plasmids at copy numbers equivalent to EBNA1, and oriP plasmids are lost from cells expressing EBNA1 or HMGA1a-DBD at indistinguishable rates (48). HMGA1a associates with chromosomes through a glycine-argine (GR) repeat protein motif termed the AT hook, which associates with DNA sequences that are rich in adenine and thymine (4). We noticed that domains A and B contain such GR-repeats that bear striking similarity to the AT hooks of cellular AT hook proteins. We have investigated whether domains A and B can function as AT hooks and have tested whether AT hook function or an association with EBP2 is required for EBNA1 to associate with cellular chromosomes and support the replication of oriP plasmids.
Our results indicate that both domains A and B of EBNA1 contain AT hooks and that AT hook activity correlates well with EBNA1's ability to associate with cellular chromosomes and stably replicate oriP plasmids. Further, by utilizing the observation that only domain B of EBNA1 associates with EBP2 and by characterizing the localization and activities of a fusion between EBP2 and the DBD of EBNA1 in human cells, we demonstrate that EBP2 is unlikely to mediate EBNA1's association with mitotic chromosomes. Our results do not eliminate the possibility that EBP2 serves as a loader to load EBNA1 onto mitotic chromosomes, without remaining a component of the final bound complex.
MATERIALS AND METHODS
Bacterial strains and plasmid purification.
All plasmids were propagated in the Escherichia coli strains DH5α, MC1061/P3, or STBL2 (Invitrogen, Carlsbad, Calif.). Plasmids used for transfection were purified on isopycnic CsCl gradients (36).
Replication reporter plasmids.
The construction of replication reporter plasmid AGP74, containing 20 EBNA1 binding sites in FR has been described previously (21). This contains a puromycin resistance gene driven by the simian virus 40 early promoter. A derivative of AGP74 was constructed that contains 30 EBNA1 binding sites in FR (AGP264). To construct this plasmid, a BssHII-Bsu36I fragment containing 10 EBNA1 binding sites from AGP212 was added to AGP74 digested with BssHII and Bsu36I.
Generation of EBNA1 derivatives, EBP2 derivatives, and the EBP2-DBD fusion.
Domain A of EBNA1 (amino acids 33 to 89) was PCR amplified with oligonucleotides AGO41 and AGO42. These oligonucleotides contain PflMI and HindIII recognition sites (underlined) at their 5′ ends: AGO41, 5′-GGATCCCCAAGTCCTGGAACCATGTCTGACGAGGGGCCAGG; and AGO42, 5′-GAATTCAAGCTTGGCCTGACTGGCCACCGTGGGTCCCTTTGCAG.
This PCR product was introduced into AGP108 digested with SfiI and HindIII to create 1A-DBD (AGP117). To generate 2A-DBD (AGP119) and 3A-DBD (AGP130), the A domain (aa 33 to 89) was PCR amplified with oligonucleotides AGO17 and AGO43. The product contains the following Bsu36I and PflMI restriction sites: AGO17, 5′-GCTTTGTCATAACAAGGTCC; and AGO43, 5′-GAATTCCCAAGTCCTGGAGTGGACCTCAAAGAAGAGG.
This PCR product was inserted into AGP117 cut with SfiI and Bsu36I to generate 2A-DBD (AGP119). To generate 3A-DBD (AGP130), the PCR product, described above, used to make AGP119 was inserted into AGP119 cut with SfiI and Bsu36I. It is important to note that, in addition to lacking domain B, 1A-DBD, 2A-DBD, and 3A-DBD also lack aa 386 to 450 of EBNA1. We and others have previously shown that this region (aa 386 to 450) of EBNA1 is not required for its function. The coding sequences for 1A-DBD, 2A-DBD, and 3A-DBD were moved into the retroviral expression vector AGP164, a derivative of pLXSN (AGP171, AGP172, and AGP173, respectively).
Harris Busch and Ben Valdez (Baylor College of Medicine, Houston, Tex.) kindly provided us with a cDNA clone of EBP2 in pcDNA3 (10). A derivative of this clone was constructed such that N-terminal methionine of EBP2 was replaced by an in-frame FLAG epitope (AGP104). Independently, wild-type EBP2 was amplified from pcDNA3-EBP2 with oligonucleotides AGO37 and AGO38 (show below): AGO37, ATTGGCCAGTCAGGCCCAACCATGGACACTCCCCCGCTCTCGGATTC; and AGO38, CCCGGGGGAAGCTTGGTGTGTTCTGTTCTTCATCTTCTC.
The PCR product was cut with SfiI and HindIII and inserted into AGP102 (48) to construct an EBP2-DBD fusion protein (AGP107). The coding sequence for this fusion protein was also moved into AGP164 to create a retroviral expression vector for EBP2-DBD (AGP178). Sequences for all of the plasmids used in the present study can be found online (http://ebv.mimnet.northwestern.edu/plasmids).
Nitrocellulose filter-binding assays to measure AT hook activity.
To produce baculo-EBNA1 (bEBNA1), a cDNA clone of the 1553 version of EBNA1 (2) was cloned into the baculovirus transfer vector pBACgus3 (Novagen). Recombinant baculovirus was generated by standard methodology and used to infect Sf9 cells. Cell supernatants were concentrated and loaded onto a Talon immobilized metal affinity chromatography column (Clontech). Eluted protein was then cleaved with enterokinase to remove the S-tag and His6 tag added by the vector. The baculovirus-expressed protein (bEBNA1) was ca. 90% pure by colloidal Coomassie staining. Domains A and B were purified as His6-tagged fusion proteins as described previously (39), after which the His6 tag was proteolytically removed. These proteins were determined to be at least 95% pure by colloidal Coomassie blue staining and used in filter-binding assays as described recently to examine the DNA-binding specificity of the Schizosaccharomyces pombe and the human origin recognition complex (ORC) (32, 55). To avoid the possibility that the binding observed was due to contaminating bacterial proteins, assays were also performed with chemically synthesized peptides corresponding to the presumptive AT hook regions of EBNA1. The domain B peptide is contained entirely within domain B, whereas the domain A peptide has 3 aa that are amino terminal to the start of domain A. Identical results were obtained with the peptides and bacterially expressed proteins. The sequences of the peptides, with the presumptive AT hook underlined, were follows: domain A hook region peptide, Ac-GPQRRGGDNHGRGRGRGRGRGGGRPG-NH2 (EBNA1 aa 30 to 55); and domain B hook region peptide, Ac-GGGRGRGGSGGRGRGGSGGRGRGGS-NH2 (EBNA1 aa 326 to 350).
Peptides were acetylated at the amino terminus and amidated at the carboxy terminus. Purified HMGA1a [HMG-I(Y)] was kindly provided by Raymond Reeves (Washington State University), and a peptide corresponding to the first two AT hooks of HMGA1a was a gift from Jonathan Leis (Northwestern University).
Filter-binding assays were conducted in a final volume of 15 μl, containing 25 mM HEPES-NaOH (pH 7.5), 50 mM NaCl, 5 mM magnesium acetate, 1 mM dithiothreitol, 0.1 mg of bovine serum albumin/ml, 125 ng of 32P-labeled DNA probe, 1 μg of poly(dA-dC)(dG-dT) as competitor. After incubation for 15 min at 25°C, the reaction volume was increased to 1 ml by the addition of wash buffer (25 mM HEPES-NaOH [pH 7.5], 50 mM NaCl, 5 mM magnesium acetate, 1 mM dithiothreitol) and then filtered through nitrocellulose filters (0.45 μm, type HA). Filters were then washed three times with 1 ml of wash buffer and dried, and the radioactivity absorbed on the filter was counted by liquid scintillation counting. The probe used for binding was poly(dA-dT), whereas poly(dG-dC), a HindIII digest of lambda, and purified oriP DNA were used as competitors. Probe and competitors were purchased from Sigma, except oriP DNA, which was excised from AGP74 as a BamHI-KpnI fragment. According to the manufacturer's specifications, the poly(dA-dT) ranged in length from 3,000 to 5,000 bp, and the poly(dG-dC) ranged in length from 800 to 2,500 bp. Binding reactions typically contained 15 ng (2.6 pmol) of domain A, 15 ng (3 pmol) of domain B, 2.5 pmol of the respective peptides, 15 ng (1.3 pmol) of HMGA1a, 2.5 pmol of the HMGA1a peptide, or 50 ng (1.3 pmol) of baculovirus expressed EBNA1 that lacks the glycine-alanine repeats.
Cell culture and generation of stable 293 cell lines.
The human EBNA1 expressing cell line 293/EBNA1 is a derivative of 293 cells (18) that stably express wild-type EBNA1. 293/1A-DBD cells are a derivative of 293 cells, which stably express through retroviral integration domain A fused to the DBD of EBNA1 (AGP171). 293/2A-DBD cells are a derivative of 293 cells, which stably express through retroviral integration two A domains fused to the DBD of EBNA1 (AGP172). 293/3A-DBD cells are a derivative of 293 cells, which stably express through retroviral integration three A domains fused to the DBD of EBNA1 (AGP173). 293/EBP2-DBD cells are a derivative of 293 cells, which stably express through retroviral integration human EBP2 fused to the DBD of EBNA1 (AGP178). All cells were propagated in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and antibiotics. G418 was supplemented for 293/EBNA1 cells at a final concentration of 200 μg/ml and supplemented at 300 μg/ml for the other 293 derivatives. All cells were grown in 37°C in a humidified 5% CO2 atmosphere.
A derivative of 293 cells that stably express the MLV Gag and Pol proteins (gp293) was used to package and propagate retroviral vectors. GP293 cells were grown in the conditions described above to a density of 50%, at which time the cells were cotransfected by the calcium phosphate method with 10 μg of AGP171, AGP172, or AGP173 and 1 μg of vesicular stomatitis virus protein G expression plasmid (AGP154) to pantropically pseudotype the packaged virus. One hundred nanograms of a simian virus 40 large T-antigen expression plasmid (AGP5) was included in the cotransfection, because large T antigen dramatically increases the titers of retroviral vectors expressed from AGP164 and its derivatives. After 16 h, the medium was replaced. After 72 h, the virus-containing medium was collected and centrifuged to pellet the suspended cells, and then 5 ml of virus-containing medium was used to infect 293 cells grown to 50% cell density in a 10-cm dish. To increase the efficiency of retroviral infection, Polybrene was added during the infection at a final concentration of 7.5 μg/ml. After 48 h, infected cells were split 1:30 and then placed under selection with 300 μg of G418/ml. Clones were obtained within 2 to 3 weeks of selection. Expression of the appropriate protein in each cell line was confirmed by immunoblot and indirect immunofluorescence (see Results).
Calcium phosphate transfection.
Cells were transfected by the calcium phosphate method as described previously (36). For colony formation assays and Southern blots, transfections were normalized by the inclusion of 1 μg of a cytomegalovirus-EGFP expression plasmid, 2145, in each transfection. Upon harvest, a fraction of the cells were profiled by using a Becton Dickinson FACScan or FACSCalibur. Transfection efficiency was measured as the fraction of GFP-expressing live cells quantified by using CellQuest software (Becton Dickinson, Franklin Lakes, N.J.).
