Abstract
To address whether human herpesvirus 8 (HHV-8) DNA in peripheral blood mononuclear cells (PBMCs) might be the product of latent or lytic infection and to shed light on sporadic detection of HHV-8 DNA in individuals seropositive for the virus, we studied the frequency of infected cells, total virus load, and virus load per infected cell in PBMCs from men coinfected with HHV-8 and human immunodeficiency virus (HIV), some of whom had Kaposi's sarcoma. The low frequencies of infected cells detected (fewer than one per million cells in some individuals) suggest that the prevalence of the virus in circulating leukocytes was underestimated in previous studies that employed more conventional sampling methods (single, small-volume specimens). Mean virus loads ranged from 3 to 330 copies per infected PBMC; these numbers can represent much higher loads in individual lytically infected cells (>103 genomes/cell) in mixtures that consist predominantly of latently (relatively few genomes) infected cells. The presence in some subjects of high HHV-8 mean genome copy numbers per infected cell, together with viral DNA being found in plasma only from subjects with positive PBMCs, supports earlier suggestions that the virus can actively replicate in PBMCs. In some individuals, mean virus loads were less than 10 genomes per infected cell, suggesting a tightly controlled purely latent state. HHV-8 genome copy numbers are substantially higher in latently infected cells derived from primary effusion lymphomas; thus, it appears that HHV-8 is able to adopt more than one latency program, perhaps analogous to the several types of Epstein-Barr virus latency.
Kaposi's sarcoma (KS) is an endothelial neoplasm characterized by intense angiogenesis, proliferation of spindle-like cells, and inflammation (20). Of the four epidemiologic forms, epidemic KS (also known as AIDS-KS) is frequently aggressive and is the neoplasm most frequently associated with human immunodeficiency virus (HIV) infection in the United States (15).
Human herpesvirus 8 (HHV-8, or KS-associated herpesvirus) was discovered in KS lesions (11) and is necessary for the development of the disease (30). Essentially all KS lesions are positive for HHV-8 DNA. Although important aspects of its biology take place in nonlymphoid cells (e.g., KS lesion spindle cells), like the other gammaherpesviruses (e.g., Epstein-Barr virus [EBV]), HHV-8 is lymphotropic. B cells are an important target for viral infection (1, 12, 17), and latency is established in lymphoid cells. Detection of viral DNA in peripheral blood mononuclear cells (PBMCs) correlates with the appearance of KS and is a useful indicator of KS risk (1, 10, 13, 36). Nonetheless, HHV-8 DNA is commonly detected in only about 50% of PBMC specimens from subjects with AIDS-KS (who are thus obviously infected with the virus). In comparison, EBV viral load in PBMCs is much more tightly linked to the development and progression of EBV-related malignancies, including nasopharyngeal carcinoma, Hodgkin's disease, posttransplantation lymphoproliferative disease (PTLD), and AIDS-related lymphoma. At the time of PTLD diagnosis, virus loads in PBMCs are 1,000- to 10,000-fold higher than those in healthy seropositive individuals (28). These and other differences noted by others (14) make it clear that although they have some similarities, the biologies of latency and persistence for EBV and HHV-8 are distinct.
Much of what we know of HHV-8 latency has been learned from study of transformed cell lines derived from HHV-8-associated primary effusion lymphomas (PELs). While this has been both convenient and informative, PEL is a very rare outcome of HHV-8 infection, and there are certain to be important differences between the biology of the virus in these cells and in vivo in HHV-8-infected individuals without PEL.
Thus there are several outstanding questions about the mechanisms by which HHV-8 persists in PBMCs and the relationship of persistence to KS pathogenesis. Does the virus in PBMCs represent lytic or latent infection? What is the balance between lytically and latently infected PBMCs? Are PBMCs (be they lytically or latently infected) the source of virus for initiating KS lesions, or do they become infected by circulating in proximity of a nexus of infection (such as a KS lesion) (2)? In individuals who have KS or are HHV-8 seropositive, but whose PBMCs are negative for viral DNA by PCR, is the virus load simply beneath the level of detection or is the virus truly absent?
We have been studying the behavior of the virus in the natural host, focusing on men infected with both HIV and HHV-8, some with KS (10, 16a). Thus far, studies by others have focused on the number of HHV-8 genomes per volume of blood or per quantity of PBMCs (4, 6, 8, 23, 33). The frequency of infected blood cells or the number of HHV-8 genomes per infected cell, fundamental metrics in answering some of the outstanding questions outlined above, has not been measured. Here we describe the results obtained with a PCR-based limiting dilution assay developed for this purpose.
MATERIALS AND METHODS
Subjects and specimens.
Thirteen HIV-1-infected men who have sex with men were included in the study. All were HHV-8 seropositive; nine had a KS history (five with active and four with prior disease). Because they are infected with both HIV-1 and HHV-8, those who never had KS are referred to as having high risk for KS. Patients were chosen from individuals participating in ongoing studies of HIV-related disease and HHV-8 (10, 31). Informed consent was obtained from subjects, and human experimentation guidelines of the U.S. Department of Health and Human Services and those of the authors' institutions were followed in the conduct of this research. PBMCs were obtained by Ficoll density gradient centrifugation of heparinized or EDTA-treated peripheral blood. For subject 12, PBMCs were obtained by leukapheresis. Residual erythrocytes in isolated PBMCs were lysed with 155 mM ammonium chloride. PBMCs were preserved in cryopreservation medium and stored in liquid nitrogen. Serum and plasma were stored at −20°C.
HHV-8 serology.
All sera were screened in duplicate wells with an enzyme-linked immunosorbent assay (ELISA) using a peptide derived from the HHV-8 open reading frame (ORF) K8.1 virion envelope protein (31). Endpoint titers were determined using twofold dilutions. Antigen preparation, plate coating, and ELISA methods were as described previously (22). The assay cutoff was the mean corrected optical density at 450 nm (OD450) of five pools of negative control sera plus 0.150 OD units. Lytic antigen immunofluorescence was done on BCBL-1 cells as previously described (24).
HIV virus load measurement.
