Abstract
Human immunodeficiency virus type 1 (HIV-1) impacts multiple lineages of hematopoietic cells, including lymphocytes and macrophages, either by direct infection or indirectly by perturbations of cell networks, leading to generalized immune deficiency. We designed a study to discover, in primary human macrophages, sentinel genetic targets that are impacted during replication over the course of 7 days by a CCR5-using virus. Expression of mRNA and proteins in virus- or mock-treated macrophages from multiple donors was evaluated. Hierarchical agglomerative cluster analysis grouped into distinct temporal expression patterns >900 known human genes that were induced or repressed at least fourfold by virus. Expression of more than one-third of the genes was induced rapidly by day 2 of infection, while other genes were induced at intermediate (day 4) or late (day 7) time points. More than 200 genes were expressed exclusively in either virus- or mock-treated macrophage cultures, independent of the donor, providing an unequivocal basis to distinguish an effect by virus. HIV-1 altered levels of mRNA and/or protein for diverse cellular programs in macrophages, including multiple genes that can contribute to a transition in the cell cycle from G1 to G2/M, in contrast to expression in mock-treated macrophages of genes that maintain G0/G1. Virus treatment activated mediators of cell cycling, including PP2A, which is impacted by Vpr, as well as GADD45 and BRCA1, potentially novel targets for HIV-1. The results identify interrelated programs conducive to optimal HIV-1 replication and expression of genes that can contribute to macrophage dysfunction.
Human immunodeficiency virus type 1 (HIV-1) pathogenesis results from perturbations in complex cellular networks leading to global immune dysfunction and a spectrum of AIDS-defining illnesses. HIV-1 infection of macrophages, which are long-lived cells, can also lead to establishment of reservoirs for virus production and spread to other cell types and compartments (2, 3, 7, 25, 27, 30, 31, 40, 82). Viral infection of macrophages produces a global impact on macrophage physiology, including alterations in the cell cycle (78, 80), chemotaxis (1, 21), levels of tumor necrosis factor alpha and other Th2-type cytokines (16, 24, 56), phagocytosis (9, 20, 45, 59), and antigen presentation (42, 51). In particular, HIV-1 or envelope gp120 activates multiple signal transducer and activator of transcription (STAT) proteins that modulate the expression of multiple receptors and/or cytokines contributing to generalized immune dysfunction in hematopoietic cells (11, 43, 66).
Discerning interactive networks through analysis of single pathways has limitations. In contrast, global analysis of gene expression profiles provides a comprehensive approach to developing an integrated picture of the complex perturbations of cellular gene expression by virus treatment. Initial genomic approaches to the study of HIV-1 infection have examined interactions between prototypic T-cell-tropic CXCR4 strains of HIV-1 and proliferating T-cell lines or primary peripheral blood mononuclear cells (PBMC) (28, 61, 63, 76) or between gp120 and PBMC or monocyte-derived macrophages (MDM) (16). We designed a unique study using the Affymetrix GeneChip system in combination with high-throughput Western blot analysis to develop a genetic profile of virus treatment by a prototypic macrophage-tropic CCR5 virus, HIV-1JR-FL, over the course of 7 days in human macrophages from multiple donors. A primary objective of using a systems biology approach was to identify sentinel genetic events that would distinguish between virus and mock treatment of macrophages. Subsequent analysis focused on the expression of networks of genes involved in cell cycle regulation (33, 44, 64, 78, 80).
MATERIALS AND METHODS
Preparation of viral stocks and infection of MDM.
A stock of HIV-1JR-FL (National Institutes of Health, AIDS Reference and Reagent Program, Rockville, Md.) was prepared in PBMC from a seronegative donor and calculated to have a titer of 104.5 50% tissue culture infective doses per ml (73). Supernatants for mock treatment of MDM cultures were prepared simultaneously from uninfected PBMC from the same seronegative donor.
Monocytes from four HIV-1- and hepatitis C virus-seronegative donors were isolated using RosetteSep (a positive-depletion monocyte enrichment method; Stem Cell Technologies, Vancouver, Canada) according to the manufacturer's protocol, which produces cell populations that are >99% CD14+ monocytes with <1% B or T cells (CD3+). MDM from four donors provided a 90% chance of detecting a twofold difference between virus- and mock-treated cultures, based on analysis of variance using a randomized complete block design, a two-sided t test, and a standard deviation of 0.5. Cells were induced to adhere to plastic at a concentration of 2 × 106 per ml and were incubated in differentiation medium comprised of Dulbecco's modified Eagle's medium (Life Technologies, Gaithersburg, Md.) with 15% human AB serum (Sigma-Aldrich Corp., Saint Louis, Mo.) and a single treatment of 10 U of granulocyte-macrophage colony-stimulating factor (Invitrogen, Carlsbad, Calif.) per ml. After 7 days, >70% of cells expressed CD4 and >80% were positive for CCR5 as measured by flow cytometry (data not shown). After differentiation, CD14+, Fcγ receptor+ macrophages were the only cell type detected; no CD3+ or CD19+ T or B lymphocytes, respectively, were detected (49, 73). MDM cultures were transferred to GM-CSF-free media and treated on day 0 with 2 ml of either HIV-1JR-FL (multiplicity of infection, ∼0.02) or mock supernatants for 24 h, washed, and cultured in GM-CSF-free media. Levels of supernatant p24 antigen on days 2, 4, and 7 were assessed using a specific enzyme-linked immunosorbent assay kit (Coulter, Hialeah, Fla.). Aliquots of cells were washed twice with phosphate-buffered saline and lysed in a nonionic detergent buffer (50 mM KCl, 10 mM Tris-Cl, pH 8.3, 2.5 mM MgCl2, 0.1 mg of gelatin/ml, 0.45% NP-40, and 0.45% Tween 20) containing proteinase K (100 μg per ml) (K buffer) for quantitation of infected cells by TaqMan real-time PCR assay using HIV-1 gag as the analytical parameter and human apoB as a genomic template control. The primers and probe used for quantitation of apoB were as follows: forward primer, 5′-TGAAGGTGGAGGACATTCCTCTA; reverse primer, 5′-CTGGAATTGCGATTTCTGGTAA; probe, VIC (Applied Biosystems)-CGAGAATCACCCTGCCAGACTTCCGT-6-carboxy-tetramethylrhodamine (TAMRA). Viral gag gene copies by TaqMan were quantified using forward primer (5′-ACATCAAGCAGCCATGCAAAT-3′), reverse primer (5′-ATCTGGCCTGGTGCAATAGG-3′), and probe (5′-6-carboxy-fluorescein (FAM)-CATCAATGAGGAAGCTGCAGAATGGGATAGA-TAMRA-3′) (19). Amplification reactions consisted of TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, Calif.), 450 nM (each) forward and reverse primers, and 125 nM probe in a total volume of 50 μl. The PCR conditions were 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Thermal cycling and data acquisition were performed using a Prism 7700 Thermal Cycler (Applied Biosystems).
RNA isolation and hybridization.
Total RNA for each sample was extracted and prepared for hybridization according to protocols in the GeneChip Expression Analysis Technical Manual (Affymetrix). Briefly, cells from virus-treated or mock-treated cultures at days 0, 2, 4, and 7 were washed two times in MDM rinse medium and resuspended in RLT RNA lysis buffer (QIAGEN, Valencia, Calif.) for total RNA isolation (RNEasy RNA isolation kit; QIAGEN). RNA aliquots (8 μg) were reverse transcribed from a (dT)24 primer into double-stranded cDNA, which served as a template for in vitro transcription to produce biotin-labeled cRNA that was fragmented under conditions of high magnesium concentration (1 mM) and heat. The quality of the cRNA was first assessed using Test 3 Genechip arrays (Affymetrix) before hybridization to HG-U95A (version 2) arrays. The arrays were stained by a streptavidin-phycoerythrin conjugate (Molecular Probes, Eugene, Oreg.) using the Affymetrix GeneChip Fluidic Station and scanned by an Agilent argon-ion laser with 488-nm emission and 570-nm detection (GeneArray Scanner). High-resolution chip images were analyzed using Microarray Suite version 4 (Affymetrix). Intensity values from each probe cell were adjusted for background and multiplied by a scaling factor to facilitate comparisons across the arrays. Images were converted into both average difference values and numerical (n-fold) change data based on comparison of virus-treated and mock-treated samples.
RNA data analysis.
