Abstract
A key end goal of gene delivery research is to develop clinically-relevant vectors that can be used to combat elusive diseases such as AIDS. Despite promising engineering strategies, efficiency and ultimately gene modulation efficacy of nonviral vectors have been hindered by numerous in vitro and in vivo barriers that have resulted in sub-viral performance. In this perspective, we concentrate on the gene delivery barriers associated with the two most common classes of nonviral vectors, cationic-based lipids and polymers. We present the existing delivery barriers and summarize current vector-specific strategies to overcome said barriers.
Keywords: Gene delivery, gene therapy, nonviral vectors, lipoplexes, polyplexes
INTRODUCTION
Gene therapy is a treatment option predicated on the underlying principle that disease can be addressed by introduction of exogenous genetic material into somatic cells of patients for the purpose of modulating gene expression of desired proteins. For example, deleterious proteins from specific pathogens can be used in genetic vaccinations to instigate cell-mediated and humoral immune responses. Thus, the delivery of genetic material, DNA and RNA, can be utilized to combat various acquired or heritable diseases and under circumstances where natural immunity is aberrant or lacking (cancer, AIDS, etc.).
Since it was first conceptualized in 1972,1 gene delivery has resulted in numerous studies but has produced a distinct shortage of licensed clinical treatments for human diseases, as most current applications are used in veterinary settings.2 Shortcomings arise due to the degradation of unprotected genetic material by extracellular enzymes that reduce the amount of gene cargo available to elicit a desired response. As a result, the scarcity of genetic material further hinders already low cellular uptake kinetics. After cellular entry, the genetic payload is subject to intracellular degradation in endosomal/lysosomal compartments. Unassisted, genetic material compartmental release is negligible, and the small fraction that may escape is exposed to further cytoplasmic degradation and poor translocation kinetics (especially with larger gene constructs). In the case of gene therapeutics that require nuclear entry for activity, crossing the nuclear membranes represents an additional barrier. A final consideration is the possible requirement of prolonged gene expression and the subsequent need for continuous administration. Regardless of the desired outcome, the presence of the aforementioned barriers necessitates the use of natural or artificial gene carrier vector systems.
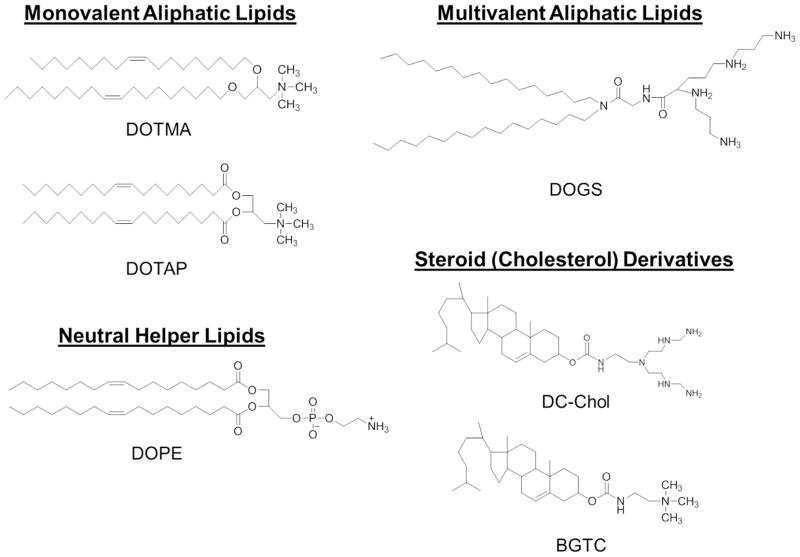
Many delivery systems have been developed to overcome specific gene delivery barriers, and vectors can be divided into two common classes: viral and nonviral systems. To conduct translational research, property requirements for gene delivery vectors include biocompatibility, biodegradability, resistance to premature degradation, non-immunogenicity, cell-specific targeting (if applicable), and modulation of gene expression for a desired period of time. From this perspective, viral vectors have resulted in unsurpassed levels of transgene transfection efficacy, but have not been utilized as an all-utility vector due to drawbacks such as nanoparticle formulation, storage difficulties, gene carrying capacity, and residual viral elements that can potentially cause insertional mutagenesis, cytotoxicity, immunogenicity, and tumorigenicity.3, 4 These limitations in viral vector delivery have given rise to the field of nonviral vectors which include biocompatible materials rationally designed and fabricated through innovative synthesis schemes. Two major categories of nonviral vectors have been developed and extensively studied: cationic lipids (CLs) and cationic polymers (CPs). The use of CLs as a synthetic carrier for gene delivery into mammalian cells was first reported with cationic N-[1-(2,3-dioleyloxy)propyl-]-N,N,N-trimethylammonium chloride (DOTMA) mixed with a neutral helper lipid, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), in 1987 by Felgner and coworkers (Figure 1).5 Delivery is accomplished by the spontaneous formation into an array of morphologically-distinct CL-nucleic acid carriers, termed lipoplexes, by means of electrostatic interactions. As of a result of their relatively high delivery efficiency, CLs have emerged as the most studied system for nonviral gene delivery.6, 7 However, limitations in fabrication reproducibility, colloidal stability, and cytotoxicity (in vitro and in vivo) have hindered expansion beyond research purposes.8
Figure 1.
Chemical structures of common cationic lipids used in transfection.
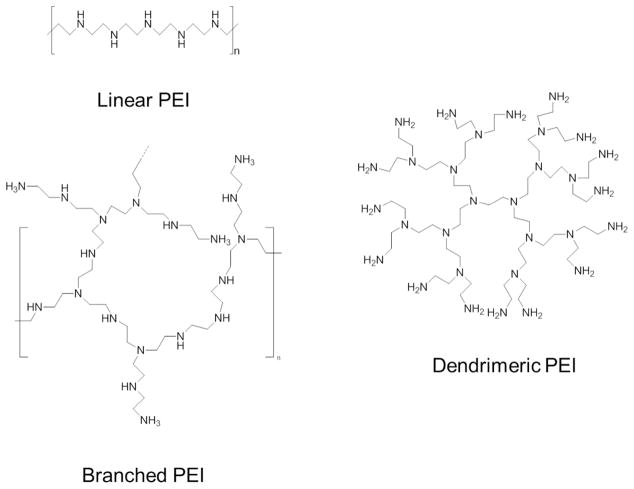
Similarly, CPs, which consists of linear, branched, and dendritic structures (Figure 2), are synthesized through various routes (block addition, living polymerization, etc.) and also form nucleic acid-loaded particles, termed polyplexes, through spontaneous electrostatic interactions. The most distinct difference between CLs and CPs is that CPs do not (normally) contain hydrophobic tail moieties and are completely soluble in water.9 Despite similarities, CPs have gained significant attention because of the flexibility in polymer chemistries that have allowed improvements to design and formulation. The improvements permit precise control of structure and surface modifications enabling use in a plethora of biomedical applications.6, 10 However, delivery efficacy still falls short of viral vector standards. Considering limitations of both nonviral systems to be profiled in this perspective, there is a need to reevaluate the status of gene delivery efforts and the challenges facing the field. Below, strategies to overcome nonviral gene delivery barriers starting with formulation and ending with gene expression/silencing are summarized.
Figure 2.
Structural variants of the PEI cationic polymer.
VECTOR DESIGN AND FORMULATION
The formation of stable CL- and CP-nucleic acid particles that possess the chemical and biophysical properties required to overcome delivery barriers is the first step and first barrier in the gene delivery process. Complexation of genetic material for the formation of lipoplexes and polyplexes proceeds by electrostatic interactions between cationic vectors and anionic nucleic acids resulting in condensation (i.e., a compacted state) of sub-micron-sized particles. These interactions result in the spontaneous formation of thermodynamically quasi-stable structures that contain numerous DNA (or RNA) molecules per nanoparticle.11 Effective particle formation sterically protects genetic payloads from nucleolytic enzymes, enhances cell permeability/uptake, and increases cytosolic mobility (as compared to naked DNA). The formulation process and the resulting structure are related to the degree of complexation and/or the amount of material being used. Complexation of genes is most easily controlled by adjusting polymer/lipid:gene weight ratios that are initially identified by nucleic acid retardation in a gel-shift assay. The identified ratios correspond to the amount of vector needed to fully complex the payload and serve as a starting point for further optimization. Gene delivery is thought to be improved by use of excessive charge due to the presence of charge reservoirs which enable nucleic acids to achieve transient yet stable interactions within the nanoparticle. Conversely, the use of excessive charge can result in undesired toxicity and binding forces that are too strong for efficient gene unpacking. Thus, desired specific structural properties for each vector type must be considered prior to synthesis to allow rational design of nanoparticles with the greatest gene delivery potential.
