Abstract
The aim of this study was to investigate the relationship between lymphocyte P-glycoprotein (P-gp) expression and genotype in vivo and the expression of lymphocyte receptors critical in the life cycle of human immunodeficiency virus type 1 (HIV-1), i.e., CD4, CCR5, and CXCR4. Using flow cytometry to quantify each membrane receptor/transporter, we demonstrate a highly significant correlation between P-gp protein expression and the expression of CXCR4 (rho = 0.874; P < 0.0001). Furthermore, confocal microscopy showed colocalized expression of CXCR4 and P-gp in the lymphocyte membrane. This significant relationship was also apparent at the mRNA level by use of reverse transcription-PCR (rho = 0.61; P < 0.005) and was present in both phytohemagglutinin-stimulated and unstimulated peripheral blood mononuclear cells. Genotypic analysis of the C3435T single-nucleotide polymorphism of P-gp confirmed significantly higher levels of P-gp in C (range, 2.45 to 11.00 relative fluorescence units [RFU])- than in T (range, 0.25 to 5.00 RFU)-homozygous individuals (P = 0.0088; 95% confidence interval [95% CI], 0.7 to 6.3 RFU). An equivalent association between CXCR4 levels and C (range, 12.7 to 44.1 RFU) versus T (range, 3 to 18.9 RFU) genotype was also demonstrated (P = 0.0019; 95% CI, 5.4 to 23.7). Functionally, although these correlates had no impact on HIV-1 production from either X4- or R5-tropic virus, expression correlated significantly with the activity of the HIV-1 protease inhibitor (PI) saquinavir for both P-gp (rho = 0.75; P = 0.0019) and CXCR4 (rho = 0.71; P = 0.0041). This study defines an association between P-gp (expression and genotype) and CXCR4 that may have implications for the selection of viral tropism and the access of drugs to protease for specific tropic types. The interplay between these two proteins may also influence the viral genotypes which escape effective chemotherapy and which therefore have the opportunity to evolve resistance to PIs.
A number of G protein-coupled CC and CXC chemokine receptors have been shown to act as human immunodeficiency virus (HIV-1) coreceptors in vitro (47, 48). CCR5 and CXCR4 are the major HIV-1 coreceptors in vivo (46). The selective use of the CCR5 and/or CXCR4 coreceptor is the predominant determinant of cellular tropism observed for different HIV-1 isolates (3, 7). CCR5 is the principal coreceptor for primary and early infection (R5 isolates). The appearance of variants that use CXCR4 or both coreceptors (X4 and R5X4 isolates) results in accelerated CD4+ T-cell loss and disease progression (6, 36), and evidence suggests that patients with higher expression of CXCR4 in lymphocytes acquire X4-tropic strains of virus more rapidly (24).
The introduction of protease inhibitors (PIs) has dramatically improved the prognosis for HIV infections. However, PIs such as saquinavir (SQV) have a variable and frequently low bioavailability (29). High dosages are often required; this has been attributed to the actions of both cytochrome P450 3A4 (8, 17) and P glycoprotein (P-gp) (15, 31). The effect of P-gp on bioavailability, combined with its expression at certain sanctuary sites such as the brain, testes, and lymphocytes, may enhance the development of PI-resistant strains of HIV.
P-gp is a member of the largest class of membrane transport proteins, designated the ATP-binding cassette (ABC) superfamily (19). A number of recent studies have also implicated P-gp in the infectivity of HIV (22, 32, 37). P-gp overexpression blocks insertion of the influenza virus fusion protein (hemagglutinin-2) into the plasma membrane (32), and this inhibits membrane fusion and infectivity. Furthermore, HIV-1 infectivity is lower in CD4+ T-cell lines, which overexpress P-gp (22). The authors concluded that P-gp expression inhibited HIV-mediated membrane fusion, as well as a subsequent step(s) in the HIV-1 life cycle. Recently, Speck et al. reported similar data for drug-selected (P-gp-overexpressing) CEM cells; the effect was reversible by verapamil (a known P-gp inhibitor), and the authors speculated that overexpression of P-gp and its localization to lipid rafts may disrupt critical protein-protein interactions because of the physical size and abundance of P-gp (37). Indeed, evidence suggests that CD4 and CXCR4 form clusters within lipid rafts that are necessary for efficient HIV infection (23). However, this does not explain the sensitivity to verapamil, and subsequently, a significant difference in the expression of CD4 and CXCR4 between CEM and drug-selected CEM cells grown in our lab was observed (27, 33).
In addition to these biochemical analyses, information on the relationship between HIV and P-gp has emerged by analysis of MDR1 single-nucleotide polymorphisms (SNPs). The C-to-T transition at position 3435 is the most extensively studied MDR1 SNP. The T allele at this position has been related to better immune recovery (9), and a trend toward virological failure with antiretroviral therapy has been reported for CC homozygotes (4). Furthermore, a nonsignificant association between this SNP and infectivity has also been reported (16). However, these observations are currently subject to substantial debate. Interestingly, the C3435T SNP does not result in an amino acid change, and as such is unlikely to influence P-gp expression directly, but may be linked to other variants that govern expression or mRNA processing.
In order to investigate the complex interactions between P-gp, HIV receptors, viral infectivity, and PI effect, we have undertaken a detailed analysis of HIV-1 receptor and coreceptor expression and of P-gp transporter expression and genotype in peripheral blood mononuclear cells (PBMC) from a cohort of healthy volunteers. Furthermore, their relationship to HIV infectivity and HIV protease inhibitor activity has been investigated.
MATERIALS AND METHODS
Materials.
Viruses HIV-IIIB (X4-tropic) and HIV JRCSF (R5-tropic), MT-4 cells, and CXCR4- and CCR5-specific antibodies were provided by the AIDS Reagent Project of the National Institute of Biological Standards and Controls (South Mimms, United Kingdom). RPMI 1640 medium, chloroform, RNase-free water, Hanks balanced salt solution, and a lectin (phytohemagglutinin [PHA]) were purchased from Sigma Chemical Co. Ltd. (Poole, United Kingdom). Fetal calf serum and Trizol were purchased from Gibco Life Technologies Ltd. (Paisley, Scotland). Immunoglobulin G2a, used as a negative control, and R-phycoerythrin (R-PE)-conjugated goat anti-mouse immunoglobulin G2a were obtained from Serotech Ltd. (Oxford, United Kingdom). Antibody UIC2 was purchased from Immunotech (Marseilles, France). CellFIX was purchased from Becton Dickinson (Oxford, United Kingdom). Isopropyl alcohol and ethanol were obtained from Fisher Scientific (Loughborough, United Kingdom). Saquinavir was a gift from Roche Ltd. (Welwyn, United Kingdom). All TaqMan primers, probes, and master mixes, as well as reverse transcription reagents, were obtained from Applied Biosystems UK (Warrington, United Kingdom), and the miniprep blood kit was purchased from QIAGEN Ltd. (Hilden, Germany). Buffy coats were supplied by the Manchester Blood Transfusion Service (Manchester, United Kingdom). p24 enzyme-linked immunosorbent assay (ELISA) kits were purchased from Bio-Rad (Hertfordshire, United Kingdom). Interleukin-2 (IL-2) was supplied by Peprotech UK Ltd. (Nottingham, United Kingdom), and the P-gp inhibitor XR9576 was a gift from Xenova (Slough, United Kingdom).