A custom relational database was designed using Microsoft (Redmond, Wash.) Access 2000 on the Windows 2000 platform. The database contained all raw data, as well as links between unique gene identifiers for mRNA and protein, functional categories, and summary of function. Functional categories of genes were based on the Gene Ontology Consortium public domain database (http://www.geneontology.org/). Cluster analysis was performed to gain an overview of temporal gene expression patterns within the data set. Prior to clustering, filters were applied so that a robust set of genes could be analyzed. Genes were first filtered to remove any transcripts that were considered absent under all conditions. A subsequent filter identified genes in infected cultures that displayed >4-fold change (induced or repressed) in hybridization intensities relative to levels in uninfected cultures. Average linkage hierarchical agglomerative clustering with uncentered correlation was performed on the median values obtained for each probe set for all donors at each time point using the software package Cluster (M. Eisen, Berkeley, Calif.). Major branches in the dendrogram were defined by correlation coefficients of >0.75 (mean, 0.85; range, 0.75 to 0.96)
To identify subsets of genes that were expressed exclusively in HIV-1 or mock-treated cultures of MDM, queries were performed within GeneSpring version 4.1 (Silicon Genetics, Redwood City, Calif.). Genes from each treated population were subjected to three high-stringency statistical filters, which required that a gene be present in all donors, vary by no more than 0.5 standard deviations from the mean expression in all donors, and be induced at least twofold relative to the globally normalized background intensity for the entire data set. The combined effect of the filters identified a set of genes that was robust to outlier, interassay, and donor variability. All genes expressed in virus-treated cultures were compared to all genes expressed in mock-treated cultures by using Venn diagrams. Unique genes that were expressed by each population were used to create lists and import them into the custom relational database.
Protein extraction and PowerBlot screening.
Monocytes (3 × 107 to 4 × 107) from each of four donors were plated in six-well plates (4 × 106 cells/well), differentiated for 7 days, and treated as previously described. Two of the four donors were assayed for both proteins and mRNA. After 48 h of virus treatment, the cells were lysed with boiling lysis buffer (10 mM Tris, pH 7.4, 1 mM sodium orthovanadate, 1% sodium dodecyl sulfate). Lysates were heated at 95°C for 30 s and passed 10 times through a 25-gauge needle to shear the DNA. Total protein yields were quantitated using the bicinchoninic acid protein assay (Pierce, Rockford, Ill.).
Immunoblot analysis of proteins was performed by BD Biosciences. Briefly, 5 mg of total protein per sample was loaded on a total of five gradient (4 to 15%) SDS-polyacrylamide gels and subsequently transferred to Immobilon-P membranes (Millipore). A total of 1,005 proteins were screened using different complex antibody cocktails. Digitally captured images of all blots from duplicate or triplicate runs of the two samples (HIVJR-FL and mock treated) were compared, and confidence values based on change and reproducibility were designated (semiquantitative). Only proteins with a confidence value of ≥3 were considered in the final analysis. Each protein was entered into our custom relational database and identified by locus link identification (ID) and Swiss Protein ID numbers, predicted and observed molecular weights, and functional categories. A query was designed in the database to link mRNA and protein data according to common locus link ID numbers.
RESULTS
CCR5-tropic viral replication and spread in MDM cultures.
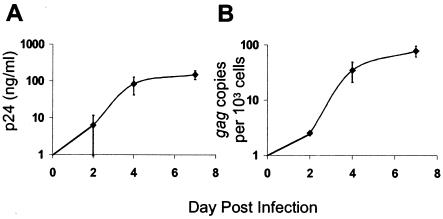
To relate gene expression to viral replication kinetics, the supernatant p24 antigen production and number of gag DNA copies were measured in MDM cultures over the course of 7 days (Fig. 1). In general, peak levels of supernatant p24 antigen (100 ng/ml) developed between days 4 and 7 following HIVJR-FL infection (Fig. 1A). Production of supernatant p24 levels paralleled virus spread based on cell-associated virus DNA copies (Fig. 1B). By 2 days posttreatment, ∼0.2% of MDM harbored virus, based on one gag copy per cell, while at day 7, ∼10% of macrophages were infected (Fig. 1B). Maximal infection of 10% of MDM by HIV-1JR-FL was consistent with other data from our laboratory and indicated that nearly 90% of macrophages in virus-treated cultures remained uninfected. An impact of virus treatment on genetic networks in MDM, including both infected and uninfected cells, was compared to gene expression by uninfected cells in mock-treated MDM cultures. A 7-day time course was analyzed to provide an overview of genetic networks during the course of spreading virus infection.
FIG. 1.
Kinetics of HIV-1 replication and spread in MDM. (A) Supernatant p24 antigen. (B) Number of gag DNA copies per 103 cells. The data are means, with error bars representing standard errors of the mean for triplicate donors.
Temporal expression of genes in HIV-1-treated MDM cultures.
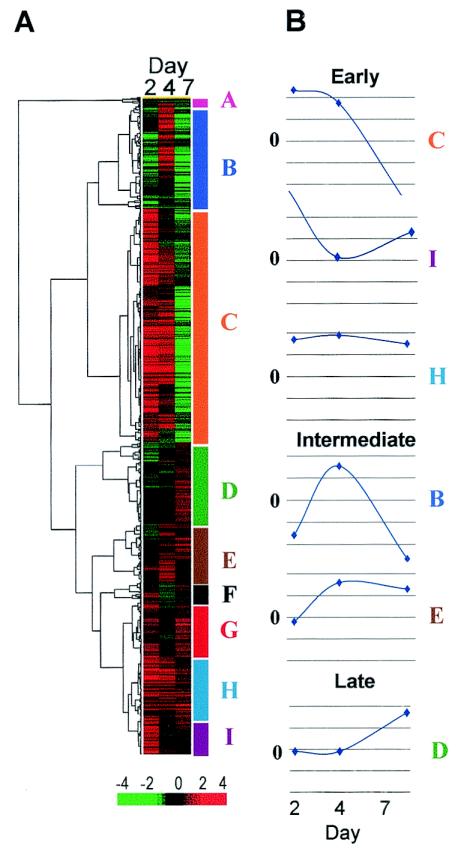
Approximately 900 genes with defined functions were induced or suppressed by >4-fold between HIV-treated and mock-treated cultures from any donor at any time point. Based on hierarchical agglomerative cluster analysis of expression patterns over time, the genes fell into nine clusters (Fig. 2A, A through I). Median induction or repression for the entire data set was ∼4-fold.
FIG. 2.
Clustering of genes perturbed by HIV-1 infection of MDM. (A) Hierarchical agglomerative clustering of ∼900 genes was based on expression patterns over time. Median induction (red) or repression (green) of expression ranged between +4 and −4 for each gene. Genes were distributed into nine cluster patterns designated A to I (color coded on the right [y] axis of the dendrogram). (B) Median induction or repression (n-fold) over time for ∼800 genes in six clusters ranged between +2- and −2-fold. Three patterns of induction of gene expression were observed: early (day 2; clusters C, H, and I), intermediate (day 4; clusters B and E), and late (day 7; cluster D).
Approximately 800 of the 900 functionally annotated genes fell into six major clusters (B, C, D, E, H, and I). Patterns of expression over time for clusters of genes were represented as waveforms that displayed the median change for the population of genes within each cluster (Fig. 2B). More than 50% of the genes (465 of 808) were induced as early as 2 days following HIV-1 treatment and were included in clusters C, H, and I (Fig. 2B). About 28% of the genes (231 of 808; clusters B and E) were induced at the intermediate day 4 time point, while 14% of the genes (112 of 808; cluster D) were induced late, by day 7 following treatment (Fig. 2B). Although the genes were classified by temporal patterns of induction of expression, distinct patterns of gene repression over time were also apparent.
Genes expressed exclusively in virus-treated or mock-treated MDM cultures.
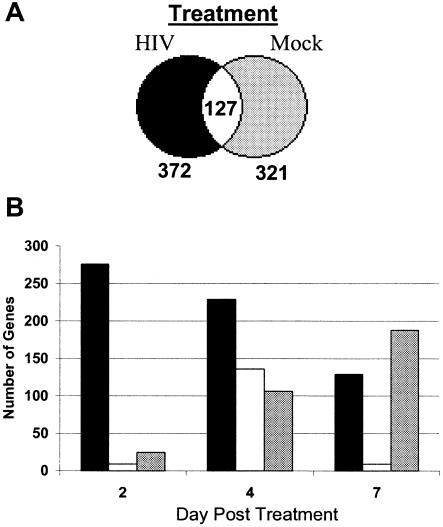
Temporal analysis required that genes be expressed in both virus- and mock-treated MDM cultures and excluded genes that were expressed only under one or the other condition. To determine if genes might be expressed exclusively in virus- or mock-treated MDM, a series of statistical filters was applied to the data set and the results were summarized using Venn diagrams (Fig. 3A). Analysis identified 372 genes that were expressed exclusively in virus-treated macrophage cultures at all time points, 321 genes that were expressed exclusively in mock-treated cultures, and 127 genes that were expressed by MDM independent of treatment.
FIG. 3.
