Abstract
The IE2 86-kDa gene product is an essential regulatory protein of human cytomegalovirus (HCMV) with several functions, including transactivation, negative autoregulation, and cell cycle regulation. In order to understand the physiological significance of each of the IE2 functions, discriminating mutants of IE2 are required that can be tested in a viral background. However, no such mutants of IE2 are available, possibly reflecting structural peculiarities of the large and ill-defined C-terminal domain of IE2. Here, we revisited the C-terminal domain by analyzing IE2 mutants for transactivation, DNA binding, autoregulation, and cell cycle regulation in parallel. We found it to contain an unexpectedly concise core domain (amino acids 450 to 544) that is defined by its absolute sensitivity to any kind of mutation. In contrast, the region adjacent to the core (amino acids 290 to 449) generally tolerates mutations much better. Although it contributes more specific sequence information to distinct IE2 activities, none of the mutations analyzed abolished any particular function. The core is demarcated from the adjacent region by the putative zinc finger region (amino acids 428 to 452). Surprisingly, the deletion of the putative zinc finger region from IE2 revealed that this region is entirely dispensable for any of the IE2 functions tested here in transfection assays. Our work supports the view that the 100 amino acids of the core domain hold the key to most functions of IE2. A systematic, high-density mutational analysis of this region may identify informative mutants discriminating between various IE2 functions that can then be tested in a viral background.
The human cytomegalovirus (HCMV), a member of the beta subgroup of herpesviruses, is characterized by its narrow host range and its prolonged replicative cycle in tissue culture. HCMV gene expression is temporally regulated, giving rise to immediate-early (IE), early, and late gene products. During the IE phase, the so-called major IE gene locus is most extensively transcribed and by different splicing events gives rise to several gene products, among which the 72-kDa IE protein (IE1) and the 86-kDa IE protein (IE2) are the most abundant. These two nuclear proteins have been extensively studied with respect to their ability to regulate transcription of numerous viral and cellular promoters (41, 54, 55). The IE2 protein is essential for viral replication and appears to play the role of a master regulator in triggering the lytic replicative cycle. Consistent with this, it has proven impossible to generate infectious HCMV progeny from mutants lacking IE2 (22, 40).
Of particular importance is the role of IE2 as a potent transcriptional activator of endogenous viral gene expression. In addition, IE2 transactivates various heterologous promoters, such as the cellular cyclin E promoter, the c-fos promoter, and the human immunodeficiency type 1 (HIV-1) long terminal repeat (LTR) (41, 54, 55). Transactivation by IE2 seems to involve both protein-protein and protein-DNA interactions. For instance, IE2 is able to directly activate basal promoters like those of the hsp70 and c-fos genes in a TATA box-dependent mechanism (20). This appears to result from a direct interaction of IE2 with components of the basal transcription complex, including the TATA-binding protein (TBP) (8, 19, 26, 27) and TFIIB (8). Protein-protein interactions also seem to be the molecular basis for the ability of IE2 to rescue the transcriptional defect in ts13 cells caused by a temperature-sensitive mutation in TAFII250, suggesting that IE2 may even exhibit TAF-like functions (35). In addition to the interaction with basal transcription factors, IE2 has also been shown to interact with upstream transcription factors, such as CREB (29), Sp-1 (36), and Egr-1 (64), and to functionally interact with the histone acetyltransferase P/CAF (6). Further, transcriptionally relevant interaction partners of IE2 include cell cycle regulatory proteins like Rb (12, 14, 18, 52). The Rb IE2 interaction correlates with the ability of IE2 to derepress a synthetic promoter via E2F-binding sites (18) and to counteract the flat cell phenotype of Rb-overexpressing SAOS-2 cells (14).
In contrast, much less is known about the role of IE2 as a DNA-binding protein in transcriptional activation. DNA binding sites for IE2 have been identified within the HCMV early UL112/113 promoter (2, 48, 50) and in the cyclin E promoter (5), where binding has been shown to contribute to transcriptional activation.
In addition to acting as a transcriptional activator, IE2 also has been shown to function as a transcriptional repressor of it's own promoter (11, 23, 26, 30, 38, 44-46, 56). Autorepression depends on direct binding of IE2 to a sequence termed the cis repression signal (CRS), which is located between the TATA box and the transcriptional start site of the major IE enhancer-promoter (10, 34, 44). IE2 binding to the CRS appears to block assembly of the transcription initiation complex by steric hindrance (32). The CRS has been identified as a 14-bp sequence element with a partially palindromic structure. It consists of two copies of the dinucleotide CG separated by an A/T-rich stretch of 10 nucleotides and can confer IE2-dependent repression to heterologous promoters in an orientation-independent but location-dependent manner (10, 34, 44). Mutation analyses of the CRS has shown that the flanking CG dinucleotides of the CRS are critical for recognition by IE2 (59) and that the correct spacing of the CG motif is more important than the specific interspersed sequence element (11). Recently, additional IE2-responsive cis-repressive sequences have been identified within the MIE promoter-enhancer (25) and also in the RSV LTR, which also can be repressed by IE2 (57). Although the ability to downregulate the MIE promoter-enhancer in transient-transfection assays can be easily demonstrated in various cell lines (23, 44, 46, 56, 57), it has been unexpectedly difficult, to date, to demonstrate DNA-binding activity in extracts from cells transfected with IE2 (25, 59), and no binding has been shown with extracts derived from cells infected with HCMV. Instead, DNA binding of IE2 has been analyzed by using IE2 expressed in and purified from bacteria (11, 15, 26, 30, 38) or in a yeast one-hybrid assay (1). This has so far hampered a direct correlation of IE2 sequence requirements for DNA binding, transactivation, and autorepression in a single test system.
A more recently discovered function of IE2 is its cell cycle regulatory activity. IE2 has been shown to block cell cycle progression around the G1/S transition in various experimental systems and cell lines (42, 61, 62), including primary fibroblasts (28, 43). At the same time, IE2 has been reported to induce the E2F-dependent gene expression program and to induce cyclin E-associated kinase activity (53, 62), thereby mimicking the arrest observed in HCMV-infected cells. Consistent with the latter finding, IE2 has also been reported to induce terminally differentiated cells to reenter the cell cycle from G0 (7, 51).
The transactivation capacity of IE2 has been attributed to its two independent acidic activation domains at the N terminus (amino acids [aa] 25 to 85) and the C terminus (aa 544 to 579) as well as on a large portion of the C terminus (aa 195 to 544) (39, 45, 56). Sequences required for the interaction with basal transcription factors also reside in the C terminus. Binding to TFIIB involves aa 290 to 542 (8), and TBP interaction is mediated by aa 290 to 504 and additionally aa 47 to 153 (8, 27). In addition, this region also seems to be involved in the interaction with promoter-specific transcription factors, as the minimal CREB-binding domain has been located between aa 290 and 410 (29). Furthermore, an overlap exists between the C-terminal transactivation domain and the regions associated with DNA binding and autorepression. Autoregulation has been shown to require aa 290 to 579 (1, 23, 45, 46, 56). The DNA-binding domain resides within this region and has been mapped to aa 346 to 579 (1, 11, 59). The N-terminal border of the DNA binding domain is currently unclear, since sequences N terminal to position 346 also seem to contribute to the DNA-binding activity (1). More recently, the C terminus of IE2 has also been suggested to be involved in mediating the cell cycle arrest function of IE2, whereas the N terminus (aa 1 to 195) was found to be dispensable (61).
Within the C terminus, particular emphasis has been attributed to a region that is located between aa 428 and 452 and that is believed to adopt a zinc finger structure (the putative zinc finger). Point mutations of either the two cysteine or histidine residues have been shown to abolish autorepression as well as DNA binding and to also affect transactivation function (5, 26, 37, 38, 57, 63). On this basis, the putative zinc finger has been considered to be of functional significance for IE2.
The overlapping domain structure of IE2 makes it difficult to study one particular activity of IE2 on the basis of a mutational analysis without affecting the others, and this would be a particular problem when analyzing IE2 mutants in a viral background. These circumstances necessitate further definition and discrimination of protein domains of IE2 responsible for specific activities of the protein in cell culture-based experimental systems prior to addressing these aspects in the viral context. Here, we define dispensable and required sequences in the C terminus of IE2 for the various IE2 activities. We show that the previously ill-defined C terminus of IE2 can be subdivided into a strictly required concise core domain and a modulating adjacent region. These two domains are demarcated by the putative zinc finger region that was found to be dispensable for all the functions of IE2 tested here. The concise core domain should qualify for a systematic mutational approach to define discriminating mutants of IE2 that can then be tested in a viral background.
MATERIALS AND METHODS
Cells.
All cell lines were maintained as monolayers in Dulbecco's modified Eagle medium supplemented with 5% newborn and 5% fetal calf serum (FCS) and 100 U of penicillin-streptomycin per ml. U-373-MG cells were purchased from Cell Lines Service (Heidelberg, Germany). Every 3 days, U373 cells were split in a 1:3 ratio and HeLa cells were split in a 1:10 ratio to maintain a subconfluent state.
Plasmids.