HIV virus loads were measured with a duplex fluorescent 5′ nuclease reverse transcription-PCR (RT-PCR; Taqman) (Ou et al., unpublished data). HIV-1 RNA was isolated from 200 μl of plasma with a QIAGEN virus kit (QIAGEN, Valencia, Calif.). An RNA-containing phage was added to the plasma to serve as an internal calibrator for RNA isolation and the subsequent RT-PCR detection. Plasma RNA was eluted in 50 μl of water, and 25 μl was used in the assay, which consisted of RT, amplification, and simultaneous detection of HIV and the internal control RNA, with a dynamic detection range extending from 457 to 1 million HIV-1 copies per ml.
CD4 counts.
CD4 counts and percentages were analyzed on whole blood by fluorescence-activated cell sorting analysis on FACScan or FACSort using TriTEST or MultiTEST reagents with MultiSET software (Becton Dickinson Biosciences, San Jose, Calif.).
DNA extraction from plasma.
DNA present in 200 μl of clarified plasma (500 × g for 15 min) was extracted with the QIAamp DNA blood mini kit (QIAGEN) after addition of 6 μg of poly(dA) as a carrier (Roche Molecular Biochemicals). Extracted DNA was eluted in 60 μl of distilled deionized UV-treated type II water. Negative and positive controls consisted of 200 μl of plasma from an HHV-8 PCR-negative blood donor, both alone and spiked with 100 copies of control plasmid containing the HHV-8 orf37 PCR target (orf37-pPICZαA), respectively.
LDA.
The limiting dilution assay (LDA) was performed as follows. As described by Trippler et al. (34), multiple replicates of serial threefold dilutions of both intact and lysed cells were analyzed in a separate series of experiments to measure both the number of cells containing one or more HHV-8 genome copies (infected-cell frequency) and the number of lysed cells needed to detect one copy on average (genome frequency). The quotient of these two frequencies provides the mean HHV-8 genome copy number per infected cell.
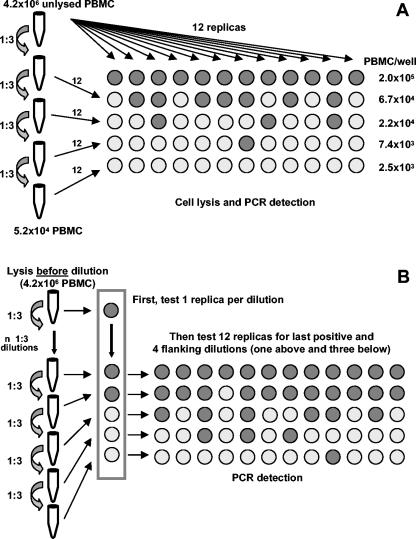
For the cell dilution method, four serial threefold dilutions of cells were made starting from a suspension of 4.2 × 106 PBMCs and 12 replicates per dilution were tested: the number of cells per well ranged from 2 × 105 to 2.5 × 103 (Fig. 1A). PBMCs were lysed in 96-well plates in a final volume of 10 μl of lysis buffer (10 mM Tris-HCl, pH 8.0, 0.5% lauryl ether, 100-mg/ml proteinase K), incubated for 2 h at 65°C, and then inactivated for 15 min at 95°C in a thermocycler (96-well GeneAmp PCR system 9700; Applied Biosystems).
FIG. 1.
LDA. (A) Cell dilution LDA for the assessment of infected-cell frequencies. (B) Cell lysate dilution LDA for the assessment of genome frequencies.
For the cell lysate dilution method (Fig. 1B), 4.2 × 106 cells were lysed in 210 μl of lysis buffer, incubated for 2 h at 65°C, and then inactivated for 15 min at 95°C in a heating block. Six 1:3 dilutions were then made in the same way as for the cell dilution method. To determine the first negative dilution, 1 of 13 replicates from each threefold dilution was tested by quantitative PCR in a preliminary experiment. The other 12 replicates were subsequently tested for the five serial dilutions surrounding and including this dilution (one above and three below the last positive dilution). Dilutions were done on ice.
Although a quantitative Taqman PCR system was used in the LDA experiments, the LDA calculations were based solely on the qualitative use of the Taqman results (positive or negative for each well).
Quantitative PCR.
A duplex quantitative Taqman PCR system was optimized for HHV-8 orf37 and cellular RNase P. The PCR primers and 6-carboxyfluorescein (FAM)-labeled probe used to quantify orf37 have been described previously (32), but a Black Hole Quencher (BHQ1; Biosearch Technologies, Novato, Calif.) was used instead of 6-carboxytetramethylrhodamine (TAMRA). The forward and reverse primers used for RNase P amplification (kindly provided by Karen McCaustland and Brian Holloway) were 5′-AGATTTGGACCTGCGAGCG-3′ and 5′-GAGCGGCTGTCTCCACAAGT-3′, respectively; the probe was 5′-TTCTGACCTGAAGGCTCTGCGCG-3′, labeled at the 5′ end with 6-carboxy-4,5-dichloro-2,7-dimethoxyfluorescein (JOE) and at the 3′ end with BHQ. Primers and probes were synthesized by standard phosphoramide chemistry techniques at the Centers for Disease Control and Prevention Biotechnology Core Facility. Amplification was performed in 50-μl reaction mixtures consisting of 2× universal PCR Mastermix (Applied Biosystems) containing Amp Erase uracil N-glycosylase (UNG), primers and probes for both HHV-8 and RNase P, and 10 μl of template DNA. To achieve an efficient duplex reaction, primer/probe concentrations for both targets were reoptimized starting from previously optimized concentrations for single-plex amplification (32). The optimized concentrations for the HHV-8 orf37 primers were 900 nM, 250 nM for the probe, and 100 nM for the RNase P primers and probe. Following activation of the UNG (2 min at 50°C) and then the polymerase (10 min at 95°C), 45 cycles (15 s at 95°C and 1 min at 60°C) were performed with an ABI Prism 7700 Sequence detector system (Applied Biosystems). Ten-fold dilutions of the orf37-pPICZαA plasmid were used to generate an HHV-8 standard curve (10,000 to 10 copies, in duplicate) or controls for single-copy and 10-copy sensitivity (1 or 10 copies in each of six replicates), by adding the plasmid dilutions to lysis buffer alone or to 2 × 105 uninfected PBMCs prior to cell lysis. The standard curve dilutions, the controls for one-copy sensitivity, and negative controls (lysed uninfected PBMCs alone and water alone) were included in each 96-well PCR plate. For the RNase P standard curve, purified human DNA was used (Roche Molecular Biochemicals, Indianapolis, Ind.) in a background of lysis buffer. For the clinical specimens, the quantity of human DNA in micrograms determined by quantitative PCR based on the standard curve was converted to cell number, assuming 6.6 pg of human DNA per diploid cell. The absence of inhibitors in the processed clinical specimens was assessed by verifying that the RNase P copy number decreased linearly along the dilution series.