HIV stocks.
Viral stocks were obtained from the AIDS Reagent Project as cell-free culture supernatants (1 ml) and expanded by passage through appropriate target cells. HIV-IIIB was added to MT-4 cells (1:10 dilution in RPMI-10% fetal calf serum; cell concentration, 0.5 × 106 · ml−1), and cells were observed microscopically for signs of viral cytopathic effects (CPE), i.e., syncytium formation. When CPE was observed, further MT-4 cells were added (10 ml; 1 × 106 · ml−1), and cultures were incubated (37°C). When the culture showed signs of CPE, it was centrifuged (5 min, 400 × g), and the supernatant fraction was frozen in aliquots until quantification of viral content by p24 ELISA.
To expand HIV JRCSF (which will not grow in laboratory-adapted cell lines), PBMC were used. Cells were isolated from buffy coats and activated with PHA (10 μg · ml−1) prior to resuspension (at 10 × 106 cells · ml−1) in lymphocyte growth medium (LGM, consisting of RPMI 1640 with 15% fetal calf serum) containing 20 IU of IL-2 · ml−1. JRCSF was added (1:10 dilution), and cultures were incubated (37°C, 5% CO2, 7 days). The culture was then centrifuged (5 min, 400 × g), and the supernatant fraction was frozen in aliquots until quantification of viral content by p24 ELISA.
Membrane protein expression in peripheral blood mononuclear cells from healthy donors.
Blood samples (60 ml) were obtained from healthy Caucasian volunteers by venopuncture, and PBMC were isolated as described previously (5). A total of 4 × 106 cells per assay were resuspended in CellFIX for assessment of P-gp, CD4, CXCR4, and CCR5 expression by flow cytometry. RNA extraction and quantification of MDR1, CXCR4, and CCR5 mRNA were performed on RNA from 10 × 106 cells suspended in Trizol. Whole blood (200 μl) was also frozen for DNA extraction and genotyping for the C3435T polymorphism in exon 26 of MDR1.
Infection of activated peripheral blood mononuclear cells with HIV-1.
To assess the relationship between membrane protein expression and the sensitivity of cells to infection, PBMC were isolated from the buffy coat or healthy donor PBMC and activated with PHA. Cells (4 × 106) were assayed for membrane protein expression by flow cytometry. Whole blood (200 μl) was kept for DNA extraction and C3435T genotyping. Cells were resuspended in LGM containing IL-2 (20 IU · ml−1) in the presence or absence of the potent and selective P-gp inhibitor XR9576 (80 nM) (25). HIV-IIIB (X4-tropic) or HIV JRCSF (R5-tropic) was added at a multiplicity of infection (MOI) of 0.001. Cells were then cultured at 37°C under 5% CO2 for 7 days, after which the supernatant fraction was assayed for viral production by p24 ELISA.
Calculation of the 50% inhibitory concentration (IC50) of saquinavir against HIV replication in peripheral blood mononuclear cells.
PBMC were isolated from buffy coat samples as described above and resuspended in LGM containing PHA (10 μg · ml−1). Following culture (37°C, 5% CO2, 72 h), cells were washed, and the cell density was adjusted to 2 × 106 · ml−1. Cells were then incubated with a range of SQV concentrations from 0 to 84 nM. IL-2 was present in the culture at 20 IU · ml−1 throughout. HIV-IIIB was then added to each well (MOI, 0.001), and negative controls were mock infected with LGM. Following incubation (37°C, 5% CO2, 24 h), cells were washed, resuspended in LGM containing IL-2, and incubated at 37°C under 5% CO2 for 6 days. The supernatant fraction was then assayed for viral content by p24 ELISA.
Flow cytometric analysis.
Flow cytometric analysis of P-gp, CXCR4, CCR5, and CD4 was carried out as previously described (10, 27). The fluorescence of the cells was plotted against the number of events, and the data were registered on a logarithmic scale prior to calculation of the median fluorescence. Surface expression was then determined by subtracting the median fluorescence of the isotype control antibody from that of the test antibody. All data are presented in relative fluorescence units (RFU) for each protein.
Confocal laser scanning microscopy.
For experimentation, 106 PBMC were incubated at room temperature for 1 h with a CXCR4-specific monoclonal antibody (12G5) or, as a negative control, with a CD11a-specific monoclonal antibody. Cells were then washed three times with Hanks balanced salt solution prior to incubation for 1 h with a fluorescein isothiocyanate-conjugated goat anti-mouse secondary antibody. Following a further three washes, cells were incubated with a P-gp-specific monoclonal antibody (UIC2) directly conjugated to PE. For microscopy, cells were adhered to coverslips previously coated with poly-l-lysine and were loaded onto a Bioptechs perfusion chamber maintained at 37°C. The cells were scanned on a Zeiss LSM Pascal confocal microscope in multichannel mode. For fluorescein isothiocyanate, fluorescence (green pseudocolor) was excited by using the 488-nm line of an argon laser and collected from an NFT 545 dichroic mirror through a 505- to 530-nm band-pass filter. For PE, fluorescence (red pseudocolor) was excited by using the 543-nm line of a helium/neon laser and collected from an NFT 545 dichroic mirror through a 560-nm long-pass filter. Laser intensity was less than 2% at all times.
mRNA quantification by real-time reverse transcription-PCR.
Quantification of mRNA transcripts for MDR1 and CXCR4 was achieved by real-time PCR using the ABI PRISM 7000 sequence detection system. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the housekeeping gene. Forty nanograms of cDNA was combined with Universal master mix, sense and antisense primers (0.4 μM each), and an oligonucleotide probe (0.2 μM) in a final volume of 20 μl. Amplification was carried out for 40 cycles with a combined annealing-extension temperature of 60°C. Primers and probes were obtained via the Assays-on-Demand (MDR1 and GAPDH) and Assays-by-Design (CXCR4) gene expression products available through the Applied Biosystems website.
C3435T genotypic analysis.
C3435T genotyping was carried out using previously validated primers and probes (home.appliedbiosystems.com). Briefly, DNA was isolated by using a QIAamp DNA mini kit and combined with the TaqMan Universal master mix and primer-probe mix. Amplification was carried out for 40 cycles with a combined annealing-extension temperature of 60°C. An allelic discrimination protocol was then carried out on an ABI PRISM 7000 sequence detection system.
Statistical analysis.
For comparison of P-gp and receptor expression, statistical analysis was carried out by Spearman's rank correlation. For comparisons between genotypes, data are presented as scatter graphs with a bar at the median. Statistical analyses were carried out by one-way analysis of variance. In all cases a P value of <0.05 was considered indicative of significance.
RESULTS
Expression of CD4, CXCR4, CCR5, and P-gp on peripheral blood mononuclear cells isolated from healthy volunteers.
Data on correlations between CD4, CXCR4, CCR5, and P-gp on PBMC from healthy donors are summarized in Table 1. P-gp expression on healthy donor PBMC (n = 21) was 2.75 RFU (range, 0.25 to 11.00 RFU). CXCR4 expression was 11.2 RFU (range, 3.00 to 44.10 RFU), and CCR5 expression was 0.23 RFU (range, 0.010 to 0.48 RFU). CD4 expression on PBMC of healthy volunteers was 0.11 RFU (range, 0.01 to 5.58 RFU).