Functional categories for genes expressed exclusively in infected or in uninfected MDM cultures. (A) Genes that were expressed only in virus-treated or mock-treated MDM cultures were identified by Venn diagram analysis. (B) Numbers of genes expressed exclusively in virus- or mock-treated cultures were identified on each day. Black bars, genes expressed uniquely in virus-treated cultures; gray bars, genes expressed uniquely in mock-treated cultures; white bars, coexpressed genes.
A second set of queries examined mutually exclusive gene expression on different days of treatment (Fig. 3B). A dramatic difference in the number of genes expressed exclusively by HIV- or mock-treated cultures was apparent as early as day 2 (Fig. 3B). The number of genes expressed exclusively in virus-treated cultures gradually decreased from 260 to 130 over 7 days (Fig. 3B). Simultaneously, the number of genes that were expressed exclusively in mock-treated cultures increased from <50 to 190 (Fig. 3B).
Functional categories of genes modulated by HIV-1.
More than 800 genes that were summarized by temporal expression (Fig. 2) were subsequently classified into 21 functional categories as defined by the Gene Ontology Consortium. About 17% of the genes (142 of 808) were classified in the energy or protein metabolism category (Table 1). More than 30% (258 of 808) were grouped into categories with related functions, such as apoptosis, cell proliferation, cytoskeletal and cell motility, signal transduction, or transcription, while 11% of the genes (89 of 808) were classified into the hematopoietic, immune response, or inflammation category.
TABLE 1.
Functional categories of genes with altered mRNA expression in MDM following HIV-1 treatment
| Gene functiona | No. of genesb
|
|||
|---|---|---|---|---|
| E | I | L | Total | |
| Adhesion and extracellular matrix | 31 | 15 | 6 | 52 |
| Apoptosis | 14 | 2 | 5 | 21 |
| Cell proliferation | 20 | 14 | 4 | 38 |
| Cell stress | 8 | 8 | 1 | 17 |
| Cell-cell signaling | 21 | 8 | 5 | 34 |
| Chromatin structure | 7 | 3 | 0 | 10 |
| Cytoskeleton and cell motility | 33 | 24 | 12 | 69 |
| DNA binding protein | 7 | 4 | 0 | 11 |
| DNA synthesis and repair | 14 | 8 | 3 | 25 |
| Energy metabolism | 42 | 25 | 14 | 81 |
| Hematopoetic | 3 | 5 | 8 | 16 |
| Hormone or growth factor | 23 | 10 | 4 | 37 |
| Immune response | 27 | 17 | 13 | 57 |
| Inflammatory and coagulation | 10 | 3 | 3 | 16 |
| Ion and solute transport | 28 | 12 | 7 | 47 |
| Protein metabolism | 31 | 20 | 10 | 61 |
| RNA binding protein | 16 | 3 | 0 | 19 |
| Signal transduction | 37 | 19 | 3 | 59 |
| Transcription | 52 | 14 | 5 | 71 |
| Tumor related | 21 | 7 | 3 | 31 |
| Vesicle related | 20 | 10 | 6 | 36 |
| Total | 465 | 231 | 112 | 808 |
Gene function was assigned based on Gene Ontology Consortium categories.
The numbers of genes within each category for each cluster are tabulated. The total number of genes for each category across all clusters is also tabulated in the final column. Only gene categories with ≥10 genes per category are included for six of the nine clusters, resulting in omission of some genes. E genes changed during the early (day 2) timepoint; I, intermediate (day 4) timepoint; L, late (day 7) time point.
Similar analysis of mutually exclusive expression profiles (Fig. 3) identified genes from multiple functional categories (data not shown). In particular, HIV-1 treatment altered the expression of genes in a subset of functional categories related to immune function, cell cycle proliferation, and cell stress. In contrast, macrophages in mock-treated cultures expressed few, if any, genes in these categories. Patterns of mutually exclusive expression of genes provided a signature for virus treatment.
HIV impacts similar genes in lymphocytes and macrophages.
To determine if similar target genes were altered by HIV-1 treatment in different cell types, we compared our data from primary macrophages with those of other studies that analyzed an impact of HIV-1 on gene expression in transformed monocytic or lymphoid cell lines (Table 2) (8, 9, 13, 18, 26, 28, 29, 31, 33, 40, 42, 47, 52, 54, 61, 63, 64, 65, 67, 79, 81, 82, 83). Twenty genes, including 4 exact matches and 16 that were similar based on function or alternate isoforms of the same gene product, were induced following virus treatment (Table 2). In addition, treatment with HIV-1 repressed 38 genes, including 11 that were exact matches and 27 that were similar by function (Table 2). For example, both CD4 mRNA levels and CD4 cell surface expression were down regulated by virus treatment in cell lines and MDM (13, 73). In addition, HIV-1 induced expression of ERF, BCL2, and IFRD2 genes involved in regulation of the cell cycle in both monocytic and lymphocytic cells, as well as macrophages (Table 2). Therefore, we focused our subsequent analysis to determine the extent to which cell cycle mediators might be impacted by virus in macrophages.
TABLE 2.
Genes induced or repressed by HIV-1
| Status | Genbank no. | Descriptiona | mRNAb | Proteinb |
|---|---|---|---|---|
| Induced | M75099 | FK506 binding proteind | 61 | |
| NM005347 | HSP70d | 77 | 77 | |
| U15655 | Ets repressor factor (ERF)d | 61 | ||
| M13995 | BCL2d | 78 | 7 | |
| AA427491 | TCR alpha, c region | 30 | ||
| D87116 | Map kinase kinase 3 | 61 | ||
| J03909 | Interferon-inducible protein (IP-30) | 61 | ||
| L27584 | Ca2+ channel B3 subunit | 61 | ||
| M13755 | Interferon-induced 17/15 kDa | 61 | ||
| M20681 | GLUT3 | 25 | ||
| M60485 | FGF receptor | 61 | 28 | |
| M61827 | Leukosialin (CD43) | 61 | 50 | |
| M62831 | Transcription factor ETR101 | 61 | ||
| M77476 | Helix-loop-helix zipper protein | 61 | ||
| S63368 | TNF receptor 75 kDa | 61 | 12 | |
| U09585 | IFRD2 | 61 | ||
| U09848 | Zinc finger protein (ZNF139) | 61 | ||
| U24576 | Breast tumor autoantigen | 61 | ||
| XM055937 | HSP27 | 77 | 77 | |
| NM001228 | Caspase 8 | 8 | 53 | |
| Repressed | D13748 | EF 4A1d | 61 | |
| J02683 | ADP/ATP carrier proteind | 61 | ||
| J03191 | Profilind | 61 | ||
| L15189 | HSP 75d | 64 | ||
| L25080 | rhoCd | 61 | ||
| M14630c | Prothymosin alphad | 61, 30 | ||
| NM000616 | CD4d | 32 | 17, 76 | |
| U90313 | GST homologd | 61 | ||
| X01060 | Transferrin receptord | 61 | 62 | |
| AR201243 | MHC-11d | 74 | 41 | |
| AA401111 | Glucose phosphate isomerase | 30 | ||
| AA455931 | Adenylosuccinate lyase | 30 | ||
| AF006082 | ARP2 | 61 | ||
| D11086 | IL-2-R gamma | 61 | 39 | |
| J03592 | ADP/ATP translocase | 61 | ||
| L00160 | Phosphoglycarate kinase | 30 | ||
| M27132 | ATP synthetase beta subunit | 61 | ||
| M84326 | ADP ribosylation factor 1 | 61 | ||
| U36764 | TGF-beta R-interacting protein 1 | 61 | ||
| U94855 | Translation initiation factor 3 | 61 | ||
| X54473 | COX Vib | 61 | ||
| X69549 | Rho GDP-dissociation inhibitor 2 | 61 | ||
| X79538 | Translation initiation factor nuk34 | 61 | ||
| Y08890 | RanGTP binding protein 5 | 61 | ||
| Z37166 | Nuclear RNA Helicase | 61 |
Regulators of cell cycle (boldface underlined). TCR, T-cell receptor; TNF, tumor necrosis factor; GST, glutathione S-transferase; MHC, major histocompatibility complex; IL-2, interleukin-2; TGF, transforming growth factor.
Numbers correspond to citation numbers of publications that have documented induction or repression of these genes at either the mRNA or protein level during HIV-1 treatment.
Addition M26708.
Exact matches based on GenBank accession number.
HIV-1 impact on genes involved in regulation of the cell cycle in MDM cultures.
Database queries were performed to focus analysis on genes in the cell cycle category that were preferentially impacted by HIV-1 in MDM. Twelve genes involved in regulating cell cycle transitions were expressed exclusively in mock-treated MDM cultures (Table 3). Seventeen other genes that are implicated in transitions through cell cycle checkpoints were expressed uniquely in HIV-treated cultures (Table 3). When ratios for expression of cell cycle genes in HIV- and mock-treated MDM cultures were evaluated, temporal patterns were apparent. Sixteen genes involved in cell cycling were induced in virus-treated cultures by day 2 (early) and subsequently were repressed or returned to near-baseline values (Table 3). Ten genes were induced exclusively by day 4 (intermediate time point), while four genes were induced by day 7 (late).