Coimmunoprecipitation of EBNA1 mutants with human EBP2.
Coimmunoprecipitation was performed by the following protocol. 293 cells were cotransfected with expression plasmids for FLAG-EBP2, along with expression plasmids for WT-EBNA1, 1A-DBD, 2A-DBD, 3A-DBD, or a derivative of WT-EBNA1 that lacks aa 386 to 450, because this region of EBNA1 is also missing from 1A-DBD, 2A-DBD, and 3A-DBD. At 2 days posttransfection, cells were harvested, washed with phosphate-buffered saline (PBS), lysed with 2 ml of immunoprecipitation buffer (20 mM Tris [pH 7.5], 250 mM NaCl, 0.5% NP-40, 0.5 mM EDTA, 0.1 mM AEBSF, and 0.5% protease inhibitor cocktail [Sigma]), and incubated on ice for 10 min. Extracts were pelleted at 50,000 × g for 20 min at 4°C. Under these conditions, EBNA1 is largely extracted away from the pelleted chromatin. Supernatants were used for coimmunoprecipitation. Then, 10 μg of K67.3 anti-EBNA1 DBD was added per 1 ml of cell extract and incubated with end-over-end mixing for 2 h at 4°C. Next, 50 μl of prewashed protein A/G-Sepharose (Santa Cruz Biochemicals) was then added to the extract and mixed end over end for 1 h at 4°C. Immunoprecipitates were washed three times with lysis buffer, mixed with 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and boiled for 5 min. Samples were then subjected to immunoblotting as described below, with membrane blocking performed in Blotto buffer (50 mM Tris [pH 7.5], 100 mM NaCl, 0.1% Tween 20, and 10% dry milk). Samples were probed by using either a monoclonal anti-EBNA1 antibody (1H4) or a monoclonal anti-FLAG antibody (M2; Sigma) as the primary antibody.
Immunoblotting.
Immunoblotting was done according to standard protocols (36). Briefly, extracts from 1.5 × 106 cells were separated on an SDS-10% polyacrylamide gel and transferred to a polyvinylidene difluoride membrane. After a blocking step for 1 h with 10% nonfat milk and 0.05% Tween 20, the membrane was treated with a rabbit polyclonal anti-EBNA1 serum diluted 1:1,000 (34) in blocking solution for 1 h at 37°C. The secondary antibody was a horseradish peroxidase-conjugated goat anti-rabbit antibody diluted 1:10,000 in blocking solution for 1 h at 37°C. Membranes were briefly washed once with 0.5% Tween 20 in PBS and once briefly with deionized water. Detection was performed by conventional chemiluminescence methods.
Metaphase chromosome spread preparation.
Isolation of metaphase chromosomes was done as described previously (51). Spreads were prepared from 293 cells and 293 cells expressing wild-type EBNA1 or mutant derivatives that were blocked in early metaphase with Colcemid (0.1 μg/ml). Cells were dropped onto poly-l-lysine-coated glass coverslips (Fisher, Hanover Park, Ill.) and dehydrated with increased ethanol washes. After PBS rehydration, slides were visualized by indirect immunofluorescence.
Indirect immunofluorescence and deconvolution microscopy.
Indirect immunofluorescence and image capture was performed similarly to that done previously (48), except that DNA was counterstained with Hoechst 33342 instead of DAPI (4′,6′-diamidino-2-phenylindole). For detection of EBP2-DBD in intact cells stalled in metaphase, 293/EBP2-DBD cells were treated with Colcemid, harvested, and seeded onto coverslips before the indirect immunofluorescence procedure. All EBNA1 derivatives were recognized by the primary rabbit polyclonal K67.3 antibody, which recognizes the EBNA1 DBD. Human EBP2 was detected by a primary mouse monoclonal antibody (a kind gift of Ben Valdez and Harris Busch). FLAG-EBP2 was expressed 2 days posttransfection of 10 μg of AGP104 and detected by a primary anti-FLAG mouse monoclonal antibody (Chemicon) at 1:2,000. The secondary antibodies used are specified with each figure. For cell imaging, an Olympus IX inverted fluorescence microscope and a Photometrix cooled charge-coupled device camera (CH350/LCCD) utilizing DeltaVision software from Applied Precision, Inc. (Seattle, Wash.) was used for data collection. Forty 100-nm optical sections (Z-sections) were obtained through the depth of the cell, and DeltaVision software (softWoRx) was used to deconvolve these images into individual layers. DeltaVision softWoRx uses a constrained iterative deconvolution algorithm to remove out-of-focus blur in fluorescence optical sections and was set for a minimum of 15 iterative cycles. Fluorescence emission was linear over a 100-fold range of emission signals within which fluorescein isothiocyanate (FITC), rhodamine, and DAPI/Hoechst 65535 signals were detected. Images were acquired with 100 NA 1.35 objective lenses either with or without 1.5× optical enhancement.
Colony formation assays to assess plasmid maintenance and partitioning.
The ability of 293 derivatives stably expressing the domain A derivatives of EBNA1 to stably partition an oriP plasmid was performed as described previously (21, 48), with transfection of AGP74 replication reporter.
Southern hybridization analysis to assess plasmid replication.
The detection of replicated, oriP plasmids in the 293 derivative cell lines was performed as previously described unless otherwise noted (21, 23, 48). To assess the dominant-negative effect of EBP2-DBD on plasmid replication by EBNA1, 10 μg of an EBP2-DBD expression plasmid containing wild-type oriP (AGP113) was transfected into 293/EBNA1 cells.
RESULTS
The A and B domains of EBNA1 specifically and directly bind AT-rich DNA in vitro.
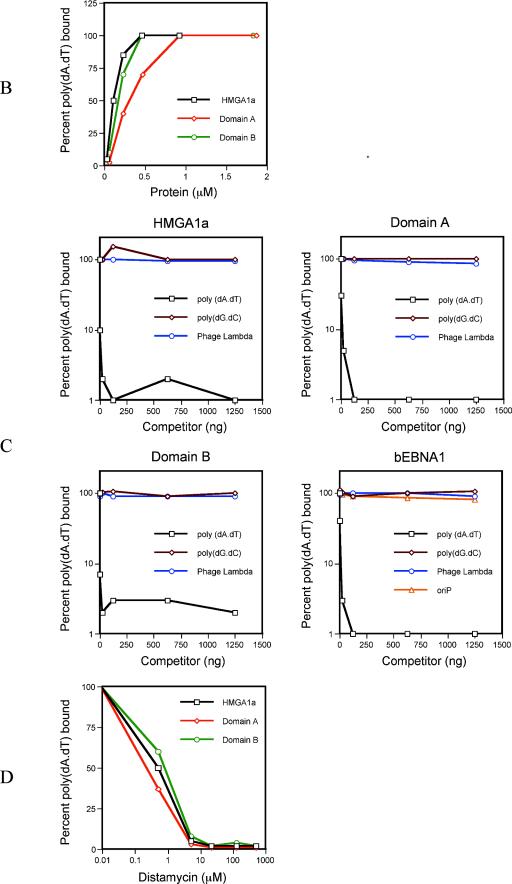
A schematic representation of EBNA1 is shown in Fig. 1A, along with sequences within domain A and domain B that are similar to AT hook motifs present within the cellular AT hook proteins HMGA1a and MeCP2/MBD2a, both of which are known to bind the AT-rich scaffold-associated regions (SARs) of mammalian chromosomes (4, 13, 52, 56). HMGA1a is known to associate with AT-rich DNA through its AT hooks (5). We therefore tested whether domains A and B of EBNA1 would function as AT hooks in vitro by using nitrocellulose filter-binding assays that have recently been used to characterize the DNA-binding specificity of the S. pombe or human ORC (12, 32, 55). Domains A and B were expressed and purified from bacteria or were chemically synthesized as peptides and then tested for their ability to bind AT-rich DNA by using labeled poly(dA-dT) as probe. As a control, binding assays were performed with purified HMGA1a or a peptide corresponding to the first two AT hooks of HMGA1a. The results of this experiment are shown in Fig. 1B. HMGA1a bound 50% of the labeled poly(dA-dT) probe (62.5 ng) at a protein concentration of 0.11 μM. Similar results were obtained by using a peptide corresponding to the first two AT hooks of HMGA1a (50% binding at a 0.5 μM concentration of peptide). Under these conditions, purified domain A and domain B also bound the poly(dA-dT) probe, such that a 0.23 μM concentration of domain A bound 50% of the probe and 0.13 μM concentration of domain B bound 50% of the probe. Moreover, 0.8 to 1.0 μM concentrations of peptides corresponding to the presumptive AT hooks of domains A and B bound 50% of the poly(dA-dT) probe.
FIG. 1.
(A) Schematic diagram of EBNA1. Domain A spans aa 33 to 89, and domain B spans aa 328 to 378. The presumptive AT hooks within domains A and B are compared to the known AT hooks of HMGA1a (GenBank accession number NM_145899) and MBD2a (GenBank accession number NP_003918). Amino acid mismatches are italicized and underlined. (B) Bacterially expressed domains A and B bind AT-rich DNA in vitro. One hundred twenty-five nanograms of labeled poly(dA-dT) probe was incubated in the presence of 1 μg of poly(dA-dC)(dG-dT), and increasing amounts of bacterially expressed HMGA1a, domain A or domain B, after which reactions were stopped and filtered through nitrocellulose. Nitrocellulose filters were washed and scintillation counted. The binding curve obtained with each protein is indicated by the legend within the graph. (C) The association between AT-rich DNA and domains A and B is specific. We incubated 0.1 μM HMGA1a, 0.2 μM domain A, 0.2 μM domain B, or 0.1 μM bEBNA1 with 125 ng of labeled poly(dA-dT) probe, 1 μg of poly(dA-dC)(dG-dT), and increasing amounts of cold poly(dA-dT), poly(dG-dC), or a HindIII digest of phage lambda as competitor DNA prior to assay by filter binding. For bEBNA1, a fragment of AGP74 that contains oriP was also used as a cold competitor. The binding curve obtained with each competitor is indicated by the legend within each graph. For all of the proteins tested, only cold poly(dA-dT) functioned as an effective competitor. (D) Distamycin A inhibits the association of HMGA1a, domain A, and domain B with poly(dA-dT). Increasing amounts of distamycin A were preincubated with the labeled poly(dA-dT) probe (125 ng) for 10 min. At this time, 0.2 μM HMGA1a, domain A, or domain B was added, along with 1 μg of poly(dA-dC)(dG-dT). Reactions were incubated for a further 20 min and then assayed by filter binding, followed by scintillation counting.