The empty expression vector pSG5-3-hemagglutinin (HA) and the expression vectors for IE2 wild type (wt), IE2 (1-544), IE2 (1-450), IE2 (195-579), and CD20 have been described previously (61). The IE2 wt-expressing plasmid contained the full-length IE2 cDNA of AD169. To create IE2 mutants with C-terminal deletions, the corresponding cDNA fragments were PCR amplified from pSG5-3HA-IE2. The following primers were used for C-terminal deletions: IE2 (1-513), 5′-CGTAAGATCTAGACTACAGGGGCAGGCACAGGTTG-3′; IE2 (1-497), 5′-CGTAAGATCTAGACTATTGGTGGGTGTGCAATTC-3′; IE2 (1-482), 5′-CGTAAGATCTA AGCGTGGATGATCATGTTG-3′. The PCR products were cloned into the BglII site of pSG5-3HA, in frame with the N-terminal HA tag. The C-terminal amino acid exchange mutants as well as the internal deletion mutants were all generated by site-directed mutagenesis (17). The following primers were used for the in vitro mutagenesis reaction: IE2 pm501, 5′-CACGCTGCCACCCCCGCGGCAGCGGCCGCGGCTGCGAACCTGTGCCTGCCC-3′; IE2 pm510-517, 5′-GGCGCTCTCAACCTGGCCGCGGCGGCCGCGGCAGCGTTTCCCAAACAGGTC-3′; IE2 pm519-526, 5′-GATGCAAAAGTTTCCCGCAGCGGCGGCCGCGGCCGCCTTCTCCACCAACCAG-3′; IE2 pm528-535, 5′-GTGCGCATCTTCTCCGCCGCCGCGGCCGCGGCCGCGCTGCCTATCTACGAG-3′; IE2 pm537-544, 5′-GGGTTCATGCTGCCTGCCGCCGCGGCCGCGGCGGCGGCCTACGCCGTGGGG-3′; IE2del 230-249, 5′-CCTTCCACCGGCAGCGGCACGCCCTTGGGCCGGCCCGATG-3′; IE2 del 260-279, 5′-GGCCCGATGAAGATAGTTCCAAATGCAGCAGTGGCGGAGG-3′; IE2 del 290-309, 5′-GTGGCGGAGGAGCATCCGTGTGCGGCCATCAGAGCAGCGG-3′; IE2 del 330-349, 5′-CGCAAGAAGAAGAGCAAACGCTGCACACCCAACGTGCAGAC-3′; IE2 del 380-399, 5′-CCAATCGCTCTCTTGAGTACTGCAAAACCATGCAGGTGAAC-3′; IE2 del 430-449, 5′-GAGGTGGATGCGGTGCGGTGTCCCGTGACACATCCACCCGAAG-3′; IE2 del 470-489, 5′-CGATGCTTGTAACGAAGGCGTCTACCGCAACATGATCATCCAC-3′. All mutations were confirmed by DNA sequencing. For transfection, plasmids were purified by CsCl ethidium bromide equilibrium centrifugation.
DNA transfections.
Cells were transfected using the calcium phosphate coprecipitation method (9). The DNA precipitates were left on the cells for 14 to 16 h. The transfection medium was then replaced with fresh medium, and cells were harvested after an additional 32 h in the case of U373 cells and 6 h in the case of HeLa cells.
For cell cycle analysis, 16 μg of effector plasmid and 4 μg of pSG5-CD20 plasmid were applied to the cells in 100-mm-diameter dishes. For reporter gene analysis, about 4 × 106 cells were seeded in 60-mm dishes. A 4.8-μg aliquot of effector plasmid and 1.6 μg of UL112/113 reporter plasmid (pHM142; kindly provided by T. Stamminger) were filled up to 8 μg with pUC19 and applied to the cells. To measure the activation of the c-fos promoter, the same protocol as for UL112/113 was used. For analysis of the autorepressor function of IE2, 2 μg of MIEP wt promoter construct (pRR55; kindly provided by J. Sinclair) and 6 μg of effector plasmid were used.
Virus infection.
Two days before infection, cells were plated at a densitiy of 1.5 × 104 cells/cm2. Stocks of the HCMV laboratory strain AD169 were used to infect the cells at a multiplicity of infection (MOI) of 5. Virus adsorption was allowed for 1 h. Mock-infected cells were exposed to an equal volume of medium containing the same serum concentration as the viral stocks.
Flow cytometry.
Forty-eight hours posttransfection, cells were harvested by trypsination and labeled with a fluorescein isothiocyanate (FITC)-conjugated anti-CD20 antibody (2H7; Pharmingen). Cells were then permeabilized in 75% ethanol, treated with RNase, and stained with propidium iodide, according to standard procedures (13). Flow cytometry was performed on a FACScan (Becton Dickinson) equipped with CellQuest software. Single cells with a FITC staining at least 20 times stronger than that of the untransfected subpopulation were gated and analyzed for DNA content as previously described (58).
Immunoblot analysis.
Cells were lysed in 1× RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris; pH 8.0) and clarified by centrifugation. The protein concentration was determined using the Bio-Rad protein assay, and immunoblot analysis was performed according to standard protocols (13). Briefly, equal amounts of proteins were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) in 8% polyacrylamide and transferred to polyvinylidene difluoride membranes. After blocking with 6% dry milk in 0.1% Tween 20-0.1 M Tris (pH 7.6)-0.15 M NaCl (TTBS), the membranes were probed with anti-HA antibody (12CA5; Boehringer). Membranes were then developed with peroxidase-conjugated anti-mouse immunoglobulin antibody (Sigma) and enhanced chemiluminescense (NEN, Perkin-Elmer Life Sciences, Inc.) as recommended by the manufacturer.
Luciferase assay.
Forty-eight hours posttransfection, medium was removed and cells were washed with phosphate-buffered saline (PBS). Cells were then scraped from the dish in 1 ml of PBS and pelleted for 1 min in a table centrifuge. The pellet was resuspended in 200 μl of luciferase-lysis buffer (Promega), vortexed, and incubated at room temperature for 1 min. Protein concentrations were determined by Bradford analysis (Bio-Rad). Luciferase activity was determined on a luminometer (Berthold, Bad Wildbach, Germany). Transfections were repeated at least three times using different DNA preparations.
CAT assay.
Cells were transfected in 60-mm dishes and harvested 18 h later. Protein concentrations were determined by Bradford analysis. Chloramphenicol acetyltransferase (CAT) activity was determined using a CAT enzyme-linked immunosorbent assay (ELISA) system (Roche) according to the manufacturer's instructions. Transfections were repeated at least three times using different DNA preparations.
Electrophoretic mobility shift assays (EMSA).
Approximately 1.5 × 106 cells per 150-mm dish were seeded the day prior to transfection. Transfections were performed by using the calcium phosphate coprecipitation method. HeLa cells were harvested about 20 h posttransfection, and U373 cells were harvested 48 h posttransfection. Virus-infected cells were harvested at 72 h postinfection (hpi). The cells were washed and harvested by scraping into PBS. Pelleted cells were resuspended in three volumes of TGA buffer (10 mM Tris [pH 8.0], 10% glycerin, 0.5 mM EDTA, 1 mM dithiothreitol [DTT]) and lysed by sonication with a sonicator Vibra cell (small tip; output control, 80%; Sonics & Materials). After centrifugation for 10 min at 20,000 × g, the supernatant was aliquoted, flash-frozen in liquid nitrogen, and stored at −80°C for later use.
To generate the CRS probe, single-stranded oligonucleotides were annealed by incubating equal amounts of each complementary strand at 95°C for 5 min, followed by gradual cooling to room temperature. The annealed products contained 5′ overhangs that were filled in and labeled with 20 μCi of [α-32P]dATP using the Klenow enzyme. The sense strand of the CRS wt probe reads 5′-GGTGGGAGGTCTATATAAGCAGAGCTCGTTTAGTGAACCGTCAGATCGCCTGGAG AC-3′. The CRS mut probe contained the sense sequence 5′-GGTGGGAGGTCTATATAAGCAGAGCctaggTAGTGAACCGTCAGATCGCCTGGAGC-3′. Italic characters indicate the CRS; lowercase indicates the CRS nut probe. For EMSA, 20 μg of protein extracts from sonicated cell extracts was preincubated in TE buffer (pH 8.0) containing 3 μg of bovine serum albumin (BSA)/μl, 5% glycerol, 1 mM MgCl2, 12.5 ng of poly(dA-dT) · poly(dA-dT)/μl and 5 ng of denatured, sonicated herring sperm DNA/μl. After 10 min, about 50 fmol (∼20,000 cpm) of the labeled probe was added, and incubation went on for another 10 min at room temperature (RT). The reaction mixture was then separated on a 4% native polyacrylamide gel containing 0.5× TBS buffer (90 mM Tris-borate, 2 mM EDTA; pH 8.0) followed by autoradiography of the dried gel. For antibody supershift experiments, 0.5 μg of one of the following antibodies was added to the preincubation reaction mixture: anti-IE antibody (9221; NEN), anti-HA antibody (16B12; Covance), antibromodeoxyuridine (anti-BrdU) antibody (3D4; Pharmingen), or anti-CD20 antibody (2H7; Pharmingen). For cold competition experiments, a 1-, 5-, 10-, 50-, or 100-fold excess of the unlabeled wt or mutant CRS duplex was added together with the labeled probe.