To further exclude the presence of PCR inhibition by the crude cell lysates, the highest cell concentration (2 × 105 PBMCs per 10 μl) was tested for each subject using the TaqMan Exogenous Internal Positive Control Reagents kit, which contains a preoptimized internal positive control (IPC). IPC (0.2 fg) was spiked into samples to distinguish true target negatives from PCR inhibition (32). The IPC DNA was detected with limiting concentrations of primers and a VIC-labeled probe. HHV-8 virus load in plasma specimens was assessed with this HHV-8/IPC duplex reaction. Data acquisition and analysis were done with sequence detector software (version 1.7; Applied Biosystems).
Statistical analysis.
For the cell dilution and cell lysate dilution methods, frequencies of infected cells and frequency of HHV-8 genomes were calculated from the percentage of negative wells at each tested dilution according to Poisson statistics. These calculations were done with the QUALITY program (Mullins Molecular Retrovirology Laboratory; http://ubik.microbiol.washington.edu/cbg/jquality.htm), which is based on a minimum chi-square method (27). Chi-square and P values were used to test goodness of fit.
RESULTS
PCR optimization and validation.
Several aspects of the PCR assays were optimized and validated. In all quantitative PCR assays, the standard curve (threshold cycle (Ct) versus the logarithm of the plasmid copy number) was linear (r2 ≥ 0.99). The slopes of the standard curves indicated that after optimization, PCR amplification efficiency was over 85% for RNase P and over 90% for HHV-8. The presence of a crude lysate corresponding to 2 × 105 PBMCs from an HHV-8 PCR-negative blood donor had negligible effects on amplification efficiency (data not shown). Moreover, low concentrations of IPC were positive for all specimens tested, further ruling out PCR inhibition. The sensitivity of the technique was tested by using dilutions of a control plasmid containing the HHV-8 orf37 target; it was possible to amplify approximately one copy of HHV-8 DNA, in a background of lysis buffer or a cell lysate corresponding to 2 × 105 PBMCs from an HHV-8 PCR-negative blood donor, from about 50% of the replicates (range, 17 to 83%); this is in agreement with the percentage of positive reactions predicted from the Poisson distribution (63%). Cell copy numbers determined by quantitative PCR for the RNase P gene were verified by counting cells with a hemacytometer (within a twofold range); copy numbers obtained from dilution series decreased in an appropriately linear manner. Negative controls (including no-template controls and PBMCs from HHV-8-negative blood donors) were never positive. Consistently negative wells in subject PBMC dilutions below the endpoints acted as further negative controls.
LDA validation.
The LDA was validated using the HHV-8-infected BCBL-1 cell line, either uninduced or tetradecanoyl phorbol acetate (TPA) induced. In the example shown in Table 1, BCBL-1 cells were washed twice and then diluted with PBMCs from an HHV-8 PCR-negative blood donor to create a controlled representation of the frequency of HHV-8-infected cells observed in KS subjects in preliminary experiments (approximately 10 or 20 BCBL-1 cells per million PBMCs); this mixture was then processed through the LDA procedures as described above. The estimated frequencies of infected cells were close to the intended input numbers; the mean viral genome loads per infected cell were 100 copies for uninduced cells and 307 copies for induced cells. P values determined by the QUALITY program were always greater than 0.05, indicating a reasonable fit of the model to the data. With 1% of uninduced cells being in the lytic state and 10% being lytic after TPA induction (as measured experimentally by indirect immunofluorescence with a rabbit antibody to HHV-8 gB, a lytic antigen), these loads correspond to approximately 80 genomes per latently infected cell and approximately 2,400 per lytically infected cell (Table 1). The value for latently infected cells is similar to previous determinations on uninduced BCBL-1 cells by quantitative PCR (70 copies) (16, 32) and is somewhat higher than observed by fluorescent in situ hybridization or Southern blot reconstruction (25 to 30 copies) (21, 32). Taking into account differences in passage lineages, that the percentage of cells induced to the lytic state at the time of harvest may differ from experiment to experiment, and the extreme dilution of infected cells into uninfected cells in our representations of in vivo infected cell frequencies (10 and 20 infected cells per million uninfected cells), the results demonstrate that the LDA methods used here are likely to give reasonable results when applied to in vivo materials.
TABLE 1.
Mean HHV-8 genome copy number per cell in uninduced and TPA-induced BCBL-1 cells
| Run or meana | No. of infected cells/106 cells
|
No. of genomes/106 cells
|
Mean no. of genomes/infected cell (G)
|
No. of genomes/cellb:
|
||||
|---|---|---|---|---|---|---|---|---|
| Uninduced | TPA induced | Uninduced | TPA induced | Uninduced | TPA induced | Latently infected (Glyt) | Lytically infected (Glat) | |
| 1 | 19 | 7 | 2,126 | 1,628 | 113 | 231 | 100 | 1,411 |
| 2 | 15 | 12 | 1,356 | 4,674 | 88 | 383 | 55 | 3,333 |
| Mean | 100 | 307 | 78 | 2,372 | ||||
Run 1 included a cell mixture at a concentration of 20 BCBL-1 cells per million PBMCs. Run 2 included a cell mixture at a concentration of 10 BCBL-1 cells per million PBMC.
Calculated for each run from the mean genomes per infected cell (G), and the observed percentages of latently and lytically infected cells in TPA-induced and uninduced BCBL-1 cultures, using the following expressions: Ginduced = 0.9 × Glat + 0.1 × Glyt and Guninduced = 0.99 × Glat + 0.01 × Glyt.
A second line of validation was obtained by comparing the results of the LDA method with virus loads determined simultaneously by the quantitative Taqman assay. Over load ranges suitable for reliable Taqman quantitation (>10 genomic copies per reaction), results from the two methods correlated well (r2 = 0.9458; data not shown).