TABLE 1.
Correlations between proteins and mRNA species analyzed in this study and expression in different MDR1 genotypes
| Species | Correlation with the following protein
|
Expressiona in the following MDR1 genotype:
|
||||
|---|---|---|---|---|---|---|
| CXCR4 | CCR5 | CD4 | CC | CT | TT | |
| P-gp | ||||||
| Protein | r = 0.87b | r = −0.22c | r = 0.44d | 4.2 | 2.7d | 1.35e |
| mRNA | r = 0.61f | NDg | ND | 1.45 | 0.97c | 0.9c |
| CXCR4 | ||||||
| Protein | r = −0.1h | r = −0.28h | 19.1 | 9.1f | 5.7f | |
| mRNA | ND | ND | 1.69 | 1.23c | 0.7f | |
| CCR5 | ||||||
| Protein | r = 0.47i | 0.25 | 0.1c | 0.12c | ||
| mRNA | ND | ND | ND | ND | ||
| CD4 | ||||||
| Protein | 0.3 | 0.25c | 0.3c | |||
| mRNA | ND | ND | ND | |||
In median RFU for protein and arbitrary units relative to GAPDH expression for mRNA.
P < 0.0001.
P > 0.05.
P < 0.05.
P < 0.01.
P < 0.005.
ND, not determined.
P > 0.5.
P < 0.5.
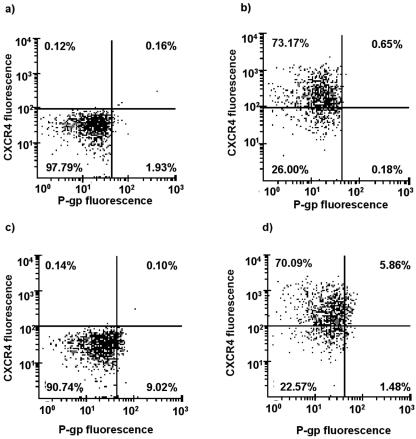
A highly significant positive correlation was observed between CXCR4 receptor expression and P-gp transporter expression (rho = 0.874; P < 0.0001) (Fig. 1a). Representative dual-color scatter plots are shown in Fig. 2, illustrating that approximately 80% of cells staining positive for P-gp also stain positive for CXCR4. There was no equivalent correlation between expression of CCR5 and P-gp (rho = −0.22; P = 0.334) (Fig. 1b). A weak correlation was observed between expression of CD4 and P-gp (rho = −0.439; P = 0.048) (Fig. 1c) and between CD4 and CCR5 expression (rho = 0.47; P = 0.032) (Fig. 1d). Analysis of P-gp and CXCR4 was repeated following activation of PBMC with PHA. Expression of P-gp on activated PBMC was 1.35 RFU (range, 0.00 to 2.60 RFU; n = 36), CXCR4 expression was 23.49 RFU (range, 3.63 to 71.96 RFU; n = 35), and CCR5 expression was 0.09 RFU (range, 0 to 0.39 RFU; n = 23). P-gp expression correlated positively with CXCR4 expression (rho = 0.519; P = 0.0016), but there was again no relationship between expression of P-gp and CCR5 on activated PBMC (rho = 0.059; P = 0.79).
FIG. 1.
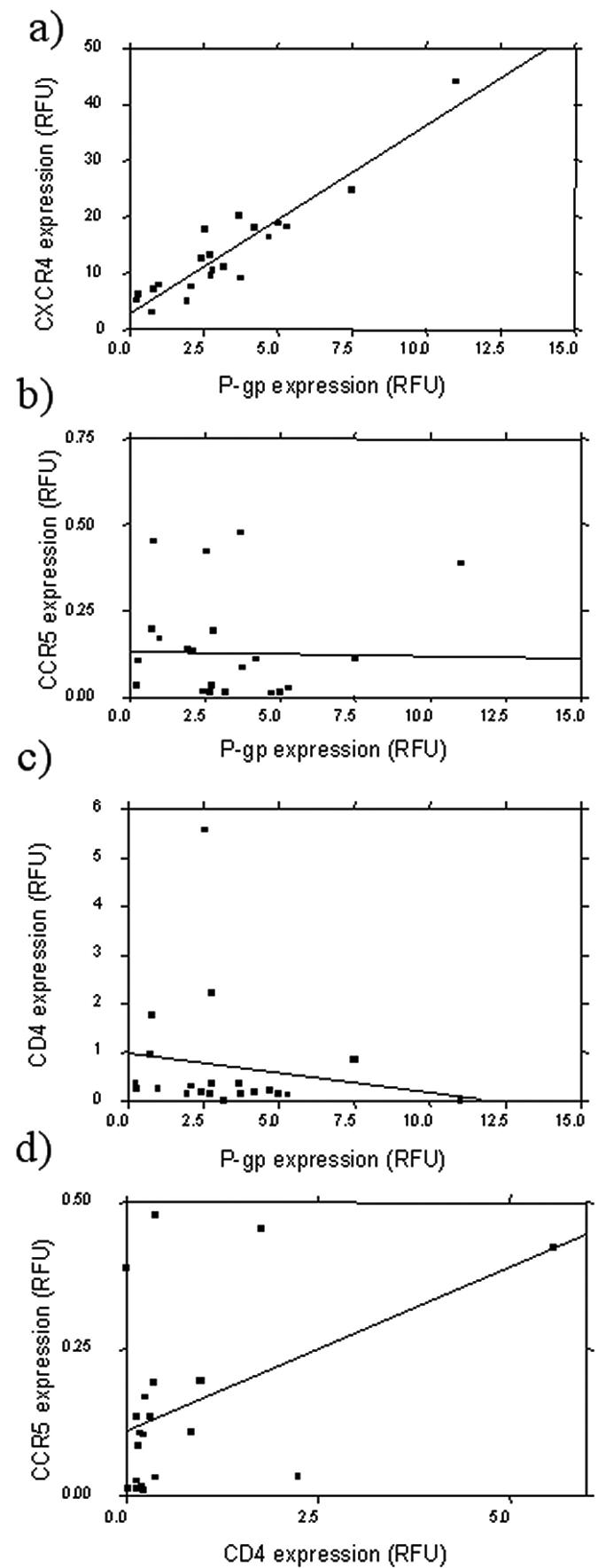
Relationship between P-gp expression and expression of (a) CXCR4, (b) CCR5, and (c) CD4 in PBMC isolated from healthy volunteers. (d) Relationship between CCR5 and CD4 in PBMC isolated from healthy volunteers. Proteins were quantified by flow cytometry, and levels are expressed in relative fluorescence units. Statistical analysis was carried out by Spearman's rank correlation. A statistically significant positive correlation was observed between P-gp and CXCR4 (n = 21; rho = 0.87; P < 0.001) and between CCR5 and CD4 (n = 21; rho = 0.47; P < 0.05). A significant inverse correlation was observed between P-gp and CD4 (n = 21; rho = −0.44; P < 0.05).
FIG. 2.