We next examined whether this association was specific for AT-rich DNA, by adding various cold competitor DNAs during the binding reaction (Fig. 1C). A 1,000-fold excess of cold poly(dG-dC) or a HindIII digest of phage lambda DNA did not compete for the association of HMGA1a (0.1 μM protein), domain A (0.2 μM protein), or domain B (0.2 μM protein) with the labeled poly(dA-dT) probe. In contrast, cold poly(dA-dT) effectively competed for the association of HMGA1a, domain A, and domain B with the labeled probe. Similar results were observed in experiments with peptides corresponding to the AT hook regions of HMGA1a and the presumptive AT hooks within domains A and B (0.2 μM peptide). When assays were performed with labeled poly(dG-dC) as probe, cold poly(dA-dT) remained an effective competitor for all three proteins tested (data not shown), indicating that domains A and B share an affinity for AT-rich DNA with HMGA1a. Experiments were also performed with baculovirus-expressed EBNA1, bEBNA1 (lacking the Gly-Ala repeats), to test whether domains A and B have AT hook activity in the context of the entire protein (Fig. 1C). bEBNA1 bound 50% of the probe at a 0.1 μM protein concentration (data not shown), and this binding was specifically competed for by poly(dA-dT) but not by lambda DNA or poly(dG-dC). Because this protein also has EBNA1's DBD, we included oriP DNA as a competitor and found that it failed to effectively compete the binding of EBNA1 for AT-rich DNA. This result confirmed that the amino terminus of EBNA1 has an affinity for AT-rich DNA.
Drugs such as distamycin A and netropsin are synthetic AT hook analogs that bind AT-rich DNA sequences in the minor groove and displace AT hook proteins from binding in the minor groove (8, 44). This property of distamycin A has been used previously to demonstrate that HMGA1a associates with AT-rich DNA in the minor groove (44). We tested whether preincubation of the poly(dA-dT) probe with distamycin A would block binding by domains A and B and included HMGA1a as a control. Interestingly, distamycin A at 5 to 10 μM, the concentration at which HMGA1a was blocked from DNA binding, inhibited both the A domain and the B domain from binding the poly(dA-dT) probe (Fig. 1D). Similar results were obtained with the HMGA1a peptide and with the peptides corresponding to the AT hook motifs within domains A and B and with bEBNA1. Netropsin also blocked the association of domain A, domain B, and HMGA1a with poly(dA-dT) (data not shown).
In summary, these data indicate that domains A and B can bind AT-rich DNA directly in a manner very similar to HMGA1a and that this association is very likely through AT hooks within domains A and B associating with the minor groove of AT-rich DNA. As such, domains A and B cannot be distinguished biochemically from the prototypic AT hook protein, HMGA1a.
Construction and expression of EBNA1 derivatives that lack domain B but contain one or more copies of domain A.
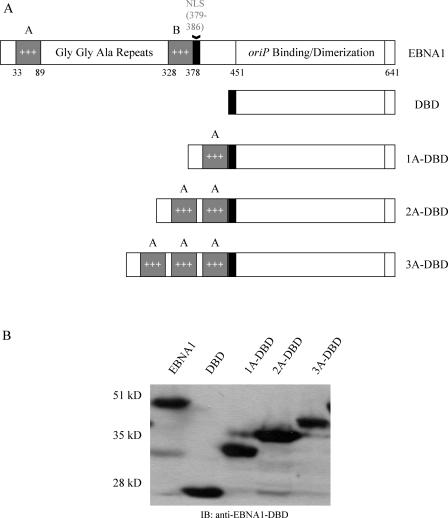
Because EBP2 associates exclusively with domain B (50), we constructed three derivatives of EBNA1 that lacked domain B and contained one (1A-DBD), two (2A-DBD), or three (3A-DBD) copies of domain A directly fused to the DBD of EBNA1. It should be noted that these derivatives all contain the nuclear localization signal (NLS) of EBNA1 (aa 379 to 386) but lack aa 386 to 450, a region that we have previously shown is not necessary for EBNA1's functions in the stable replication and partitioning of oriP plasmids (48). 1A-DBD, 2A-DBD, and 3A-DBD are schematically represented in Fig. 2A. The individual A domains in proteins that contained multiple copies of domain A were separated by a short peptide linker. Derivatives of 293 cells were created by retroviral transduction that stably expressed each derivative of EBNA1, and expression was confirmed through immunoblot analysis (Fig. 2B). All of the derivatives migrated at the expected sizes.
FIG. 2.
(A) Schematic representation of EBNA1 and derivatives lacking domain B but containing one or more copies of domain A. The three A domain derivatives contained one, two, or three copies of domain A (aa 33 to 89) fused to the EBNA1 NLS (aa 379 to 386), and the DBD (aa 451 to 641). In derivatives that contain two or more A domains, adjacent A domains are separated by a short peptide linker with the sequence QSWS. The version of EBNA1 used here contains only five copies of the Gly-Gly-Ala repeat between domains A and B but functions like wild-type EBNA1 (2, 48). Retroviral vectors expressing EBNA1, DBD, 1A-DBD, 2A-DBD, and 3A-DBD were used to transduce 293 cells and establish cell lines that stably express each of these proteins. (B) An immunoblot analysis of clones of 293 cells that express the proteins described above. Cell extracts were prepared from 293 cell clones and immunoblotted with a rabbit polyclonal antibody that recognizes the DBD of EBNA1. The cell line used is indicated above each lane. All of the derivatives migrated at their expected mobilities relative to wild-type EBNA1.
Domain B is required for EBNA1 to associate with EBP2 but not for EBNA1 to associate with metaphase chromosomes.
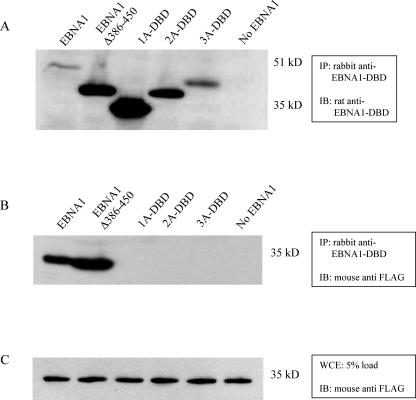
Next, we sought to confirm that 1A-DBD, 2A-DBD, and 3A-DBD no longer associated with EBP2, since they lacked domain B (50). We examined this by coimmunoprecipitation, such that FLAG-EBP2 was transiently overexpressed in 293 cells, along with wild-type EBNA1, 1A-DBD, 2A-DBD, 3A-DBD, or a derivative of wild-type EBNA1 that lacks aa 386 to 450 (EBNA1Δ386-450) (Fig. 3). Whole-cell extracts were immunoprecipitated with a rabbit polyclonal antibody to EBNA1's DBD. Immunoprecipitates were immunoblotted with a rat monoclonal antibody to EBNA1 (Fig. 3A) or a mouse monoclonal anti-FLAG antibody to detect EBP2 (Fig. 3B). As can be seen in Fig. 3B, EBP2 was readily coimmunoprecipitated by derivatives of EBNA1 that contained domain B but not by 1A-DBD, 2A-DBD, or 3A-DBD. A total of 5% of the extracts used for immunoprecipitation were directly examined for the expression of FLAG-EBP2. This analysis, shown in Fig. 3C, confirmed that FLAG-EBP2 was expressed approximately equivalently in all of the transfections. An immunoblot performed against EBNA1 confirmed that all of the derivatives of EBNA1 were immunoprecipitated with the polyclonal antibody. In our experience, deletion of aa 386 to 450 increases the expression of EBNA1 in transient transfection, which is why EBNA1Δ386-450, 1A-DBD, 2A-DBD, and 3A-DBD are expressed at higher levels than EBNA1 (Fig. 3A).
FIG. 3.
Domain B is required for EBNA1 to associate with the cellular protein EBP2. 293 cells were transiently transfected with a FLAG-EBP2 expression plasmid and expression plasmids for the indicated derivative of EBNA1 (indicated above each lane) or with just the FLAG-EBP2 expression plasmid (lane No EBNA1). Extracts were prepared as described in the text and immunoprecipitated with a rabbit polyclonal antibody to the DBD of EBNA1. Immunoprecipitates were analyzed by SDS-PAGE and then immunoblotted with a rat monoclonal antibody to EBNA1 (A) or a mouse monoclonal antibody to FLAG-EBP2 (B). Only EBNA1 and EBNA1Δ386-450, which both contain domain B, were observed to immunoprecipitate FLAG-EBP2. (C) An immunoblot against FLAG-EBP2 with a 5% load of whole-cell extract from each of 293 cell cotransfections, indicating that the expression of FLAG-EBP2 was approximately equal in all transfections. The antibodies used for immunoprecipitation (IP) or immunoblotting (IB) are indicated adjacent to each blot. The positions of molecular mass markers on the SDS-PAGE gels are also indicated in kilodaltons.
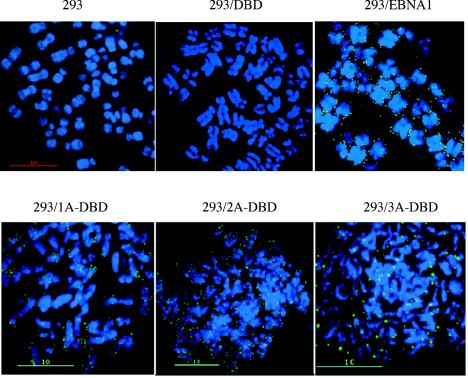
These results confirm those reported previously (50) that domain B is absolutely required for an association of EBNA1 with EBP2. We next examined whether this association was essential for EBNA1 to bind metaphase chromosomes. 293, 293/DBD, 293/EBNA1, 293/1A-DBD, 293/2A-DBD, and 293/3A-DBD cells were stalled in early metaphase through Colcemid treatment (0.1 μg/ml) for 16 h and used to isolate metaphase spreads that were subjected to indirect immunofluorescence to detect whether 1A-DBD, 2A-DBD, and 3A-DBD associated with metaphase chromosomes. This analysis is shown in Fig. 4. Chromosomes isolated from 293/EBNA1 cells showed discrete, punctate staining on metaphase chromosomes similar to what we have described previously for EBNA1 and HMGA1a-DBD (48). No chromosome-associated staining was observed in chromosomes isolated from 293/DBD cells or from the negative-control 293 cells. All three derivatives that lacked domain B showed a staining pattern on metaphase chromosomes very similar to wild-type EBNA1, although the expression levels of these proteins differed.
FIG. 4.
1A-DBD, 2A-DBD, and 3A-DBD bind metaphase chromosomes. Metaphase chromosomes were isolated from 293, 293/DBD, 293/EBNA1, 293/1A-DBD, 293/2A-DBD, and 293/3A-DBD cells stalled in mitosis by Colcemid treatment. Indirect immunofluorescence was performed by using the K67.3 rabbit polyclonal antibody against the DBD of EBNA1. Images of individual layers (40 z-sections of 100 nm each) were captured using a ×100 objective, with ×1.5 optical enhancement, and deconvolved by using softWoRx. The cell line used for metaphase chromosome isolation is indicated above each panel. All three proteins were observed to localize to metaphase chromosomes in discrete punctate spots that resemble those observed previously with wild-type EBNA1 or HMGA1a-DBD (48).