Cationic Lipids
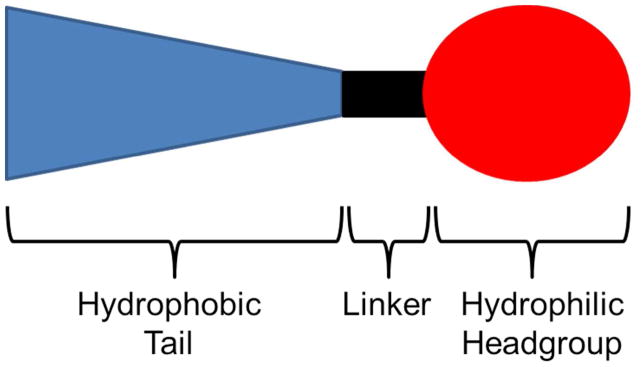
The chemical structure of CLs determines the physical characteristics of the lipoplex and is an essential factor in the determination of final gene delivery and cytotoxicity.12 CL structure can be subdivided into three sections: a positively charged, hydrophilic headgroup; a hydrophobic anchor group (tail); and a linker group that joins the polar and non-polar regions (Figure 3). Lipoplex formation requires a headgroup that can sustain a cationic charge at physiological pH. In early CLs, the cationic charge was localized on either primary, secondary, or tertiary amines or quaternary ammonium salts; however, the choice of headgroup has expanded to include natural architectures and functionalities with established nucleic acid binding capabilities as well as non-amino-based moieties. Thus, before discussing more extensively studied amine-based headgroups, it is worth briefly considering alternatives that have shown a nucleic acid binding capacity. For example, Floch and coworkers demonstrated that use of phosphorus or arsenic atoms instead of nitrogen, which incorporate phosphonate linkers, convey cationic charge and result in improved transfection efficiency and reduced cytotoxicity.13
Figure 3.
Three basic structural domains of cationic lipids: hydrophobic tail, linker, and headgroup.
The majority of CLs, however, utilize nitrogen atoms within the hydrophilic headgroup, as there appears to be a relationship between gene delivery efficiency and hydration of amino-based headgroups.14 Charge conferral usually proceeds via the protonation of a monovalent amine group; alternatively, multivalent amino groups form lipoplexes with greater surface charge density and are generally believed to enable higher levels of nucleic acid binding and delivery. Of note, monovalent headgroup dehydration by incorporation of hydroxyalkyl chains capable of forming hydrogen bonds with neighboring headgroups (reducing available space for water inclusion) can promote inverted hexagonal phases, thus, facilitating lipid mixing and subsequent gene delivery. Accordingly, hydroxylation of DOTMA and the ester-linked variant 1,2-dioleoyloxy-3-(trimethylammonium)-propane (DOTAP) to yield 1,2-dioleoyl-3-dimethyl-hydroxyethyl ammonium bromide (DORIE) and 1,2-dioleoyloxypropyl-3-dimethyl-hydroxyethyl ammonium chloride (DORI), respectively, significantly improved gene delivery.15, 16 Furthermore, incorporation of two or more hydroxyethyl moieties in the headgroup conveyed increased gene delivery and decreased cytotoxicity.17 However, other studies concluded that the relationship between the hydroxyethyl headgroup incorporation and transfection efficiency is related to the overall lipoplex charge ratio.18 A slightly positive correlation exists between gene delivery efficiency and +/− charge ratios. That is, with increasing cationic charge, delivery efficiency and cytotoxicity generally increase concomitantly. Shortly after the development of DOTMA, multivalent headgroup CLs emerged, reported with the synthesis of dioctadecylamidoglycylspermin (DOGS), with elevated nucleic acid delivery capability (Figure 1). The use of DOGS demonstrated high initial transfection ability, without requiring helper lipids, both in vitro and in vivo. This unexpected observation promoted further studies that analyzed the impact upon transfection efficiency and cytotoxicity related to headgroup shape (linear, branched, globular, T-shaped), length, and the separation distance between two consecutive nitrogen atoms in a polyamine.19, 20 Despite increased gene delivery and gene compaction, multivalent CLs may result in overly strong nucleic acid binding and the formation of micelles contributing to increased cytotoxicity.21 Regardless of charge conferral type, an imbalance between the cross-sectional area of the headgroup and the hydrophobic tail will result in cone-shaped lipids which have a propensity to form unstable lipoplexes and fuse with anionic vesicles. Despite concerns over complex instability, a cone-shape CL architecture is presumed to result in improved gene delivery (in vitro) as increased fusion between the cationic lipoplexes and the endosomal membranes leads to efficient nucleic acid release into the cytoplasm.22, 23 For a more in-depth look into CL headgroup design readers are directed to other detailed reviews.14, 24
The hydrophobic anchor (tail) domain is an accompanying non-polar hydrocarbon region of CLs made up of two types of moieties, aliphatic chains and steroid domains. The lipoplex phase transition temperature, bilayer fluidity, overall stability, gene delivery potential, and associated cytotoxicity are significantly dependent upon structural variations within the hydrophobic region. These include such variations to domain length, type of chemical bonds, and the relative hydrocarbon chain position.25 Furthermore, commonly utilized aliphatic chains are typically linear and either fully saturated or mono-unsaturated (oleyl, lauryl, myristyl, palmityl, stearyl moieties).26 CLs commonly have one to four hydrocarbon chains, with studies demonstrating that incorporation of aliphatic chains with different numbers can boost transfection efficiency, presumably by promoting endosomal escape.5 However, double-chained CLs, such as DOTMA and DOTAP, are among the most actively researched structures. To this end, double-tailed CLs have the ability to form lamellar phases without the need for a helper lipid and thus lead to increases in gene delivery. The lack of studies that use single- and tripled-tailed CLs is, generally speaking, due to their surfactant nature that leads to micelle formation and subsequent increases in cytotoxicity relative to their double-tailed counterparts. Aliphatic chain length and degree of saturation affect CL gene delivery and cytotoxicity efficiency. Using a series of hydroxylethyl quaternary ammonium lipids with myristol, palmitoly, stearoyl, and oleoyl chains, Felgner and coworkers established a chain length-activity correlation of C14:0>C18:1>C16:0>C18:0.16 This trend has since been corroborated leading to the general consensus that gene delivery efficiency is correlated with decreases in chain length.27 Beyond chain length, the vast majority of studies report the use of unsaturated chains, particularly C18:1 oleyl, to promote the best transfection. Interestingly, substitution of the cis-double bonds of the oleyl tail with C—C triple bonds led to an increase of gene delivery and reduced cytotoxicity in both in vitro and in vivo models.28 As for the second type of moiety used for hydrophobic domains, steroids are common alternatives because of their rigidity, biodegradability, and fusion activity. Cholesterol is the most commonly encountered steroid. It is noteworthy in lipoplex formation because of its internal cyclic structure conferring a high degree of stability, exemplified by a BGTC/DOPE/DNA lipoplex resistant to shear forces involved in nebulization.29 The aforementioned improvements to gene delivery and cytotoxicity by the use of shorter, unsaturated hydrophobic chains are presumably caused by increases in lipoplex stability and membrane fluidity at physiological conditions. Increased lipoplex stability improves the protection of nucleic acids from degradation during inter- and intracellular trafficking. At the same time, these modifications result in the shifting of the lipid transition conditions towards the edge of physiologically-relevant temperatures (36 °C) and pH (~7), hence preventing over-stability and excessive rigidity that may diminish desired cell/endosomal membrane—lipoplex interactions.
The linker bond connects the hydrophobic and hydrophilic moieties of CLs and mediates stability. A balanced choice must be made between a desired vector’s persistence and subsequent cytotoxicity which is presumably related to the half-life within the cell. Common linkages are ether, ester, amide, and carbamate groups, with no particular bond emerging as a consistent optimum in structure-activity studies. However, it is generally recognized that ether bonds, such as DOTMA, render better gene delivery efficiency. They are particularly stable and, as such, are expected to be more toxic (further explained below) than ester-linked lipids (DOTAP), which are more easily cleaved within the cell. Carbamate and amide linkers, by contrast, are expected to serve as a reasonable balance between CL gene delivery efficiency and cytotoxicity and are therefore used in DC-Chol and BGTC.14 Interest has also been placed on the development of nucleic acid release by means of stimuli-triggered decomplexation and biodegradation mechanisms through the incorporation of functional group linkers. Thus, linkers have been developed that are sensitive to acidic pH, changes in redox potential, enzymes, and more recently, light exposure.30 The use of cleavable linker types should be emphasized; as their use may lead to reduced cytotoxicity and elevated gene delivery potential as compared to more chemically-stable variants.31
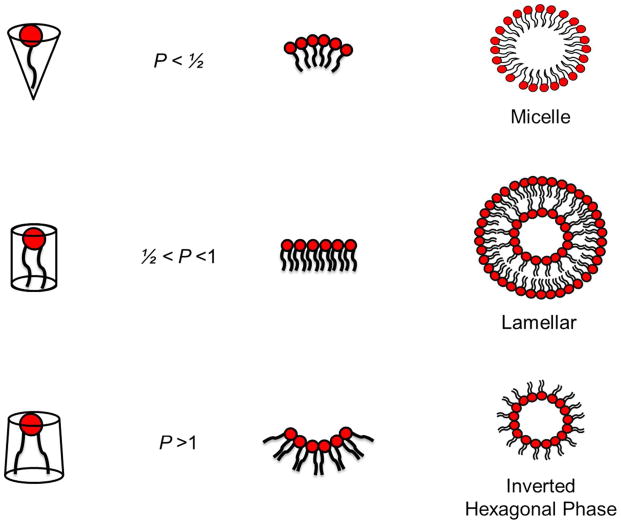
Colloidal stability and the formation of lipoplexes with desired morphology can be aided by the use of neutrally-charged helper lipids such as DOPE and 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), and these are often employed with other CLs in order to gain higher gene delivery efficiencies. An important property of CLs with regards to vector stability is overall geometry. Like any cationic vector, when suspended in an aqueous environment, CLs can adopt a number of structural phases, such as the micellar, lamellar, and inverted hexagonal phase (Figure 4). The specific phase adopted can be best predicted by a factor known as the packing parameter, , defined as the ratio of the hydrophobic tail volume, v, and the product of the hydrophilic headgroup area, a, and the hydrophobic tail length, Lc (Figure 4).32 When P exceeds the value of 1 (i.e., the area occupied by the hydrophobic region is larger than headgroup), the CL will adopt the inverted hexagonal phase, which is a bilayer destabilizing structure. However, when P is less than ½, the CL monomer will be cone-like shaped and will assemble into micellar phases. Thus, when P possesses a value between ½ and 1, CL monomers will adopt lamellar phases
Figure 4.