Subject characteristics.
Of the 13 subjects, 5 had active KS lesions, 4 previously had KS lesions, and 4 never had KS but were considered at high risk because they were infected with both HIV and HHV-8. Characteristics of these subjects are summarized in Tables 2 and 3. Subjects 7 and 8 were seen at two visits 3 and 2 months apart, respectively. All subjects were on highly active antiretroviral therapy (HAART) except for subject 8. Most subjects with KS lesions were on doxorubicin therapy, but none of them was being treated with agents that have significant in vitro activity against HHV-8. KS subjects had lower median CD4 percentages and higher median HHV-8 ORF K8.1 antibody endpoint titers (14 versus 22 and 3,200 versus 400, respectively), but too few subjects were analyzed to reliably assess whether the medians were statistically different.
TABLE 2.
Characteristics of and LDA results for HHV-8 PCR-negative subjects
| Group | KS status | CD4 count in cells/μl (%) | HIV load (no. of copies/ml) | HIV treatmenta | KS treatment | Antiherpes treatment | No. of HHV-8 genomes/106 PBMCb | HHV-8 antibody titer |
|---|---|---|---|---|---|---|---|---|
| KS subjects | ||||||||
| 1 | Cutaneous (progressing) | 613 (47) | 565 | d4T, 3TC, NVR | Doxorubicin | No | <0.21 | 6,400 |
| <0.15c | ||||||||
| 2 | Inactive unresolved cutaneous KS | 93 (7) | 1,266 | NFV, Combivir | No | ACV | <0.93 >0.58d | 3,200 |
| 3 | Resolved KS, no current lesions | 435 (12) | 226 | NFV, combivir | No | No | <1.00 | 400 |
| Subjects at risk for KS | ||||||||
| 4 | No KS | 558 (25) | 719 | NFV, NVR, Combivir | No | No | <1.20 0.34c | 400 |
| 5 | No KS | 201 (15) | 1.1 × 104 | ddl, d4T, NFV, NVR | No | No | <0.67 | 400 |
| 6 | No KS | 434 (18) | Undetectable | Combivir, EFV | No | No | <0.37 | 6,400 |
d4T, stavudine; 3TC, lamivudine; NVR, nevirapine; NFV, nelfinavir; combivir, zidovudine and lamivudine; ACV, acyclovir; ddl, didanosine; EFV, efavirenz.
The less than signs indicate the upper bound of virus load in individuals in whom virus DNA was not detected.
Result when the number of replicates was increased from 12 to 22, which led to a positive result for subject 4.
Result when the number of replicates was increased from 12 to 17.
TABLE 3.
Clinical characteristics, frequency of HHV-8-infected PBMCs, and virus load per positive cell in HHV-8 PCR-positive subjectsa
| Groupb | KS status | CD4 count in cells/μl (%) | HIV load (no. of copies/ml) | HIV treatment | KS treatment | Antiherpes treatment | No. of HHV-8-infected cells/106 PBMCs | No. of HHV-8 genomes/106 PBMCs | No. of HHV-8 genomes/infected cellc | No. of HHV-8 genomes/200 μl of plasma | HHV-8 antibody titer | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KS subjects | |||||||||||||
| 7a | Cutaneous (progressing) | 207 (13) | 1,041 | AZT, ddl, Kaletra | Doxo | ACV | 29 | 169 | 5.8 | 832 | 51,200 | ||
| 7b | Cutaneous (progressing) and oropharyngeal | 225 (12) | <500 | AZT, ddl, Kaletra | Doxo | ACV | 5.4 | 522 | 97 | 51 | 51,200 | ||
| 8a | Cutaneous (progressing) | 732 (29) | 2.1 × 106 | No | Doxo | No | 0.84c | 278 | 331d | ND | 25,600 | ||
| 8b | Cutaneous (progressing) and oropharyngeal | 342 (21) | 6.4 × 105 | No | No | No | 4.8 | 826 | 172 | 46 | 25,600 | ||
| 9 | Cutaneous (improving) | 251 (14) | 772 | NFV, combivir | Doxo | No | 4.5 | 12 | 2.7 | ND | 800 | ||
| 10 | No current lesions (cutaneous 2 yr before) | 961 (47) | 170 | IND, combivir | No | No | 1.1 | 210 | 191 | ND | 1,600 | ||
| 11 | No current lesions (transient oral 2 yr before) | 7 (1) | 7.3 × 106 | AZT, tenofovir, amprenavir | No | No | NS | 13 | NS | ND | 400 | ||
| 12 | No current lesions (transient oral 7 yr before) | 410 (19) | 1.2 × 104 | d4T, 3TC, NLF | No | No | 18 | 1,090 | 61 | 5.3 | 3,200 | ||
| Subjects at risk for KS
|
|||||||||||||
| 13 | No KS | 296 (29) | Und. | ddl, RTV, NVR, SAQ | No | No | 3.3c | 592 | 179c | ND | 400 | ||
AZT, zidovudine; ddl, didanosine; Kaletra, lopinavir/ritonavir; doxo, doxorubicin; ACV, acyclovir; NFV, nelfinavir; Combivir, zidovudine/lamivudine; IND, indinavir; RTV, ritonavir; SAQ, saquinavir; ND, not detected; Und., undetectable; NS, insufficient specimen.
For subjects 7 and 8, specimens were obtained at two time points, 3 and 2 months apart, respectively.
Mean genomes per infected cell.
Calculations based on 1 dilution only.
Frequency of infected PBMCs and virus load in HHV-8-infected subjects.
Six of the 13 subjects were PCR negative in the cell lysate dilution LDA (Table 2); thus the cell dilution LDA was not performed for them. Three of these subjects had KS, and the other three were at high risk. The frequency of the HHV-8 genome in these subjects was thus less than 0.21 to 1.2 per million PBMCs. This value is also the maximum estimated frequency of infected cells, since the number of infected cells cannot exceed the genome copy number. For subjects 1 and 4, enough cells were available to repeat the assay with 22 instead of 12 replicates; for subject 2, the assay was repeated with 17 replicates. In these assays, subjects 1 and 2 remained negative, with lowered maximum frequencies, but subject 4 tested positive with a frequency of 0.34 genomes per million PBMCs.