From this analysis, we conclude that although domain B is absolutely required for an association of EBNA1 with EBP2, it is not essential for EBNA1 to associate with metaphase chromosomes. As a corollary, binding EBP2 cannot be the only mechanism by which EBNA1 associates with metaphase chromosomes. However, is this binding, in the absence of EBP2, sufficient to support the stable replication and partitioning of oriP plasmids?
1A-DBD, 2A-DBD, and 3A-DBD support the replication and partitioning of oriP plasmids.
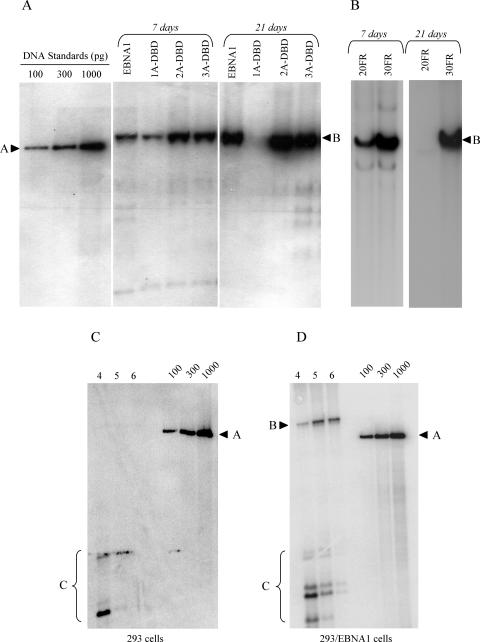
Our previous results led us to hypothesize that metaphase chromosome tethering is a requirement for the replication of oriP plasmids by EBNA1 in mammalian cells (48). Because 1A-DBD, 2A-DBD, and 3A-DBD bound to metaphase chromosomes without binding EBP2, we tested whether they were still competent to replicate an oriP plasmid. For this, the oriP replication reporter plasmid, AGP74, was transfected into 293/EBNA1, 293/1A-DBD, 293/2A-DBD, and 293/3A-DBD cells. Hirt DNAs were isolated 1 and 3 weeks posttransfection, digested with DpnI, linearized with XbaI, and quantified by Southern blotting (Fig. 5 and Table 1). By 7 days posttransfection, replicated oriP reporter was readily detected in all cell types, which reinforces our previous conclusion that metaphase chromosome association is required for the replication of oriP plasmids (48). However, at 3 weeks posttransfection, the levels of replicated oriP reporter plasmid detected from 2A-DBD and 3A-DBD were similar to that observed with wild-type EBNA1 (Fig. 5 and Table 1). In contrast, little to no replicated oriP reporter plasmid was observed with 1A-DBD at 3 weeks posttransfection, a finding consistent with previous results obtained in C33A cells (50). Our results with 2A-DBD and 3A-DBD suggested that the ability of EBNA1 to stably replicate an oriP plasmid was dependent upon the affinity of EBNA1 for metaphase chromosomes. Wild-type EBNA1 associates via two regions—A and B—as does 2A-DBD. We speculated that we might be able to rescue the stable replication defect in 1A-DBD by increasing the number of EBNA1 binding sites on the replication reporter plasmid. Simplistically, we felt this would increase the amount of 1A-DBD on the plasmid and therefore increase the probability that the replication reporter would associate with metaphase chromosomes. Thus, we constructed a derivative of AGP74 which has an FR containing 30 EBNA1 binding sites (AGP264) and tested the ability of 1A-DBD to support the stable replication of this novel replication reporter plasmid. As shown in Fig. 5B and Table 1, when there are 30 EBNA1 binding sites in FR, 1A-DBD supports the stable replication of an oriP plasmid, supporting our simple model.
FIG. 5.
EBNA1 derivatives 1A-DBD, 2A-DBD, and 3A-DBD support the episomal replication of oriP plasmids. (A) The oriP replication reporter plasmid, AGP74 (20FR), was transfected into 293/EBNA1, 293/1A-DBD, 293/2A-DBD, or 293/3A-DBD cells, along with a GFP expression plasmid. Two days after transfection, live transfected cells were recovered and subjected to puromycin selection for an additional 5 days (total = 7 days) or 19 days (total = 21 days). At this time, episomal DNAs were recovered, digested with DpnI, linearized, and quantified by Southern blotting. The cell line transfected is indicated above each lane, as is the time point at which episomal plasmids were recovered. The migration of standards is indicated by an arrowhead and “A,” and the linearized plasmid DNA is indicated by an arrowhead and “B.” Copy numbers of replicated plasmids per cell at each time point from three replicates of this experiment are indicated in Table 1. 1A-DBD only supported the replication of AGP74 (20FR) up to 7 days posttransfection but not at the later time point. 2A-DBD and 3A-DBD supported replication of this plasmid at both time points. (B) The replication defect in 1A-DBD is compensated by a plasmid, AGP264, that contains 30 EBNA1-binding sites in FR. AGP74 (20FR) or AGP264 (30FR) were transfected into 293/1A-DBD cells, which were processed as in panel A. Replicated episomal DNAs were detected for both plasmids at 7 days posttransfection, but only for AGP264 (30FR) at 21 days posttransfection, indicating that increasing the number of EBNA1 binding sites in FR compensated for the defect in 1A-DBD. Copy numbers for replicated plasmids per cell at each time point from two replicates of this experiment are indicated in Table 1. As a control for DpnI digestion, AGP74 was transfected into 293 (C) or 293/EBNA1 cells (D). Hirt DNAs were recovered at 4, 5, or 6 days posttransfection, digested with DpnI, linearized, and quantified by Southern blot. The DpnI fragments are indicated by “C,” whereas DpnI-resistant DNAs and markers are indicated as previously.
TABLE 1.
Average copy number of DpnI-resistant replication reporter plasmids 7 and 21 days posttransfection in 293 cells expressing 1A-DBD, 2A-DBD, 3A-DBD, or EBNA1
| Cell line transfected | Replication reporter plasmida (mean no. of plasmids ± SD)
|
|||
|---|---|---|---|---|
| AGP74 (20 FR) at:
|
AGP264 (30 FR)b at:
|
|||
| 7 days | 21 days | 7 days | 21 days | |
| 293/1A-DBD | 11 ± 3 | 1.1 ± 0.7 | 34 ± 11 | 40 ± 8 |
| 293/2A-DBD | 35 ± 16 | 43 ± 17 | ND | ND |
| 293/3A-DBD | 42 ± 8 | 37 ± 12 | ND | ND |
| 293/EBNA1 | 18 ± 7 | 27 ± 15 | ND | ND |
Numbers of episomal DpnI-resistant plasmids detected at the indicated time point after transfection in the presence of puromycin selection. The numbers represent the means and standard deviations of three experiments, except as noted.
Numbers represent the means and standard deviations of two experiments. ND, not done.
Domains A and B both have AT hook activity in vitro like HMGA1a, but only domain B associates with EBP2. Our results examining the stable replication of oriP plasmids indicate that this activity of EBNA1 correlates well with its AT hook activity. However, are the plasmids that are stably replicated by 1A-DBD, 2A-DBD, and 3A-DBD partitioned or does partitioning require an association with EBP2?
We therefore tested whether oriP plasmids were partitioned efficiently by the 1A-DBD, 2A-DBD, and 3A-DBD proteins in comparison to wild-type EBNA1 by using colony formation assays. Such assays test the ability of replicated plasmids to be distributed as a single cell proceeds to form a colony of cells. If partitioning is dependent upon an association with EBP2, all three proteins should not perform well in this assay. On the other hand, our previous results with HMGA1a-DBD (which has three AT hooks) suggested that partitioning correlated with AT hook activity. If this were also true for EBNA1, then we would expect that 1A-DBD would partition plasmid inefficiently (with only one AT hook region), whereas 2A-DBD and 3A-DBD should partition plasmids with an efficiency that is close to that of wild-type EBNA1. Colony formation assays were performed by using AGP74 as described in Materials and Methods, and the number of transfected, surviving, cells that proceeded to form colonies that were ≥2 mm in size were scored 21 days posttransfection. These data are presented in Table 2. 1A-DBD is very ineffective at partitioning but substantially above background for the assay. In contrast, 2A-DBD and 3A-DBD are approximately as efficient as wild-type EBNA1.
TABLE 2.
Colony formation efficiency of AGP74 (pPUR-oriP) in 293 cells that stably express the 1A-DBD, 2A-DBD, and 3A-DBD derivatives of EBNA1
| Cell line | No. of puromycin-resistant coloniesa |
|---|---|
| 293 | 1 ± 1 |
| 293/EBNA1 | 1,222.4 ± 75.5 |
| 293/1A-DBD | 118.6 ± 28.9 |
| 293/2A-DBD | 750.5 ± 68.2 |
| 293/3A-DBD | 951 ± 117.4 |
Data represent the number of puromycin-resistant colonies formed per 2 × 105 live, GFP- positive cells plated 48 h posttransfection. Selection was initiated 4 days posttransfection and continued for 17 days. Each cell line was transfected at least twice. At least two plates were counted from each transfection. Only colonies that were ≥2 mm were counted.
Contributions of EBP2 to the replication of oriP plasmids in human cells.
We and others have shown that the entire amino terminus of EBNA1 can be functionally replaced by cellular proteins that associate with metaphase chromosomes (24, 48). This strategy has also been employed successfully to study the association of LANA protein of KSHV with chromosomes (49). Because EBP2 is proposed to mediate EBNA1's association with metaphase chromosomes, we tested whether the amino terminus of EBNA1 could be functionally replaced by EBP2. This new fusion, EBP2-DBD, is schematically depicted in Fig. 6A. It contains full-length wild-type EBP2 (without any tags), fused to aa 379 to 386 of EBNA1 (the NLS) and then aa 451 to 640 (the DBD). We constructed a derivative of 293 cells that expressed this fusion, 293/EBP2-DBD, via retroviral transduction and confirmed expression by immunoblot (data not shown).
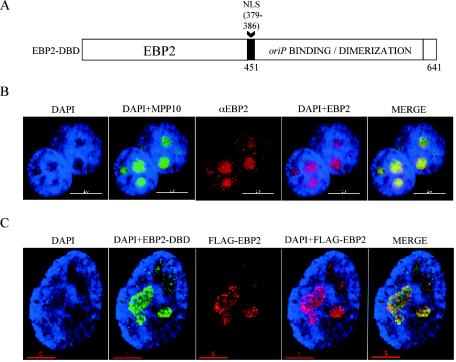
FIG. 6.