Representative scatter plots obtained from dual-color flow cytometry of PBMC stained with (a) isotypically matched negative-control antibodies and with (b) CXCR4-specific, (c) P-gp-specific, and (d) P-gp- and CXCR4-specific antibodies. Values in each quartile indicate number of events within the region.
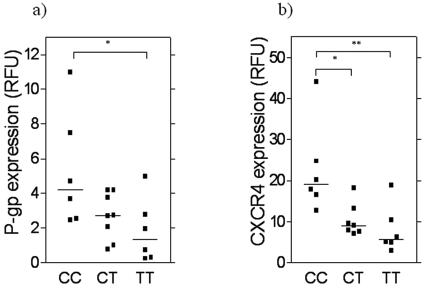
P-gp expression in PBMC isolated from individuals homozygous for the C allele (n = 6), heterozygous (n = 7), and homozygous for the T allele (n = 6) at position 3435 of MDR1 was 4.2 (range, 2.45 to 11.00), 2.7 (range, 0.8 to 4.2), and 1.35 (range, 0.25 to 5.00) RFU, respectively (Fig. 3a). P-gp expression was significantly higher in CC individuals than in CT (P = 0.0195; 95% confidence interval [95% CI], 0.2 to 5.5) and TT (P = 0.0088; 95% CI, 0.7 to 6.3) individuals. When CXCR4 expression on PBMC was assessed in the same genotype groups, values were 19.1 RFU (range, 12.7 to 44.1 RFU) in CC individuals, 9.1 RFU (range, 7.15 to 18.15 RFU) in CT individuals, and 5.7 RFU (range, 3 to 18.9 RFU) in TT individuals (Fig. 3b). Expression was again significantly higher in CC individuals than in CT (P = 0.0045; 95% CI, 3.5 to 21.1) and TT (P = 0.0019; 95% CI, 5.4 to 23.7) individuals.
FIG. 3.
Expression of (a) P-gp and (b) CXCR4 in relation to C3435T genotype in healthy volunteers (n = 6 CC, 7 CT, 6 TT). Expression was determined by flow cytometry and is expressed in relative fluorescence units for each protein and genotype by TaqMan allelic discrimination. Statistical analysis was carried out by one-way analysis of variance (*, P < 0.05; **, P < 0.01).
No significant differences in expression of CCR5 or CD4 were observed between C3435T genotypes (data not shown).
Localization of P-gp and CXCR4 by confocal laser scanning microscopy.
Confocal laser scanning microscopy revealed P-gp and CXCR4 to be expressed on the same cells within the lymphocyte population (Fig. 4b and c). Furthermore, expression of these proteins was colocalized within individual cells (Fig. 4d). This was not the case for P-gp and CD11a (Fig. 4e to h).
FIG. 4.
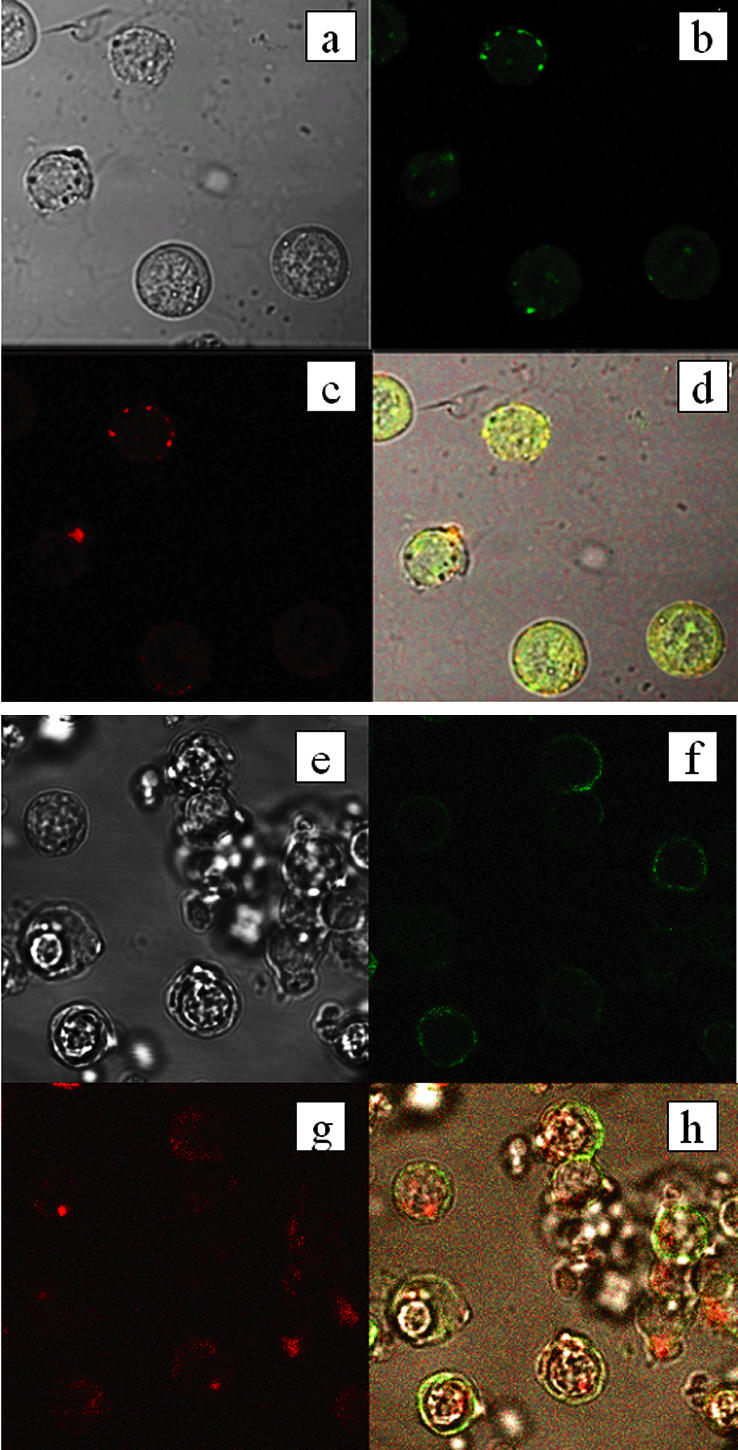
Combined fluorescence and bright-field images of peripheral blood mononuclear cells. (a and e) Bright-field images of collections of cells. (b and c) Localization of fluorescence of anti-CXCR4 and anti-P-gp antibodies, respectively, in the cells shown in panel a. (d) Superimposition of the three images in panels a to c, with areas of colocalization in yellow. (f and g) Localization of fluorescence of anti-CD11a and anti-P-gp antibodies, respectively, in the cells shown in panel e. (h) Superimposition of the three images in panels e to g, with areas of colocalization in yellow.
Relative expression of CXCR4 and MDR1 mRNAs in peripheral blood mononuclear cells isolated from healthy volunteers.
Data on the expression of CXCR4 and MDR1 mRNA in PBMC, expressed relative to GAPDH expression in arbitrary units, are summarized in Table 1. In PBMC isolated from healthy donors (n = 19), MDR1 expression was 1.0 arbitrary unit (range, 0.16 to 2.95) and CXCR4 expression was 1.16 arbitrary units (range, 0.43 to 2.41). A significant positive correlation was observed between the MDR1/GAPDH ratio and the CXCR4/GAPDH ratio (rho = 0.61; P = 0.005) (Fig. 5).