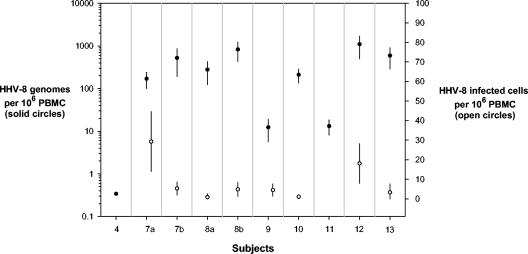
The cell dilution LDA was performed on samples from the seven PCR-positive subjects (six KS subjects and one subject at high KS risk) (Table 3). Infected cell frequencies ranged from 0.84 to 29 per million PBMCs (median, 4.7), and genome frequencies ranged from 12 to 1,090 (median, 278) per million PBMCs (Fig. 2). Virus loads ranged from 2.7 to 331 copies per infected PBMC (median, 134 copies). For subject 12, sufficient PBMCs were available to perform the LDA on B cells and non-B PBMCs obtained by immunomagnetic sorting with an antibody to CD19. HHV-8 DNA was almost exclusively detected in B cells, at a level (2.5 genomes per infected cell) consistent with a low-copy-number latent infection (data not shown). The highest HHV-8 virus load per positive cell was seen in a subject (patient 8) with progressing cutaneous KS (specimen 8a), who later developed oropharynx involvement (specimen 8b). This subject was not on HAART therapy and had falling CD4 counts, which could have contributed to these results.
FIG. 2.
Frequency of HHV-8 genome and frequency of infected cells in PCR-positive subjects with or at risk for KS. Error bars represent 95% confidence intervals (for some points, the confidence intervals are smaller than the plotted symbol). The measured frequencies were based on testing 12 replicates in each of five serial dilutions (22 replicas for subject 4), among which three to four dilutions showed percentages of negative wells between 0 and 100% and therefore were suitable for Poisson analysis. For subject 10, calculations for infected cell frequency are based only on the two informative dilutions. For the three subjects with the lowest frequency of infected cells (patients 8a and 13) and genomes (patient 4), only the lowest dilution was positive.
Rates of detection of HHV-8 DNA in PBMCs and plasma.
Three of five subjects with active KS lesions, three of four subjects with prior KS, and one (two if a subject is included for whom 22 instead of 12 replicate PCRs was done) of four subjects at high KS risk were positive by PCR in PBMCs. When plasma from the same visits was tested, only two of the five subjects with KS lesions and one of the four subjects with past KS were positive by PCR (Table 3). The lower detection rate in plasma than in PBMCs is probably due at least in part to a sample volume issue: while 200 μl of plasma was tested (representing approximately 0.4 ml of blood), the PBMCs tested in the 12 replicate reactions represented a total blood volume of 0.4 to 2.1 ml. In some studies reporting high detection rates in plasma or serum, virus particles were concentrated by centrifugation prior to DNA extraction (6, 36).
DISCUSSION
Methods used thus far for quantitative PCR of HHV-8 in PBMCs have given information only on the net number of HHV-8 genomes in the total cellular lysate sampled (expressed as either copies per microgram of total DNA or copies per million cells), without regard to the percentage of the cell population actually infected (3-5, 9, 16, 23, 26, 33). Because latently infected cells can be expected to have many fewer viral genomes than lytically infected cells, some insights into the biology of HHV-8 infections of PBMCs can be gleaned from the frequency and mean viral load of infected cells. Here we developed a method for measuring the frequency of both HHV-8-infected cells and HHV-8 genomes among PBMCs, which enabled computation of the mean virus load per infected cell.
HHV-8 genome and infected-cell frequencies in PBMCs.
When measured as net HHV-8 genome copy numbers per bulk quantity of PBMCs, the frequency of HHV-8 genomes in KS subjects with a positive PCR result ranged from 12 to 1,090 copies per 106 PBMCs (median of 244), in agreement with other studies that used quantitative PCR (3, 4, 26). In EBV-infected individuals with PTLD, these virus genome loads would be considered low (29). Even though EBV-infected lymphoblasts proliferate in lymph nodes and circulating memory B cells are only latently infected, PTLD patients have EBV genome frequencies of 5,000 to 50,000 copies per 106 PBMCs at the time of diagnosis (19, 28). Furthermore, the frequency of infected cells in PCR-positive KS subjects (0.84 to 29 per million PBMCs) is low enough to overlap with the higher end of the range of frequencies of EBV latently infected cells in healthy carriers (0.1 to 24 per million PBMCs) (37), again emphasizing the biological differences between the two human gammaherpesviruses. Explanations for this relatively low frequency of infected cells in blood of persons with KS may be that although HHV-8 is a lymphotropic virus, infected lymphocytes may be localized mainly in lymphoid tissues rather than circulating in peripheral blood, or other tissues might harbor the virus. Relevant to this hypothesis, the closely related murine gammaherpesvirus 68 (murid herpesvirus 4) can establish a latent infection in the spleen, with infected-cell frequencies among splenocytes that vary depending on the stage of infection (35).
Consistent with studies in which detection of HHV-8 DNA in PBMCs correlated with the appearance of KS (1, 13, 26, 36), among PCR-positive subjects, the range of infected-cell frequencies for KS subjects was higher than that in subjects at high KS risk (0.84 to 29 per million PBMCs versus 0.34 to 3.3 per million PBMCs), although the number of subjects included in our study was too small to claim this as a definitive result. Likewise, the frequency of detecting HHV-8 DNA in PBMCs was higher in subjects with active or prior KS lesions than in subjects at high KS risk (6 of 9 versus 1 of 4 under our standard LDA conditions). In both of the subjects in whom KS progressed to the oropharynx between consecutive visits, HHV-8 genome copy numbers in their blood increased. In one subject, this was associated with an increased number of virus genomes per positive cell, and in the other, it was associated with an increase in the frequency of infected cells in PBMCs. Interestingly, KS spindle-like cells can be detected in the PBMC fraction of peripheral blood of HIV-positive patients with or at risk for KS, a possible explanation for the multifocal nature of this disease (7). However, the third subject with progressing KS had fewer than 1 HHV-8 genome per 6 million PBMCs, consistent with prior observations that KS lesions are not always associated with high virus levels in PBMCs (4, 5, 16). In sum, while there is an association in our data and in data from others, peripheral blood virus load is much less tightly linked to pathogenesis for HHV-8 and KS than for EBV and PTLD.