TABLE 3.
Differentially expressed genes involved in cell cycling
| Unique genesa
|
Cluster analysisb
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mock treated
|
Treated
|
Early
|
Intermediate
|
Late
|
|||||
| GenBank no. | Description | GenBank no. | Description | GenBank no. | Description | GenBank no. | Description | GenBank no. | Description |
| AF053305 | BUB1L | M25753 | CCNB | D43969 | AML1 | M14752 | ABL | Y11284 | AFX |
| AL080146 | CCNB2 | U47414 | CCNG2 | U77629 | ASCL2 | AF045581 | BRCA1 | U82759 | HOX1 |
| X05360 | CDC2 | U65410 | MAD2L | L36844 | INK4B | M29551 | Calcineurin A2 | A1018098 | Integrin Beta 1 |
| U02570 | CDC42-GAP | AF053306 | MAD3L | X06292 | C-FES | X66364 | CDK5 | M26682 | Rhombotin-1 |
| L29218 | CDC-like 2 | X98263 | MPP-6 | X86371 | HUGL1 | AF049703 | ESE2 | ||
| AI304854 | CDKN1B | L07590 | PP2A B | U59464 | PTC1 | L06895 | MAX-BP | ||
| AF048731 | Cyclin T2 | M69199 | G0/G1 Switch | Y11306 | TCF4 | AB010419 | MTG16 | ||
| Z36714 | FBX1 | X71440 | CDK3 | AJ000414 | CIP4 | AF059681 | AIK3 | ||
| L13698 | GAS1 | X61123 | BTG1 | AF038198 | Chordin | L19267 | DM9 | ||
| U26727 | INK4 | M29551 | Calcineurin A2 | X70991 | MADER | M15881 | Uromodulin | ||
| X66363 | PCTAIRE1 | L25876 | CIP2 | S72043 | Metallothionein III | ||||
| D38305 | TROB1 | X06292 | C-FES | U33822 | TAX BP 181 | ||||
| X55504 | NOP120 | M31523 | TCF3 | ||||||
| X66363 | PCTAIRE1 | Z47363 | TCF7 | ||||||
| AB014561 | RB-BP | X66362 | PCTAIRE3 | ||||||
| Z11773 | ZNF187 | AF071538 | PDEF | ||||||
| D84557 | MCM6 | ||||||||
Specific genes unique to either virus- or mock-treated cultures are grouped.
Genes were clustered into groups based on whether they are induced at early, intermediate, or late time points posttreatment.
Qualitative identification of genes involved in cell cycle regulation checkpoints.
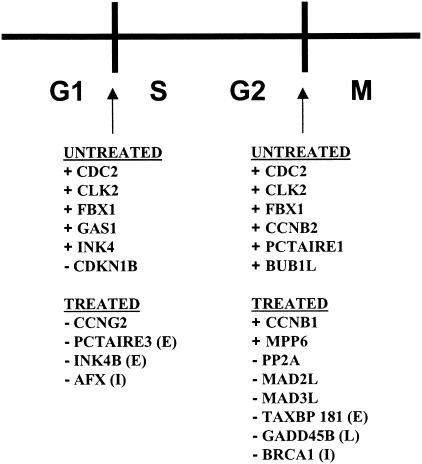
A number of genes involved in regulating G1/S or G2/M checkpoints in the cell cycle were detected exclusively in mock-treated MDM cultures (Fig. 4). For example, CDC2, CDKN1B, and GAS1 genes are implicated in G1/S transition, while CDC2 and PCTAIRE1 (a gene homologous to the CDC2 gene), CDC2-like kinase (CLK2), and cyclin B2 (CCNB2) genes are required for G2/M transition. In contrast, none of these genes was expressed in virus-treated cultures. Expression by only four genes that are involved in the maintenance of G1 (G1/S transitioning) was detected uniquely in virus-treated cultures. Most of the genes uniquely expressed following virus treatment, including protein phosphatase 2A (PP2A), breast cancer 1 (BRCA1), and growth arrest and DNA-damage-inducible (GADD45) genes, were involved in arresting cell cycling at the G2/M checkpoint (Fig. 4).
FIG. 4.
Impact of HIV-1 infection on cell cycle genes in MDM. Schematic of the cell cycle, with G1, S, G2, and M indicated. Genes expressed differentially by cluster analysis or mutually exclusive analysis in mock- or virus-treated MDM cultures are listed below the G1/S and G2/M checkpoints. Genes marked + promote progression through cell cycle checkpoints. Genes marked − inhibit progression through cell cycle checkpoints. Time points at which genes were detected are indicated in parentheses: early (E), intermediate (I), and late (L).
Altered protein expression in macrophages treated with a CCR-5-tropic virus.
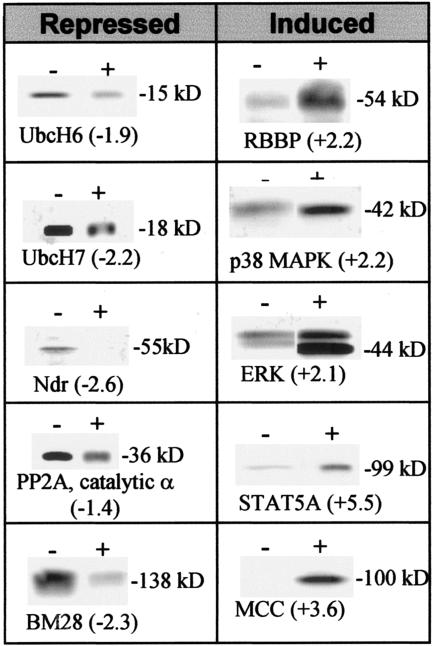
To determine the effect(s) on protein expression in MDM of treatment with a CCR5 strain of HIV-1, >1,000 proteins from virus- or mock-treated cultures were assayed and ranked based on changes. High-throughput Western blot screening with monoclonal antibodies evaluated levels rather than posttranslational modifications, including phosphorylation, that can be critical determinants of function for some proteins. Overall, 300 proteins had detectable differences of ≥1.5-fold in expression in virus-treated MDM from each donor, including ∼50 proteins that can have a functional role in regulation of the cell cycle or cell proliferation. A subset of proteins is presented in Fig. 5. For example, ubiquitination protein (UbcH6 and UbcH7) levels were reduced, while the levels of retinoblastoma binding protein 4 (RBBP) and interferon-induced protein kinases (p38 MAPK and ERK) and a related signal transducer and activator of transcription factor, STAT 5A, were induced. Cell cycle control was modified following virus treatment of macrophages through increased levels of MCC (a negative regulator of cell cycling) (15, 69), in combination with repression of PP2A catalytic alpha subunit, the nuclear Dbf2-related (Ndr) family of protein serine-threonine kinases (negative regulators of CDK1 required for completion of the cell cycle) (70), and BM28 (a protein associated with cell division) (72).
FIG. 5.
Altered protein expression in MDM. Expression levels of representative proteins determined by Powerblot analysis from mock (−)- or HIVJRFL (+)-treated monocyte-derived macrophages. Digitally captured images of all blots from duplicate runs were semiquantitated for changes compared to relative controls (indicated in parentheses). Proteins with changes of >1.5 were confirmed on duplicate verification blots.
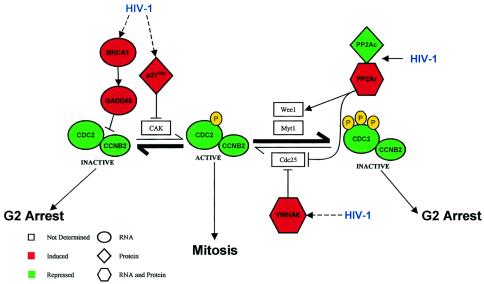
The overall effects of HIV-1 in MDM on specific networks of cellular mRNA and proteins that promote transition from G1 to G2 from our data are integrated in Fig. 6. Global effects include G2 arrest and suppression of mitosis through two pathways. One pathway, as expected, involved the PP2A complex, presumably through a Vpr-dependent mechanism (22, 23, 33). Both induction of the PP2A regulatory subunit and repression of the catalytic subunit can promote the enzymatic activity of Wee1 and inhibit the phosphatase CDC25 (41), resulting in hyperphosphorylation of CDC2 that leads to G2/M arrest (46, 53). Inactivation of CDC25 prevents dephosphorylation of CDC2, thus inhibiting mitosis (32, 41). Induction of mRNA and protein levels of tyrosine 3-monooxygenase/tryptophan 5-monooxygenase-activation protein, epsilon polypeptide (YWHAE), which blocks CDC25, also promotes G2 arrest (Fig. 6). The alternate pathway involves the nonphosphorylated form of the CDC2/CCNB2 complex. HIV-1 induces BRCA1 and subsequently GADD45 mRNA levels, which results in the inactivation of CDC2 and G2/M arrest (38, 77). BRCA1 is a downstream target of Rad3-related protein (ATR) phosphorylation, and HIV-1 Vpr activates ATR (5, 60, 84). In addition, HIV-1 induces a CDK inhibitor, p21Cip1/Waf1 protein, which blocks the CDK-activating kinase (CAK) phosphorylation of the cyclin D/CDC2 complex (10, 71), promoting G2 arrest. Activation of ERKs by phosphorylation can induce p21Cip1/Waf1 (6, 58), and activation of MEK/ERK is reported to occur in T-cell lines following infection with HIV-1 (57).