FIG. 5.
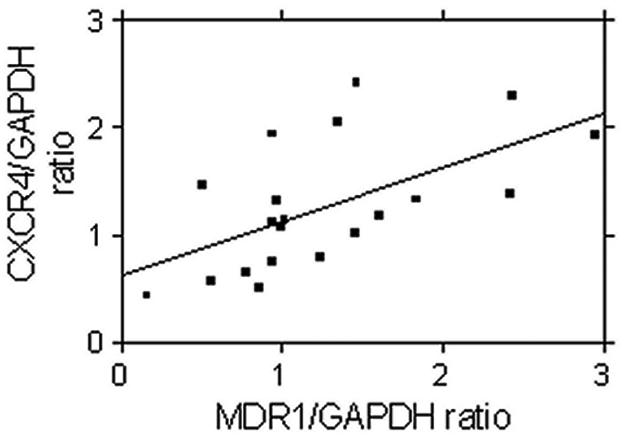
Relationship between MDR1 expression and expression of CXCR4 mRNA in PBMC isolated from healthy volunteers. Transcripts were quantified by real-time reverse transcription-PCR, and statistical analysis was carried out by Spearman's rank correlation. A statistically significant correlation was observed between MDR1 and CXCR4 (n = 21; rho = 0.61; P < 0.005).
MDR1 expression for CC individuals (n = 6), CT individuals (n = 7), and TT individuals (n = 6) was compared. Expression of MDR1 mRNA from individuals with CC, CT, and TT genotypes was 1.45 (range, 0.97 to 2.43), 0.97 (range, 0.51 to 2.95), and 0.90 (range, 0.16 to 2.41) arbitrary units, respectively. CXCR4 mRNA expression was 1.69 (range, 1.02 to 2.41) arbitrary units in PBMC isolated from CC individuals, 1.23 (range, 0.57 to 1.94) arbitrary units for CT individuals, and 0.70 (range, 0.43 to 1.38) arbitrary units for TT individuals. Expression of CXCR4 mRNA in CC individuals was significantly higher than that in TT individuals (P = 0.0027; 95% CI, 0.3 to 1.5).
Relationship between P-gp or chemokine receptor expression and viral replication in activated peripheral blood mononuclear cells.
To determine the influence of membrane proteins on HIV replication, activated PBMC were infected with either HIV-IIIB (X4-tropic) or JRCSF (R5-tropic), and p24 recovery was measured after 7 days. Median HIV-IIIB (n = 24) and HIV JRCSF (n = 24) p24 recoveries were 61,210 (range, 20,040 to 263,400) and 27,704 (range, 3,727 to 2,335,000) pg · ml−1, respectively. No significant correlation was observed between HIV-IIIB p24 recovery and expression of CXCR4 (rho = 0.22; P = 0.30; n = 23) or P-gp (rho = 0.39; P = 0.06; n = 23). Results were similar for HIV-JRCSF p24 recovery compared to expression of CXCR4 (rho = 0.40; P = 0.06; n = 23) and P-gp (rho = 0.29; P = 0.17; n = 23).
When P-gp activity was blocked by addition of XR9576, there was no significant difference between p24 recovery for HIV-IIIB (median, 129,045 [range, 15,782 to 1,351,481] pg · ml−1; P = 0.19) or HIV-JRCSF (median, 32,382 [range, 4,894 to 2,169,319] pg · ml−1; P = 0.24) and p24 recovery in the absence of XR9576.
Relationship between membrane-bound protein expression and the SQV IC50 and IC90 against viral replication in activated peripheral blood mononuclear cells.
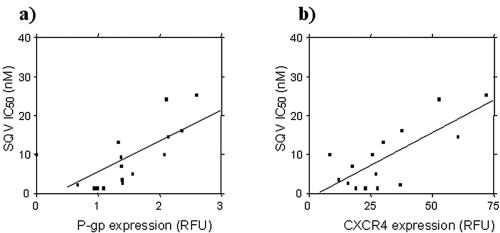
The IC50 and IC90 of SQV against HIV-IIIB in activated PBMC were 8.05 nM (range, 1.3 to 25.2 nM; n = 16) and 11.97 nM (range, 1.3 to 69.18 nM; n = 16), respectively. The SQV IC50 and IC90 correlated significantly with both P-gp expression (for the IC50, rho = 0.83, P = 0.0002, and n = 15; for the IC90, rho = 0.75, P = 0.0019, and n = 15) (Fig. 6a) and CXCR4 expression (for the IC50, rho = 0.57, P = 0.028, and n = 15; for the IC90, rho = 0.71, P = 0.0041, and n = 15) (Fig. 6b) on PBMC.
FIG. 6.
Relationship between expression of (a) P-gp (n = 15; rho = 0.71; P = 0.0002) or (b) CXCR4 (n = 15; rho = 0.57; P = 0.028) and the SQV IC50 against HIV-IIIB in activated PBMC. Proteins were quantified by flow cytometry and are expressed in relative fluorescence units. IC50 was calculated as described in Materials and Methods. Statistical analysis was carried out by Spearman's rank correlation.
DISCUSSION
In this study we have utilized healthy donor PBMC as a model to investigate the effects of P-gp, receptor, and coreceptor expression on HIV susceptibility. In contrast to CEM and CEMVBL cells (27, 33), a strong positive correlation of P-gp and CXCR4 expression on the surfaces of PBMC was observed, with 80% of P-gp-expressing PBMC also expressing CXCR4 (Fig. 2), suggesting limitations to the cell line model. The two proteins were further shown to colocalize on the surfaces of individual cells. This relationship was also observed at the mRNA level, and expression of both P-gp protein and P-gp mRNA was related to the C3435T genotype in the following order (from highest to lowest): CC, CT, TT. This relationship between P-gp expression and genotype is in agreement with the findings of previous studies (9, 14). However, a novel finding was that the coexpression of P-gp and CXCR4 was also reflected in relation to C3435T genotype, with CXCR4 protein and mRNA also expressed in the order (from highest to lowest) of CC, CT, and TT.
The mechanism by which P-gp expression and CXCR4 expression are linked is unclear, although correlation at the level of mRNA suggests that the mechanism may be in part transcriptional. Examination of a 1-kb sequence upstream of the CXCR4 gene indicates putative recognition sites for a number of transcription factors, including AP-1, NF-κB, SP-1, C/EBPβ, and NF-Y. Indeed, NF-κB has recently been shown to regulate CXCR4 via these sequences (13), and all of these proteins have been shown to influence P-gp expression via sequences in the MDR1 promoter. It is possible that interindividual variability in the activity of one or more of these nuclear proteins may account for differential regulation of the two genes under baseline conditions. However, this is speculative, and given the apparent lack of biological plausibility, it is now imperative that the relationship between CXCR4 expression and MDR1 genotype be examined in larger cohorts.
CD4 expression in PBMC reflects the pattern observed in CEM and CEMVBL cells (27, 33), with increased P-gp expression correlating with decreased CD4 expression. However, although this relationship in PBMC was statistically significant, it was not as striking as that observed between P-gp and CXCR4; it appears to be the result of a few outliers expressing extremely high levels of CD4. Indeed, CD4 expression was independent of the C3435T genotype. This finding does, however, suggest that the relationship observed between P-gp and CXCR4 expression is specific to these proteins and is not a reflection of general coordination of membrane surface proteins.