Schematic representation of the packing efficiency of cationic lipids and their resulting phase structure. A packing parameter, , less than ½ confers a cone-like shaped structure that assembles into micellar phases. Values between ½ and 1 allow formation of the lamellar phase (bilayers). For P > 1, the negative curvature leads to the formation of the inverted hexagonal phase.
Stability of lipoplexes has further been observed to be a function of medium sodium chloride concentration, pH, and DNA concentration, as the formation of nanoparticles is presumably aided by the expulsion of both nucleic acid and lipid counter ions as a driving force.33 In addition, lipoplexes can be decorated with functional elements for steric stabilization and shielding from extracellular environments (using poly(ethylene glycol) (PEG), for example). Each condition requires specific optimization during synthesis and formulation and is prone to trial-and-error-based experiments.
Cytotoxicity of lipoplexes is mostly associated with the cationic nature of its hydrophilic headgroup. CLs become toxic by presumably interacting with enzymes such as protein kinase C (PKC); however, the reason for this effect is unclear.12 Quaternary ammonium groups were shown to be more toxic than their tertiary amine counterparts.34 To circumvent these problems, the headgroup cationic charge distribution can be delocalized into heterocyclic rings, such imidazolium or pyridinium heads,35–37 which has been reported to display higher gene delivery and lower cytotoxicity when compared to traditional amino-based headgroups.38, 39 As outlined above, ether linkers render better gene delivery; however, these linkages result in cytotoxicity due to strong stability effects that result in a lack of biodegradability. CLs synthesized with ester bonds, such as DOTAP, were reported to be more biodegradable and less cytotoxic.40–43 However, ester or amide linkers are likely to readily degrade in physiological circulation conditions. To address these concerns, carbamate-linked CLs allow acid-catalyzed hydrolysis and are stable in neutral conditions, as demonstrated by improved cytotoxicity when compared to other linkages. In addition, studies have indicated that an increase in linker segment length leads to decreased cytotoxicity in vitro.44 Lastly, the effect of the hydrophobic chain on cytotoxicity has not been thoroughly addressed; however, single-tailed aliphatic chains are more toxic and less efficient than their double-tailed counterparts, due to the propensity towards micelle formation.45, 46
Cationic Polymers
To precisely design a CP that can properly counter-balance gene delivery and cytotoxicity, five major structural features must be considered that include charge density (number of cationic charges per polymer backbone), charge center type, polymer molecular weight, grafting of functionalities, and colloidal stability. Charge density within the polymer backbone can be precisely controlled through use of sophisticated polymer chemistry techniques. Specifically, successful synthesis of controlled charge density additions has been reported using atom transfer radical polymerization (ATRP),47, 48 reversible addition-fragmentation chain-transfer polymerization (RAFT),49, 50 ring-opening polymerization,51, 52 click-chemistry52–54, and other various chemical procedures such as Michael additions.55–59 These strategies are reproducible and can facilitate synthesis of well-defined charge density polymers. Several groups have reported that at least six to eight charges per single polymer chain are required for effective nucleic acid condensation.60–62
Prior to implementation of synthetic schemes, selection of the type of charge center to result in desired properties must be addressed. Traditionally, polymer chemists have utilized different forms of nitrogen centers (primary, secondary, tertiary, and quaternary) as the cationic source. It has been reported that complexation ability proceeds in order of quaternary>tertiary>secondary>primary.63, 64 However, gene delivery has resulted in contradictory results. Quaternary amines on pectin resulted in higher levels of gene delivery as compared to other amine types.63 Using a dextran backbone, Xu and coworkers observed the same complexation trends noted above, but for gene delivery, tertiary amines recorded the highest levels.64 This discrepancy was explained by a loss of buffering capacity leading to reduced endosomal escape (see later discussion related to the “proton sponge effect”) and associated cytotoxicity with quaternized amines.
Once synthesis has started, desired polymer molecular weights (MW) should be taken into consideration because, aside from the expected effects upon complexation ability (i.e., increased MW and number of cationic units per polymer chain causing tighter binding), gene delivery is positively correlated with increasing molecular weights. Yet, despite increases in gene delivery, increasing levels of cytotoxicity also generally accompany increased CP MW.65 For example, Neoh and coworkers synthesized variable chain length poly((2-dimethyl amino)ethyl methacrylate) (pDMAEMA)-based copolymers by ATRP.64 Enhanced plasmid DNA (pDNA) complexation was achieved with increasing cationic chain lengths, eventually resulting in higher transfection efficiency. Corroborative results were observed utilizing different polymer systems such as polyethylenimine,66 polycyclooctene,64, 67 and poly(beta amino esters).68 Other studies that demonstrated elevated complexation and transfection efficiency with increased molecular weight, however, were also accompanied by increases in cytotoxicity despite usage of the same N/P ratios (the amount of polymeric nitrogen compared to nucleic acid phosphate).69 Increased cytotoxicity was assumed to be caused by enhanced CP hydrophobic properties resulting from high molecular weights, which led to increased cell membrane damage.
Polymers are traditionally designed to incorporate specified functionalities to address a specific intracellular barrier, but outcomes have been mixed, with some CPs losing delivery efficacy and others performing as well as, or even slightly better than, the best commercial agents.7, 70 In summary, efforts to improve complexation and subsequent gene delivery have resulted in the synthesis of CPs with amine groups intermittently interchanged with thiol moieties,71 surface attachment of pendant hydroxyl groups, 51, 72 backbone alkylation,73 and imidazole grafting (upon a chitosan backbone).35 In addition to improved complexation, imidazole-functionalized CPs possessed pKa values of ~6, which is expected to facilitate enhanced gene delivery by polyplex escape from endosomal compartments (to be further explained below). The aforementioned functionalizations offer a variety of approaches to enhance or influence complexation ability; however, excessive complexation is inhibitory to the unpacking of gene cargo in the cytoplasm. Thus, researchers have attempted to address this issue through use of stimuli-response approaches. For example, Kataoka and coworkers synthesized charge-conversional polymers possessing ternary cross-linked polyplexes, exhibiting high complexation at neutral pHs but weak complexation at acidic pHs.74 Such properties can also be conferred by incorporating acid-labile linkers such as imine,75 hydrazone,76 ketal,77 derivatized maleamate,74, 78 and acetal groups.79
Colloid stability of polyplexes is ultimately the rate-limiting step for CP preparation. Poor stability can result in premature gene unpacking, degradation, aggregation, and clearance both in vitro and in vivo. The consensus strategy to improve colloidal stability is by the introduction of hydrophilic layers, which offer steric shielding effects to minimize undesired non-specific interactions. Commonly used hydrophilic layers include: PEG, poly(N-(2-hydroxyproyl)methacrylamide (PHPMA), poly(glucopyranose), and zwitterionic polymers. PEGylation is one of the most extensively studied hydrophilic functionalizations for improving colloidal stability and reducing aggregation.7 Successful gene delivery studies have been completed using structures such as PEG-b-cationic polymers46, 80, 81 and cationic polymers-g-PEG.82–85 However, a contradictory report indicates that PEGylation without efficient release of the PEG moiety results in a lower surface charges and a subsequent decrease in both cellular uptake and gene delivery.86 Thus, PEGylated CPs were synthesized with acid-labile or disulfide linkages which are designed to degrade upon exposure to acidic conditions and, as a result, improve gene delivery.75, 87 Due to oxidation concerns of PEG moieties, the use of zwitterion groups in CPs have been utilized to provide high resistance to nonspecific protein adsorption from human serum while maintaining gene transfection.88, 89
Lastly, polymer biocompatibility/cytotoxicity is strongly associated with biodegradability of the underlying polymeric backbone.90 CP biodegradability is directed by the use of hydrolysable backbones and disulfide linkages. For most CPs, the polymeric backbone is designed to be degraded under physiological conditions, followed by elimination from the human body through renal clearance. Towards this end, various hydrolysable bonds have been explored including esters, amides, phosphoesters, and urethane bonds, resulting in the formation of biodegradable polymers such as aliphatic polyesters, poly(L-lysine), polyphosphoesters, and polyurethanes, respectively.55, 82, 91 Of these polymers, aliphatic polyesters demonstrated greatest biocompatibility and biodegradability, exemplified by FDA approval of polylactide (PLA).92 As a result, Cheng and coworkers synthesized a cationic variant (CPLA) through ring-opening polymerization (ROP) and thiol-ene functionalization and utilized polyplexes for safe delivery of small interfering RNA (siRNA) and pDNA to prostate cancer cells and macrophage and fibroblast cells, respectively.52, 53 As compared with commercial gene delivery vectors (Fugene 6), CPLA variants facilitated enhanced transfection efficiencies, even in the presence of physiologically-relevant levels of fetal bovine serum (data not published), due in part to the polymer’s high complexation ability and biodegradability. To similar effect, disulfide linkages were introduced to polymer backbones and crossing-linkages which promoted cleavage into small fragments by exposure to reductive intracellular environments through glutathione (GSH)-mediated thiol-disulfide exchange reactions.65 Accordingly, incorporations of disulfide bonds significantly reduced the cytotoxicity of non-degradable high molecular weight polymers and were an effective approach for such polymers to release their gene payloads in cytoplasm.93 Pun and coworkers corroborate these previous observations through synthesis of HPMA-oligolysine polymers containing either bioreducible or non-bioreducible linkers between polymer backbones and cationic moieties.94 By proper experimental design, a positive correlation between bioreducible linker inclusion on cell cytotoxicity and transfection efficiency was clearly identified.