Prevalence of HHV-8 in PBMCs.
Rates for detecting HHV-8 DNA by PCR in PBMCs from AIDS-KS patients in America and Europe in the HAART era range from 30 to 70% (3-5, 9, 10, 18). Our results make it clear that HHV-8 DNA detection rates in PBMCs are dependent on the number of cells used for DNA extraction.
Subjects with KS or at risk for KS who had the lowest detected frequencies of infected cells harbored one HHV-8-infected cell per 8 or 20 μg of DNA (obtained from approximately 0.8 or 2 ml of blood, respectively); according to the Poisson distribution, even larger volumes would need to be sampled to achieve reliable detection. In patients with higher numbers of genomes per infected cell, the detection of one HHV-8 genome would require less human DNA per PCR, as long as the initial number of cells processed for DNA extraction was sufficient to contain at least one infected cell. To obtain reliable detection in patients with low loads per infected cell (e.g., patient 9 with three copies per infected cell), a large number of cells would need to be processed and a large quantity of this material would need to be assayed by PCR.
Thus, the likelihood of detecting the HHV-8 genome is dependent on the number of cells lysed initially, the virus copy number per infected cell, and the total amount of DNA sampled by PCR. In some reports with low rates of HHV-8 DNA detection in PBMCs from AIDS-KS, small amounts of DNA (0.2 to 3.3 μg, representing 3 × 104 to 5 × 105 PBMCs) were used per PCR, and multiple specimens were not analyzed to increase the amount of DNA being sampled (3-5, 9, 10, 13, 18, 26). In one carefully described example, DNA was extracted from 3 million PBMCs, of which 2 μg was used per PCR; the detection rate was 50% among 16 AIDS-KS patients (9). Prior to this work, we would have considered that to be both a large cell sample and a large amount of DNA to test by PCR. Unfortunately, according to our results, there would have been too few PBMCs in the bulk lysate and too few replicates to obtain a positive PCR result in some of our PCR-positive subjects. The number of cells originally lysed for DNA extraction, a critical variable needed to compare rates of detection reported from different studies, is seldom described (including in our own studies) (10). Here, we started from a lysate of 4.2 ×106 PBMCs and tested up to 7.4 μg per patient for those who were PCR positive. Even when these large amounts of DNA were used (or even larger quantities in the retests of subjects 1 and 2), three of nine KS subjects in our study were negative by PCR. Subject 4 went from negative to positive when the number of replicates was increased from 12 to 22, raising the possibility that other subjects with negative results would test positive if an even larger number of cells were sampled. Thus, HHV-8 PCR results obtained with more conventional sampling methods (single, small-volume specimens) are likely to have underestimated the prevalence of the virus in PBMCs.
Evidence for both lytic and latent infection in PBMCs.
The measured mean virus loads per infected cell should not be interpreted as a homogeneous distribution of genomes in infected cells. Our results make it clear that there is almost certainly a sharp polarization between the copy number per latently infected cell and per lytically infected cell. The ratio between numbers of cells in these states is likely to vary over time. For example, induction of lytic HHV-8 replication in BCBL-1 cells (all of which harbor the virus) resulted in only a threefold increase in the mean net virus load (from 100 to 307 HHV-8 genomes per infected cell), even though lytically infected cells harbor on the order of 2,400 viral genomes per cell. Thus, even low elevations of the mean virus load, such as the threefold elevation seen in our BCBL-1 experiments, can represent substantial active virus replication in a small subset of cells. Among our study subjects, we saw up to 70-fold differences in mean virus loads per positive cell and hypothesize that (i) levels seen in some individuals of fewer than 10 genomic copies per infected cell represent tightly regulated latent infections and (ii) the highest mean virus loads (the data do not support defining a clear threshold) are likely to indicate appreciable lytic virus replication in PBMCs. The meaning of the intermediate values cannot be discerned at present. Interestingly, three of the eight subjects with PCR-positive PBMCs (and none of the subjects with PCR-negative PBMCs) were simultaneously PCR positive in their plasma, further supporting the idea that active virus replication was taking place, in agreement with other studies (12, 25). We cannot exclude the possibility that free virions in plasma may arise from noncirculating infected cells such as epithelial cells or vessel endothelial cells. However, all patients with detectable virus in plasma had positive PBMCs as well; free virions were not detected in any of the subjects with PCR-negative PBMCs.
The final point is that the lowest viral copy numbers per infected cell we observed (approximately three and six genomes per infected cell in patients 9 and 7) are substantially less than the copy number in latently infected BCBL-1 cells. This suggests that viral genome copy number is regulated differently in nontransformed cells during in vivo latency than in PEL cells. Thus, in analogy with EBV, HHV-8 may employ more than one latency program. Because of their low infected-cell frequencies, this will be particularly difficult to study in healthy, immunocompetent individuals. It is easy to forget that such individuals represent the predominant HHV-8-infected host phenotype.
In conclusion, our results suggest the following. (i) While laborious, the PCR-LDA method described here has clear value in monitoring HHV-8 activity and will be of use in studies of KS progression in a larger patient group. (ii) Standard methods for sampling PBMCs lead to underreporting of HHV-8 presence in circulating leukocytes. (iii) HHV-8 can persist in circulating latently infected B cells and is likely to undergo lytic replication in PBMCs; this may contribute to KS pathogenesis by delivering circulating infectious virus into tissues. (iv) In vivo HHV-8 latency may be regulated differently from latency in transformed cells. (v) Complementary approaches, such as further analysis of latent and lytic transcripts in PBMCs (25) and quantitative assessments of whether virus genomes are linear or circular (12), as well as assessment of the ratio of latent to lytically infected cells over time, will ultimately be needed to more fully understand the biology of HHV-8 in circulating leukocytes.
Acknowledgments
This study was supported in part by a fellowship from Generalitat de Catalunya (AGAUR), Spain, and by funding from the CDC Opportunistic Infections Working Group.