We failed to show a correlation between CCR5 and P-gp expression in the cell membrane. However, as CCR5 expression is known to be low on PBMC (the majority of CCR5 is expressed on macrophages), it cannot be ruled out that in many individuals CCR5 protein expression is below the limit of detection by the assay employed in this study. For this reason, CCR5 expression was not investigated at the mRNA level. We are currently investigating whether a relationship between CCR5 and P-gp exists in monocyte-derived macrophages.
In order to further investigate the implications of membrane-bound protein expression on PBMC for cell-virus interactions, activated PBMC were assessed for expression of P-gp, CXCR4, and CCR5 and were infected with either an X4- or an R5-tropic virus. The protein expression relationships observed in nonactivated PBMC were maintained following activation: a strong correlation was observed between P-gp and CXCR4 expression, and CCR5 expression did not correlate with that of P-gp. These findings again suggest a common regulatory mechanism for P-gp and CXCR4, requiring further investigation.
Following 7 days of infection with HIV-IIIB (X4-tropic) or HIV-JRCSF (R5-tropic), there was large intraindividual variation in the amount of virus produced by PBMC, with a ca. 10-fold difference between the highest and lowest values obtained for HIV-IIIB and a 1,000-fold difference for HIV-JRCSF.
Increased expression of CXCR4 and CCR5 has recently been shown to enhance infection by X4- and R5-tropic strains of HIV (40). Similarly, a weak correlation was observed between either CXCR4 or P-gp expression and HIV-IIIB p24 recovery from PBMC in our study. However, when P-gp function was blocked by addition of XR9576, a potent and specific P-gp inhibitor, no effect was observed on HIV-IIIB p24 recovery, suggesting either that CXCR4 is the determinant of HIV-IIIB recovery and P-gp is merely a “bystander” as a result of coregulation of expression or that P-gp does not act directly on HIV infectivity. This is in contrast to the findings of Speck et al., who showed a reversal of infectivity by use of the P-gp inhibitor verapamil (37). This discrepancy may be explained by the observation that verapamil has numerous effects on cellular processes in lymphocytes. These include effects on Ca2+ signaling, other transporters (2, 11), immunologically important proteins (41, 45), and membrane proteins (43, 44). However, a similar effect was observed by Lee et al. using both quinidine and PSC 833 as P-gp inhibitors (22). JRCSF production also correlated weakly with expression of P-gp and CXCR4, and addition of XR9576 to inhibit P-gp activity had no effect on p24 recovery.
Finally, there was a significant correlation between the expression of P-gp or CXCR4 and the concentration of SQV required to inhibit viral replication in PBMC. Because chemokine receptors are not thought to be involved in drug transport, it would seem likely that the increased IC50 of SQV in cells expressing high levels of membrane-bound proteins was due to decreased intracellular drug concentrations as a result of active efflux by P-gp. In vivo studies have not demonstrated a correlation between P-gp expression on lymphocytes and response to antiviral therapy including a PI (1, 26), although Fellay et al. noted increased immune reconstitution in HIV patients with the TT genotype (and therefore lower P-gp expression) at position 3435 in the MDR1 gene (9). This finding is also in contrast to previous studies reporting no significant differences in the IC90 of either SQV, ritonavir, indinavir, or nelfinavir between P-gp-overexpressing cells and their parental cell line (39). The authors suggested that intracellular drug concentrations would be affected by P-gp efflux only at high extracellular concentrations in excess of those required to inhibit viral replication. Our findings with PBMC suggest this is not the case, but which of these methods more accurately predicts the in vivo scenario is unclear at this time.
Our findings have a number of potential clinical implications. First, the correlation between P-gp expression and the IC50 of saquinavir suggests an important role for this transporter in HIV therapy. Second, since P-gp expression and CXCR4 expression correlate positively, it is possible that the relatively rapid emergence of more pathogenic X4-tropic strains of HIV that has been observed in patients with high lymphocyte CXCR4 expression (24) may be exacerbated by the increased IC50 of SQV against viruses in these cells. That is, in patients on therapy, higher CXCR4 expression may lead to increased CXCR4-dependent infection and concomitant higher P-gp expression, rendering the viruses within these cells less sensitive to drugs. This may facilitate the replication, and thereby speed the emergence, of X4-tropic viruses, a phenomenon that is related to accelerated disease progression.
The role of X4-tropic viruses in hastening disease progression has been the subject of debate but is supported by the clinical observation that X4-tropic strains of virus are temporally associated with a decline in CD4+ T-cell numbers and with AIDS (6, 20, 33, 35, 38, 42). Also, X4 and X4/R5 strains (experimental strains as well as primary isolates) deplete CD4+ cells in vitro (activated PBMC, ex vivo lymphoid tissue, and noninflammatory human spleen tissue), whereas the effects of R5 strains on the CD4/CD8 ratio are much less marked despite similar rates of replication (12, 21, 28, 34). Furthermore, this CD4-selective toxicity is eliminated in these models by compounds that block CXCR4 (34). Finally, in support of our hypothesis, clinical data suggesting that antiretroviral treatment may create an environment for the emergence of CXCR4 tropism are now beginning to emerge (18, 30).
In summary, a correlation between P glycoprotein and CXCR4 expression that influences virus production and the IC50 of saquinavir exists in PBMC. This is likely to be important for established antiretroviral drugs that are known to be substrates for P-gp as well as for new-generation compounds that target these coreceptors as well as fusion. The simultaneous higher levels of P-gp and CXCR4 may therefore have implications for the efficacy of both protease inhibitors and fusion inhibitors when given in combination. Further work to fully characterize this relationship with respect to HIV is now necessary, and studies are currently under way to investigate whether a similar relationship is observed on the surfaces of various cellular subsets and on PBMC isolated from HIV-positive individuals.
Acknowledgments
The support of the British Society for Antimicrobial Chemotherapy (BSAC) is gratefully acknowledged. A. Owen is funded by BSAC. B. Chandler is funded by BBSRC and Roche. P. G. Bray is funded by BBSRC.