FIG. 6.
Summary of impacts of a CCR5 strain of HIV-1 on cell cycle regulators in MDM. Progression through G2 is regulated by phosphorylation states of the CDC2-cyclin B2 complex. Reversible phosphorylation reactions of the CDC2-cyclin B2 complex determine active (phosphorylated) or subsequent inactive (hyperphosphorylated) states. Hyperphosphorylated CDC2-cyclin B2 complex can be reactivated through dephosphorylation catalyzed by CDC25. HIV-1 exposure induces the expression of cell cycle regulators (PP2A, BRCA1/GADD45, p21Cip1, and YWHAE) involved in arresting MDM in G2. Inactive CDC2-cyclin B2 complex is promoted in HIV-1-treated cells (boldface arrows). Red indicates factors that were induced, and green indicates factors that were suppressed. Ovals, diamonds, and hexagons indicate whether the differential expression was detected at the level of mRNA, protein, or both, respectively.
DISCUSSION
The overall impact of HIV-1 on gene expression patterns in MDM is global and complex. Perturbations by virus clearly distinguished between HIV-1 and mock-treated cultures even though <0.2% of MDM treated with virus were infected by day 2 and only 10% were infected by day 7. The limited frequency of infected macrophages ex vivo resembles restricted HIV-1 infection of lymphoid tissue macrophages or alveolar macrophages in vivo (12, 50, 62). The number of infected MDM was estimated based on the premise of one viral DNA copy per cell, although cells in vivo can be infected by more than one virus (39). Even if estimates of infected MDM in our study exceed the actual numbers of cells that harbor virus, interpretation of the data would remain the same. Even though low levels of supernatant p24 antigen or cell-associated viral DNA were detected 2 days following HIV-1 treatment, expression of >400 genes at the mRNA or protein level changed relative to mock treatment. In addition, mutually exclusive expression at day 2 of ∼250 genes distinguished virus- from mock-treated MDM. The number of genes in macrophages altered by treatment with a CCR5 strain of HIV-1 in our analyses was similar to the number of genes in primary peripheral blood mononuclear cells or MDM that were altered by CCR5 gp120 treatment (16) but greater than the number of genes in lymphocytic or monocytic cell lines identified by treatment with CXCR4 strains of HIV-1 (28, 61, 63). Differences between primary cells and cell lines could reflect methodologies for measuring changes in mRNA or the types of viruses, as well as the possibility that transformed cell lines could have elevated basal levels of expression by genes that are altered in primary cells by HIV-1.
The number of genes expressed exclusively in mock-treated MDM increased over time, suggesting that macrophages in culture are dynamic rather than static. Increases in the number of genes expressed in the absence of virus could reflect actual changes induced by culture conditions or apparent changes reflecting down regulation of genes over time in virus-treated macrophages. Loss of cell viability is an unlikely explanation, because macrophage viability is >90% for 14 to 21 days following differentiation and infection (73) in contrast to rapid depletion of CD4 T lymphocytes (74). Either way, the net effect would be an increase in the number of genes expressed exclusively in mock-treated MDM over time. Clear differences between HIV-treated and mock-treated cells occur on day 2, which corresponds to the completion of first-round replication. The results agree with our earlier data showing that signal cascades are initiated by early virus-cell interactions (43). Expression of mutually exclusive genes over the course of the study provided sentinel biomarkers that characterized the effects of virus treatment on macrophage populations and distinguished unequivocally between mock- and virus-treated macrophages.
Using a systems biological approach, we identified substantial changes in mRNA and protein levels within the HIV-1-treated cultures that might occur exclusively in the small subset of macrophages that were infected directly, in the uninfected bystander cells, or in both types of cells. The possibility of residual CD4+ T lymphocytes in the MDM cultures was ruled out by flow cytometry and an inability of MDM cultures to support entry or replication by non-macrophage-tropic viruses (75). A difference between direct and bystander effects of virus in macrophage cultures at early time points would be difficult to evaluate, because so few macrophages were infected and strategies to separate infected from uninfected macrophages could alter mRNA or protein profiles. Nonetheless, the fact that a distinct subset of genes was detected in mock-treated MDM cultures suggested that uninfected cells not exposed to virus differed from uninfected cells in virus-treated MDM. Bystander effects could be mediated by cytokine expression from infected cells that would influence neighboring uninfected cells in an exocrine fashion. Alternatively, soluble viral components, such as gp120, could mediate changes in signal transduction cascades that impact downstream gene expression in the absence of infection (14, 17, 36, 43, 65).
A number of genes that were reported in other studies as induced or repressed at the mRNA or protein level in monocytic or lymphocytic cell lines infected by CXCR4 strains of HIV-1 were also detected in our biological systems approach involving primary macrophages treated by a CCR5 strain of HIV-1 (8, 13, 28, 29, 47, 61, 78, 79, 81, 86). Similarities among genes that were induced or repressed during treatment of primary human macrophages or cell lines by CCR5 or CXCR4 strains of virus, respectively, suggest that elements of virus-host cell interactions are highly conserved regardless of the viral envelope, the coreceptor preference, or even the cell type infected, which could have implications for developing novel therapeutics.
Normal macrophage differentiation involves G1 arrest (83), which would explain preferential expression of genes mediating the maintenance of G1 phase in mock-treated MDM in our studies. Even though differentiated cells are typically defined as nondividing or nonproliferating, differentiated cells can maintain a capacity to shift in their cell-cycling state following adequate stimulation (48). Our data are consistent with studies that indicate alteration of the cell cycle stage in macrophages by HIV-1 infection and suggest that the intracellular environment of cells no longer in G1, rather than cell division itself, plays a role in virus replication in nondividing cells, such as macrophages (33, 64, 80). G2/M arrest in lymphocytes is critical for optimal gene expression by long terminal repeats (4, 85), while transition from G1 to G2 in MDM correlates with enhanced HIV-1 replication (35, 68). An ability by HIV-1 to mediate transition from G1 to G2 could contribute to the fitness of viruses in MDM (34), although G2-dependent enhancement of long-terminal-repeat-mediated gene expression in macrophages is unclear. The theme of virus-induced cell cycle arrest, specifically in G2/M, is not unique to HIV-1. For example, reovirus and herpes simplex virus also mediate G2/M arrest via the CDC2-cyclin B pathway (23, 37, 55). A virus-dependent transition from G1/S to G2/M in macrophages could impact MDM physiology and function and contribute to immune dysfunction. The potential for more extensive involvement with the cell cycle by HIV-1, beyond the known role of Vpr, warrants further examination.
Patterns of induced or repressed genes identified within virus-treated MDM cultures have dynamic implications for a cascade of downstream effects that can alter cellular physiology. Our studies focused on global differential gene expression induced in association with the viral replication cycle, similar to the global effects associated with gp120-macrophage interactions (16, 78). In contrast to approaches that analyze single genes or pathways, global analysis provides an overall view of networks and interactions between known and novel genes and can provide insight into macrophage physiology and immune deficiency.
Acknowledgments
We are grateful to Maria Elena Bottazzi and Keith Peden for critical comments and insights into the data. We gratefully acknowledge the joint UF Shands Cancer Center/Interdisciplinary Center for Biotechnology Research Microarray Core Facility and the University of Florida Genetics Institute Microarray Core Facility for help in preparing samples, performing experiments, and analysis of data. The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-1JR-FL (catalog no. 394) from Irvin Chen.
Research was supported in part by PHS R01 awards HD32259, AI28571, HL58005, AI47723, T32 AR07603, and T32 CA09126; the Children's Miracle Network; and a University of Florida Presidential Fellowship.