EXTRACELLULAR BARRIERS
In vitro Delivery
The first step in performing translational research of nonviral vectors is in vitro gene delivery determination to identify and remove non-effective vectors. Once a lipid or polymer has been designed and formulated, a general consideration for all future (in vivo) studies is the quality of DNA/RNA available for complexation. High quality (pure), intact pDNA or RNA is important for achieving high performance transfections, and can be obtained through established protocols or commercial vendors such as Elim Biopharm. The formation of lipoplexes and polyplexes is then conducted in the absence of serum, as certain serum proteins can interfere with or prevent stable complex formation.
With the formation of quality vectors, only cell culture considerations remain. The most obvious barrier is the choice of cell type, as various cells, such is stem cells, are notoriously difficult to transfect with nonviral methods. There is typically an overarching theme associated with in vitro studies towards eventual application, such as cancer treatment, and thus, there exist a number of immortalized and primary cells to assess this future goal. Along with the proper vector design and formation, cell plating density is another important in vitro consideration. Optimal cell density for transfection varies for different cell types and applications, but for adherent cell lines, generally 70–90% confluency at the time of transfection or 5 × 105 to 2 × 106 cells/mL provides good results. Lipids as a class are known to increase cell permeability, and if antibiotics are used in transfection media, their simultaneous transport into cells can result in cytotoxicity and lower gene delivery. Thus, for lipoplexes, antibiotics are removed or reduced when plating for transfection. Finally, aggregation of vectors can result in false-positive results that will lead to contradictory outcomes in vivo. When using adherent cells, aggregated/sedimented vectors are still able to interact with cells, and thus may exhibit gene delivery. However, when this same vector is applied to suspension cells or in vivo, the aggregation will result in poor transfection by means of sedimentation and clearance, respectively. Unlike other gene delivery challenges, the aforementioned barriers can be manipulated by the end user, and thus, are easy to overcome with proper experimental design.
In vivo Delivery
The most important and most difficult challenge in gene therapy is the issue of in vivo delivery. Not only must a vector evade the reticuloendothelial system (RES) after systemic administration, but it must also cross several barriers prior to entering target cells. The first consideration is the selection of an administration route for prepared vectors. General methods include intravenous injection, topical application, mucosal administration, and oral delivery.3 Once introduced into circulation, vectors are subject to serum inactivation, enzymatic degradation, complement-mediated clearance, and RES recognition. Neutral polyplexes in physiological salt concentrations rapidly form large aggregates that result in ineffective gene delivery and associated cytotoxicity through mechanisms such as lung embolization. In contrast, positively charged particles are easily maintained in solution; however, studies have shown that aggregate size is time dependent95 and expedited by adsorption of serum albumin and other blood proteins which can cause further aggregation and clearance. As introduced above, most undesired interactions can be remedied by decoration of the vector surface with shielding moieties such as PEG, HPMA, sugars, and proteins.7 The shielding effect depends on the hydrophilic functionalization’s molecular mass, grafting density, and method of attachment. Surface PEG coating (i.e., stealth particles) sterically hinders the interactions of blood components and degradative enzymes with the vector surface, thus, preventing opsonization and recognition by phagocytic cells (e.g., macrophages, dendritic cells, and monocytes) and resulting in prolonged circulation in the blood.96
Targeting ligands are frequently grafted on the vector particle surface to guide cell specificity and enhance cellular uptake by receptor-mediated endocytosis. Commonly used targeting ligands include peptides or proteins,97 antibodies,98 cell penetrating peptides,99 sugars,100 and other small molecule ligands.101 In the case of tumors and inflammatory tissue, accumulation at the target site can be achieved by taking advantage of the increased transport of macromolecules across the tumor endothelium attributed to its leaky and discontinuous vascular structure and poor lymphatic drainage.3 This phenomena is referred to as the “enhanced permeability and retention” (EPR) effect. Normally, penetration of macromolecules and nanoparticles are enhanced by prolonged exposure, thus, those particles that have been PEGylated are not rapidly cleared from circulation and will have an increased chance to encounter and pool in the leaky tumor vasculature. However, size restrictions have been identified with relation to specific cancerous tissue, as not all tumors are equally leaky, and for tumors with less porous vasculatures, small nanoparticle sizes (less than 30 nm) are desirable.102 Conversely, for most tumors the optimal mean nanoparticle size has been identified as 100 nm; whereas, particles greater than or equal to 400 nm do not easily enter tumoral capillary gaps. Studies have also shown that particles smaller than 70 nm are able to access parenchymal cells in the liver after crossing sinusoidal space (liver blood vessels).103 Unfortunately, despite the need for smaller particles, reduced sizes are prone to renal excretion through the glomeruli in the kidney.3
A major challenge regardless of nanoparticle type is circumvention of the host immune response. When combined with unmethylated CpG-containing nucleic acid compositions present in the gene payload, nanoparticles can stimulate acute inflammatory responses in the host. This is followed by subsequent production of cytokines and clearance of transfected cells. These factors help contribute to the observed treatment-related cytotoxicity and shortened duration of gene expression. The inflammatory response is mediated by toll-like receptors (TLRs) TLR3, TLR7, TLR8, and TLR9 recognizing double stranded RNA, single stranded RNA in endosomes, G-rich oligonucleotides, and unmethylated CpG sequences, respectively. Additionally, delivery aspects such as chemical and physical properties of the vector, pharmacokinetics, biodistribution of nucleic acids, and route of administration are known to be determinants for specific immune reactions.104
INTRACELLULAR BARRIERS
Uptake Mechanisms
It is generally accepted that cell-surface interactions are driven by electrostatic differences between nanoparticles (cationic) and the cell surface (anionic), to which transmembrane proteins are considered the major binding site. Once semi-stable interactions have been established, nanoparticles are likely to redistribute over the cell surface to participate in a variety of uptake pathways. A comprehensive analysis of nanoparticle uptake mechanism has been presented by various reviews, and thus, will only be briefly covered here.96, 105
Receptor-mediated endocytosis is the most common route of nanoparticle uptake, and encompasses a variety of entry pathways such as clathrin- and caveolae-mediated endocytosis, macropinocytosis, and pathways that are both clathrin- and caveolae-independent. An additional uptake mechanism, phagocytosis, is available to specialty cells, such as macrophages and dendritic cells. Each route is distinguished from the other by particle sizes that are preferred and the involvement of specific proteins mediating endocytic entry. The particle size requirements for each uptake mechanism varies with cell type, but for most cells, endocytosis is on the order of 200 nm or less, macropinocytosis is on the order of 500 nm, and phagocytosis is capable of accommodating sizes up to 10 μm.52
Vesicle Fate and Compartmental Escape
Although most cells will internalize vectors, only a small fraction (<2%) of nucleic acid escapes the endocytic vesicle.106 No matter the uptake route, nanoparticles will be localized within endocytic vesicles, which represent a hostile environment known to facilitate nucleic acid degradation. The first vesicle, termed the early endosome, slowly matures by fusing with other sorting vesicles (from which material can be recycled back to the cell surface by exocytosis) and is eventually trafficked to a late endosome. Transition from early to late endosome is marked by the rapid acidification (pH 5–6) due to the action of the ATPase proton-pump enzyme. Sequential trafficking to the lysosome leads to further acidification (pH ~4.5) and the activation of various degradative enzymes. At this point, those nucleic acids that are unable to escape into the cytoplasm are rapidly degraded. Recent studies have observed that nanoparticles become trapped in the late endosome/multivesicular late endosome and are recycled through multiple pathways; consequently, gene delivery efficiency may be improved by designing delivery vectors that can escape the recycling pathway.107
Several strategies, often vector-specific, are employed to facilitate nucleic acid release. Lipoplexes release their cargo into the cytosol by a lipid mixing mechanism, which presumably involves fusion of carrier lipids with the endosomal membrane, where local perturbations are formed that allow nucleic acid release. The proposed underlying mechanism is that CLs, with or without helper lipids, form nonbilayer structures that participate in the lipoplex-induced flip-flop of negatively-charged phospholipids from the cytoplasmic to the inner face of the endosome (Figure 5). This phenomenon is followed by the formation of charge-neutral ion pairs with the lipoplex, thereby destabilizing the endosomal lamellar membrane organization and causing dissociation of the nucleic acids into the cytosol.
Figure 5.
Schematic representation of classical and proposed lipid mixing mechanisms. a) Classical lipid mixing mechanism was believed to proceed by membrane fusion of the lipoplex and host endosomal membranes followed by exchange of negatively charged phospholipids from the cytoplasmic to the inner face of the endosome. Membrane destabilization and nucleic acid release occur as a result. b) Conversely, lipid mixing may function by an altered mechanism where lipoplexes that are in close proximity to the inner-endosomal membrane lose lipid molecules through dispersal or degradation. Release of lipid molecules permits the passive release of nucleic acids, followed by subsequent cytoplasmic transport mediated by transient pore formation resulting from integration of freed lipids.