REFERENCES
- 1.Agrati, C., F. Poccia, S. Topino, P. Narciso, C. Selva, L. P. Pucillo, G. D'Offizi, G. Antonelli, F. Bellomi, O. Turriziani, and F. Bambacioni. 2003. P-glycoprotein expression by peripheral blood mononuclear cells from human immunodeficiency virus-infected patients is independent from response to highly active antiretroviral therapy. Clin. Diagn. Lab. Immunol. 10:191-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aszalos, A., K. Thompson, J. J. Yin, and D. D. Ross. 1999. Combinations of P-glycoprotein blockers, verapamil, PSC833, and cremophor act differently on the multidrug resistance associated protein (MRP) and on P-glycoprotein (Pgp). Anticancer Res. 19:1053-1064. [PubMed] [Google Scholar]
- 3.Berger, E. A. 1998. HIV entry and tropism. When one receptor is not enough. Adv. Exp. Med. Biol. 452:151-157. [PubMed] [Google Scholar]
- 4.Brumme, Z. L., W. W. Dong, K. J. Chan, R. S. Hogg, J. S. Montaner, M. V. O'Shaughnessy, and P. R. Harrigan. 2003. Influence of polymorphisms within the CX3CR1 and MDR-1 genes on initial antiretroviral therapy response. AIDS 17:201-208. [DOI] [PubMed] [Google Scholar]
- 5.Chandler, B., L. Almond, J. Ford, A. Owen, P. Hoggard, S. Khoo, and D. Back. 2003. The effects of protease inhibitors and nonnucleoside reverse transcriptase inhibitors on P-glycoprotein expression in peripheral blood mononuclear cells in vitro. J. Acquir. Immune Defic. Syndr. 33:551-556. [DOI] [PubMed] [Google Scholar]
- 6.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dragic, T. 2001. An overview of the determinants of CCR5 and CXCR4 co-receptor function. J. Gen. Virol. 82:1807-1814. [DOI] [PubMed] [Google Scholar]
- 8.Eagling, V. A., H. Wiltshire, I. W. Whitcombe, and D. J. Back. 2002. CYP3A4-mediated hepatic metabolism of the HIV-1 protease inhibitor saquinavir in vitro. Xenobiotica 32:1-17. [DOI] [PubMed] [Google Scholar]
- 9.Fellay, J., C. Marzolini, E. R. Meaden, D. J. Back, T. Buclin, J. P. Chave, L. A. Decosterd, H. Furrer, M. Opravil, G. Pantaleo, D. Retelska, L. Ruiz, A. H. Schinkel, P. Vernazza, C. B. Eap, and A. Telenti. 2002. Response to antiretroviral treatment in HIV-1-infected individuals with allelic variants of the multidrug resistance transporter 1: a pharmacogenetics study. Lancet 359:30-36. [DOI] [PubMed] [Google Scholar]
- 10.Ford, J., P. G. Hoggard, A. Owen, S. H. Khoo, and D. J. Back. 2003. A simplified approach to determining P-glycoprotein expression in peripheral blood mononuclear cell subsets. J. Immunol. Methods 274:129-137. [DOI] [PubMed] [Google Scholar]
- 11.Germann, U. A., P. J. Ford, D. Shlyakhter, V. S. Mason, and M. W. Harding. 1997. Chemosensitization and drug accumulation effects of VX-710, verapamil, cyclosporin A, MS-209 and GF120918 in multidrug resistant HL60/ADR cells expressing the multidrug resistance-associated protein MRP. Anticancer Drugs 8:141-155. [DOI] [PubMed] [Google Scholar]
- 12.Grivel, J. C., M. L. Penn, D. A. Eckstein, B. Schramm, R. F. Speck, N. W. Abbey, B. Herndier, L. Margolis, and M. A. Goldsmith. 2000. Human immunodeficiency virus type 1 coreceptor preferences determine target T-cell depletion and cellular tropism in human lymphoid tissue. J. Virol. 74:5347-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helbig, G., K. W. Christopherson II, P. Bhat-Nakshatri, S. Kumar, H. Kishimoto, K. D. Miller, H. E. Broxmeyer, and H. Nakshatri. 2003. NF-κB promotes breast cancer cell migration and metastasis by inducing the expression of the chemokine receptor CXCR4. J. Biol. Chem. 278:21631-21638. [DOI] [PubMed] [Google Scholar]
- 14.Hitzl, M., S. Drescher, H. van der Kuip, E. Schaffeler, J. Fischer, M. Schwab, M. Eichelbaum, and M. F. Fromm. 2001. The C3435T mutation in the human MDR1 gene is associated with altered efflux of the P-glycoprotein substrate rhodamine 123 from CD56+ natural killer cells. Pharmacogenetics 11:293-298. [DOI] [PubMed] [Google Scholar]
- 15.Huisman, M. T., J. W. Smit, H. R. Wiltshire, R. M. Hoetelmans, J. H. Beijnen, and A. H. Schinkel. 2001. P-glycoprotein limits oral availability, brain, and fetal penetration of saquinavir even with high doses of ritonavir. Mol. Pharmacol. 59:806-813. [DOI] [PubMed] [Google Scholar]
- 16.Ifergan, I., N. F. Bernard, J. Bruneau, M. Alary, C. M. Tsoukas, and M. Roger. 2002. Allele frequency of three functionally active polymorphisms of the MDR-1 gene in high-risk HIV-negative and HIV-positive Caucasians. AIDS 16:2340-2342. [DOI] [PubMed] [Google Scholar]
- 17.Inaba, T., N. E. Fischer, D. S. Riddick, D. J. Stewart, and T. Hidaka. 1997. HIV protease inhibitors, saquinavir, indinavir and ritonavir: inhibition of CYP3A4-mediated metabolism of testosterone and benzoxazinorifamycin, KRM-1648, in human liver microsomes. Toxicol. Lett. 93:215-219. [DOI] [PubMed] [Google Scholar]
- 18.Johnston, E. R., L. S. Zijenah, S. Mutetwa, R. Kantor, C. Kittinunvorakoon, and D. A. Katzenstein. 2003. High frequency of syncytium-inducing and CXCR4-tropic viruses among human immunodeficiency virus type 1 subtype C-infected patients receiving antiretroviral treatment. J. Virol. 77:7682-7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, P. M., and A. M. George. 2000. Symmetry and structure in P-glycoprotein and ABC transporters: what goes around comes around. Eur. J. Biochem. 267:5298-5305. [DOI] [PubMed] [Google Scholar]
- 20.Koot, M., I. P. Keet, A. H. Vos, R. E. de Goede, M. T. Roos, R. A. Coutinho, F. Miedema, P. T. Schellekens, and M. Tersmette. 1993. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann. Intern. Med. 118:681-688. [DOI] [PubMed] [Google Scholar]
- 21.Kreisberg, J. F., D. Kwa, B. Schramm, V. Trautner, R. Connor, H. Schuitemaker, J. I. Mullins, A. B. van't Wout, and M. A. Goldsmith. 2001. Cytopathicity of human immunodeficiency virus type 1 primary isolates depends on coreceptor usage and not patient disease status. J. Virol. 75:8842-8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, C. G., M. Ramachandra, K. T. Jeang, M. A. Martin, I. Pastan, and M. M. Gottesman. 2000. Effect of ABC transporters on HIV-1 infection: inhibition of virus production by the MDR1 transporter. FASEB J. 14:516-522. [DOI] [PubMed] [Google Scholar]
- 23.Manes, S., G. del Real, R. A. Lacalle, P. Lucas, C. Gomez-Mouton, S. Sanchez-Palomino, R. Delgado, J. Alcami, E. Mira, and A. C. Martinez. 2000. Membrane raft microdomains mediate lateral assemblies required for HIV-1 infection. EMBO Rep. 1:190-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manetti, R., L. Cosmi, G. Galli, F. Annunziato, M. Mazzetti, S. Romagnani, and E. Maggi. 2000. Enhanced expression of the CXCR4 co-receptor in HIV-1-infected individuals correlates with the emergence of syncytia-inducing strains. Cytokines Cell Mol. Ther. 6:19-24. [DOI] [PubMed] [Google Scholar]