RESULTS
In order to define more closely the structural interdependencies for the various functions of IE2, we set up a structure-function analysis of this protein that allowed us to (semi)quantitatively study cell cycle regulation, transactivation, and autorepression in HCMV-permissive U373 cells that transiently express the IE2 protein.
Mapping of IE2 protein domains involved in cell cycle regulation and interdependencies with other IE2 activities.
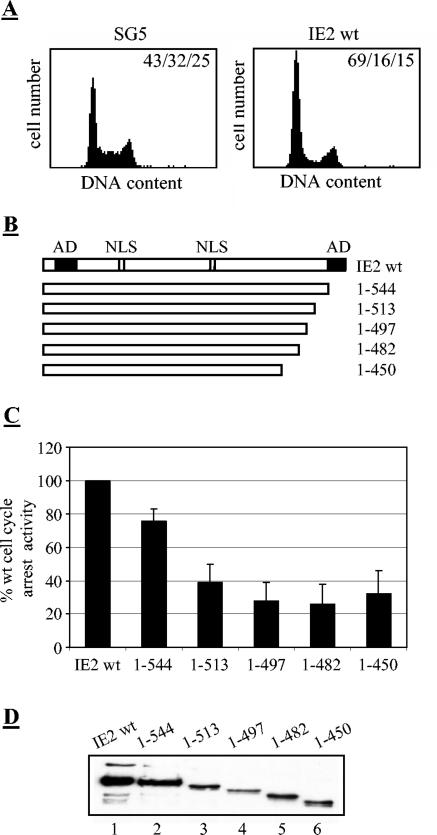
In a previous study, our investigators showed that a mutant with the last 37 aa (1 to 544) deleted was still able to induce a cell cycle arrest, whereas a mutant that had a deletion down to position 450 (1 to 450) failed to do so (61). In order to more precisely determine the C-terminal border of the cell cycle arrest domain, additional C-terminal deletion mutants between aa 450 and 544 were constructed and analyzed. The IE2-mediated cell cycle arrest occurs around the G1/S transition period. In U373 cells, the majority of cells are arrested immediately after S phase entry, but the bulk of cellular DNA remains unreplicated (62). Therefore, fluorescence-activated cell sorter (FACS) analysis after propidium iodide (PI) staining shows an increase in what is generally known as the G1 peak but which, due to the low resolution of the PI-based method, also contains the IE2-arrested early S phase cells (62) (Fig. 1A). However, for monitoring the cell cycle arrest activity of IE2, this method is entirely sufficient and we will refer to this cell cycle block as an arrest at the G1/S transition. For comparative studies, the relative difference between IE2-expressing cells arrested at the G1/S transition (69%) and control cells in G1 (43%) was set at 100%, and the relative difference between IE2 mutants and control cells was determined accordingly. As shown in Fig. 1C, deletion of a further 30 aa (1 to 513) severely impinged upon the cell cycle arrest domain, with even further deletions reducing the arrest activity down to 25% of the wt level. Since this level was close to the limitation of the assay, those mutants were considered to be inactive for cell cycle regulation. Also, the relative loss of cell cycle arrest activity could not be explained by differences in the expression levels of mutant IE2 species (Fig. 1D). In addition, we tested the capacity of the mutant IE2 proteins to autorepress and to transactivate. As expected and consistent with similar mutants tested by other laboratories in the past (23, 45, 56), all of these mutants were completely inactive (data not shown). Therefore, we conclude from this first set of experiments that, with respect to the C terminus, the functional domains mediating transactivation, autoregulation, and cell cycle regulation are not identical but do overlap to a great extent.
FIG. 1.
Requirements of C-terminal sequences for the cell cycle arrest activity of IE2. U373 cells were transiently transfected with plasmids expressing IE2 wt protein or C-terminal deletion mutants as indicated. Cells were harvested 46 h post transfection. (A) Cell cycle arrest activity of the wt IE2 protein. Cells were harvested after transfection, stained for CD20 expression and the DNA content was analyzed by Flow cytometry. The percentage of cells in the G0-G1/S/G2-M compartments is indicated. (B) Schematic representation of the IE2 deletion constructs used. The designation of the mutants indicates the amino acid residues (aa) of the IE2 protein present. The independent activation domains (AD) and the nuclear localisation signals (NLS) of IE2 are indicated. (C) Cell cycle arrest activity of C-terminal deletion mutants. The DNA content of transfected cells was analyzed by flow cytometry. The relative increase of the population arrested by IE2 at G1/S transition compared to the G1 population of control transfected cells was set at 100% with activities of the indicated IE2 mutants determined accordingly. The bar chart gives the mean value obtained in at least 3 independent experiments. Standard deviations are indicated by error bars. (D) Equal amounts of extracts from U373-cells expressing HA-tagged forms of full-length IE2 or the indicated IE2 deletion mutants were separated by SDS-PAGE and analyzed by immunoblotting using an antibody recognizing the HA epitope.
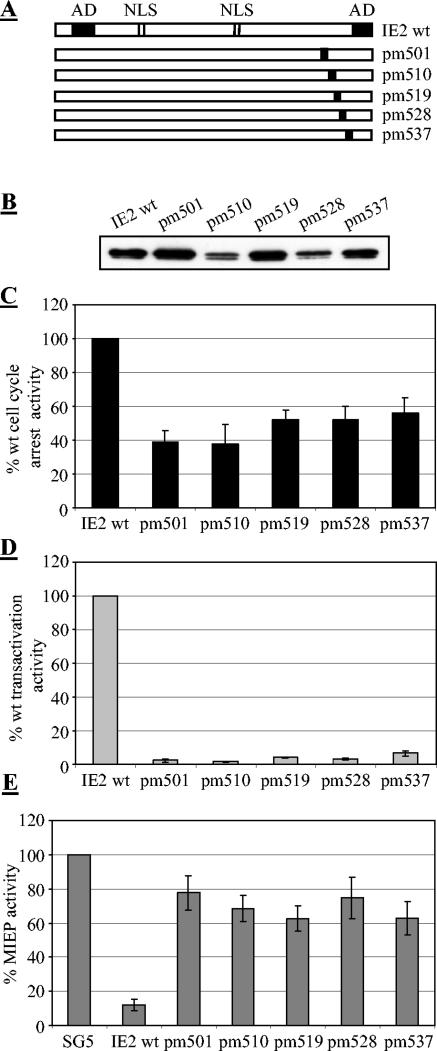
To analyze the C-terminal region in more detail, we constructed five IE2 mutants, each of which contained an exchange of seven consecutive amino acids to alanines (Fig. 2A). All mutants and IE2 wt were expressed at comparable levels (Fig. 2B) and were tested for their ability to arrest the cell cycle as well as to transactivate and to autoregulate. Since transactivation by IE2 is mediated by different mechanisms, it was monitored on two independent reporter systems, namely, the c-fos promoter, as an example of a validated cellular target (4, 20), and the UL112/113 promoter, as a well-studied HCMV-derived early target (2, 29, 49, 50). Since we did not observe any significant differences in the regulation of these two promoters by IE2, only the results from transactivation of the UL112/113 promoter are presented, although all IE2 mutants were tested on both reporter constructs. The activation of these promoters by IE2, which typically was about 50- to 60-fold (data not shown), was set to 100%, allowing us to compare the IE2 mutants accordingly. The ability of IE2 to repress its own promoter was measured by using a construct containing the MIE promoter-enhancer from −671 to +52 relative to the transcription start site upstream of a CAT reporter gene (47). MIEP activity was typically repressed fivefold in IE2-transfected cells (data not shown) and was set at 100%, allowing a direct comparison with the activities mediated by IE2 mutants. As can be seen from Fig. 2C to E, all functions were severely affected by each of the five mutations, indicating that the different activities have overlapping sequence requirements between aa 501 and 544 that cannot be discriminated, even by this more subtle type of mutational analysis.
FIG. 2.
Cell cycle arrest, transactivation and autoregulation functions have overlapping sequence requirements in the C terminus of IE2. (A) Schematic representation of the amino acid exchange mutants used. Seven consecutive amino acids were substituted by alanine residues. The position of the first exchanged residue is indicated. (B) U373 cells were transfected as described in Fig. 1. The expression levels of the indicated proteins were analyzed by immunobloting using an anti-HA antibody. (C) Cell cycle arrest activity of wt and mutant IE2 proteins. Cell cycle distribution was determined as described in Fig. 1. (D) Transactivation of the UL112/113 promoter by wt and mutant IE2 proteins. The transactivation of the UL112/113 promoter by IE2 in transiently transfected U373 cells was compaired to control transfected cells and set at 100%. The activity of mutant IE2 proteins was determined accordingly. (E) Autorepressor function of wt and mutant IE2 proteins. The autoregulation function was analyzed by transfecting U373 cells with a MIE promoter-reporter construct together with IE2 or the indicated IE2 mutants. Control transfected cells received the reporter construct together with the pSG5 plasmid. Cells were harvested 46 h posttransfection and CAT activity was determined as described in Materials and Methods. The promoter activity seen in control transfected cells was set at 100% and the promoter activity in wt or mutant IE2 transfected cells was determined accordingly. The bar chart gives the average of 4 independent experiments, the standard deviation is indicated by error bars.