We thank Karen McCaustland and Brian Holloway for developing and optimizing the RNase P single-plex reaction and their assistance with optimization of the duplex TaqMan PCR, Eng-Chun Mar for providing the orf37-pPICZαA plasmid, Kay Radford and Tim Bailiff for performing HHV-8 serology and measuring HIV virus loads, Lee Lam for performing CD4 counts, David O. Willer from Sam Speck's laboratory for sharing their limiting dilution assay method, and Clifford J. Gunthel and the Grady Infectious Diseases Clinic staff for their cooperation and support with specimen collection.
REFERENCES
- 1.Ambroziak, J. A., D. J. Blackbourn, B. G. Herndier, R. G. Glogau, J. H. Gullett, A. R. McDonald, E. T. Lennette, and J. A. Levy. 1995. Herpes-like sequences in HIV-infected and uninfected Kaposi's sarcoma patients. Science 268:582. [DOI] [PubMed] [Google Scholar]
- 2.Antman, K., and Y. Chang. 2000. Kaposi's sarcoma. N. Engl. J. Med. 342:1027-1038. [DOI] [PubMed] [Google Scholar]
- 3.Boivin, G., S. Cote, N. Cloutier, Y. Abed, M. Maguigad, and J. Routy. 2002. Quantification of human herpesvirus 8 by real-time PCR in blood fractions of AIDS patients with Kaposi's sarcoma and multicentric Castleman's disease. J. Med. Virol. 68:399-403. [DOI] [PubMed] [Google Scholar]
- 4.Boivin, G., A. Gaudreau, and J. P. Routy. 2000. Evaluation of the human herpesvirus 8 DNA load in blood and Kaposi's sarcoma skin lesions from AIDS patients on highly active antiretroviral therapy. AIDS 14:1907-1910. [DOI] [PubMed] [Google Scholar]
- 5.Boivin, G., A. Gaudreau, E. Toma, R. Lalonde, J.-P. Routy, G. Murray, J. Handfield, and M. G. Bergeron. 1999. Human herpesvirus 8 DNA load in leukocytes of human immunodeficiency virus-infected subjects: correlation with the presence of Kaposi's sarcoma and response to anticytomegalovirus therapy. Antimicrob. Agents Chemother. 43:377-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broccolo, F., G. Locatelli, L. Sarmati, S. Pierpaoli, F. Veglia, M. Andreoni, S. Buttò, B. Ensoli, P. Lusso, and M. S. Malnati. 2002. Calibrated real-time PCR assay for quantitation of human herpesvirus 8 DNA in biological fluids. J. Clin. Microbiol. 40:4652-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browning, P. J., J. Sechler, M. Kaplan, R. Washington, R. Gendelman, R. Yarchoan, B. Ensoli, and R. C. Gallo. 1994. Identification and culture of Kaposi's sarcoma-like spindle cells from the peripheral blood of human immunodeficiency virus-1-infected individuals and normal controls. Blood 84:2711-2720. [PubMed] [Google Scholar]
- 8.Campbell, T. B., M. Borok, L. Gwanzura, S. MaWhinney, I. E. White, B. Ndemera, I. Gudza, L. Fitzpatrick, and R. T. Schooley. 2000. Relationship of human herpesvirus 8 peripheral blood virus load and Kaposi's sarcoma clinical stage. AIDS 14:2109-2116. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, T. B., L. Fitzpatrick, S. MaWhinney, X. Zhang, and R. T. Schooley. 1999. Human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) infection in men receiving treatment for HIV-1 infection. J. Acquir. Immune Defic. Syndr. 22:333-340. [DOI] [PubMed] [Google Scholar]
- 10.Cannon, M. J., S. C. Dollard, J. B. Black, B. R. Edlin, C. Hannah, S. E. Hogan, M. M. Patel, H. W. Jaffe, M. K. Offermann, T. J. Spira, P. E. Pellett, and C. J. Gunthel. 2003. Risk factors for Kaposi's sarcoma in men seropositive for both human herpesvirus 8 and human immunodeficiency virus. AIDS 17:215-222. [DOI] [PubMed] [Google Scholar]
- 11.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 12.Decker, L. L., P. Shankar, G. Khan, R. B. Freeman, B. J. Dezube, J. Lieberman, and D. A. Thorley-Lawson. 1996. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J. Exp. Med. 184:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engels, E. A., R. J. Biggar, V. A. Marshall, M. A. Walters, C. J. Gamache, D. Whitby, and J. J. Goedert. 2003. Detection and quantification of Kaposi's sarcoma-associated herpesvirus to predict AIDS-associated Kaposi's sarcoma. AIDS 17:1847-1851. [DOI] [PubMed] [Google Scholar]
- 14.Grundhoff, A., and D. Ganem. 2004. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi's sarcoma pathogenesis. J. Clin. Investig. 113:124-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iscovich, J., P. Boffetta, S. Franceschi, E. Azizi, and R. Sarid. 2000. Classic Kaposi sarcoma: epidemiology and risk factors. Cancer 88:500-517. [PubMed] [Google Scholar]
- 16.Lallemand, F., N. Desire, W. Rozenbaum, J.-C. Nicolas, and V. Marechal. 2000. Quantitative analysis of human herpesvirus 8 viral load using a real-time PCR assay. J. Clin. Microbiol. 38:1404-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Laney, A. S. S. C. Dollard, H. W. Jaffe, M. K. Offermann, T. J. Spira, C. J. Gunthel, P. E. Pellett, and M. J. Cannon. 2004. Repeated measures study of human herpesvirus 8 (HHV-8) DNA and antibodies in men seropositive for both HHV-8 and HIV. AIDS 18:1819-1826. [DOI] [PubMed]
- 17.Mesri, E. A., E. Cesarman, L. Arvanitakis, S. Rafii, M. A. Moore, D. N. Posnett, D. M. Knowles, and A. S. Asch. 1996. Human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus is a new transmissible virus that infects B cells. J. Exp. Med. 183:2385-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min, J., and D. A. Katzenstein. 1999. Detection of Kaposi's sarcoma-associated herpesvirus in peripheral blood cells in human immunodeficiency virus infection: association with Kaposi's sarcoma, CD4 cell count, and HIV RNA levels. AIDS Res. Hum. Retrovir. 15:51-55. [DOI] [PubMed] [Google Scholar]