(A) Schematic representation of the EBP2-DBD fusion protein in which the EBNA1 amino terminus (aa 1 to 450) is replaced by human EBP2. EBNA1's NLS (aa 379 to 386) was added to the fusion junction. A retrovirus vector expressing EBP2-DBD was used to transduce 293 cells and create a 293 derivative cell line that stably expresses this protein (293/EBP2-DBD). (B) Endogenous EBP2 localizes to nucleoli in interphase human cells. The stain or antibody used is indicated above each panel. HeLa cells were transfected with an expression plasmid for the nucleolar protein MPP10 fused to GFP to locate nucleoli (57). MPP10-GFP largely localizes to DAPI-negative regions (compare panel DAPI to panel DAPI+MPP10). EBP2 largely colocalizes with MPP10 (compare panel DAPI+MPP10 to panels DAPI+EBP2 and MERGE). This colocalization indicates that the majority of EBP2 is localized to nucleoli as observed previously (10). (C) EBP2-DBD colocalizes with EBP2 in 293 cells. 293/EBP2-DBD cells were transfected with an expression plasmid for FLAG-EBP2, and interphase cells were visualized by using antibodies against the DBD (FITC) or FLAG-EBP2 (rhodamine). EBP2-DBD and FLAG-EBP are largely localized to regions of the nucleus that stain poorly with DAPI (compare panel DAPI to panels DAPI+EBP-DBD and DAPI+FLAG-EBP2). Also, EBP2-DBD localizes similarly to EBP2 within cells (compare panel DAPI+EBP2-DBD to panels DAPI+FLAG-EBP2 and MERGE).
To characterize the EBP2-DBD protein, we compared its intracellular localization relative to native EBP2. p40/EBP2 was first reported by Busch and coworkers to be a strictly nucleolar protein (10) in HeLa cells, although other analyses in HeLa and other cells suggest that it is both nucleolar and associated with chromatin (26, 58). We therefore performed experiments in both HeLa and 293 cells, the results of which are shown in Fig. 6B. HeLa cells were transfected with GFP-tagged MPP10 (a nucleolar marker) (57). Interphase cells were then processed for indirect immunofluorescence by using deconvolution (or confocal) microscopy with a monoclonal antibody to native EBP2. As shown in Fig. 6B, GFP-MPP10 stained several nucleoli per cell, specifically coinciding with where DAPI staining was weakest inside nuclei (see Fig. 6B, the first two panels). Consistent with the previous characterization of EBP2 (10), endogenous EBP2 was found to be strictly nuclear and heavily enriched within nucleoli (see Fig. 6B, the third and fourth panels). The final merged image in Fig. 6B shows a close colocalization of native EBP2 and GFP-MPP10 within the nucleoli of HeLa cells. Identical results were obtained for EBP2 localization in 293 cells (data not shown). We could not examine the localization of EBP2-DBD by using the monoclonal antibody to EBP2, since it would detect both native and fused proteins. We therefore transfected 293/EBP2-DBD cells with FLAG-EBP2. The localization of FLAG-EBP2 was examined by using a monoclonal antibody to the FLAG epitope, and localization of EBP2-DBD was examined by using polyclonal antibodies to the DBD (Fig. 6C). As shown in the second panel, EBP2-DBD, like endogenous EBP2, was specifically enriched within DAPI-negative nucleoli, as was FLAG-EBP2. There was almost complete overlap between the localization of EBP2-DBD with FLAG-EBP2 (Fig. 6C, last panel). Our results indicate that in interphase HeLa and 293 cells, EBP2, FLAG-EBP2, and EBP2-DBD are all localized to nucleoli, with very little to no chromatin staining, a finding consistent with observations published previously for EBP2/p40, in which no chromatin staining was observed (10).
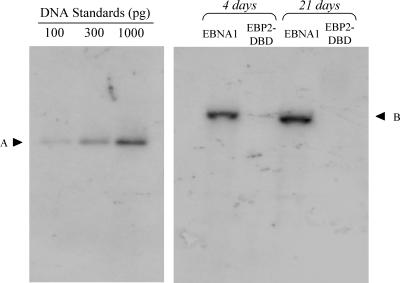
We tested whether EBP2-DBD could support the replication of oriP plasmids by transfecting 293/EBP2-DBD cells with the oriP replication reporter plasmid, 1033 (28). DNA's were extracted, digested with DpnI to remove unreplicated DNA, and examined by Southern blot for episomal plasmids 4 and 21 days posttransfection in the absence of drug selection (Fig. 7 and Table 3). Plasmid 1033 replicated robustly in 293/EBNA1 cells. In contrast, replicated plasmid 1033 DNA was present at substantially reduced levels by 4 days posttransfection and could not be detected by 21 days posttransfection in 293/EBP2-DBD cells. We have previously shown that another fusion protein that also does not support transient replication of oriP plasmids, HMG1-DBD, functions as a dominant-negative inhibitor of wild-type EBNA1. We tested whether EBP2-DBD would function as such an inhibitor by examining the replication of an EBP2-DBD expression plasmid that also contains oriP (AGP113) in 293/EBNA1 cells. No replicated DNA was detected at 4 days posttransfection or beyond, confirming that EBP2-DBD can bind oriP but does not functionally support the replication or partitioning of oriP plasmids (data not shown).
FIG. 7.
EBP2-DBD does not support the replication of oriP plasmids in human cells. (A) An oriP replication reporter plasmid (i.e., plasmid 1033) was transfected into 293/EBNA1 and 293/EBP2-DBD cells, after which cells were propagated in the absence of selection and harvested 4 or 21 days posttransfection. The transfected cell line is indicated above each lane, and the time point is given above each set of lanes. DNAs were extracted and digested with DpnI, linearized with XbaI, and quantified by Southern analysis with a probe generated from plasmid 1033 digested with HincII. The amounts of standards loaded are indicated above each lane, and their electrophoretic mobilities are indicated by “A.” “B” indicates the electrophoretic mobility of the linearized replicated plasmid 1033. Quantified results from two repetitions of this experiment are given in Table 3.
TABLE 3.
Average copy number of DpnI-resistant oriP replication reporter plasmid (i.e., plasmid 1033) at 4 and 21 days posttransfection in 293 cells expressing EBP2-DBD or EBNA1
| Days posttransfection | Cell line transfecteda (mean copy no. ± SD)
|
|
|---|---|---|
| 293/EBNA1 | 293/EBP2-DBD | |
| 4 | 32 ± 13 | 2 ± 2 |
| 21 | 23 ± 5 | <0.5 |
Data represent the means and standard deviations of two experiments. Copy numbers are corrected for transfection efficiency. The average transfection efficiency was 81%, as determined by flow cytometry for a cotransfected GFP expression plasmid.
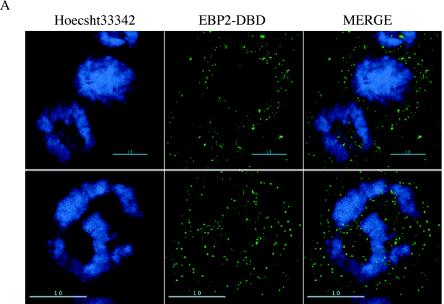
Our previous results indicated that even transient replication of oriP plasmids correlated with an association between EBNA1 and metaphase chromosomes (48). We therefore tested whether EBP2-DBD, as well as native EBP2, associated with metaphase chromosomes. This analysis was performed by using deconvolution microscopy with a ×100 objective lens, with or without ×1.5 optical enhancement, and 40 z-sections of 100 nm (Fig. 8). 293/EBP2-DBD cells were blocked with Colcemid (0.1 μg/ml) and prepared for whole-cell immunofluorescence by using an antibody against the EBNA1 DBD. Images were deconvolved and are shown with the condensed chromatin in focus (Fig. 8A). As can be seen in the images of multiple cells, there is no overlap between EBP2-DBD and condensed chromatin in the same geometric plane. This is indicated by the almost perfect exclusion of the FITC (EBP2-DBD) signal from anywhere the DAPI signal is present. This indicates that EBP2-DBD does not bind condensed chromatin within intact cells.
FIG. 8.
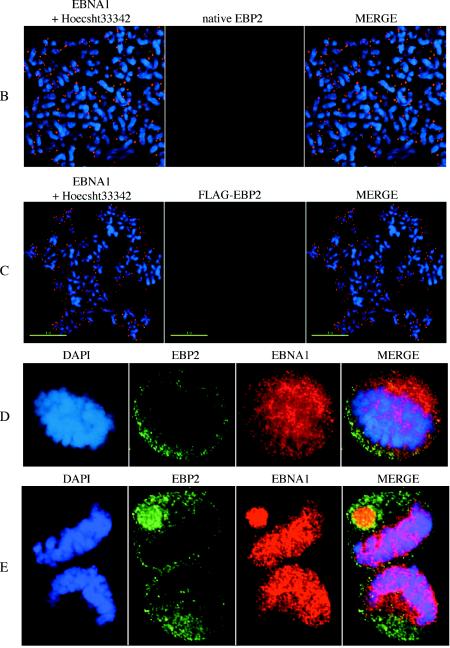
EBP2-DBD and EBP2 do not associate with condensed mitotic chromatin within cells or with isolated metaphase chromosomes. (A) 293/EBP2-DBD cells were treated with Colcemid and subjected to whole-cell immunofluorescence with the K67.3 antibody to the DBD of EBNA1. In these images, the cells are mitotic, the nuclear envelope has broken down, but the cell membrane is intact. EBP2-DBD is visualized with a FITC-conjugated secondary antibody, whereas condensed mitotic chromatin is counterstained with DAPI. The upper row shows deconvolved images from three cells visualized with a ×100 objective lens, indicating that EBP2-DBD stains wherever mitotic chromatin is absent. The lower row shows a single cell also taken with a ×100 objective lens, but with 1.5× optical enhancement from an independent experiment, also indicating an exclusion of EBP2-DBD from condensed mitotic chromatin within intact cells, although the protein can be detected readily within such cells. (B) Endogenous EBP2 is not bound to metaphase chromosomes isolated from 293/EBNA1 cells. Metaphase spreads were recovered from 293/EBNA1 cells and subjected to coimmunofluorescence with K67.3 (rhodamine-conjugated secondary antibody) and a monoclonal antibody against endogenous human EBP2 (FITC-conjugated secondary antibody). As previously observed (48), EBNA1 (red) coats metaphase chromosomes in a distinct, punctate manner; EBP2 (green) is not found on the isolated metaphase chromosomes. (C) Overexpressed FLAG-EBP2 is not bound to isolated metaphase chromosomes. Metaphase spreads were recovered from 293/EBNA1 cells transiently transfected with a FLAG-EBP2 expression plasmid and subjected to coimmunofluorescence as in panel B. EBNA1 was detected and visualized as described above, whereas FLAG-EBP2 was detected by using a monoclonal anti-FLAG antibody (Chemicon) and visualized by using an FITC-conjugated secondary antibody. Even when overexpressed, EBP2 was not detected on metaphase chromosomes. (D) Localization of EBP2 and EBNA1 within a cell in early metaphase isolated by mitotic shake-off. 293/EBNA1 cells in mitosis were isolated by shake-off, fixed on slides, and stained with a monoclonal antibody against endogenous EBP2, followed by an FITC-conjugated secondary antibody, and the K67.3 rabbit polyclonal antibody against EBNA1 (rhodamine-conjugated secondary). (E) Localization of EBP2 and EBNA1 within a cell in late anaphase. 293/EBNA1 cells were isolated and processed as in panel D.