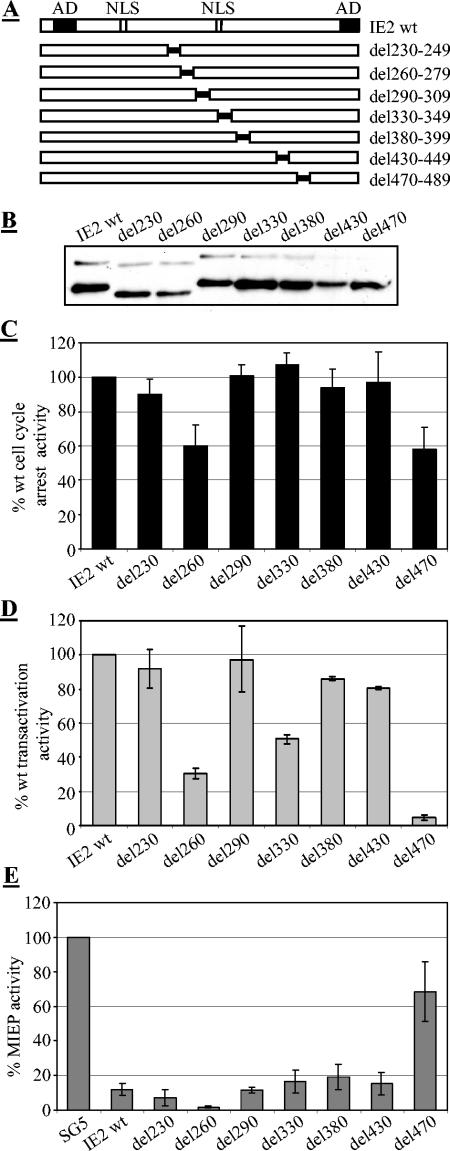
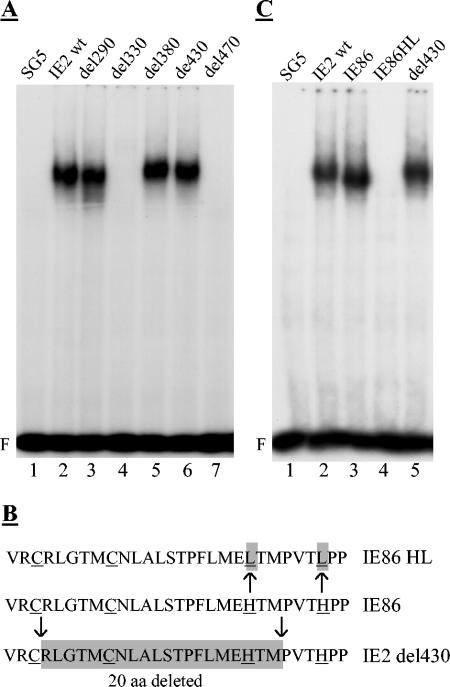
The above data suggest that the IE2 C terminus, which has previously been found to be essential for transactivation and autoregulation (11, 23, 39, 45, 56, 61, 63) is, to a great extent, also indispensable for cell cycle regulation. Next, we set out to define the N-terminal border of this domain to then analyze the region adjacent to it for its contribution to the various activities of IE2 studied here. In order to address this point, 20-aa deletions were introduced into the central part of IE2 (Fig. 3A). The deletion of the serine-rich stretch of residues resulted in faster-migrating IE2 proteins on SDS-PAGE (Fig. 3B, del230 and del260), which has previously also been noted by others (21, 52). Importantly, however, all mutants were expressed to similar levels, such that differences in the activities of IE2 mutants could not simply be allocated to differences in protein expression. The N-terminal border of the invariably required C-terminal region of IE2 (which from now on we will refer to as the core) appears to reside between aa 450 and 470, since del470 was still strongly impaired in all three functions tested but del430 had approximately wt activity in these functions (Fig. 3C and D). Two short stretches seemed to modulate transcriptional activation. Both del260 and del330 were reduced by at least 50% in their abilities to activate the UL112/113 promoter. Interestingly, del330 was fully capable of arresting cells. This suggested that in contrast to the core, different regions of the central portion of IE2 (between aa 260 and 449) can contribute independent modulating activities to transactivation and cell cycle regulation. Unexpectedly, none of the mutants N terminal to del470 impaired the autoregulatory function of IE2 (Fig. 3E). Of note, autorepression by these mutants was still dependent on an intact CRS element (data not shown), indicating that the downregulation of the MIEP by these mutants was still specific. While these results do not suggest that the central portion N terminal to the core is generally dispensable for autoregulation (1, 42), they do suggest that many sequence elements within this region indeed are dispensable, which again clearly contrasts this region from the core. In particular del430, which lacks the putative zinc finger region, is entirely dispensable for IE2 autoregulation (and transactivation).
FIG. 3.
Internal deletion mutants identify a consice core domain and a modulating adjacent region in IE2. U373 cells were transfected as described in Fig. 1 and 2. (A) Schematic of the internal deletion mutants with 20 consecutive amino acids deleted as indicated. (B) Expression levels of the mutant proteins by immunoblot analysis using an anti-HA antibody. (C) Cell cycle arrest activity of wt and mutant IE2 proteins. Cell cycle distribution was determined as described in Fig. 1. (D) Transactivation of the UL112/113 promoter by wt IE2 and mutant proteins. Transactivation function was determined as described in Fig. 2. (E) Autorepressor function of wt IE2 and mutant proteins. The autoregulatory function was determined as described in Fig. 2. Relative activities were determined as described in Fig. 2.
IE2 derived from HCMV-infected or transiently transfected cells can bind to the CRS in a standard DNA binding assay.
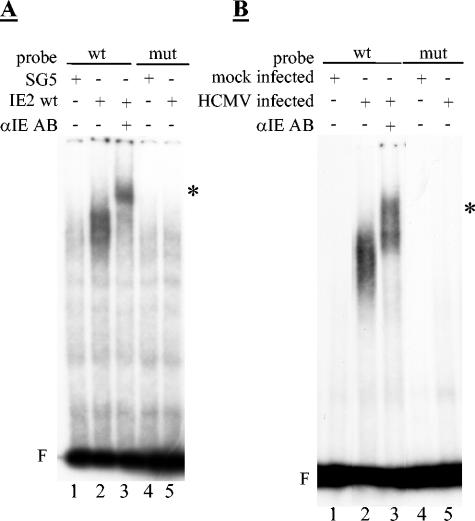
The above findings also imply that neither the putative zinc finger nor the strict integrity of the region adjacent to the core are required for DNA binding of IE2. Since this was contrary to expectations (1, 23, 26, 38, 59), we set out to directly test DNA binding of the IE2 mutants. To this end, we aimed at assaying the IE2 DNA-binding activity in both extracts prepared from IE2-transfected and HCMV-infected cells rather than the binding activity of IE2 prepared from bacteria. This would allow us to build on the most genuine DNA-binding conditions of IE2 and to directly correlate DNA binding and autoregulation of IE2 in a single test system.
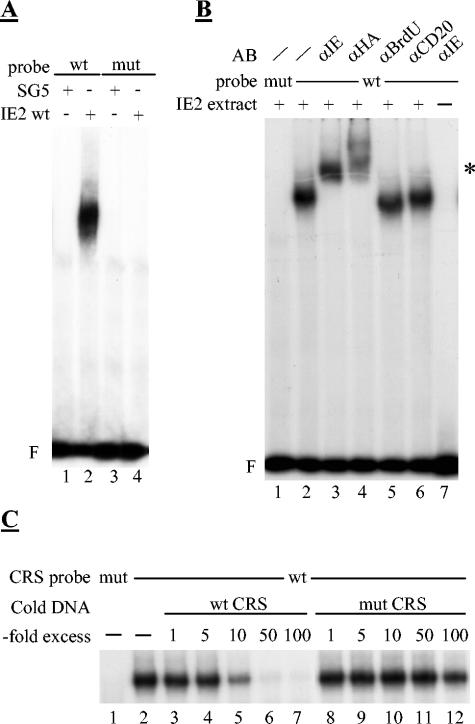
In order to set up the assay system, IE2-transfected HeLa cells were initially used, because of their high transfection efficiency. Extracts were prepared from transfected cells by sonication in a low-salt buffer, leaving out detergents completely. Keeping to this protocol allowed us to observe a mobility shift of the CRS probe when using extracts from IE2-transfected but not from control-transfected cells (Fig. 4A, lanes 1 and 2). This shift in mobility was not seen when we used a mutated CRS element (lanes 3 and 4), suggesting that the binding of IE2 was specific for its known target site. Addition of antibodies recognizing either IE2 or directed against the HA epitope fused to IE2 resulted in a supershift of the observed complex, indicating that IE2 was physically present in this complex rather than inducing a cellular protein binding to the CRS (Fig. 4B, lanes 3 and 4). In contrast, two different nonspecific antibodies did not result in a supershift (lanes 5 and 6). Also, the observed DNA binding of IE2 was specific for the CRS, since a 10- to 50-fold excess of a cold competitor (wt CRS) but not of a nonspecific competitor (mutant CRS) could compete for DNA binding (Fig. 4C). Consistent with previously published data on the DNA binding of bacterially expressed IE2 (11), we could only observe DNA binding of IE2 in the presence of poly(dA-dT) · (dA-dT) as a nonspecific competitor, whereas addition of poly(dI-dC) · (dI-dC) even at low concentrations completely abolished binding (data not shown). However, in contrast to previously published results (59), we did not observe a dependency on dephosphorylating IE2 prior to the DNA-binding assay (data not shown).