REFERENCES
- 1.Albini, A., R. Benelli, D. Giunciuglio, T. Cai, G. Mariani, S. Ferrini, and D. M. Noonan. 1998. Identification of a novel domain of HIV tat involved in monocyte chemotaxis. J. Biol. Chem. 273:15895-15900. [DOI] [PubMed] [Google Scholar]
- 2.Aquaro, S., E. Balestra, A. Cenci, M. Francesconi, R. Calio, and C. F. Perno. 1997. HIV infection in macrophage: role of long-lived cells and related therapeutical strategies. J. Biol. Regul. Homeost. Agents 11:69-73. [PubMed] [Google Scholar]
- 3.Aquaro, S., R. Calio, J. Balzarini, M. C. Bellocchi, E. Garaci, and C. F. Perno. 2002. Macrophages and HIV infection: therapeutical approaches toward this strategic virus reservoir. Antivir. Res. 55:209-225. [DOI] [PubMed] [Google Scholar]
- 4.Argyropoulos, C., G. C. Nikiforidis, M. Theodoropoulou, P. Adamopoulos, S. Boubali, T. N. Georgakopoulos, F. Paliogianni, A. G. Papavassiliou, and A. Mouzaki. 2004. Mining microarray data to identify transcription factors expressed in naive resting but not activated T lymphocytes. Genes Immun. 5:16-25. [DOI] [PubMed] [Google Scholar]
- 5.Arlt, M. F., A. M. Casper, and T. W. Glover. 2003. Common fragile sites. Cytogenet. Genome Res. 100:92-100. [DOI] [PubMed] [Google Scholar]
- 6.Auer, K. L., J. S. Park, P. Seth, R. J. Coffey, G. Darlington, A. Abo, M. McMahon, R. A. Depinho, P. B. Fisher, and P. Dent. 1998. Prolonged activation of the mitogen-activated protein kinase pathway promotes DNA synthesis in primary hepatocytes from p21Cip-1/WAF1-null mice, but not in hepatocytes from p16INK4a-null mice. Biochem. J. 336:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balestra, E., C. F. Perno, S. Aquaro, S. Panti, A. Bertoli, M. Piacentini, F. Forbici, R. D'Arrigo, R. Calio, and E. Garaci. 2001. Macrophages: a crucial reservoir for human immunodeficiency virus in the body. J. Biol. Regul. Homeost. Agents 15:272-276. [PubMed] [Google Scholar]
- 8.Bartz, S. R., and M. Emerman. 1999. Human immunodeficiency virus type 1 Tat induces apoptosis and increases sensitivity to apoptotic signals by up-regulating FLICE/caspase-8. J. Virol. 73:1956-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biggs, B. A., M. Hewish, S. Kent, K. Hayes, and S. M. Crowe. 1995. HIV-1 infection of human macrophages impairs phagocytosis and killing of Toxoplasma gondii. J. Immunol. 154:6132-6139. [PubMed] [Google Scholar]
- 10.Bottazzi, M. E., X. Zhu, R. M. Bohmer, and R. K. Assoian. 1999. Regulation of p21(cip1) expression by growth factors and the extracellular matrix reveals a role for transient ERK activity in G1 phase. J. Cell Biol. 146:1255-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bovolenta, C., L. Camorali, M. Mauri, S. Ghezzi, S. Nozza, G. Tambussi, A. Lazzarin, and G. Poli. 2001. Expression and activation of a C-terminal truncated isoform of STAT5 (STAT5 Delta) following interleukin 2 administration or AZT monotherapy in HIV-infected individuals. Clin. Immunol. 99:75-81. [DOI] [PubMed] [Google Scholar]
- 12.Brodie, S. J. 2000. Nonlymphoid reservoirs of HIV replication in children with chronic-progressive disease. J. Leukoc. Biol. 68:351-359. [PubMed] [Google Scholar]
- 13.Brooks, D. G., and J. A. Zack. 2002. Effect of latent human immunodeficiency virus infection on cell surface phenotype. J. Virol. 76:1673-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cefai, D., P. Debre, M. Kaczorek, T. Idziorek, B. Autran, and G. Bismuth. 1990. Human immunodeficiency virus-1 glycoproteins gp120 and gp160 specifically inhibit the CD3/T cell-antigen receptor phosphoinositide transduction pathway. J. Clin. Investig. 86:2117-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan, G. K., and T. J. Yen. 2003. The mitotic checkpoint: a signaling pathway that allows a single unattached kinetochore to inhibit mitotic exit. Prog. Cell Cycle Res. 5:431-439. [PubMed] [Google Scholar]
- 16.Cicala, C., J. Arthos, S. M. Selig, G. Dennis, Jr., D. A. Hosack, D. Van Ryk, M. L. Spangler, T. D. Steenbeke, P. Khazanie, N. Gupta, J. Yang, M. Daucher, R. A. Lempicki, and A. S. Fauci. 2002. HIV envelope induces a cascade of cell signals in non-proliferating target cells that favor virus replication. Proc. Natl. Acad. Sci. USA 99:9380-9385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cicala, C., J. Arthos, A. Rubbert, S. Selig, K. Wildt, O. J. Cohen, and A. S. Fauci. 2000. HIV-1 envelope induces activation of Caspase-3 and cleavage of focal adhesion kinase in primary human CD4+ T cells. Proc. Natl. Acad. Sci. USA 97:1178-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark, E., F. Santiago, L. Deng, S. Chong, C. de La Fuente, L. Wang, P. Fu, D. Stein, T. Denny, V. Lanka, F. Mozafari, T. Okamoto, and F. Kashanchi. 2000. Loss of G1/S checkpoint in human immunodeficiency virus type 1-infected cells is associated with a lack of cyclin-dependent kinase inhibitor p21/Waf1. J. Virol. 74:5040-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cota, M., M. Mengozzi, E. Vicenzi, P. Panina-Bordignon, F. Sinigaglia, P. Transidico, S. Sozzani, A. Mantovani, and G. Poli. 2000. Selective inhibition of HIV replication in primary macrophages but not T lymphocytes by macrophage-derived chemokine. Proc. Natl. Acad. Sci. USA 97:9162-9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crowe, S. M., N. J. Vardaxis, S. J. Kent, A. L. Maerz, M. J. Hewish, M. S. McGrath, and J. Mills. 1994. HIV infection of monocyte-derived macrophages in vitro reduces phagocytosis of Candida albicans. J. Leukoc. Biol. 56:318-327. [DOI] [PubMed] [Google Scholar]
- 21.Del Sorbo, L., A. DeMartino, L. Biancone, B. Bussolati, P. G. Conaldi, A. Toniolo, and G. Camussi. 1999. The synthesis of platelet-activating factor modulates chemotaxis of monocytes induced by HIV-1 Tat. Eur. J. Immunol. 29:1513-1521. [DOI] [PubMed] [Google Scholar]
- 22.Elder, R. T., Z. Benko, and Y. Zhao. 2002. HIV-1 VPR modulates cell cycle G2/M transition through an alternative cellular mechanism other than the classic mitotic checkpoints. Front. Biosci. 7:D349-D357. [DOI] [PubMed] [Google Scholar]
- 23.Elder, R. T., M. Yu, M. Chen, X. Zhu, M. Yanagida, and Y. Zhao. 2001. HIV-1 Vpr induces cell cycle G2 arrest in fission yeast (Schizosaccharomyces pombe) through a pathway involving regulatory and catalytic subunits of PP2A and acting on both Wee1 and CDC25. Virology 287:359-370. [DOI] [PubMed] [Google Scholar]
- 24.Fantuzzi, L., L. Conti, M. C. Gauzzi, P. Eid, M. Del Corno, B. Varano, I. Canini, F. Belardelli, and S. Gessani. 2000. Regulation of chemokine/cytokine network during in vitro differentiation and HIV-1 infection of human monocytes: possible importance in the pathogenesis of AIDS. J. Leukoc. Biol. 68:391-399. [PubMed] [Google Scholar]
- 25.Fischer-Smith, T., S. Croul, A. E. Sverstiuk, C. Capini, D. L'Heureux, E. G. Regulier, M. W. Richardson, S. Amini, S. Morgello, K. Khalili, and J. Rappaport. 2001. CNS invasion by CD14+/CD16+ peripheral blood-derived monocytes in HIV dementia: perivascular accumulation and reservoir of HIV infection. J. Neurovirol. 7:528-541. [DOI] [PubMed] [Google Scholar]
- 26.Furtado, M. R., D. S. Callaway, J. P. Phair, K. J. Kunstman, J. L. Stanton, C. A. Macken, A. S. Perelson, and S. M. Wolinsky. 1999. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N. Engl. J. Med. 340:1614-1622. [DOI] [PubMed] [Google Scholar]