We next examined whether endogenous EBP2 was localized to metaphase chromosomes in cells that expressed EBNA1 (Fig. 8B). For this, metaphase spreads were prepared from 293/EBNA1 cells stalled in early metaphase with Colcemid, followed by indirect immunofluorescence against both EBNA1 (rhodamine) and EBP2 (FITC). Because the FITC channel of our camera is more sensitive than the rhodamine channel, we decided to detect EBNA1 by using a rhodamine-conjugated secondary antibody (red) and EBP2 with an FITC-conjugated secondary antibody (green) for this analysis. As observed previously, EBNA1 stained metaphase chromosomes as discrete punctate dots, which overlapped greatly with the Hoechst-stained metaphase chromosomes (48). In stark contrast, despite our ability to readily stain EBP2 in the nucleoli of interphase cells (Fig. 6), we failed to observe any signal on isolated metaphase chromosomes (Fig. 8B). This result is inconsistent with those obtained with a GFP-EBP2 fusion (26) but is consistent with those reported previously for native EBP2 in HeLa cells (10). Possible reasons for the inconsistency are discussed below. We also observed that EBP2 failed to stain metaphase chromosomes prepared from 293 cells (data not shown).
We were concerned that our localization studies were performed with the same monoclonal antibody as those performed previously (10), although this antibody does work to detect EBP2 in nucleoli (Fig. 6) (10). To eliminate the possibility that this monoclonal antibody does not detect EBP2 bound to metaphase chromosomes, we transfected FLAG-EBP2 into 293/EBNA1 cells, isolated metaphase chromosomes, and proceeded to detect EBNA1 and FLAG-EBP2 by indirect immunofluorescence (Fig. 8C), with rabbit polyclonal antibodies to EBNA1 and a monoclonal anti-FLAG antibody. We failed to observe any association between EBP2 and metaphase chromosomes, although the association of EBNA1 with such chromosomes was clearly evident (Fig. 8C).
EBP2 is crucial for the formation of ribosomes in yeast, and it is likely to have that same function in mammalian cells. As such it has to be partitioned into both daughter cells, or one daughter will not survive. We were concerned that the methods we used to stall cells in mitosis, drop swollen cells on slides, or fix chromosomes masked prevented the detection of EBP2. To test this possibility, we examined the localization of EBNA1 and EBP2 in 293/EBNA1 cells obtained by mitotic shake-off without any drug treatment. For this purpose, we grew >108 cells and recovered ca. 107 cells in various stages of mitosis. Such intact cells were fixed onto coverslips and then processed for indirect immunofluorescence with antibodies to EBNA1 and EBP2. An example of a cell in early metaphase is shown in Fig. 8D. We observed that although EBNA1 was distributed throughout the cell and is not especially concentrated on the condensed DNA, most of the EBP2 was on the periphery directed away from the condensed DNA. An example of a cell in late anaphase is shown in Fig. 8E. Here it can be seen that most, but not all, of the EBNA1 staining coincides with the condensed DNA. The EBP2 signal is about equivalent in what will become the two daughter cells but is not associated with DNA. In our examination of more than 200 such mitotic cells, we did not find any cells in which EBP2 was associated with mitotic DNA. However, it is very likely that EBP2 is partitioned equally. Examining the FITC signal with softWoRx indicates that it is often approximately equal in the two daughters just prior to cytokinesis. Our studies on such intact cells also indicated that EBNA1 and EBP2 do often colocalize, but when they colocalize it is not on condensed chromosomes.
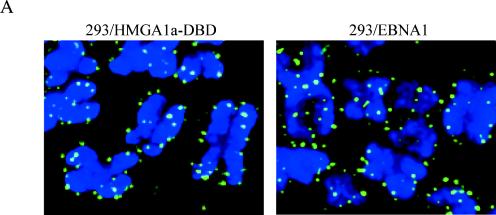
In summary, we observed that EBP2 and EBP2-DBD are localized to nucleoli. We observe that neither protein associates with metaphase chromosomes, either in intact cells (Fig. 8A, D, and E) or in metaphase spreads (Fig. 8B and C). Consistent with this, EBP2-DBD fails to support the robust replication of an oriP plasmid, in contrast to HMGA1a-DBD, a protein that does bind metaphase chromosomes (48) and whose staining on such chromosomes closely resembles that of EBNA1 (Fig. 9A).
FIG. 9.
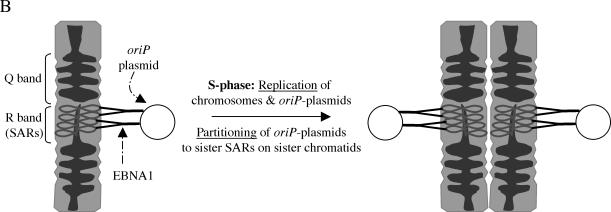
Model for the partitioning of oriP plasmids on a per-replicon basis. (A) High-resolution indirect immunofluorescence comparing the localization of wild-type EBNA1 and HMGA1a-DBD on metaphase chromosomes by using an antibody to the DBD of EBNA1. The localizations of both proteins were similar; many pairs of sister chromatids contained an equal number of dots for both proteins. In several instances, the dots were symmetrically positioned on both sister chromatids. (B) Model of a portion of a chromosome based on the models constructed by Laemmli and coworkers (20, 45, 46). For convenience we have depicted a metaphase chromosome with Q and R bands containing AT-rich SARs. Laemmli and coworkers have demonstrated that sequences present as SARs on interphase chromosomes are present in R bands in metaphase chromosomes (45, 46). We propose that EBNA1 or HMGA1a-DBD tethers oriP plasmids to SARs that are present relatively infrequently compared to other sequences on chromosomes. Upon S phase, when there is replication of chromosomes, we hypothesize that the replicated oriP plasmids are partitioned between the sister SARs on sister chromatids, by a distribution of EBNA1 or HMGA1a-DBD to each sister SAR. Key to this model is that the actual partitioning event is concomitant with replication and occurs during S phase. The plasmids remain tethered to sister chromatids and piggyback on the sisters during mitosis (9).
DISCUSSION
The latent genome of EBV is stably maintained as an episome within cells, a process that requires the viral protein EBNA1. EBNA1 maintains EBV genomes or oriP plasmids in proliferating cells by tethering them to host chromosomes. The genome or oriP maintenance activity of EBNA1 requires the amino-terminal domain of EBNA1 that is known to associate with chromatin. This association requires two positively charged regions within the amino terminus that termed “A” (aa 33 to 89) and “B” (aa 328 to 378). Region B associates with a cellular nucleolar protein (p40), originally cloned by Busch and coworkers (10), and reisolated as an EBNA1-interacting protein and termed EBP2 (50). In genetic experiments, it has been shown that expression of human EBP2 permits budding yeast that express EBNA1 to retain yeast ARS/FR plasmids through multiple cell cycles (27), although partitioning was not directly addressed. Using flat-field indirect immunofluorescence microscopy, an association between a GFP-EBP2 fusion with human metaphase chromosomes has been observed at a ×630 resolution (26). On this basis it has been proposed that EBP2 mediates EBNA1's association with metaphase chromosomes. These studies intrigued us for several reasons. First, EBP2 associated exclusively with domain B of EBNA1, but not with domain A, whereas studies by others suggested that EBNA1 derivatives lacking either domain B or domain A were equivalent in function (34). Second, the localization of GFP-EBP2 (26) differs from that originally observed (10) and also differs from that reported for the yeast homolog of EBP2 (54). Third, our own studies examining the localization of EBNA1 on metaphase chromosomes indicated that it is bound in a punctate manner at discrete locations (Fig. 4, 8, and 9) (48) and does not uniformly coat chromosomes as reported for GFP-EBP2 (26). Finally, studies by Marechal et al. with fusions of domain A and domain B with GFP indicated that each could associate with metaphase chromosomes in a punctate manner (37), a result we interpreted to indicate minimally that EBNA1 has EBP2-dependent and -independent modes of chromosome association.
Recent work by us and others has elucidated the requirements of metaphase chromosome association for the replication and partitioning functions of EBNA1 (24, 48). In both studies, the entire amino terminus of EBNA1 (both domains A and B) was replaced with cellular proteins that associate with metaphase chromosomes, histone H1 and HMGA1a (24, 48). Both of these fusion proteins support the long-term replication of oriP plasmids. For one of these proteins, HMGA1a-DBD, we have determined that its activity in the replication of oriP plasmids is indistinguishable from wild-type EBNA1 (48). In addition, the rate of loss for oriP plasmids in human cell lines that express the HMGA1a-DBD protein is mathematically indistinguishable from the rate of loss of the same plasmids in matched cell lines that express wild-type EBNA1. This observation suggested to us that the amino terminus of EBNA1 and HMGA1a acted via similar molecular mechanisms. A corollary to this argument implies that this molecular mechanism is also shared by histone H1. One property that histone H1 and HMGA1a share with EBNA1 is that they associate with metaphase chromosomes. It appeared unlikely to us that this property alone explained the ability of EBNA1 to support the stable replication and partitioning of oriP plasmids because a fusion between histone H3 and the DBD of EBNA1 does associate with metaphase chromosomes but does not support the transient or stable replication of oriP plasmids (A. Balasubramaniam and A. Aiyar, unpublished results). Detailed studies on the association of histone H1 and HMGA1a with metaphase chromosomes indicate that these two proteins share another property. Both proteins bind AT-rich sequences with high affinity, although the protein motifs are different (16, 17, 25, 38, 46): histone H1 through a motif that is now called the histone H1-fold and HMGA1a through a motif called the AT hook (43). Using giant chromosomes from Drosophila, or the Indian muntjac, Laemmli and coworkers have demonstrated that metaphase chromosomes are not uniformly condensed (41, 46). Rather, these chromosomes consist of highly condensed regions (Q bands) connected by AT-rich R bands that are not condensed (20, 45, 46). These AT-rich R bands have been termed by Laemmli as SARs. How does this relate to the distribution of histone H1 or HMGA1a on metaphase chromosomes? The groups of Reeves and Laemmli have together shown that HMGA1a and histone H1 are largely or entirely localized to these SARs on metaphase chromosomes (25, 46, 52, 63), such that they stain in a highly punctate manner (16, 25, 46) that is indistinguishable from the punctate staining of EBNA1 on metaphase chromosomes. That histone H1 and HMGA1a have a high affinity for the same regions on chromosomes is also indicated by studies demonstrating that HMGA1a can compete with histone H1 for binding to SARs in vitro (63). Thus far, the only cellular proteins that have functionally replaced the amino terminus of EBNA1 (24, 48) preferentially associate with AT-rich DNA and are localized to SARs on metaphase chromosomes.
We therefore sought to determine whether the amino terminus of EBNA1 preferentially associated with AT-rich DNA and if this association correlated with the stable replication of oriP plasmids. Sequence analysis indicated that domains A and B of EBNA1 shared the GR repeats found in many AT hook proteins (4), including HMGA1a. Other members of this family, such as ARBD or MBD2a, have much longer GR repeats, such as those found in EBNA1. Our experiments indicated that both domains have a high affinity for AT-rich DNA. Further, the binding is displaced by the minor groove binding drugs distamycin and netropsin (data not shown), indicating that the association occurs via interactions in the minor groove, as it does for HMGA1a. Full-length EBNA1 (that lacks the Gly-Ala repeats) expressed in baculovirus also had AT hook activity comparable to purified domains A and B or peptides containing the AT hook region.