FIG. 4.
IE2 produced in transiently transfected HeLa cells specifically binds to the CRS element. DNA-binding of IE2 as assayed by electro mobility shift assays (EMSA; F = free probe; * = supershift after antibody incubation) (A) Autoradiograph image of an EMSA experiment with cell extracts derived from transiently transfected HeLa cells using wild-type (wt) and mutant (mut) probes of the CRS. HeLa cells were transfected with an IE2 expression plasmid or the corresponding control vector pSG5 as indicated. 24 h post transfection cells were harvested and extracts prepared by sonication of cells as described in Materials and Methods. 20 μg of protein extracts were used for the binding reaction. (B) IE2 is physically present in the retarded protein/DNA complex. The EMSA was performed as described in Materials and Methods. Where indicated, 0.5 μg of the indicated antibodies (AB) were added to the reaction mixture prior to addition of the probe. (C) IE2 specifically binds to the CRS binding site. The EMSA was performed as described in Materials and Methods. The specificity of the binding was analyzed by adding the indicated excess of cold specific (wt CRS) or nonspecific (mut CRS) competitor DNAs. The image only shows the retarded IE2/DNA complexes.
In order to allow the detection of IE2 DNA binding in HCMV-infected cells, we next turned to U373 cells. First, we analyzed extracts from IE2-transfected cells and found the same binding characteristics of IE2 as seen with HeLa cell extracts (Fig. 5A). However, the signal was somewhat weaker, which most likely reflected the lower transfection rate of U373 cells, resulting in an estimated fivefold-lower expression level of IE2 in U373 cells (data not shown).
FIG. 5.
IE2 produced in HCMV-infected U373 cells can specifically bind to the CRS element. Autoradiograph image of an EMSA performed with cell extracts derived from either IE2-transfected or HCMV-infected U373 cells using wt and mut CRS probes. Where indicated an IE-specific antibody was added to the reaction mixture prior to the incubation with the labeled probe resulting in a supershifted IE2/DNA complex (*). (A) U373 cells were transfected as described in Materials and Methods and harvested 46 h later. The EMSA was performed as described in Fig. 4. (B) U373 cells were infected with HCMV as described in Materials and Methods and harvested at 72 hpi. Cell extracts were prepared and the EMSA was performed as described in Fig. 4.
We then asked whether IE2 DNA binding could also be detected under these conditions in extracts from HCMV-infected cells. As shown in Fig. 5B, the virally produced IE2 was also found to bind to the CRS with the same specificity as IE2 derived from transfected U373 or HeLa cells. The slightly drawn-out appearance of the complex might have been due to the fact that the infected cell extracts contained IE2 variants like the IE2 40-kDa protein (data not shown), which is known to retain DNA-binding activity. Alternatively, the complex might have contained higher-order DNA-IE2 complexes or other viral proteins in addition to IE2. Importantly, however, an IE-specific antibody resulted in a supershift of the entire signal (lane 3), indicating that immunoreactive IE2 is part of each retarded DNA-protein complex. Thus, our experimental conditions used to analyze IE2-DNA interactions in transfected HeLa and U373 cell extracts appeared to faithfully reflect the IE2-binding characteristics found in infected cell extracts.
The DNA-binding activity of IE2 requires an intact C-terminal core but is not necessary for the cell cycle arrest function.
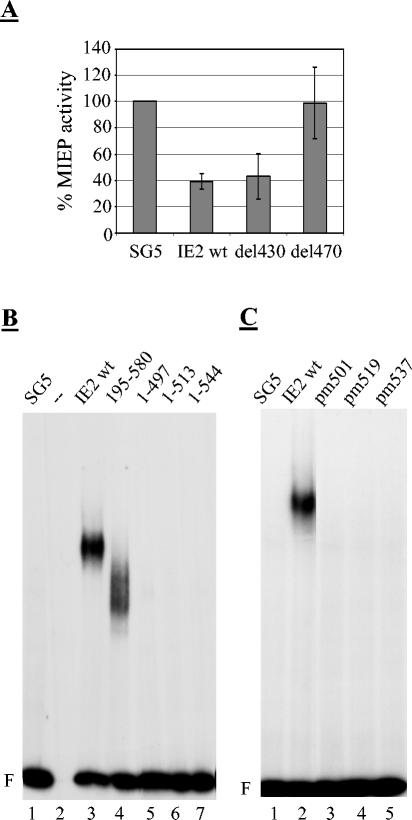
We next wished to correlate IE2-DNA binding with the other activities of IE2 characterized in the previous experiments, particularly the cell cycle regulatory and the autorepression functions of IE2. Given the comparable quality of IE2-DNA binding in HeLa and U373 cells, we decided to perform the experiments in HeLa cells, which showed the stronger IE2-DNA interaction. Importantly, the autoregulatory activity of IE2 was readily observed in HeLa cells (Fig. 6A), which was consistent with previous results obtained in nonpermissive cells (44, 57). Also, the IE2 mutants del430 and del470 led to similar results in HeLa cells (columns 4 and 5) and U373 cells (Fig. 3E), demonstrating that the preserved autorepressor activity of del430 is not specific for U373 cells and suggesting that HeLa cells can be used to correlate DNA binding and autoregulation by IE2 and its mutant derivatives. We found that the smallest truncation of the C terminus impinged on the ability of IE2 to bind to the CRS (Fig. 6B, lanes 5 to 7), which is consistent with previous studies performed in yeast (1) or with bacterially expressed IE2 (11). In contrast, deletion of the first 195 aa from the N terminus of IE2 resulted in a protein that could still bind to the CRS (lane 4). Since IE2 (1 to 544), which was defective in DNA binding, was still capable of blocking cell cycle progression (Fig. 1), these results show that the cell cycle regulatory activity of IE2 does not depend on its ability to bind DNA. In agreement with this deletion analysis, the amino acid exchange mutants resulted in a complete loss of DNA binding, indicating that the integrity of the core is also a prerequisite for DNA binding (Fig. 6C). This is consistent with the structural requirements for autoregulation (Fig. 2).
FIG. 6.
The cell cycle arrest activity of IE2 does not require DNA binding. (A) Autorepression of the MIE promoter/enhancer by IE2 in HeLa cells. HeLa cells were transfected with the indicated effector plasmids and a MIE promoter/enhancer reporter construct. 24 h post transfection cells were harvested and promoter activity was determined as described in Materials and Methods and Fig. 2. (B) DNA binding activity of N- and C-terminal deletion mutants. Autoradiograph image of an EMSA with cell extracts derived from transiently transfected HeLa cells. Cells were transfected with expression plasmids as indicated. After 24 h, cells were harvested and cell extracts were prepared as described in Materials and Methods. The EMSA was performed as described in Fig. 4. (C) DNA binding activity of amino acid exchange mutants.
Sequences in addition to the core domain contribute to DNA binding by IE2, but the putative zinc finger is dispensable.
We next examined the IE2 mutants with internal deletions in the region adjacent to the core for their ability to bind to the CRS in transfected cell extracts. Previous studies had given somehow conflicting results with respect to sequence requirements in this region. Whereas in one study the C-terminal region from aa 346 to 579 had been shown to be necessary and sufficient for DNA binding in a glutathione S-transferase (GST)-IE2 fusion protein (11), an internal deletion mutant eliminating aa 313 to 346 was found not to bind to DNA in a yeast one-hybrid assay (1). More consistently, point mutations of both the two cysteine (5, 26) or histidine (38) residues within the putative zinc finger of IE2 abolished DNA binding, indicating the need for integrity of this element.
As pointed out above, all our internal deletion mutants located between aa 290 and 449 were still able to autorepress the MIE promoter-enhancer, indicating that they can also still bind to DNA, and with one exception this is what we found (Fig. 7A). del470, which lies most C terminal in this series of mutants and which was defective in all other activities tested in this study, is also not capable of binding to the CRS (lane 7), again indicating that this mutant still resides within the highly sensitive C-terminal core. In contrast del430, which eliminates the putative zinc finger of IE2, still strongly bound to the CRS (lane 6). This was consistent with the above data, demonstrating that this mutant retains the ability to autorepress (and to transactivate) (Fig. 3). Together these results further support the view that the homogenous C-terminal core per se does not extend N terminally beyond aa 450. Also consistent with this, the adjacent mutant (del 380) has wt activity in DNA binding (lane 5). Unexpectedly however, we found that del330, although retaining full autorepressor activity, displayed a grossly reduced DNA-binding activity (lane 4). However, it should be mentioned that in contrast to any of the series of the C-terminal deletion mutants (Fig. 6), we have consistently noted a remaining but very weak DNA-binding activity of del330 on long exposures of autoradiographs (data not shown). This may indicate that del330 is grossly impaired but not completely unable to bind to DNA under bandshift conditions. At the same time, this result is consistent with the above-mentioned published work demonstrating that a deletion outside the minimal DNA-binding domain can affect the interaction of IE2 with the CRS (1, 11).
FIG. 7.