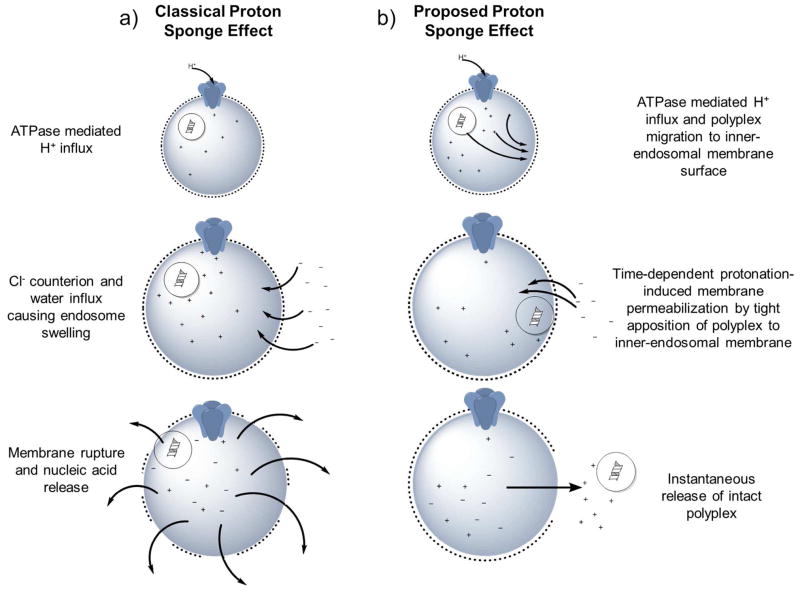
CPs in contrast are believed to release their cargo by the so-called “proton sponge effect” (Figure 6). Cationic polyplexes that contain a large number of titratable secondary and tertiary amines confer pKa values between physiological and lysosomal pH (typically 5.5–6). Thus, upon endolysosomal acidification, the amines become protonated and promote the influx of additional H+ ions and the concomitant influx of Cl− counterions. In order to compensate for increased ion uptake, vesicles introduce additional water molecules, causing osmotic swelling and eventual rupture.
Figure 6.
Schematic representation of classical and proposed proton-sponge mechanisms. a) Classical polyplex-induced proton sponge effect proceeds by the increase of protons due to protonation of amines in the polyplex and ATPase-mediated H+ influx. This is followed by concomitant influx of both counter-ions (Cl−) and water into endocytic vesicles, resulting in osmotic-induced lysis. However, recent studies support an alternative mechanism b) where nucleic acid release relies on a time-depend protonation-induced membrane permeabilization by tight binding of the polyplex and the inner endosomal membrane.
Despite being widely cited, numerous reports have questioned details of the proton sponge mechanism.66, 108, 109 These studies comment on the lack of direct evidence for polyplex-mediated compartmental escape by pH buffering. An alternative mechanism (Figure 6b) has been proposed that suggests escape relies on time-dependent protonation-induced membrane permeabilization by tight apposition of the polyplex and the inner-endosomal membrane. This mechanism was corroborated by Zuhorn and coworkers, where neither complete rupture of endosomes nor release of intact polyplexes (and lipoplexes) into the cytosol was observed.110 Rather, they suggest that at the time of endosomal escape, an almost instantaneous and complete discharge of nucleic acid and remnants of the carrier occurs for polyplexes. In contrast, they observed lipoplexes promote the formation of multiple transient pores, where nucleic acids were more gradually transferred into the cytosol (Figure 5b), which contradicts the accepted “lipid-mixing” paradigm. The last major observation of the study was the frequency of escape, where as few as one and upwards of four/five particles per cell (average of 1–2/cell) contribute to release of nucleic acids from the endosome and subsequent accumulation in the nucleus. The low cellular uptake and even lower endosomal release was unexpected, but was later confirmed by a recent study that utilized colloidal-gold particles conjugated to siRNA to monitor the uptake and release of lipid nanoparticles.106 Based upon observations and computational modeling, the authors estimate that escape of siRNA from the endosome occurs at 1–2% efficiency and only during a limited time-frame when the nanoparticles reside in a vesicle that shares both early and late endosome characteristics.
Other strategies have been utilized to overcome the endosomal barrier. Concurrent chloroquine treatment of cells at the time of transfection is known to buffer pH in acidic vesicles, resulting in improved gene delivery of polyplexes. This strategy and others like it are impractical for in vivo gene delivery and thus are limited to bench-scale experiments. In contrast, fusogenic viral or endosomal lytic peptides can be attached to nanoparticles to provide endosomal escape. These peptides are typically pH-sensitive and biologically-derived. For example, listeriolysin O from Listeria monocytogenes is a pore-forming peptide that is used extensively with bacterial-based gene vectors.111, 112 Alternatively, Li and coworkers developed a lipid-calcium-phosphate nanoparticle (LCP) that achieved efficient siRNA delivery by LCP endosomal disassemble.113 The disassembly resulted in the CaP core dissolving in acidic conditions, causing osmotic pressure-induce vesicle rupture.
Nucleic Acid Translocation
After endosomal escape and cytoplasmic release, nucleic acids should be delivered to their target intracellular compartment to obtain desired biological activity. It can be assumed that for cytoplasmic activity-based nucleic acids, such as siRNA, this translocation barrier is nonexistent. However, the mobility of larger molecules, such as pDNA, is extremely low in the cytoplasm, making the payload susceptible to cytoplasmic nucleases.110, 114 It has been reported that the half-life of naked pDNA in the cytoplasm is 50–90 min.115 Previously, researchers believed that upon escape from the endosome, remnants of nanoparticles may protect larger nucleic acid constructs from degradation. However, recent studies disproved these assumptions by observations of empty lipoplexes that were anchored to/continuous with the endosomal membrane, and free polymer being expelled into the cytosol with no accompanied translocation away from the endosomal vesicles at the time of nucleic acid release.110
The determining factor for the nucleic acid mobility rate through the cytoplasm is the size and spherical structure of the molecule, with circular pDNA being faster than linear structures.116 The state of DNA compaction with reference to gene delivery is poorly understood, but tight compaction may result in increased mobility and stability against cytoplasmic nucleases. Nuclear expression requires overcoming an addition intracellular barrier, transnuclear passage. For dividing cells, nucleic acids can enter the nucleus during nuclear membrane breakdown; however, in non-dividing cells, gene cargo must cross the membrane via the nuclear pore complex (NPC) which only permits entry of molecules up to 9 nm in size and <40 kDa by free diffusion.117 In the case of larger macromolecules, transferal is energy-dependent.118 Introduction of specific sequences, termed Nuclear Localization Sequences (NLS), to nucleic acids can promote the attachment of proteins that are expected to enter the nucleus and mediate transnuclear transport through the NPC.119, 120 Despite noted success121, attachment of NLS-recognition proteins can affect biological expression/activity of pDNA120 and attempts at ex vivo attachment require a complex synthesis mechanism.122 Overcoming nuclear translocation is poorly studied and requires more extensive research to investigate potential solutions.
TYPES OF NUCLEIC ACIDS
Successful therapy is accomplished by delivery of one of three mechanism-exclusive sets of nucleic acid payloads to either the cytoplasm or nucleus. The first set consists of the delivery of pDNA and other larger DNA-based constructs (bacterial artificial chromosomes123 and human artificial chromosomes124) to the nucleus, facilitating subsequent expression of a transgene. Similarly, antisense oligonucleotides (AONs), which are 15–30 base pair (bp) constructs, also require entry into the nucleus (and the cytosol in rare occurrences), but in contrast, they prevent the expression of a target gene by hybridizing with host complimentary mRNA. 3, 125 The last set is composed of siRNAs, short hairpin RNA (shRNA) and microRNA (miRNA), that participate in RNA interference (RNAi) mechanisms. Upon delivery into the cytoplasm, these small RNA-based therapeutics operate by a sequence-specific hybridization to their complementary target mRNA, thus, blocking further translation of a desired protein and facilitating target mRNA cleavage and degradation.
Expression Systems
Plasmid DNA expression systems contain a transgene that encodes for a specific protein, that is borne from circular, double-stranded DNA, ranging in size from a few hundred basepairs (bp) to several thousand bp (Table 3). Following nuclear uptake, DNA transcription is driven by a designed promoter, followed by translation by the host’s native machinery. In addition, pDNA can contain other regulatory signals that play an important role in gene expression such as enhancer sequences and splicing and polyadenylation sites.2 Promoter sequences can be engineered to include strong or cell-specific expression; viral-based promoters, such as the cytomegalovirus (CMV) promoter, are the most commonly used. Additionally, selection of proper enhancers can drastically improve transcription efficiency.126, 127 Other elements include a prokaryotic antibiotic resistance gene, origin of replication, and the remainder of the bacterial-derived plasmid backbone (BB). These additional features have been attributed to the failure in vivo of pDNA-based expression systems to achieve efficient and sustained activity.128 It has been proposed that silencing of pDNA transgenes begins when 1 kb or more of genetic material is outside of the transcriptional unit.129 To address this concern, minicircle DNA (MC) vectors, which are devoid of BBs, have resulted in sustained and elevated transgene expression in quiescent tissues in vivo as compared to tradition pDNA.128, 130, 131 Even though protocols have been established for MC production,132 the experimental scheme is still tedious and substantially more complicated than traditional pDNA preparation. Recently, an improved expression vector, mini-intronic plasmid (MIP), was created by Kay and coworkers.128 They report that placement of the bacterial origin of replication and an antibiotic-free selectable marker as an intron in the transgene expression cassette results in elevated gene delivery compared to both pDNA and MC while still maintaining the advantage of facile propagation and purification. Other expression systems such as bacterial and human artificial chromosomes have also been used in gene delivery.123, 124.
Table 3.