Because both domains A and B had AT hook function, but only domain B associated with EBP2, we could test whether the association of EBNA1 with metaphase chromosomes, and its ability to support the transient and stable replication of oriP-plasmids better correlated with AT hook function or an association with EBP2. For this we constructed versions of EBNA1 that had one or more copies of domain A (but lacked domain B) or a derivative of EBNA1 in which the entire amino terminus was replaced with EBP2. Our studies clearly indicate that any version of EBNA1 that contains at least two copies of domain A supports stable replication of wild-type oriP (which has 20 binding sites for EBNA1 in FR). Curiously, a derivative with just a single copy of domain A also supports the stable replication of oriP plasmids that contain 30 EBNA1 binding sites in FR. This result leads us to hypothesize that the tethering of EBV episomes with metaphase chromosomes is dependent upon two associations: the first is between the amino terminus of EBNA1 and the chromosome, and the second is between the carboxy terminus of EBNA1 and the EBV episome. The former is modulated by the number of copies of domain A (and presumably domain B) in EBNA1, and the latter is modulated by the number of EBNA1-binding sites on the EBV episome. Although 1A-DBD presumably associates with chromosomes less well and therefore does not stably replicate a wild-type oriP-plasmid, increasing the number of binding sites in FR from 20 to 30 provides additional A domains that can tether the plasmid to chromosomes, rescuing the defect. Put more simply, increasing the amount of 1A-DBD on the episome increases the avidity of the plasmid-protein complex for chromosomes without changing the affinity of each 1A-DBD for chromatin per se. Our results with 1A-DBD closely parallel those of Mackey and Sugden, who observed that this protein was approximately half as functional as wild-type EBNA1 and equivalent in function to 1B-DBD (34).
Our results with the EBP2-DBD fusion are intriguing. This fusion localizes precisely as wild-type EBP2 in cells. It is primarily nucleolar, and we did not find any association of either protein with metaphase chromosomes similar to the original observations (10). We have examined the localization of EBP2 within cells obtained by mitotic shake-off, and these results confirm our observations that it does not associate with condensed chromosomes. There are two differences in the methods used by us and those used by others (26, 58). We used deconvolution or confocal microscopy instead of flat-field microscopy and higher-powered objectives (×100 and ×150). That we failed to observe EBP2 localized to metaphase chromosomes is underscored by the inability of the EBP2-DBD fusion protein to support the transient or stable replication of oriP plasmids and that this protein, which is localized to nucleoli, acts as a potent dominant-negative of wild-type EBNA1. This result is similar to what we have demonstrated previously with another protein (HMG1-DBD) (48). These results are consistent with our previous hypothesis that nuclear localization by itself is not sufficient for the transient replication of oriP plasmids; rather, metaphase chromosome localization is required for both transient and stable replication of these episomes (48).
How do we reconcile our observation that EBP2 does not colocalize with EBNA1 to metaphase chromosomes with genetic data that expression of human EBP2 increases the retention of ARS/FR plasmids in budding yeast? There are at least two possibilities. The simplest explanation is that, although AT hook proteins such as HMGA1a, bind SARs on metaphase chromosomes, it is possible that their binding is mediated by a “loading protein” that doesn't remain a part of the bound complex. EBP2 may function as such a loader for EBNA1. If it does, there have to be other such loaders as well, because derivatives of EBNA1 that lack domain B continue to bind metaphase chromosomes and stably replicate oriP plasmids. Another possible explanation for the genetic observations in budding yeast may lie in the fundamental differences in mitosis between budding yeast and human cells. The nuclear envelope is not dissolved in budding yeast during mitosis, and there is typically a single large nucleolus that occupies about half the nucleus (62). The contents of this large nucleolus are retained within the nucleus during mitosis (on account of the nuclear membrane not dissolving during mitosis). Therefore, a complex of EBP2 and EBNA1 may permit the efficient retention of ARS/FR plasmids within this large nucleolus during interphase, and the random retention of its contents during mitosis. Such retention and distribution of ARS plasmids is not hitherto unknown. ARS plasmids with Sir4p binding sites are retained more efficiently than plasmids with an ARS alone (3), although Sir4 does not function in the partitioning of chromosomes. It should also be noted that although ARS plasmids containing Sir4 sites are retained more efficiently in budding cells, they are not partitioned equally when tested by partitioning assays (3, 22, 30).
The observation that the amino terminus of EBNA1 has AT hooks probably has implications for both the replication and partitioning of EBV episomes. It is clear that the replication of these episomes is ORC dependent, although the mechanism by which ORC is loaded onto oriP remains enigmatic (15, 47). Kelly and coworkers have recently purified human ORC and tested its DNA-binding specificity. Although this complex lacks AT hooks that the ORC from S. pombe has (12, 32), it still associates preferentially with AT-rich DNA when tested with synthetic substrates and in an assay identical to the one we used (55). We believe that an association between EBNA1 and ORC may be first mediated by their common localization to AT-rich DNA within cells, such as SARs. This suggestion explains two observations. The first is that the DBD alone of EBNA1 does not support even the transient replication of oriP plasmids (28). The second is that fusing proteins that bind AT-rich DNA, such as histone H1 or HMGA1a, to the DBD of EBNA1 restores the replication of oriP plasmids (24, 48).
Are EBV episomes partitioned equally and how might the AT hooks of EBNA1 contribute to this process? In studies conducted 15 years ago, Sugden and Warren monitored the loss of Hygr and G418r oriP plasmids present at different copy numbers in the same cell and found that both the selected and the unselected plasmids were lost at the same rate (53). When these data are analyzed by using mathematical models created to understand the partitioning of plasmids (42), it best fits a model in which plasmids are largely, if not entirely, partitioned equally on a per-replicon basis. Random partitioning of a plasmid pool would have resulted in the low-copy unselected plasmid being diluted out very rapidly (within five to eight passages) by the high-copy selected plasmid. Equal partitioning also fits the observations of Delecluse et al., who observed by fluorescence in situ hybridization analysis that episomal EBV often appeared as a pair of dots on paired sister chromatids in metaphase spreads (14). How might the AT hooks of EBNA1 facilitate an equal partitioning event that occurs for each replicon? Although we have no direct evidence that EBNA1 associates with SARs on metaphase chromosomes as HMGA1a does, the distribution of EBNA1 and HMGA1a-DBD on metaphase chromosomes cannot be distinguished, and the maintenance of oriP plasmids by these two proteins is mathematically identical. If EBNA1 were to localize to a SAR on an interphase chromosome, it is possible that after S phase, EBNA1, along with the replicated daughter oriP plasmid, is distributed to the sister SAR on the sister chromatid. The indirect immunofluorescence analysis of some of our metaphase spreads indicates that the distribution of EBNA1 “dots” on some sister chromatids is approximately equal, an observation that is also true for HMGA1a-DBD (see Fig. 9A). In this model that is presented schematically in Fig. 9B, the per-replicon partitioning event is simply the distribution of EBNA1 to the sister SAR on the newly replicated sister chromatid. Replication of DNA may be sufficient to very transiently displace EBNA1 bound to a SAR and permit reassociation with SARs on both sister chromatids. However, there is at least one other possibility. The Saccharomyces cerevisiae 2μm plasmids in budding yeast are partitioned on a per replicon basis via a counting event that involves cohesin cleavage, although these plasmids are not centromeric (40). On human chromosomes cohesins are now known to be loaded via the NuRD complex (19), which has a high affinity for AT-rich sequences (59). It is possible that cohesins are loaded onto oriP plasmids by virtue of EBNA1 and the NuRD complex being colocalized to AT-rich sequences and that the actual per-replicon partitioning event requires cohesin cleavage, much as occurs for 2μm plasmids.
Our results that EBNA1 associates with chromosomes via AT hooks may also have implications for the replication and partitioning of Kaposi's sarcoma herpesvirus. Kaye and coworkers have demonstrated that the N terminus of LANA is essential for the stable replication of KSHV episomes (6). Krithivas et al. have shown that this region of LANA associates with MeCP2 (also called MBD2a) (31). In addition to binding methyl CpG, MeCP2/MBD2a is an AT hook protein that is known to associate with SARs (4, 13, 56). Further, expression of MeCP2 in mouse cells facilitates the association of LANA with chromosomes in mouse cells, suggesting that LANA associates with chromosomes indirectly but through an AT hook-mediated tether (31). Finally, similar to EBNA1, the N terminus of LANA can be replaced with histone H1 but not with cellular DNA-binding proteins that associate with metaphase chromosomes but do not preferentially associate with AT-rich sequences, such as histone H2b (49).
In summary, we have determined that the amino terminus of EBNA1 has two regions that have AT hook activity. We have demonstrated this activity strongly correlates with the ability of EBNA1 to associate with metaphase chromosomes and to support the replication and partitioning of oriP plasmids. Our results provide a molecular mechanism that explains how EBNA1 tethers EBV genomes to cellular chromosomes, a process that is essential for the replication and partitioning of viral genomes in cells latently infected by EBV.
Acknowledgments
We thank Leslie Elbert for constructing the EBP2-DBD fusion and Scott Battle for constructing the 1A-DBD, 2A-DBD, and 3A-DBD derivatives of EBNA1. We thank Raymond Reeves for providing HMGA1a protein and for discussions on structural similarities between EBNA1 and HMGA1a. We thank Harris Busch and Ben Valdez for generously providing a cDNA clone of EBP2/p40 and for monoclonal antisera against EBP2. We thank Jonathan Leis for helping us design the filter-binding assays and Pat Spear for providing the filter apparatus. We thank Sui Huang for providing reagents against nucleolar proteins and for discussions on the structure and function of nucleoli in yeast and human cells. We thank Pat Spear and Hank Seifert for critically reading the manuscript. Fluorescence-activated cell sorting analyses were performed at the Immunobiology Center of Northwestern University.
J.S. is a predoctoral trainee supported by the Carcinogenesis Training Grant (T32 CA09560). A.A. is supported by an award from the National Cancer Institute.