The IE2 zinc finger region is dispensable for DNA binding. Autoradiograph image of EMSAs performed with cell extracts derived from transiently transfected HeLa cells. Cells were transfected with expression plasmids as indicated and processed as described in Fig. 4. (A) DNA binding activity of the indicated internal deletion mutants (B) Sequence comparison of wt IE2, a prototype zinc finger point mutant (IE86HL) (38, 63) and del430. The strategy (arrows) and point mutated (IE86HL) or deleted (IE2del430) residues (shaded boxes) are indicated. (C) DNA binding activity of del430 and IE86HL compared to their respective maternal wild-type proteins (IE2wt and IE86).
How can one reconcile the seemingly contradictory results of del430 and the zinc finger point mutants in DNA binding and autorepression? In Fig. 7B, the amino acid sequence of del430 is compared to that of the well-established point mutant IE86HL (38, 63). Whereas in the del430 used in this study the putative zinc finger between the outermost cysteine and histidine residues is nearly completely excised in the prototypic zinc finger mutant, the two histidine residues are exchanged for leucines. This suggested to us that the amino acid exchanges within IE86HL may exhibit a dominant constraint on the protein rather than reflecting the absolute requirement of this structural feature. This notion is consistent with the result of a direct comparison of the DNA-binding activities of del430 and IE86HL. Figure 7C confirms that when these two mutants were analyzed in parallel using the same experimental system, both of them behaved as previously described. This result showed that the differences in DNA binding between del430 and IE86HL are real and do not depend on the experimental systems in use. More importantly, however, the results obviously show that the putative zinc finger is indeed required neither for DNA binding nor for autoregulation, transactivation, or cell cycle regulation by IE2.
DISCUSSION
The mutational analysis of the functionally important but ill-defined, large C-terminal domain of IE2 presented here aimed at determining sequence requirements and structural interdependencies for distinct IE2 functions, namely, transactivation, autorepression, and cell cycle regulation.
We also included in our studies an analysis of the DNA-binding activity of IE2 to the CRS, which we were able to demonstrate by using extracts from IE2-transfected and also from HCMV-infected cells. As outlined in the introduction, DNA binding of IE2 has so far typically been analyzed by GST fusion proteins expressed in and purified from bacteria (11, 30, 31, 37, 38, 59). Only more recently has one study reported on a weak interaction of IE2 with the CRS, where IE2 was purified from transfected nonpermissive human lung cancer cells via a heparin-Sepharose column (25). Other studies have suggested that DNA binding was dependent on limiting posttranslational modifications (59) and that DNA binding by IE2 could only be observed in yeast (1) or cellular extracts that were phosphatase treated (59). However, our study does not confirm these limiting requirements for the IE2-DNA interaction. There are several reasons why, in contrast to previous studies, we were able to show a strong interaction of IE2 with DNA. First, extracts were prepared in a low-salt buffer by sonication of cells using an optimized protocol in the absence of detergents. Second, BSA was included in the binding reaction, and that has been shown to stabilize protein-DNA interactions of minor groove binding proteins like TBP (27, 33). Third, we were using the CRS within an extended 60-bp DNA fragment, which further stabilized protein-DNA interactions.
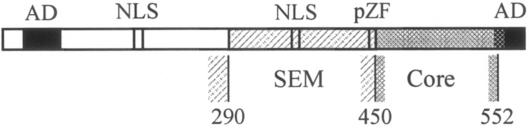
Consistent with previous studies (11, 23, 39, 45, 56, 61, 63), our data demonstrate that IE2 contains a C-terminal domain that plays an essential role in all activities tested. However, by using internal deletion and amino acid exchange mutants, we showed that this domain appears much more concise than previously thought. This region, which we refer to as the “Core” (Fig. 8), is localized between aa 450 and 544. We have not been able to generate any mutation in the core without severely impinging on any of the analyzed IE2 activities. The Core does exclude the furthest C-terminal sequences of the protein. Although the last 37 aa were found to be strictly required for transactivation (45), autorepression (23, 45, 56), and DNA binding (11), they are not essential for cell cycle regulation by IE2. This is an important feature, since it demonstrates that IE2 does not require its DNA binding activity to block cell cycle progression. In addition, it supports the notion that transactivation is not required for the IE2 cell cycle regulatory activity which has previously also been concluded by using an independent transactivation-deficient IE2 mutant (61). The core is likely to extend even a little further to the C terminus but not beyond aa 552, since a mutant with the last 28 aa of IE2 deleted (1 to 552) has been shown to autoregulate and to bind DNA (37). The most C-terminal part of IE2 is likely to form a domain independent of the Core, since the last 35 aa have been shown to contain a transferable transactivation domain (45). Thus, at the C terminus the Core seems to border to an independent transactivation domain that can be physically separated from it without affecting its function.
FIG. 8.
The Core and SEM domains of the IE2 C terminus. The C terminus of IE2 contains a concise core domain (Core) that is invariably required for cell cycle arrest, transactivation, autorepression and in vitro DNA binding. C-terminally the Core extends at least up to aa 544 but more likely up to aa 552. The most C-terminal sequences are essential for transactivation only and the last 34 amino acids have been shown to contain an independent transactivation domain (AD) (45) The N-terminal border of the Core that excludes the putative zinc finger (pZF) lies between aa 450 and 470. Outside the core domain, the C terminus of IE2 is less complex. Within the “Specific and Essential Modulator” (SEM) distinct activities of IE2 appear to have different sequence requirements but in this study no single mutation was identified that abolishes any of the IE2 activities examined (see text). C-terminally the SEM borders to the Core. On the N-terminal side, the border is as yet unclear. The extreme N-terminal sequences (aa25-85) are again only involved in transactivation, whereas for cell cycle arrest, autorepression and DNA binding aa 1-195 are dispensable.
The N-terminal border of the Core resides between aa 450 and 470. Although the Core appears to be nonsufficient to perform any of the IE2 functions independently of its N-terminally flanking part, there is a clear difference between these two regions. In contrast to the core, the N-terminally flanking region tolerates well “internal” manipulations at various positions (Fig. 2), suggesting that these two regions can be considered two separate functional and/or structural components of IE2. In support of this view, different mutations within the flanking region affect distinct IE2 activities, which contrasts with the findings for the core, where all mutations severely affected all functions tested. For instance, del330 was grossly impaired in its DNA-binding capacity and only moderately affected in its transactivation function but retained full cell cycle regulatory and autorepressor functions. Deletion of aa 260 to 279 impaired transactivation and cell cycle regulatory activities of IE2 without affecting autorepression. These results show that different activities of IE2 have different sequence requirements within this region. However, none of the internal deletions appear to completely eliminate any of the IE2 functions (see the discussion of del330, below). Therefore, we consider this region flanking the core at its N terminus as a specific and essential modulator (SEM) for the various activities of IE2 (Fig. 8).
None of the internal deletion mutants within the SEM impaired the ability of IE2 to autorepress the MIE promoter-enhancer. Since DNA binding is considered a prerequisite for autorepression, it was somewhat unexpected to find a strongly impaired DNA-binding activity associated with del330 that on the other hand perfectly well autorepressed the MIEP. As stated above, we consistently saw a very weak interaction between del330 and the CRS in in vitro bandshift assays, but the retarded complex was diffuse rather than a well-defined signal, as obtained with the full-length IE2 protein. Importantly, the del330 autorepressor activity still depended on the integrity of the CRS (data not shown). This leaves two possible explanations. On the one hand, del330 may be sufficient to bind the CRS only when in vivo it is part of a stabilizing complex with cellular factors and/or an intact promoter structure, whereas del330 is grossly impaired to assemble into a sufficiently stable complex with an isolated CRS element in in vitro bandshift assays. Alternatively, del330 might have indeed lost its ability to bind DNA in vivo, too. But in vivo, the DNA-binding domain of IE2 may not be strictly required for autorepression as long as the CRS per se adds a limiting (e.g., a structural) component in cis to facilitate the formation of a functional transcriptional repressor complex that in the presence of del330 is stabilized purely on the basis of protein-protein interactions. Given our aforementioned observation of a weak DNA interaction of del330, we favor the first possibility. This would also be reminiscent of other minor groove-binding proteins, like TBP (for review, see reference 16). A TBP-DNA interaction cannot readily be observed in DNA bandshift experiments in vitro, although the TATA box is a rate-limiting DNA-binding element for transcription in vivo. On the other hand, a TFIID complex, which besides TBP contains several associated factors, is significantly more stable on DNA. Thus, our data are most consistent with the view that the 330-349 element adds a possibly structural component to IE2 that is nonessential in vivo but necessary for detecting a stable IE2-CRS interaction in vitro. This view is further supported by the finding that those sequences deleted in the del330 mutant are outside the minimal DNA-binding domain defined with GST-IE2 fusion proteins (11). Yet, when aa 313 to 346 were deleted from the full-length protein, this mutant was unable to bind to the CRS in a yeast one-hybrid assay (1). Together these findings support the view that the 330-349 element is not absolutely essential for DNA binding but rather modulates it under certain conditions.