- 19.Miyashita, E., B. Yang, D. H. Crawford, and D. A. Thorley-Lawson. 1995. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell 80:593-601. [DOI] [PubMed] [Google Scholar]
- 20.Moore, P. S., and Y. Chang. 2001. Kaposi's sarcoma-associated herpesvirus, p. 2803-2833. In D. M. Knipe, P. M. Howley, B. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 21.O'Neill, E., J. L. Douglas, M.-L. Chien, and J. V. Garcia. 1997. Open reading frame 26 of human herpesvirus 8 encodes a tetradecanoyl phorbol acetate- and butyrate-inducible 32-kilodalton protein expressed in a body cavity-based lymphoma cell line. J. Virol. 71:4791-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pau, C. P., S. Lee-Thomas, W. Auwanit, J. George, C. Y. Ou, B. S. Parekh, T. Granade, D. Holloman, S. Phillips, and G. Schochetman. 1993. Highly specific V3 peptide enzyme immunoassay for serotyping HIV-1 specimens from Thailand. AIDS 7:337-340. [DOI] [PubMed] [Google Scholar]
- 23.Pellet, C., S. Chevret, L. Blum, C. Gauville, M. Hurault, F. Agbalika, C. P. D. Lascoux, P. Morel, F. Calvo, and C. Lebbe. 2001. Virologic and immunologic parameters that predict clinical response of AIDS-associated Kaposi's sarcoma. J. Investig. Dermatol. 117:858-863. [DOI] [PubMed] [Google Scholar]
- 24.Pellett, P. E., D. J. Wright, E. A. Engels, D. V. Ablashi, S. C. Dollard, B. Forghani, S. A. Glynn, J. J. Goedert, F. J. Jenkins, T.-H. Lee, F. Neipel, D. Todd, D. Whitby, G. Nemo, M. P. Busch et al. 2002. Multicenter comparison of serologic assays and estimation of Human herpesvirus 8 seroprevalence in US blood donors. Transfusion (Paris) 43:1260-1268. [DOI] [PubMed] [Google Scholar]
- 25.Polstra, A., J. Goudsmit, and M. Cornelissen. 2003. Latent and lytic HHV-8 mRNA expression in PBMCs and Kaposi's sarcoma skin biopsies of AIDS Kaposi's sarcoma patients. J. Med. Virol. 70:624-627. [DOI] [PubMed] [Google Scholar]
- 26.Quinlivan, E., C. Zhang, P. Stewart, C. Komoltri, M. Davis, and R. Wehbie. 2002. Elevated virus loads of Kaposi's sarcoma-associated human herpesvirus 8 predict Kaposi's sarcoma disease progression, but elevated levels of human immunodeficiency virus type 1 do not. J. Infect. Dis. 185:1736-1744. [DOI] [PubMed] [Google Scholar]
- 27.Rodrigo, A., P. Goracke, K. Rowhanian, and J. Mullins. 1997. Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res. Hum. Retrovir. 13:737-742. [DOI] [PubMed] [Google Scholar]
- 28.Rowe, D. T., L. Qu, J. Reyes, N. Jabbour, E. Yunis, P. Putnam, S. Todo, and M. Green. 1997. Use of quantitative competitive PCR to measure Epstein-Barr virus genome load in peripheral blood of pediatric transplant patients with lymphoproliferative disorders. J. Clin. Microbiol. 35:1612-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rowe, D. T., S. Webber, E. Schauer, J. Reyes, and M. Green. 2001. Epstein-Barr virus load monitoring: its role in the prevention and management of post-transplant lymphoproliferative disease. Transplant Infect. Dis. 3:79-87. [DOI] [PubMed] [Google Scholar]
- 30.Sarid, R., S. J. Olsen, and P. S. Moore. 1999. Kaposi's sarcoma-associated herpesvirus: epidemiology, virology, and molecular biology. Adv. Virus Res. 52:139-232. [DOI] [PubMed] [Google Scholar]
- 31.Spira, T. J., L. Lam, S. C. Dollard, Y.-X. Meng, C. P. Pau, J. B. Black, D. Burns, B. Cooper, M. Hamid, J. Huong, K. Kite-Powell, and P. E. Pellett. 2000. Comparison of serologic assays and PCR for diagnosis of human herpesvirus 8 infection. J. Clin. Microbiol. 38:2174-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stamey, F. R., M. M. Patel, B. P. Holloway, and P. E. Pellett. 2001. Quantitative, fluorogenic probe PCR assay for detection of human herpesvirus 8 DNA in clinical specimens. J. Clin. Microbiol. 39:3537-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tedeschi, R., M. Enbom, E. Bidoli, A. Linde, P. De Paoli, and J. Dillner. 2001. Viral load of human herpesvirus 8 in peripheral blood of human immunodeficiency virus-infected patients with Kaposi's sarcoma. J. Clin. Microbiol. 39:4269-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trippler, M., K. Meyer zum Buschenfelde, and G. Gerken. 1999. HBV viral load within subpopulations of peripheral blood mononuclear cells in HBV infection using limiting dilution PCR. J. Virol. Methods 78:129-147. [DOI] [PubMed] [Google Scholar]
- 35.Weck, K. E., S. S. Kim, H. W. Virgin IV, and S. H. Speck. 1999. B cells regulate murine gammaherpesvirus 68 latency. J. Virol. 73:4651-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitby, D., M. R. Howard, M. Tenant-Flowers, N. S. Brink, A. Copas, C. Bishoff, T. Hatzioannou, F. E. A. Suggett, D. M. Aldam, A. S. Denton, R. F. Miller, I. V. D. Weller, R. A. Weiss, R. S. Tedder, and T. F. Schultz. 1995. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet 346:799-802. [DOI] [PubMed] [Google Scholar]
- 37.Yang, J., Q. Tao, I. W. Flinn, P. G. Murray, L. E. Post, H. Ma, S. Piantadosi, M. A. Caligiuri, and R. F. Ambinder. 2000. Characterization of Epstein-Barr virus-infected B cells in patients with posttransplantion lymphoproliferative disease: disappearance after rituximab therapy does not predict clinical response. Blood 96:4055-4063. [PubMed] [Google Scholar]