- 27.Garbuglia, A. R., M. Zaccarelli, S. Calcaterra, G. Cappiello, R. Marini, and A. Benedetto. 2001. Dynamics of viral load in plasma and HIV DNA in lymphocytes during highly active antiretroviral therapy (HAART): high viral burden in macrophages after 1 year of treatment. J. Chemother. 13:188-194. [DOI] [PubMed] [Google Scholar]
- 28.Geiss, G. K., R. E. Bumgarner, M. C. An, M. B. Agy, A. B. 't Wout, E. Hammersmark, V. S. Carter, D. Upchurch, J. I. Mullins, and M. G. Katze. 2000. Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology 266:8-16. [DOI] [PubMed] [Google Scholar]
- 29.Geleziunas, R., S. Bour, and M. A. Wainberg. 1994. HIV-1 associated down-modulation of CD4 gene expression is differentially restricted in lymphocytic and monocytic cell lines. J. Leukoc. Biol. 55:589-595. [DOI] [PubMed] [Google Scholar]
- 30.Gendelman, H. E., J. M. Orenstein, L. M. Baca, B. Weiser, H. Burger, D. C. Kalter, and M. S. Meltzer. 1989. The macrophage in the persistence and pathogenesis of HIV infection. AIDS 3:475-495. [DOI] [PubMed] [Google Scholar]
- 31.Gendelman, H. E., J. M. Orenstein, M. A. Martin, C. Ferrua, R. Mitra, T. Phipps, L. A. Wahl, H. C. Lane, A. S. Fauci, and D. S. Burke. 1988. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J. Exp. Med. 167:1428-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goh, W. C., N. Manel, and M. Emerman. 2004. The human immunodeficiency virus Vpr protein binds Cdc25C: implications for G2 arrest. Virology 318:337-349. [DOI] [PubMed] [Google Scholar]
- 33.Goh, W. C., M. E. Rogel, C. M. Kinsey, S. F. Michael, P. N. Fultz, M. A. Nowak, B. H. Hahn, and M. Emerman. 1998. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat. Med. 4:65-71. [DOI] [PubMed] [Google Scholar]
- 34.Goodenow, M. M., S. L. Rose, D. L. Tuttle, and J. W. Sleasman. 2003. HIV-1 fitness and macrophages. J. Leukoc. Biol. 74:657-666. [DOI] [PubMed] [Google Scholar]
- 35.Gummuluru, S., and M. Emerman. 1999. Cell cycle- and Vpr-mediated regulation of human immunodeficiency virus type 1 expression in primary and transformed T-cell lines. J. Virol. 73:5422-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta, S., S. Aggarwal, C. Kim, and S. Gollapudi. 1994. Human immunodeficiency virus-1 recombinant gp120 induces changes in protein kinase C isozymes—a preliminary report. Int. J. Immunopharmacol. 16:197-204. [DOI] [PubMed] [Google Scholar]
- 37.Hobbs, W. E., and N. A. DeLuca. 1999. Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J. Virol. 73:8245-8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin, S., M. J. Antinore, F. D. Lung, X. Dong, H. Zhao, F. Fan, A. B. Colchagie, P. Blanck, P. P. Roller, A. J. Fornace, Jr., and Q. Zhan. 2000. The GADD45 inhibition of Cdc2 kinase correlates with GADD45-mediated growth suppression. J. Biol. Chem. 275:16602-16608. [DOI] [PubMed] [Google Scholar]
- 39.Jung, A., R. Maier, J. P. Vartanian, G. Bocharov, V. Jung, U. Fischer, E. Meese, S. Wain-Hobson, and A. Meyerhans. 2002. Multiply infected spleen cells in HIV patients. Nature 418:144. [DOI] [PubMed] [Google Scholar]
- 40.Khati, M., W. James, and S. Gordon. 2001. HIV-macrophage interactions at the cellular and molecular level. Arch. Immunol. Ther. Exp. 49:367-378. [PubMed] [Google Scholar]
- 41.Kinoshita, N., H. Yamano, H. Niwa, T. Yoshida, and M. Yanagida. 1993. Negative regulation of mitosis by the fission yeast protein phosphatase ppa2. Genes Dev. 7:1059-1071. [DOI] [PubMed] [Google Scholar]
- 42.Knight, S. C., and S. E. Macatonia. 1991. Effect of HIV on antigen presentation by dendritic cells and macrophages. Res. Virol. 142:123-128. [DOI] [PubMed] [Google Scholar]
- 43.Kohler, J. J., D. L. Tuttle, C. R. Coberley, J. W. Sleasman, and M. M. Goodenow. 2003. Human immunodeficiency virus type 1 (HIV-1) induces activation of multiple STATs in CD4+ cells of lymphocyte or monocyte/macrophage lineages. J. Leukoc. Biol. 73:407-416. [DOI] [PubMed] [Google Scholar]
- 44.Kootstra, N. A., B. M. Zwart, and H. Schuitemaker. 2000. Diminished human immunodeficiency virus type 1 reverse transcription and nuclear transport in primary macrophages arrested in early G1 phase of the cell cycle. J. Virol. 74:1712-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koziel, H., Q. Eichbaum, B. A. Kruskal, P. Pinkston, R. A. Rogers, M. Y. Armstrong, F. F. Richards, R. M. Rose, and R. A. Ezekowitz. 1998. Reduced binding and phagocytosis of Pneumocystis carinii by alveolar macrophages from persons infected with HIV-1 correlates with mannose receptor downregulation. J. Clin. Investig. 102:1332-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krek, W., and E. A. Nigg. 1991. Mutations of p34cdc2 phosphorylation sites induce premature mitotic events in HeLa cells: evidence for a double block to p34cdc2 kinase activation in vertebrates. EMBO J. 10:3331-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuhn, L., A. Coutsoudis, D. Moodley, N. Mngqundaniso, D. Trabattoni, G. M. Shearer, M. Clerici, and H. M. Coovadia. 2001. Interferon-gamma and interleukin-10 production among HIV-1-infected and uninfected infants of HIV-1-infected mothers. Pediatr. Res. 50:412-416. [DOI] [PubMed] [Google Scholar]
- 48.Latella, L., A. Sacco, D. Pajalunga, M. Tiainen, D. Macera, M. D'Angelo, A. Felici, A. Sacchi, and M. Crescenzi. 2001. Reconstitution of cyclin D1-associated kinase activity drives terminally differentiated cells into the cell cycle. Mol. Cell. Biol. 21:5631-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lathey, J. L., D. Brambilla, M. M. Goodenow, M. Nokta, S. Rasheed, E. B. Siwak, J. W. Bremer, D. D. Huang, Y. Yi, P. S. Reichelderfer, and R. G. Collman. 2000. Co-receptor usage was more predictive than NSI/SI phenotype for HIV replication in macrophages: is NSI/SI phenotyping sufficient? J. Leukoc. Biol. 68:324-330. [PubMed] [Google Scholar]
- 50.Lewin, S. R., J. Kirihara, S. Sonza, L. Irving, J. Mills, and S. M. Crowe. 1998. HIV-1 DNA and mRNA concentrations are similar in peripheral blood monocytes and alveolar macrophages in HIV-1-infected individuals. AIDS 12:719-727. [DOI] [PubMed] [Google Scholar]
- 51.Macatonia, S. E., M. Gompels, A. J. Pinching, S. Patterson, and S. C. Knight. 1992. Antigen-presentation by macrophages but not by dendritic cells in human immunodeficiency virus (HIV) infection. Immunology 75:576-581. [PMC free article] [PubMed] [Google Scholar]
- 52.Michie, C. A., A. McLean, C. Alcock, and P. C. Beverley. 1992. Lifespan of human lymphocyte subsets defined by CD45 isoforms. Nature 360:264-265. [DOI] [PubMed] [Google Scholar]
- 53.Norbury, C., J. Blow, and P. Nurse. 1991. Regulatory phosphorylation of the p34cdc2 protein kinase in vertebrates. EMBO J. 10:3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piedimonte, G., D. Corsi, M. Paiardini, G. Cannavo, R. Ientile, I. Picerno, M. Montroni, G. Silvestri, and M. Magnani. 1999. Unscheduled cyclin B expression and p34 CDC2 activation in T lymphocytes from HIV-infected patients. AIDS 13:1159-1164. [DOI] [PubMed] [Google Scholar]
- 55.Poggioli, G. J., R. L. DeBiasi, R. Bickel, R. Jotte, A. Spalding, G. L. Johnson, and K. L. Tyler. 2002. Reovirus-induced alterations in gene expression related to cell cycle regulation. J. Virol. 76:2585-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poli, G., and A. S. Fauci. 1992. The role of monocyte/macrophages and cytokines in the pathogenesis of HIV infection. Pathobiology 60:246-251. [DOI] [PubMed] [Google Scholar]