Nucleic acid examples used for gene delivery experiments.
| Characteristic | pDNA | MC | MIP | AON | siRNA | shRNA | miRNA |
|---|---|---|---|---|---|---|---|
| Shape |
|
|
|
|
|
||
| Molecular Weight | ~1.5–20 kb | ~2–6 kb | ~2–4 kb | 8–50 nucleotides | 19–25 bp | Stem: 25–29 bp Loop: 4–23 nucleotides |
21–24 nucleotides |
| Synthesis/Propagation | Bacterial propagation and purification. MC requires additional restriction enzyme and ligase reactions | Synthetic, however, siRNA has been produced by bacterial propagation and purification | |||||
| Mechanism of Activity | Nuclear localization followed by transgene expression under engineered promotors (cell specific, viral, etc.) | Steric-translation-blocker and mRNA maturation interference | RNAi mechanism | ||||
| Immune Response | TLR9 | TLR9 (limited) | TLR9 (rare) | TLR9 (if construct contains CpG motifs) | TLR 3/7/8/9, retinoic acid-inducible gene 1 (RIG)-like receptor, protein kinase receptor, helicases | ||
| Advantages | Controlled expression, nonreplicative, encodes/ex presses only gene of interest | All pDNA advantages in addition to limited bacterial sequences, limited immune response, limited transgene silencing | All pDNA advantages in addition to lowest amount of bacterial sequences and transgene silencing. Highest transgene expression activity | No nuclear barrier (except AONs), insertional mutagenesis is circumvented, requires lower amount of carrier | |||
| Disadvantages | Contains bacterial backbone and antibiotic resistance markers. Requires nuclear entry and experiences transgene silencing | More complicated propagation and lower yields | Requires creation of special bacterial propagation strains | Complicated and more expensive synthesis, cannot be used to express a gene of interest, short term activity, off-target effects. Nuclear barrier can exist for the application of AONs | |||
| Peak Activity Timeframe | 1–10 days | 1–60+ days | 1–60+ days | Less than 7 days, unless expressed from an expression vector | |||
Nuclear Inhibition
AON-based techniques achieve suppression/elimination of genetic material by using synthetic nucleotide sequences (DNA, RNA, and chemical analogues) that will bind the mRNA produced by the target gene and inactivate it (Table 3). Alternatively, it can target a splicing site on pre-mRNA and modify the exon content.133 The approach has been heavily researched for various cancers, diabetes, arthritis, and other inflammatory-related diseases, and therapies are now beginning to emerge such as FDA-approved fomivirsen, vitravene, and mipomersen.2
The design of AONs is considerably easier than expression-based systems due to the advantages of lower delivery requirements, cytoplasmic and nuclear activity, and the omission of enhancer and promoter sequences. However, these nucleic acids are limited to the expression reduction of only endogenous genes and require extensive screening and design to overcome off-target effects.
Cytoplasmic Inhibition
Gene modulation that occurs in the cytoplasm proceeds by RNAi mechanisms and in limited cases by AONs. In the RNAi process, siRNA are the most encountered class (Table 3). This set of nucleic acids features a double-stranded RNA molecule ranging in size from 19–25 bp that possess a characteristic 2-nucleotide overhang at both 3′ locations. The overhangs allow recognition and sequential formation of large protein complexes, called the RNA-induced silencing complex (RISC), where the sense strand of the siRNA molecule is degraded by Argo-2. Thus, the antisense strand-RISC complex specifically cleaves complimentary mRNA. Unlike other RNA-based therapeutics that require chemical synthesis methods, recent studies have demonstrated the feasibility to produce siRNAs by bacterial propagation methods (only slightly more complex than traditional pDNA purification).134
By contrast, shRNA molecules are one continuous single strand of nucleic acid, ranging in size from 25–29 bases, which contain a loop of 4–23 nucleotides (Table 3). Similar to siRNA, shRNA also induce gene silencing through RNAi mechanisms; however, shRNA require processing by the Dicer protein to remove the loop to produce siRNA.135–137 Outside nanoparticle delivery strategies, engineered E. coli were used to produce and carry shRNA for bactofection studies.138 The last set of cytoplasmic molecules are miRNA, which are single-stranded, non-coding nucleic acids, ranging in size from 20–24 nucleotides, that bind partially to complementary sites on their target mRNA. The miRNA-induced silencing operates by affecting the stability of target mRNAs or blocking translation.136 If the miRNA has perfect complementarity to the target mRNA, it will degrade the transcript similar to siRNA.
Each of the small RNA-based approaches can additionally be utilized by delivery of pDNA or other expression systems to generate endogenous RNAi molecules. This route circumvents the price of synthesis, but reintroduces all the barriers associated with pDNA delivery. However, effective introduction by means of pDNA and variants (MCs and MIPs) can lead to a sustained down regulation of a target gene.
CONCLUSION
Gene therapy holds the promise of treating genetic diseases by introduction of exogenous transgenes into specific cells or by the introduction of therapeutic agents that can inhibit expression of target endogenous genes using approaches such as antisense oligonucleotides (AON) or RNAi-based molecules (siRNA, shRNA, and miRNA). Gene therapy has been studied for the treatment and vaccination of numerous diseases including cancer, viral infections, and genetic diseases. The successful transfer of nucleic acids (e.g., plasmid DNA, bacterial/human artificial chromosomes, AON, siRNA, shRNA, etc.) into cells is greatly aided by efficient carriers to overcome extracellular barriers that include enzymatic degradation and rapid clearance after administration. Presently, viral vectors are the most efficient carrier system; however, immunogenicity, tumorigenicity, inflammatory reactions, and formulation and scalability issues limit their clinical use. Consequently, nonviral vectors that combine tunable properties with facile synthesis and particle formation are alternatives. The ideal nonviral carrier should be non-cytotoxic and yet ensure that the DNA/RNA cargo survives the various extra- and intracellular environments while facilitating release and subsequent translocation to the appropriate cellular compartment. To date, the barriers of gene delivery have limited the effectiveness of nonviral-based gene therapeutics, precluding translational research. Thus, this perspective discusses the gene delivery barriers and potential strategies that have been employed to allow for successful eventual application.
Table 1.
Cationic lipid design parameters that influence gene delivery.
| Important Factor | Function | Desired Characteristics | Points of Design | Design Considerations |
|---|---|---|---|---|
| Hydrophilic Headgroup |
|
|
Charge
Total length |
Cationic charge is positively correlated with gene delivery efficacy; however, excessive cationic charge can result in decreased gene delivery due to overly tight nucleic acid binding, or increased associated cytotoxicity. Less obvious trends are presented with amine separation length and total length. Although, large head groups result in a less effective lipoplex morphology (micelle). |
| Hydrophobic Tail Group | Determines lipoplex:
|
|
Shape
|
The most successful vectors employed are usually monounsaturated; however, studies indicate that additional degrees of unsaturation improve gene delivery. Studies have shown that shorter tails are also correlated with increased gene delivery. Additionally, the most important consideration is the ratio of area of the tail group to the head (packing efficiency), which determines the morphology (lamellar is the best). |
| Linker |
|
|
Aliphatics Oxygen containing
|
Overly stable linkers result in lipids that don’t easily degrade and elevated levels of cytotoxicity. Conversely, easily cleaved bonds result in lipids that do not reside long enough to reach the target cell or facilitate endosomal release. A balance between lipid persistence and cytotoxicity must be determined. Thus, carbamate and ester bonds represent the majority of successful vectors. |
Table 2.
Cationic polymer design parameters that influence gene delivery.
| Important Factor | Function | Desired Characteristics | Points of Design | Design Considerations |
|---|---|---|---|---|
| Charge Density | Determines degree of interaction with nucleic acids and related to cytotoxicity | Balance of cationic charge |
Charge
|
Total charge density is the determining factor when complexation of anionic material is needed. Polymeric charge density control can be precisely controlled through sophisticated synthesis techniques which allow users to tailor polymers to desired cationic charges. |
| Charge Center Type | Confers charge and related to charge density (complexation degree) | Possess pKa that facilitate release of nucleic acids at desired locations |
Nitrogen containing
|
Studies indicate that complexation proceeds in the order of quaternary>tertiary>secondary>primary. However, contradictory findings have been reported. |
| Molecular Weight |
|
|
Length Heterogeneity of MW (PDI) |
Gene delivery is positively correlated with increasing molecular weights. Although, increasing molecular weight is usually accompanied by increases in cytotoxicity, believed to arise from enhanced CP hydrophobic properties that result in increased cell membrane damage. |
| Functionality Grafting | Incorporation of functionalities that are designed to address specific gene delivery barriers | Dependent upon desired functionality |
Polyplex stability
|
Functionalization can be achieved by a number of methods to meet any desired needs. However, outcomes have been mixed and screening is required before any additions can be implemented therapeutically. |
Acknowledgments
Funding Sources
The authors recognize support from NIH award AI088485.
The authors thank a SUNY-Buffalo Schomburg fellowship (CHJ) for financial support.
Footnotes
The authors declare no competing financial interest.