REFERENCES
- 1.Adams, A. 1987. Replication of latent Epstein-Barr virus genomes in Raji cells. J. Virol. 61:1743-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiyar, A., and B. Sugden. 1998. Fusions between Epstein-Barr viral nuclear antigen-1 of Epstein-Barr virus and the large T-antigen of simian virus 40 replicate their cognate origins. J. Biol. Chem. 273:33073-33081. [DOI] [PubMed] [Google Scholar]
- 3.Ansari, A., and M. R. Gartenberg. 1997. The yeast silent information regulator Sir4p anchors and partitions plasmids. Mol. Cell. Biol. 17:7061-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aravind, L., and D. Landsman. 1998. AT hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 26:4413-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banks, G. C., B. Mohr, and R. Reeves. 1999. The HMG-I(Y) A.T-hook peptide motif confers DNA-binding specificity to a structured chimeric protein. J. Biol. Chem. 274:16536-16544. [DOI] [PubMed] [Google Scholar]
- 6.Barbera, A. J., M. E. Ballestas, and K. M. Kaye. 2004. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 N terminus is essential for chromosome association, DNA replication, and episome persistence. J. Virol. 78:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bashaw, J. M., and J. L. Yates. 2001. Replication from oriP of Epstein-Barr virus requires exact spacing of two bound dimers of EBNA1 which bend DNA. J. Virol. 75:10603-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beckerbauer, L., J. J. Tepe, J. Cullison, R. Reeves, and R. M. Williams. 2000. FR900482 class of anti-tumor drugs cross-links oncoprotein HMG I/Y to DNA in vivo. Chem. Biol. 7:805-812. [DOI] [PubMed] [Google Scholar]
- 9.Calos, M. P. 1998. Stability without a centromere. Proc. Natl. Acad. Sci. USA 95:4084-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterjee, A., J. W. Freeman, and H. Busch. 1987. Identification and partial characterization of a Mr 40,000 nucleolar antigen associated with cell proliferation. Cancer Res. 47:1123-1129. [PubMed] [Google Scholar]
- 11.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 98:10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chuang, R. Y., and T. J. Kelly. 1999. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT hooks. Proc. Natl. Acad. Sci. USA 96:2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cross, S. H., R. R. Meehan, X. Nan, and A. Bird. 1997. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat. Genet. 16:256-259. [DOI] [PubMed] [Google Scholar]
- 14.Delecluse, H. J., S. Bartnizke, W. Hammerschmidt, J. Bullerdiek, and G. W. Bornkamm. 1993. Episomal and integrated copies of Epstein-Barr virus coexist in Burkitt lymphoma cell lines. J. Virol. 67:1292-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 16.Disney, J. E., K. R. Johnson, N. S. Magnuson, S. R. Sylvester, and R. Reeves. 1989. High-mobility group protein HMG-I localizes to G/Q- and C-bands of human and mouse chromosomes. J. Cell Biol. 109:1975-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elton, T. S., M. S. Nissen, and R. Reeves. 1987. Specific A. T DNA sequence binding of RP-HPLC purified HMG-I. Biochem. Biophys. Res. Commun. 143:260-265. [DOI] [PubMed] [Google Scholar]
- 18.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 19.Hakimi, M. A., D. A. Bochar, J. A. Schmiesing, Y. Dong, O. G. Barak, D. W. Speicher, K. Yokomori, and R. Shiekhattar. 2002. A chromatin remodeling complex that loads cohesin onto human chromosomes. Nature 418:994-998. [DOI] [PubMed] [Google Scholar]
- 20.Hart, C. M., and U. K. Laemmli. 1998. Facilitation of chromatin dynamics by SARs. Curr. Opin. Genet. Dev. 8:519-525. [DOI] [PubMed] [Google Scholar]
- 21.Hebner, C., J. Lasanen, S. Battle, and A. Aiyar. 2003. The spacing between adjacent binding sites in the family of repeats (FR) affects the functions of Epstein-Barr nuclear antigen 1 (EBNA1) in transcription activation and stable plasmid maintenance. Virology 311:263-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hieter, P., C. Mann, M. Snyder, and R. W. Davis. 1985. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell 40:381-392. [DOI] [PubMed] [Google Scholar]
- 23.Hubert, W. G., T. Kanaya, and L. A. Laimins. 1999. DNA replication of human papillomavirus type 31 is modulated by elements of the upstream regulatory region that lie 5′ of the minimal origin. J. Virol. 73:1835-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hung, S. C., M. S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 98:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izaurralde, E., E. Kas, and U. K. Laemmli. 1989. Highly preferential nucleation of histone H1 assembly on scaffold-associated regions. J. Mol. Biol. 210:573-585. [DOI] [PubMed] [Google Scholar]
- 26.Kapoor, P., and L. Frappier. 2003. EBNA1 partitions Epstein-Barr virus plasmids in yeast cells by attaching to human EBNA1-binding protein 2 on mitotic chromosomes. J. Virol. 77:6946-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapoor, P., K. Shire, and L. Frappier. 2001. Reconstitution of Epstein-Barr virus-based plasmid partitioning in budding yeast. EMBO J. 20:222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirchmaier, A. L., and B. Sugden. 1997. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. J. Virol. 71:1766-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirchmaier, A. L., and B. Sugden. 1995. Plasmid maintenance of derivatives of oriP of Epstein-Barr virus. J. Virol. 69:1280-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koshland, D., J. C. Kent, and L. H. Hartwell. 1985. Genetic analysis of the mitotic transmission of minichromosomes. Cell 40:393-403. [DOI] [PubMed] [Google Scholar]
- 31.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, J. K., K. Y. Moon, Y. Jiang, and J. Hurwitz. 2001. The Schizosaccharomyces pombe origin recognition complex interacts with multiple AT-rich regions of the replication origin DNA by means of the AT hook domains of the spOrc4 protein. Proc. Natl. Acad. Sci. USA 98:13589-13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackey, D., T. Middleton, and B. Sugden. 1995. Multiple regions within EBNA1 can link DNAs. J. Virol. 69:6199-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mackey, D., and B. Sugden. 1999. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol. Cell. Biol. 19:3349-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackey, D., and B. Sugden. 1997. Studies on the mechanism of DNA linking by Epstein-Barr virus nuclear antigen 1. J. Biol. Chem. 272:29873-29879. [DOI] [PubMed] [Google Scholar]
- 36.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Piolot, M. Coppey-Moisan, and J. C. Nicolas. 1999. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 73:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marekov, L. N., G. Angelov, and B. Beltchev. 1978. Studies on the preferential binding of histone H1 to AT-rich DNA. Biochimie 60:1347-1349. [DOI] [PubMed] [Google Scholar]
- 39.Mayo, K., M. L. Vana, J. McDermott, D. Huseby, J. Leis, and E. Barklis. 2002. Analysis of Rous sarcoma virus capsid protein variants assembled on lipid monolayers. J. Mol. Biol. 316:667-678. [DOI] [PubMed] [Google Scholar]
- 40.Mehta, S., X. M. Yang, C. S. Chan, M. J. Dobson, M. Jayaram, and S. Velmurugan. 2002. The 2 micron plasmid purloins the yeast cohesin complex: a mechanism for coupling plasmid partitioning and chromosome segregation? J. Cell Biol. 158:625-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mirkovitch, J., S. M. Gasser, and U. K. Laemmli. 1988. Scaffold attachment of DNA loops in metaphase chromosomes. J. Mol. Biol. 200:101-109. [DOI] [PubMed] [Google Scholar]
- 42.Murray, A. W., and J. W. Szostak. 1983. Pedigree analysis of plasmid segregation in yeast. Cell 34:961-970. [DOI] [PubMed] [Google Scholar]
- 43.Reeves, R., and L. Beckerbauer. 2001. HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim. Biophys. Acta 1519:13-29. [DOI] [PubMed] [Google Scholar]
- 44.Reeves, R., and M. S. Nissen. 1990. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins: a novel peptide motif for recognizing DNA structure. J. Biol. Chem. 265:8573-8582. [PubMed] [Google Scholar]
- 45.Saitoh, Y., and U. K. Laemmli. 1993. From the chromosomal loops and the scaffold to the classic bands of metaphase chromosomes. Cold Spring Harbor Symp. Quant. Biol. 58:755-765. [DOI] [PubMed] [Google Scholar]
- 46.Saitoh, Y., and U. K. Laemmli. 1994. Metaphase chromosome structure: bands arise from a differential folding path of the highly AT-rich scaffold. Cell 76:609-622. [DOI] [PubMed] [Google Scholar]
- 47.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sears, J., J. Kolman, G. M. Wahl, and A. Aiyar. 2003. Metaphase chromosome tethering is necessary for the DNA synthesis and maintenance of oriP plasmids but is insufficient for transcription activation by Epstein-Barr nuclear antigen 1. J. Virol. 77:11767-11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shinohara, H., M. Fukushi, M. Higuchi, M. Oie, O. Hoshi, T. Ushiki, J. Hayashi, and M. Fujii. 2002. Chromosome binding site of latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus is essential for persistent episome maintenance and is functionally replaced by histone H1. J. Virol. 76:12917-12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shire, K., D. F. Ceccarelli, T. M. Avolio-Hunter, and L. Frappier. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J. Virol. 73:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spector, D. L., R. D. Goldman, and L. A. Leinwand. 1998. Cells: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 52.Strick, R., and U. K. Laemmli. 1995. SARs are cis DNA elements of chromosome dynamics: synthesis of a SAR repressor protein. Cell 83:1137-1148. [DOI] [PubMed] [Google Scholar]
- 53.Sugden, B., and N. Warren. 1988. Plasmid origin of replication of Epstein-Barr virus, oriP, does not limit replication in cis. Mol. Biol. Med. 5:85-94. [PubMed] [Google Scholar]
- 54.Tsujii, R., K. Miyoshi, A. Tsuno, Y. Matsui, A. Toh-e, T. Miyakawa, and K. Mizuta. 2000. Ebp2p, yeast homologue of a human protein that interacts with Epstein-Barr virus nuclear antigen 1, is required for pre-rRNA processing and ribosomal subunit assembly. Genes Cells 5:543-553. [DOI] [PubMed] [Google Scholar]
- 55.Vashee, S., C. Cvetic, W. Lu, P. Simancek, T. J. Kelly, and J. C. Walter. 2003. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 17:1894-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weitzel, J. M., H. Buhrmester, and W. H. Stratling. 1997. Chicken MAR-binding protein ARBP is homologous to rat methyl-CpG-binding protein MeCP2. Mol. Cell. Biol. 17:5656-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Westendorf, J. M., K. N. Konstantinov, S. Wormsley, M. D. Shu, N. Matsumoto-Taniura, F. Pirollet, F. G. Klier, L. Gerace, and S. J. Baserga. 1998. M phase phosphoprotein 10 is a human U3 small nucleolar ribonucleoprotein component. Mol. Biol. Cell 9:437-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, H., D. F. Ceccarelli, and L. Frappier. 2000. The DNA segregation mechanism of Epstein-Barr virus nuclear antigen 1. EMBO Rep. 1:140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yasui, D., M. Miyano, S. Cai, P. Varga-Weisz, and T. Kohwi-Shigematsu. 2002. SATB1 targets chromatin remodeling to regulate genes over long distances. Nature 419:641-645. [DOI] [PubMed] [Google Scholar]
- 60.Yates, J., N. Warren, D. Reisman, and B. Sugden. 1984. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. USA 81:3806-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yates, J. L., and N. Guan. 1991. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J. Virol. 65:483-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zanchin, N. I., and D. S. Goldfarb. 1999. Nip7p interacts with Nop8p, an essential nucleolar protein required for 60S ribosome biogenesis, and the exosome subunit Rrp43p. Mol. Cell. Biol. 19:1518-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao, K., E. Kas, E. Gonzalez, and U. K. Laemmli. 1993. SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG-I/Y is enriched in H1-depleted chromatin. EMBO J. 12:3237-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]