In the same studies mentioned above (1, 59), some IE2 mutants were described that lacked sequences overlapping with deleted regions in the mutants used here. For instance, Ahn et al. found that dimerization of IE2 requires sequences between aa 388 and 404 and consequently mutants lacking this region neither bound to DNA nor were autorepressed (1). In the work presented here, however, a deletion lacking aa 380 to 399 bound DNA perfectly well and also autorepressed. Generally speaking, our analysis suggests that the region corresponding to the SEM, but not the core, might be less vulnerable than previous studies suggested (1, 59). A possible explanation for the observed differences could be the different experimental systems used. We performed all our experiments in U373-MG cells, which are HCMV permissive, and HeLa cells, whereas in the other studies either (monkey kidney-derived) Vero cells, yeast, or bacterially derived GST-IE2 fusion proteins were used. Posttranslational modifications (apparently required for the functionality of IE2) and cofactors are likely to differ considerably between these systems and hence may explain the observed differences. In addition, although less likely, it cannot be excluded that the minor differences in the primary sequence of the IE2 proteins derived from AD169 (used here) and Town (used in the other studies) might be responsible for the observed differences. The view of an invariably required concise core and its adjacent more-variably organized SEM is also supported by the most recent analysis of IE2 mutants in a viral background. There, an IE2 mutant deleted for aa 356 to 359 was significantly less affected in transactivation than a mutant lacking aa 501 to 511 (60).
del260 showed a reduction in transactivation and cell cycle arrest without affecting autorepression. This mutation lies within a region of IE2 that has been shown to be highly phosphorylated (21). It contains eight consecutive serine residues, whereas IE2 derived from the Town strain contains only seven serines in this region. The additional serine has been implicated in an increased transcriptional activity of IE2 derived from AD169 (3). Therefore, it is not surprising that a nearly complete deletion of this serine cluster per se will influence the transactivation capacity. Also, these results are consistent with previous data obtained with the internal deletion mutant delta 136 to 290 (24, 52). The del260 to 279 was also affected in the cell cycle arrest function. This was somewhat surprising, since a larger deletion reaching from aa 136 to 290 did not impair cell cycle arrest function (61). This may be explained if this region were to influence the IE2 overall structure rather than primarily add a specific sequence requirement to the cell cycle arrest activity of IE2. Thus, in a dominant manner, deletion of a smaller fragment (aa 260 to 279) could impose a greater negative effect than deleting a larger fragment of IE2. Consistent with this, Harel et al. reported that mutating threonine 233 and serine 234 had a complex effect on the overall phosphorylation of IE2, possibly altering the conformation of the protein (21). Of note, del260 was still able to autorepress the MIE promoter-enhancer. Therefore, this mutation is unlikely to impose a dominant structural constraint on the core but, rather, appears to influence structural features of the SEM itself or of positioning the core and SEM relative to each other.
Particular emphasis has previously been attributed to the putative zinc finger of IE2 that is located between aa 428 and 452. Two different mutants have been described in the past which have either the two cysteine residues (5, 26, 63) or two histidine residues (38, 63) exchanged. Both of these mutants are unable to bind DNA and to autoregulate (5, 26, 38), although some transactivating activity may have been retained (5, 26). In the del430 mutant used here, the putative zinc finger has nearly been completely deleted (Fig. 7) and, rather surprisingly, this mutant still autoregulates, still binds DNA, and also is able to transactivate both the c-fos and the UL112/113 promoters. At the same time, however, we could also confirm that the prototype zinc finger mutant (IE86HL [38, 63]), which was tested in parallel in our DNA-binding assay, could not bind to the CRS anymore. This shows that the alteration of the zinc finger by point mutations has a greater effect on IE2 function than deleting the entire element, suggesting that the point-mutated zinc finger imposes an intramolecular dominant constraint on IE2. Alternatively, the zinc finger may be part of a switchable protein domain that negatively regulates DNA binding of IE2. In that case, destroying the switch by a point mutation could have the opposite effect of deleting the regulatory domain. Anyway, more importantly the analysis showed that the zinc finger per se is neither required for DNA binding nor for autoregulation or transactivation. Consistent with this conclusion, this motif is not well conserved among IE2 proteins from different CMVs (11). Thus, these data further support the view that the invariably required core does not reach N terminally beyond aa 450 and that it can be clearly separated from the SEM. Most recently, the putative zinc finger region has also been mutated in the viral context by deleting the left-hand cysteine residues plus their intervening sequences. This approach most closely resembles one of the aforementioned amino acid exchange mutants (26, 63) within this region, and the deletion was indeed found to have profound effects on IE2 functions (60). However, the data presented here suggest that the observed loss of function likely results from an intramolecular constraint and that it does not necessarily indicate the specific requirement of this region for IE2 function.
In order to address the physiological significance of distinct IE2 functions, it should be most helpful to first delineate discriminating sequence requirements, to allow the introduction of informative mutations of IE2 into the viral background. The Core appears to be of utmost importance for any of the IE2 functions tested here, and our data suggest that the invariably required region is a lot more concise than previously thought. In order to identify discriminating loss-of-function mutations of IE2, a systematic high-density mutational approach of the invariably required region may be necessary and, by focusing on the Core, such an approach should be a manageable undertaking.
Acknowledgments
We wish to thank Thomas Stamminger and John Sinclair for reagents.
This work was supported by the DFG Klinische Forschergruppe “Pädiatrische Molekularbiologie” (TP2) to C.H. (GA167/6-2).
REFERENCES
- 1.Ahn, J. H., C. J. Chiou, and G. S. Hayward. 1998. Evaluation and mapping of the DNA binding and oligomerization domains of the IE2 regulatory protein of human cytomegalovirus using yeast one and two hybrid interaction assays. Gene 210:25-36. [DOI] [PubMed] [Google Scholar]
- 2.Arlt, H., D. Lang, S. Gebert, and T. Stamminger. 1994. Identification of binding sites for the 86-kilodalton IE2 protein of human cytomegalovirus within an IE2-responsive viral early promoter. J. Virol. 68:4117-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrasa, M. I., N. Harel, Y. Yu, and J. C. Alwine. 2003. Strain variations in single amino acids of the 86-kilodalton human cytomegalovirus major immediate-early protein (IE2) affect its functional and biochemical properties: implications of dynamic protein conformation. J. Virol. 77:4760-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boldogh, I., S. AbuBakar, and T. Albrecht. 1990. Activation of proto-oncogenes: an immediate early event in human cytomegalovirus infection. Science 247:561-564. [DOI] [PubMed] [Google Scholar]
- 5.Bresnahan, W. A., T. Albrecht, and E. A. Thompson. 1998. The cyclin E promoter is activated by human cytomegalovirus 86-kDa immediate early protein. J. Biol. Chem. 273:22075-22082. [DOI] [PubMed] [Google Scholar]
- 6.Bryant, L. A., P. Mixon, M. Davidson, A. J. Bannister, T. Kouzarides, and J. H. Sinclair. 2000. The human cytomegalovirus 86-kilodalton major immediate-early protein interacts physically and functionally with histone acetyltransferase P/CAF. J. Virol. 74:7230-7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castillo, J. P., A. D. Yurochko, and T. F. Kowalik. 2000. Role of human cytomegalovirus immediate-early proteins in cell growth control. J. Virol. 74:8028-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caswell, R., C. Hagemeier, C. J. Chiou, G. Hayward, T. Kouzarides, and J. Sinclair. 1993. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J. Gen. Virol. 74:2691-2698. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C., O. H. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherrington, J. M., E. L. Khoury, and E. S. Mocarski. 1991. Human cytomegalovirus IE2 negatively regulates alpha gene expression via a short target sequence near the transcription start site. J. Virol. 65:887-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiou, C. J., J. Zong, I. Waheed, and G. S. Hayward. 1993. Identification and mapping of dimerization and DNA-binding domains in the C terminus of the IE2 regulatory protein of human cytomegalovirus. J. Virol. 67:6201-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi, K. S., S. J. Kim, and S. Kim. 1995. The retinoblastoma gene product negatively regulates transcriptional activation mediated by the human cytomegalovirus IE2 protein. Virology 208:450-456. [DOI] [PubMed] [Google Scholar]
- 13.Coligan, J. E., A. M. Kruisbeek, D. H. Margulies, E. M. Shevach, and W. Strober. 1991. Current protocols in immunology. John Wiley & Sons, New York, N.Y.