References
- 1.Friedmann T, Roblin R. Gene therapy for human genetic disease? Science. 1972;175(4025):949–55. doi: 10.1126/science.175.4025.949. [DOI] [PubMed] [Google Scholar]
- 2.Elsabahy M, Nazarali A, Foldvari M. Non-viral nucleic acid delivery: key challenges and future directions. Current drug delivery. 2011;8(3):235–44. doi: 10.2174/156720111795256174. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Satterlee A, Huang L. In vivo gene delivery by nonviral vectors: overcoming hurdles? Mol Ther. 2012;20(7):1298–304. doi: 10.1038/mt.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vannucci L, Lai M, Chiuppesi F, Ceccherini-Nelli L, Pistello M. Viral vectors: a look back and ahead on gene transfer technology. The new microbiologica. 2013;36(1):1–22. [PubMed] [Google Scholar]
- 5.Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, Northrop JP, Ringold GM, Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci U S A. 1987;84(21):7413–7. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mintzer MA, Simanek EE. Nonviral vectors for gene delivery. Chemical reviews. 2009;109(2):259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 7.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4(7):581–93. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 8.Tros de Ilarduya C, Sun Y, Duzgunes N. Gene delivery by lipoplexes and polyplexes. Eur J Pharm Sci. 2010;40(3):159–70. doi: 10.1016/j.ejps.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 9.Elouahabi A, Ruysschaert JM. Formation and intracellular trafficking of lipoplexes and polyplexes. Mol Ther. 2005;11(3):336–47. doi: 10.1016/j.ymthe.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Samal SK, Dash M, Van Vlierberghe S, Kaplan DL, Chiellini E, van Blitterswijk C, Moroni L, Dubruel P. Cationic polymers and their therapeutic potential. Chemical Society Reviews. 2012;41(21):7147–7194. doi: 10.1039/c2cs35094g. [DOI] [PubMed] [Google Scholar]
- 11.Bloomfield VA. DNA condensation by multivalent cations. Biopolymers. 1997;44(3):269–82. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 12.Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release. 2006;114(1):100–9. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Guenin E, Herve AC, Floch VV, Loisel S, Yaouanc JJ, Clement JC, Ferec C, des Abbayes H. Cationic Phosphonolipids Containing Quaternary Phosphonium and Arsonium Groups for DNA Transfection with Good Efficiency and Low Cellular Toxicity**. Angewandte Chemie. 2000;39(3):629–631. [PubMed] [Google Scholar]
- 14.Martin B, Sainlos M, Aissaoui A, Oudrhiri N, Hauchecorne M, Vigneron JP, Lehn JM, Lehn P. The design of cationic lipids for gene delivery. Current pharmaceutical design. 2005;11(3):375–94. doi: 10.2174/1381612053382133. [DOI] [PubMed] [Google Scholar]
- 15.Bennett MJ, Aberle AM, Balasubramaniam RP, Malone JG, Malone RW, Nantz MH. Cationic lipid-mediated gene delivery to murine lung: correlation of lipid hydration with in vivo transfection activity. Journal of medicinal chemistry. 1997;40(25):4069–78. doi: 10.1021/jm970155q. [DOI] [PubMed] [Google Scholar]
- 16.Felgner JH, Kumar R, Sridhar CN, Wheeler CJ, Tsai YJ, Border R, Ramsey P, Martin M, Felgner PL. Enhanced gene delivery and mechanism studies with a novel series of cationic lipid formulations. J Biol Chem. 1994;269(4):2550–61. [PubMed] [Google Scholar]
- 17.Banerjee R, Mahidhar YV, Chaudhuri A, Gopal V, Rao NM. Design, synthesis, and transfection biology of novel cationic glycolipids for use in liposomal gene delivery. Journal of medicinal chemistry. 2001;44(24):4176–85. doi: 10.1021/jm000466s. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh YK, Visweswariah SS, Bhattacharya S. Nature of linkage between the cationic headgroup and cholesteryl skeleton controls gene transfection efficiency. Febs Lett. 2000;473(3):341–344. doi: 10.1016/s0014-5793(00)01558-1. [DOI] [PubMed] [Google Scholar]
- 19.Byk G, Dubertret C, Escriou V, Frederic M, Jaslin G, Rangara R, Pitard B, Crouzet J, Wils P, Schwartz B, Scherman D. Synthesis, activity, and structure--activity relationship studies of novel cationic lipids for DNA transfer. Journal of medicinal chemistry. 1998;41(2):229–35. doi: 10.1021/jm9704964. [DOI] [PubMed] [Google Scholar]
- 20.Fujiwara T, Hasegawa S, Hirashima N, Nakanishi M, Ohwada T. Gene transfection activities of amphiphilic steroid-polyamine conjugates. Biochimica et biophysica acta. 2000;1468(1–2):396–402. doi: 10.1016/s0005-2736(00)00278-9. [DOI] [PubMed] [Google Scholar]
- 21.Pedroso de Lima MC, Neves S, Filipe A, Duzgunes N, Simoes S. Cationic liposomes for gene delivery: from biophysics to biological applications. Curr Med Chem. 2003;10(14):1221–31. doi: 10.2174/0929867033457430. [DOI] [PubMed] [Google Scholar]
- 22.Xu Y, Szoka FC., Jr Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry-Us. 1996;35(18):5616–23. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- 23.Hasegawa S, Hirashima N, Nakanishi M. Comparative study of transfection efficiency of cationic cholesterols mediated by liposomes-based gene delivery. Bioorganic & medicinal chemistry letters. 2002;12(9):1299–302. doi: 10.1016/s0960-894x(02)00119-1. [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharya S, Bajaj A. Advances in gene delivery through molecular design of cationic lipids. Chem Commun (Camb) 2009;(31):4632–56. doi: 10.1039/b900666b. [DOI] [PubMed] [Google Scholar]
- 25.Zhi D, Zhang S, Wang B, Zhao Y, Yang B, Yu S. Transfection efficiency of cationic lipids with different hydrophobic domains in gene delivery. Bioconjug Chem. 2010;21(4):563–77. doi: 10.1021/bc900393r. [DOI] [PubMed] [Google Scholar]
- 26.Niculescu-Duvaz D, Heyes J, Springer CJ. Structure-activity relationship in cationic lipid mediated gene transfection. Curr Med Chem. 2003;10(14):1233–61. doi: 10.2174/0929867033457476. [DOI] [PubMed] [Google Scholar]
- 27.Liu D, Qiao W, Li Z, Chen Y, Cui X, Li K, Yu L, Yan K, Zhu L, Guo Y, Cheng L. Structure-function relationship research of glycerol backbone-based cationic lipids for gene delivery. Chemical Biology & Drug Design. 2008;71(4):336–44. doi: 10.1111/j.1747-0285.2008.00644.x. [DOI] [PubMed] [Google Scholar]
- 28.Fletcher S, Ahmad A, Perouzel E, Heron A, Miller AD, Jorgensen MR. In vivo studies of dialkynoyl analogues of DOTAP demonstrate improved gene transfer efficiency of cationic liposomes in mouse lung. Journal of Medicinal Chemistry. 2006;49(1):349–57. doi: 10.1021/jm0507227. [DOI] [PubMed] [Google Scholar]
- 29.Densmore CL, Giddings TH, Waldrep JC, Kinsey BM, Knight V. Gene transfer by guanidinium-cholesterol: dioleoylphosphatidyl-ethanolamine liposome-DNA complexes in aerosol. J Gene Med. 1999;1(4):251–64. doi: 10.1002/(SICI)1521-2254(199907/08)1:4<251::AID-JGM43>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 30.Nagasaki T, Taniguchi A, Tamagaki S. Photoenhancement of transfection efficiency using novel cationic lipids having a photocleavable spacer. Bioconjug Chem. 2003;14(3):513–6. doi: 10.1021/bc0256603. [DOI] [PubMed] [Google Scholar]
- 31.Guo X, Szoka FC., Jr Chemical approaches to triggerable lipid vesicles for drug and gene delivery. Acc Chem Res. 2003;36(5):335–41. doi: 10.1021/ar9703241. [DOI] [PubMed] [Google Scholar]
- 32.Hsu WL, Chen HL, Liou W, Lin HK, Liu WL. Mesomorphic complexes of DNA with the mixtures of a cationic surfactant and a neutral lipid. Langmuir. 2005;21(21):9426–31. doi: 10.1021/la051863e. [DOI] [PubMed] [Google Scholar]
- 33.Tranchant I, Thompson B, Nicolazzi C, Mignet N, Scherman D. Physicochemical optimisation of plasmid delivery by cationic lipids. The Journal of Gene Medicine. 2004;6(S1):S24–S35. doi: 10.1002/jgm.509. [DOI] [PubMed] [Google Scholar]
- 34.Bottega R, Epand RM. Inhibition of protein kinase C by cationic amphiphiles. Biochemistry-US. 1992;31(37):9025–30. doi: 10.1021/bi00152a045. [DOI] [PubMed] [Google Scholar]
- 35.Solodin I, Brown CS, Bruno MS, Chow CY, Jang EH, Debs RJ, Heath TD. A novel series of amphiphilic imidazolinium compounds for in vitro and in vivo gene delivery. Biochemistry-Us. 1995;34(41):13537–44. doi: 10.1021/bi00041a033. [DOI] [PubMed] [Google Scholar]
- 36.van der Woude I, Wagenaar A, Meekel AA, ter Beest MB, Ruiters MH, Engberts JB, Hoekstra D. Novel pyridinium surfactants for efficient, nontoxic in vitro gene delivery. Proc Natl Acad Sci U S A. 1997;94(4):1160–5. doi: 10.1073/pnas.94.4.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roosjen A, Šmisterová J, Driessen C, Anders Joachim T, Wagenaar A, Hoekstra D, Hulst R, Engberts Jan BFN. Synthesis and Characteristics of Biodegradable Pyridinium Amphiphiles Used for in vitro DNA Delivery. European Journal of Organic Chemistry. 2002;2002(7):1271–1277. [Google Scholar]