- 25.Mistry, P., A. J. Stewart, W. Dangerfield, S. Okiji, C. Liddle, D. Bootle, J. A. Plumb, D. Templeton, and P. Charlton. 2001. In vitro and in vivo reversal of P-glycoprotein-mediated multidrug resistance by a novel potent modulator, XR9576. Cancer Res. 61:749-758. [PubMed] [Google Scholar]
- 26.Nasi, M., V. Borghi, M. Pinti, C. Bellodi, E. Lugli, S. Maffei, L. Troiano, L. Richeldi, C. Mussini, R. Esposito, and A. Cossarizza. 2003. MDR1 C3435T genetic polymorphism does not influence the response to antiretroviral therapy in drug-naive HIV-positive patients. AIDS 17:1696-1698. [DOI] [PubMed] [Google Scholar]
- 27.Owen, A., B. Chandler, J. Ford, S. H. Khoo, and D. J. Back. 2003. Differential expression of HIV co-receptors between CEM, CEMVBL and CEME1000 cells. J. Infect. Dis. 187:874-876. [DOI] [PubMed] [Google Scholar]
- 28.Penn, M. L., J. C. Grivel, B. Schramm, M. A. Goldsmith, and L. Margolis. 1999. CXCR4 utilization is sufficient to trigger CD4+ T cell depletion in HIV-1-infected human lymphoid tissue. Proc. Natl. Acad. Sci. USA 96:663-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry, C. M., and S. Noble. 1998. Saquinavir soft-gel capsule formulation. A review of its use in patients with HIV infection. Drugs 55:461-486. [DOI] [PubMed] [Google Scholar]
- 30.Pierdominici, M., A. Giovannetti, F. Ensoli, F. Mazzetta, M. Marziali, M. R. De Cristofaro, D. Santini-Muratori, W. Leti, and F. Aiuti. 2002. Changes in CCR5 and CXCR4 expression and beta-chemokine production in HIV-1-infected patients treated with highly active antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 29:122-131. [DOI] [PubMed] [Google Scholar]
- 31.Profit, L., V. A. Eagling, and D. J. Back. 1999. Modulation of P-glycoprotein function in human lymphocytes and Caco-2 cell monolayers by HIV-1 protease inhibitors. AIDS 13:1623-1627. [DOI] [PubMed] [Google Scholar]
- 32.Raviv, Y., A. Puri, and R. Blumenthal. 2000. P-glycoprotein-overexpressing multidrug-resistant cells are resistant to infection by enveloped viruses that enter via the plasma membrane. FASEB J. 14:511-515. [DOI] [PubMed] [Google Scholar]
- 33.Scarlatti, G., E. Tresoldi, A. Bjorndal, R. Fredriksson, C. Colognesi, H. K. Deng, M. S. Malnati, A. Plebani, A. G. Siccardi, D. R. Littman, E. M. Fenyo, and P. Lusso. 1997. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat. Med. 3:1259-1265. [DOI] [PubMed] [Google Scholar]
- 34.Schramm, B., M. L. Penn, R. F. Speck, S. Y. Chan, E. De Clercq, D. Schols, R. I. Connor, and M. A. Goldsmith. 2000. Viral entry through CXCR4 is a pathogenic factor and therapeutic target in human immunodeficiency virus type 1 disease. J. Virol. 74:184-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. de Goede, R. P. van Steenwijk, J. M. Lange, J. K. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simmons, G., D. Wilkinson, J. D. Reeves, M. T. Dittmar, S. Beddows, J. Weber, G. Carnegie, U. Desselberger, P. W. Gray, R. A. Weiss, and P. R. Clapham. 1996. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic, and most can use either Lestr or CCR5 as coreceptors for virus entry. J. Virol. 70:8355-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Speck, R. R., X. F. Yu, J. Hildreth, and C. Flexner. 2002. Differential effects of P-glycoprotein and multidrug resistance protein-1 on productive human immunodeficiency virus infection. J. Infect. Dis. 186:332-340. [DOI] [PubMed] [Google Scholar]
- 38.Spijkerman, I., F. de Wolf, M. Langendam, H. Schuitemaker, and R. Coutinho. 1998. Emergence of syncytium-inducing human immunodeficiency virus type 1 variants coincides with a transient increase in viral RNA level and is an independent predictor for progression to AIDS. J. Infect. Dis. 178:397-403. [DOI] [PubMed] [Google Scholar]
- 39.Srinivas, R. V., D. Middlemas, P. Flynn, and A. Fridland. 1998. Human immunodeficiency virus protease inhibitors serve as substrates for multidrug transporter proteins MDR1 and MRP1 but retain antiviral efficacy in cell lines expressing these transporters. Antimicrob. Agents Chemother. 42:3157-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steele, A. D., E. E. Henderson, and T. J. Rogers. 2003. Mu-opioid modulation of HIV-1 coreceptor expression and HIV-1 replication. Virology 309:99-107. [DOI] [PubMed] [Google Scholar]
- 41.Walz, G., B. Zanker, C. Barth, K. J. Wieder, S. C. Clark, and T. B. Strom. 1990. Transcriptional modulation of human IL-6 gene expression by verapamil. J. Immunol. 144:4242-4248. [PubMed] [Google Scholar]
- 42.Xiao, L., D. L. Rudolph, S. M. Owen, T. J. Spira, and R. B. Lal. 1998. Adaptation to promiscuous usage of CC and CXC-chemokine coreceptors in vivo correlates with HIV-1 disease progression. AIDS 12:F137-F143. [DOI] [PubMed] [Google Scholar]
- 43.Yamaguchi, M., M. Kuzume, H. Nakano, and K. Kumada. 1998. Verapamil suppressed lymphocyte adhesion to vascular endothelial cells via selective inhibition of VCAM-1 expression. Transplant. Proc. 30:2955. [DOI] [PubMed] [Google Scholar]
- 44.Yamaguchi, M., H. Suwa, M. Miyasaka, and K. Kumada. 1997. Selective inhibition of vascular cell adhesion molecule-1 expression by verapamil in human vascular endothelial cells. Transplantation 63:759-764. [DOI] [PubMed] [Google Scholar]
- 45.Zanker, B., G. Walz, K. J. Wieder, M. Moscovitch-Lopatin, B. R. Smith, and T. B. Strom. 1989. Verapamil selectively inhibits expression of interleukin-2 messenger RNA in mitogen activated mononuclear blood cells. Transplant. Proc. 21:85-87. [PubMed] [Google Scholar]
- 46.Zhang, Y., B. Lou, R. B. Lal, A. Gettie, P. A. Marx, and J. P. Moore. 2000. Use of inhibitors to evaluate coreceptor usage by simian and simian/human immunodeficiency viruses and human immunodeficiency virus type 2 in primary cells. J. Virol. 74:6893-6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Y. J., T. Dragic, Y. Cao, L. Kostrikis, D. S. Kwon, D. R. Littman, V. N. KewalRamani, and J. P. Moore. 1998. Use of coreceptors other than CCR5 by non-syncytium-inducing adult and pediatric isolates of human immunodeficiency virus type 1 is rare in vitro. J. Virol. 72:9337-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, Y. J., and J. P. Moore. 1999. Will multiple coreceptors need to be targeted by inhibitors of human immunodeficiency virus type 1 entry? J. Virol. 73:3443-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]