- 14.Fortunato, E. A., M. H. Sommer, K. Yoder, and D. H. Spector. 1997. Identification of domains within the human cytomegalovirus major immediate-early 86-kilodalton protein and the retinoblastoma protein required for physical and functional interaction with each other. J. Virol. 71:8176-8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furnari, B. A., E. Poma, T. F. Kowalik, S. M. Huong, and E. S. Huang. 1993. Human cytomegalovirus immediate-early gene 2 protein interacts with itself and with several novel cellular proteins. J. Virol. 67:4981-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenblatt, J. 1992. Transcription: riding high on the TATA box. Nature 360:16-17. [DOI] [PubMed] [Google Scholar]
- 17.Hagemeier, C. 1996. Site-directed mutagenesis using a uracil-containing phagemid template. Methods Mol. Biol. 57:45-54. [DOI] [PubMed] [Google Scholar]
- 18.Hagemeier, C., R. Caswell, G. Hayhurst, J. Sinclair, and T. Kouzarides. 1994. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 13:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagemeier, C., S. Walker, R. Caswell, T. Kouzarides, and J. Sinclair. 1992. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J. Virol. 66:4452-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagemeier, C., S. M. Walker, P. J. Sissons, and J. H. Sinclair. 1992. The 72K IE1 and 80K IE2 proteins of human cytomegalovirus independently trans-activate the c-fos, c-myc and hsp70 promoters via basal promoter elements. J. Gen. Virol. 73:2385-2393. [DOI] [PubMed] [Google Scholar]
- 21.Harel, N. Y., and J. C. Alwine. 1998. Phosphorylation of the human cytomegalovirus 86-kilodalton immediate-early protein IE2. J. Virol. 72:5481-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heider, J. A., W. A. Bresnahan, and T. E. Shenk. 2002. Construction of a rationally designed human cytomegalovirus variant encoding a temperature-sensitive immediate-early 2 protein. Proc. Natl. Acad. Sci. USA 99:3141-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermiston, T. W., C. L. Malone, and M. F. Stinski. 1990. Human cytomegalovirus immediate-early two protein region involved in negative regulation of the major immediate-early promoter. J. Virol. 64:3532-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofmann, H., S. Floss, and T. Stamminger. 2000. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 74:2510-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, C. H., and J. Y. Chen. 2002. Identification of additional IE2-p86-responsive cis-repressive sequences within the human cytomegalovirus major immediate early gene promoter. J. Biomed. Sci. 9:460-470. [DOI] [PubMed] [Google Scholar]
- 26.Jupp, R., S. Hoffmann, A. Depto, R. M. Stenberg, P. Ghazal, and J. A. Nelson. 1993. Direct interaction of the human cytomegalovirus IE86 protein with the cis repression signal does not preclude TBP from binding to the TATA box. J. Virol. 67:5595-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jupp, R., S. Hoffmann, R. M. Stenberg, J. A. Nelson, and P. Ghazal. 1993. Human cytomegalovirus IE86 protein interacts with promoter-bound TATA-binding protein via a specific region distinct from the autorepression domain. J. Virol. 67:7539-7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kronschnabl, M., M. Marschall, and T. Stamminger. 2002. Efficient and tightly regulated expression systems for the human cytomegalovirus major transactivator protein IE2p86 in permissive cells. Virus Res. 83:89-102. [DOI] [PubMed] [Google Scholar]
- 29.Lang, D., S. Gebert, H. Arlt, and T. Stamminger. 1995. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J. Virol. 69:6030-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang, D., and T. Stamminger. 1993. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE-1/2 enhancer-promoter. J. Virol. 67:323-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lang, D., and T. Stamminger. 1994. Minor groove contacts are essential for an interaction of the human cytomegalovirus IE2 protein with its DNA target. Nucleic Acids Res. 22:3331-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, G., J. Wu, P. Luu, P. Ghazal, and O. Flores. 1996. Inhibition of the association of RNA polymerase II with the preinitiation complex by a viral transcriptional repressor. Proc. Natl. Acad. Sci. USA 93:2570-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liebermann, P. M., and A. J. Berk. 1991. The Zta trans-activator protein stabilizes TFIID association with promoter DNA by direct protein-protein interaction. Genes Dev. 5:2441-2454. [DOI] [PubMed] [Google Scholar]
- 34.Liu, B., T. W. Hermiston, and M. F. Stinski. 1991. A cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J. Virol. 65:897-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukac, D. M., N. Y. Harel, N. Tanese, and J. C. Alwine. 1997. TAF-like functions of human cytomegalovirus immediate-early proteins. J. Virol. 71:7227-7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukac, D. M., J. R. Manuppello, and J. C. Alwine. 1994. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J. Virol. 68:5184-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macias, M. P., L. Huang, P. E. Lashmit, and M. F. Stinski. 1996. Cellular or viral protein binding to a cytomegalovirus promoter transcription initiation site: effects on transcription. J. Virol. 70:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macias, M. P., and M. F. Stinski. 1993. An in vitro system for human cytomegalovirus immediate early 2 protein (IE2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate early promoter. Proc. Natl. Acad. Sci. USA 90:707-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malone, C. L., D. H. Vesole, and M. F. Stinski. 1990. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J. Virol. 64:1498-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. A. Knipe, P. M. Howley, D. Griffin, R. Lamb, M. Martin, and S. Straus (ed.), Field's virology, Xth ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 42.Murphy, E. A., D. N. Streblow, J. A. Nelson, and M. F. Stinski. 2000. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J. Virol. 74:7108-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noris, E., C. Zannetti, A. Demurtas, J. Sinclair, M. De Andrea, M. Gariglio, and S. Landolfo. 2002. Cell cycle arrest by human cytomegalovirus 86-kilodalton IE2 protein resembles premature senescence. J. Virol. 76:12135-12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pizzorno, M. C., and G. S. Hayward. 1990. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J. Virol. 64:6154-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pizzorno, M. C., M. A. Mullen, Y. N. Chang, and G. S. Hayward. 1991. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J. Virol. 65:3839-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pizzorno, M. C., P. O'Hare, L. Sha, R. L. LaFemina, and G. S. Hayward. 1988. trans-activation and autoregulation of gene expression by the immediate-early region 2 gene products of human cytomegalovirus. J. Virol. 62:1167-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prosch, S., R. Wuttke, D. H. Kruger, and H. D. Volk. 2002. NF-κB—a potential therapeutic target for inhibition of human cytomegalovirus (re)activation? Biol. Chem. 383:1601-1609. [DOI] [PubMed] [Google Scholar]
- 48.Rodems, S. M., C. L. Clark, and D. H. Spector. 1998. Separate DNA elements containing ATF/CREB and IE86 binding sites differentially regulate the human cytomegalovirus UL112-113 promoter at early and late times in the infection. J. Virol. 72:2697-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwartz, R., B. Helmich, and D. H. Spector. 1996. CREB and CREB-binding proteins play an important role in the IE2 86-kilodalton protein-mediated transactivation of the human cytomegalovirus 2.2-kilobase RNA promoter. J. Virol. 70:6955-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz, R., M. H. Sommer, A. Scully, and D. H. Spector. 1994. Site-specific binding of the human cytomegalovirus IE2 86-kilodalton protein to an early gene promoter. J. Virol. 68:5613-5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sinclair, J., J. Baillie, L. Bryant, and R. Caswell. 2000. Human cytomegalovirus mediates cell cycle progression through G1 into early S phase in terminally differentiated cells. J. Gen. Virol. 81:1553-1565. [DOI] [PubMed] [Google Scholar]
- 52.Sommer, M. H., A. L. Scully, and D. H. Spector. 1994. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J. Virol. 68:6223-6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song, Y. J., and M. F. Stinski. 2002. Effect of the human cytomegalovirus IE86 protein on expression of E2F-responsive genes: a DNA microarray analysis. Proc. Natl. Acad. Sci. USA 99:2836-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spector, D. H. 1996. Activation and regulation of human cytomegalovirus early genes. Intervirology 39:361-377. [DOI] [PubMed] [Google Scholar]
- 55.Stenberg, R. M. 1996. The human cytomegalovirus major immediate-early gene. Intervirology 39:343-349. [DOI] [PubMed] [Google Scholar]
- 56.Stenberg, R. M., J. Fortney, S. W. Barlow, B. P. Magrane, J. A. Nelson, and P. Ghazal. 1990. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J. Virol. 64:1556-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsai, H. L., G. H. Kou, F. M. Tang, C. W. Wu, and Y. S. Lin. 1997. Negative regulation of a heterologous promoter by human cytomegalovirus immediate-early protein IE2. Virology 238:372-379. [DOI] [PubMed] [Google Scholar]
- 58.van den Heuvel, S., and E. Harlow. 1993. Distinct roles for cyclin-dependent kinases in cell cycle control. Science 262:2050-2054. [DOI] [PubMed] [Google Scholar]
- 59.Waheed, I., C. J. Chiou, J. H. Ahn, and G. S. Hayward. 1998. Binding of the human cytomegalovirus 80-kDa immediate-early protein (IE2) to minor groove A/T-rich sequences bounded by CG dinucleotides is regulated by protein oligomerization and phosphorylation. Virology 252:235-257. [DOI] [PubMed] [Google Scholar]
- 60.White, E. A., C. L. Clark, V. Sanchez, and D. H. Spector. 2004. Small internal deletions in the human cytomegalovirus IE2 gene result in nonviable recombinant viruses with differential defects in viral gene expression. J. Virol. 78:1817-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiebusch, L., and C. Hagemeier. 1999. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J. Virol. 73:9274-9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiebusch, L., and C. Hagemeier. 2001. The human cytomegalovirus immediate early 2 protein dissociates cellular DNA synthesis from cyclin-dependent kinase activation. EMBO J. 20:1086-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yeung, K. C., C. M. Stoltzfus, and M. F. Stinski. 1993. Mutations of the human cytomegalovirus immediate-early 2 protein defines regions and amino acid motifs important in transactivation of transcription from the HIV-1 LTR promoter. Virology 195:786-792. [DOI] [PubMed] [Google Scholar]
- 64.Yoo, Y. D., C. J. Chiou, K. S. Choi, Y. Yi, S. Michelson, S. Kim, G. S. Hayward, and S. J. Kim. 1996. The IE2 regulatory protein of human cytomegalovirus induces expression of the human transforming growth factor β1 gene through an Egr-1 binding site. J. Virol. 70:7062-7070. [DOI] [PMC free article] [PubMed] [Google Scholar]