- 57.Popik, W., J. E. Hesselgesser, and P. M. Pitha. 1998. Binding of human immunodeficiency virus type 1 to CD4 and CXCR4 receptors differentially regulates expression of inflammatory genes and activates the MEK/ERK signaling pathway. J. Virol. 72:6406-6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pumiglia, K. M., and S. J. Decker. 1997. Cell cycle arrest mediated by the MEK/mitogen-activated protein kinase pathway. Proc. Natl. Acad. Sci. USA 94:448-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reardon, C. C., S. J. Kim, R. P. Wagner, H. Koziel, and H. Kornfeld. 1996. Phagocytosis and growth inhibition of Cryptococcus neoformans by human alveolar macrophages: effects of HIV-1 infection. AIDS 10:613-618. [DOI] [PubMed] [Google Scholar]
- 60.Roshal, M., B. Kim, Y. Zhu, P. Nghiem, and V. Planelles. 2003. Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J. Biol. Chem. 278:25879-25886. [DOI] [PubMed] [Google Scholar]
- 61.Ryo, A., Y. Suzuki, K. Ichiyama, T. Wakatsuki, N. Kondoh, A. Hada, M. Yamamoto, and N. Yamamoto. 1999. Serial analysis of gene expression in HIV-1-infected T cell lines. FEBS Lett. 462:182-186. [DOI] [PubMed] [Google Scholar]
- 62.Schacker, T., S. Little, E. Connick, K. Gebhard, Z. Q. Zhang, J. Krieger, J. Pryor, D. Havlir, J. K. Wong, R. T. Schooley, D. Richman, L. Corey, and A. T. Haase. 2001. Productive infection of T cells in lymphoid tissues during primary and early human immunodeficiency virus infection. J. Infect. Dis. 183:555-562. [DOI] [PubMed] [Google Scholar]
- 63.Scheuring, U. J., J. Corbeil, D. E. Mosier, and A. N. Theofilopoulos. 1998. Early modification of host cell gene expression induced by HIV-1. AIDS 12:563-570. [DOI] [PubMed] [Google Scholar]
- 64.Schuitemaker, H., N. A. Kootstra, R. A. Fouchier, B. Hooibrink, and F. Miedema. 1994. Productive HIV-1 infection of macrophages restricted to the cell fraction with proliferative capacity. EMBO J. 13:5929-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Selliah, N., and T. H. Finkel. 2001. HIV-1 NL4-3, but not IIIB, inhibits JAK3/STAT5 activation in CD4+ T cells. Virology 286:412-421. [DOI] [PubMed] [Google Scholar]
- 66.Shrikant, P., D. J. Benos, L. P. Tang, and E. N. Benveniste. 1996. HIV glycoprotein 120 enhances intercellular adhesion molecule-1 gene expression in glial cells. Involvement of Janus kinase/signal transducer and activator of transcription and protein kinase C signaling pathways. J. Immunol. 156:1307-1314. [PubMed] [Google Scholar]
- 67.Sleasman, J. W., L. F. Aleixo, A. Morton, S. Skoda-Smith, and M. M. Goodenow. 1996. CD4+ memory T cells are the predominant population of HIV-1-infected lymphocytes in neonates and children. AIDS 10:1477-1484. [DOI] [PubMed] [Google Scholar]
- 68.Subbramanian, R. A., A. Kessous-Elbaz, R. Lodge, J. Forget, X. J. Yao, D. Bergeron, and E. A. Cohen. 1998. Human immunodeficiency virus type 1 Vpr is a positive regulator of viral transcription and infectivity in primary human macrophages. J. Exp. Med. 187:1103-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sudakin, V., G. K. Chan, and T. J. Yen. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154:925-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tamaskovic, R., S. J. Bichsel, and B. A. Hemmings. 2003. NDR family of AGC kinases—essential regulators of the cell cycle and morphogenesis. FEBS Lett. 546:73-80. [DOI] [PubMed] [Google Scholar]
- 71.Taylor, W. R., and G. R. Stark. 2001. Regulation of the G2/M transition by p53. Oncogene 20:1803-1815. [DOI] [PubMed] [Google Scholar]
- 72.Todorov, I. T., R. Pepperkok, R. N. Philipova, S. E. Kearsey, W. Ansorge, and D. Werner. 1994. A human nuclear protein with sequence homology to a family of early S phase proteins is required for entry into S phase and for cell division. J. Cell Sci. 107:253-265. [DOI] [PubMed] [Google Scholar]
- 73.Tuttle, D. L., C. B. Anders, M. J. Aquino-De Jesus, P. P. Poole, S. L. Lamers, D. R. Briggs, S. M. Pomeroy, L. Alexander, K. W. Peden, W. A. Andiman, J. W. Sleasman, and M. M. Goodenow. 2002. Increased replication of non-syncytium-inducing HIV type 1 isolates in monocyte-derived macrophages is linked to advanced disease in infected children. AIDS Res. Hum. Retrovir. 18:353-362. [DOI] [PubMed] [Google Scholar]
- 74.Tuttle, D. L., C. R. Coberley, X. Xie, Z. C. Kou, J. W. Sleasman, and M. M. Goodenow. 2004. Effects of human immunodeficiency virus type 1 infection on CCR5 and CXCR4 coreceptor expression on CD4 T lymphocyte subsets in infants and adolescents. AIDS Res. Hum. Retrovir. 20:305-313. [DOI] [PubMed] [Google Scholar]
- 75.Tuttle, D. L., J. K. Harrison, C. Anders, J. W. Sleasman, and M. M. Goodenow. 1998. Expression of CCR5 increases during monocyte differentiation and directly mediates macrophage susceptibility to infection by human immunodeficiency virus type 1. J. Virol. 72:4962-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vahey, M. T., M. E. Nau, L. L. Jagodzinski, J. Yalley-Ogunro, M. Taubman, N. L. Michael, and M. G. Lewis. 2002. Impact of viral infection on the gene expression profiles of proliferating normal human peripheral blood mononuclear cells infected with HIV type 1 RF. AIDS Res. Hum. Retrovir. 18:179-192. [DOI] [PubMed] [Google Scholar]
- 77.Vairapandi, M., A. G. Balliet, B. Hoffman, and D. A. Liebermann. 2002. GADD45b and GADD45g are cdc2/cyclinB1 kinase inhibitors with a role in S and G2/M cell cycle checkpoints induced by genotoxic stress. J. Cell Physiol. 192:327-338. [DOI] [PubMed] [Google Scholar]
- 78.Wahl, S. M., T. Greenwell-Wild, G. Peng, G. Ma, J. M. Orenstein, and N. Vazquez. 2003. Viral and host cofactors facilitate HIV-1 replication in macrophages. J. Leukoc. Biol. 74:726-735. [DOI] [PubMed] [Google Scholar]
- 79.Wainberg, Z., M. Oliveira, S. Lerner, Y. Tao, and B. G. Brenner. 1997. Modulation of stress protein (hsp27 and hsp70) expression in CD4+ lymphocytic cells following acute infection with human immunodeficiency virus type-1. Virology 233:364-373. [DOI] [PubMed] [Google Scholar]
- 80.Wang, X., and D. E. Lewis. 2001. CD86 expression correlates with amounts of HIV produced by macrophages in vitro. J. Leukoc. Biol. 69:405-413. [PubMed] [Google Scholar]
- 81.Wang, Z., G. F. Morris, J. C. Reed, G. D. Kelly, and C. B. Morris. 1999. Activation of Bcl-2 promoter-directed gene expression by the human immunodeficiency virus type-1 Tat protein. Virology 257:502-510. [DOI] [PubMed] [Google Scholar]
- 82.Williams, K. C., S. Corey, S. V. Westmoreland, D. Pauley, H. Knight, C. deBakker, X. Alvarez, and A. A. Lackner. 2001. Perivascular macrophages are the primary cell type productively infected by simian immunodeficiency virus in the brains of macaques: implications for the neuropathogenesis of AIDS. J. Exp. Med. 193:905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xaus, J., M. Comalada, A. F. Valledor, M. Cardo, C. Herrero, C. Soler, J. Lloberas, and A. Celada. 2001. Molecular mechanisms involved in macrophage survival, proliferation, activation or apoptosis. Immunobiology 204:543-550. [DOI] [PubMed] [Google Scholar]
- 84.Yarden, R. I., S. Pardo-Reoyo, M. Sgagias, K. H. Cowan, and L. C. Brody. 2002. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat. Genet. 30:285-289. [DOI] [PubMed] [Google Scholar]
- 85.Zack, J. A. 1995. The role of the cell cycle in HIV-1 infection. Adv. Exp. Med. Biol. 374:27-31. [DOI] [PubMed] [Google Scholar]
- 86.Zhang, L., B. Ramratnam, K. Tenner-Racz, Y. He, M. Vesanen, S. Lewin, A. Talal, P. Racz, A. S. Perelson, B. T. Korber, M. Markowitz, and D. D. Ho. 1999. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N. Engl. J. Med. 340:1605-1613. [DOI] [PubMed] [Google Scholar]