- 38.Ilies MA, Johnson BH, Makori F, Miller A, Seitz WA, Thompson EB, Balaban AT. Pyridinium cationic lipids in gene delivery: an in vitro and in vivo comparison of transfection efficiency versus a tetraalkylammonium congener. Archives of biochemistry and biophysics. 2005;435(1):217–26. doi: 10.1016/j.abb.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 39.Ilies MA, Seitz WA, Johnson BH, Ezell EL, Miller AL, Thompson EB, Balaban AT. Lipophilic pyrylium salts in the synthesis of efficient pyridinium-based cationic lipids, gemini surfactants, and lipophilic oligomers for gene delivery. Journal of medicinal chemistry. 2006;49(13):3872–87. doi: 10.1021/jm0601755. [DOI] [PubMed] [Google Scholar]
- 40.Freedland SJ, Malone RW, Borchers HM, Zadourian Z, Malone JG, Bennett MJ, Nantz MH, Li JH, Gumerlock PH, Erickson KL. Toxicity of cationic lipid-ribozyme complexes in human prostate tumor cells can mimic ribozyme activity. Biochemical and molecular medicine. 1996;59(2):144–53. doi: 10.1006/bmme.1996.0080. [DOI] [PubMed] [Google Scholar]
- 41.Leventis R, Silvius JR. Interactions of mammalian cells with lipid dispersions containing novel metabolizable cationic amphiphiles. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1990;1023(1):124–132. doi: 10.1016/0005-2736(90)90017-i. [DOI] [PubMed] [Google Scholar]
- 42.Farhood H, Bottega R, Epand RM, Huang L. Effect of cationic cholesterol derivatives on gene transfer and protein kinase C activity. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1992;1111(2):239–246. doi: 10.1016/0005-2736(92)90316-e. [DOI] [PubMed] [Google Scholar]
- 43.Choi JS, Lee EJ, Jang HS, Park JS. New cationic liposomes for gene transfer into mammalian cells with high efficiency and low toxicity. Bioconjug Chem. 2001;12(1):108–13. doi: 10.1021/bc000081o. [DOI] [PubMed] [Google Scholar]
- 44.Floch V, Loisel S, Guenin E, Herve AC, Clement JC, Yaouanc JJ, des Abbayes H, Ferec C. Cation substitution in cationic phosphonolipids: a new concept to improve transfection activity and decrease cellular toxicity. Journal of medicinal chemistry. 2000;43(24):4617–28. doi: 10.1021/jm000006z. [DOI] [PubMed] [Google Scholar]
- 45.Pinnaduwage P, Schmitt L, Huang L. Use of a quaternary ammonium detergent in liposome mediated DNA transfection of mouse L-cells. Biochimica et biophysica acta. 1989;985(1):33–7. doi: 10.1016/0005-2736(89)90099-0. [DOI] [PubMed] [Google Scholar]
- 46.Tang F, Hughes JA. Synthesis of a single-tailed cationic lipid and investigation of its transfection. J Control Release. 1999;62(3):345–58. doi: 10.1016/s0168-3659(99)00158-3. [DOI] [PubMed] [Google Scholar]
- 47.Wang ZH, Li WB, Ma J, Tang GP, Yang WT, Xu FJ. Functionalized Nonionic Dextran Backbones by Atom Transfer Radical Polymerization for Efficient Gene Delivery. Macromolecules. 2010;44(2):230–239. [Google Scholar]
- 48.Ping Y, Liu CD, Tang GP, Li JS, Li J, Yang WT, Xu FJ. Functionalization of Chitosan via Atom Transfer Radical Polymerization for Gene Delivery. Adv Funct Mater. 2010;20(18):3106–3116. [Google Scholar]
- 49.Zhu C, Zheng M, Meng F, Mickler FM, Ruthardt N, Zhu X, Zhong Z. Reversibly shielded DNA polyplexes based on bioreducible PDMAEMA-SS-PEG-SS-PDMAEMA triblock copolymers mediate markedly enhanced nonviral gene transfection. Biomacromolecules. 2012;13(3):769–78. doi: 10.1021/bm201693j. [DOI] [PubMed] [Google Scholar]
- 50.Li H, Cortez MA, Phillips HR, Wu Y, Reineke TM. Poly(2-deoxy-2-methacrylamido glucopyranose)-b-Poly(methacrylate amine)s: Optimization of Diblock Glycopolycations for Nucleic Acid Delivery. ACS Macro Letters. 2013;2(3):230–235. doi: 10.1021/mz300660t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang QF, Yi WJ, Wang B, Zhang J, Ren L, Chen QM, Guo L, Yu XQ. Linear polycations by ring-opening polymerization as non-viral gene delivery vectors. Biomaterials. 2013;34(21):5391–401. doi: 10.1016/j.biomaterials.2013.03.083. [DOI] [PubMed] [Google Scholar]
- 52.Jones CH, Chen CK, Jiang M, Fang L, Cheng C, Pfeifer BA. Synthesis of cationic polylactides with tunable charge densities as nanocarriers for effective gene delivery. Mol Pharm. 2013;10(3):1138–45. doi: 10.1021/mp300666s. [DOI] [PubMed] [Google Scholar]
- 53.Chen CK, Law WC, Aalinkeel R, Nair B, Kopwitthaya A, Mahajan SD, Reynolds JL, Zou J, Schwartz SA, Prasad PN, Cheng C. Well-defined degradable cationic polylactide as nanocarrier for the delivery of siRNA to silence angiogenesis in prostate cancer. Adv Healthc Mater. 2012;1(6):751–61. doi: 10.1002/adhm.201200094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu Y, Chen CK, Law WC, Mok J, Zou J, Prasad PN, Cheng C. Well-Defined Degradable Brush Polymer-Drug Conjugates for Sustained Delivery of Paclitaxel. Mol Pharm. 2012 doi: 10.1021/mp3004868. [DOI] [PubMed] [Google Scholar]
- 55.Lynn DM, Langer R. Degradable Poly(β-amino esters): Synthesis, Characterization, and Self-Assembly with Plasmid DNA. J Am Chem Soc. 2000;122:10761–10768. [Google Scholar]
- 56.Green JJ, Zugates GT, Langer R, Anderson DG. Poly(beta-amino esters): procedures for synthesis and gene delivery. Methods in molecular biology. 2009;480:53–63. doi: 10.1007/978-1-59745-429-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sunshine JC, Sunshine SB, Bhutto I, Handa JT, Green JJ. Poly(beta-amino ester)-nanoparticle mediated transfection of retinal pigment epithelial cells in vitro and in vivo. Plos One. 2012;7(5):e37543. doi: 10.1371/journal.pone.0037543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sunshine JC, Peng DY, Green JJ. Uptake and Transfection with Polymeric Nanoparticles Are Dependent on Polymer End-Group Structure, but Largely Independent of Nanoparticle Physical and Chemical Properties. Mol Pharm. 2012 doi: 10.1021/mp3004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fields RJ, Cheng CJ, Quijano E, Weller C, Kristofik N, Duong N, Hoimes C, Egan ME, Saltzman WM. Surface modified poly(beta amino ester)-containing nanoparticles for plasmid DNA delivery. J Control Release. 2012;164(1):41–8. doi: 10.1016/j.jconrel.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wadhwa MS, Collard WT, Adami RC, McKenzie DL, Rice KG. Peptide-Mediated Gene Delivery: Influence of Peptide Structure on Gene Expression. Bioconjugate Chem. 1997;8(1):81–88. doi: 10.1021/bc960079q. [DOI] [PubMed] [Google Scholar]
- 61.Plank C, Tang MX, Wolfe AR, Szoka FC., Jr Branched cationic peptides for gene delivery: role of type and number of cationic residues in formation and in vitro activity of DNA polyplexes. Human gene therapy. 1999;10(2):319–32. doi: 10.1089/10430349950019101. [DOI] [PubMed] [Google Scholar]
- 62.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67(5):598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 63.Katav T, Liu L, Traitel T, Goldbart R, Wolfson M, Kost J. Modified pectin-based carrier for gene delivery: cellular barriers in gene delivery course. J Control Release. 2008;130(2):183–91. doi: 10.1016/j.jconrel.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 64.Xu FJ, Li H, Li J, Zhang Z, Kang ET, Neoh KG. Pentablock copolymers of poly(ethylene glycol), poly((2-dimethyl amino)ethyl methacrylate) and poly(2-hydroxyethyl methacrylate) from consecutive atom transfer radical polymerizations for non-viral gene delivery. Biomaterials. 2008;29(20):3023–33. doi: 10.1016/j.biomaterials.2008.03.041. [DOI] [PubMed] [Google Scholar]
- 65.Chu DSH, Schellinger JG, Shi J, Convertine AJ, Stayton PS, Pun SH. Application of Living Free Radical Polymerization for Nucleic Acid Delivery. Accounts Chem Res. 2012;45(7):1089–1099. doi: 10.1021/ar200242z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Godbey WT, Wu KK, Mikos AG. Size matters: molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. J Biomed Mater Res. 1999;45(3):268–75. doi: 10.1002/(sici)1097-4636(19990605)45:3<268::aid-jbm15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 67.Johnson RN, Chu DS, Shi J, Schellinger JG, Carlson PM, Pun SH. HPMA-oligolysine copolymers for gene delivery: optimization of peptide length and polymer molecular weight. J Control Release. 2011;155(2):303–11. doi: 10.1016/j.jconrel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eltoukhy AA, Siegwart DJ, Alabi CA, Rajan JS, Langer R, Anderson DG. Effect of molecular weight of amine end-modified poly(beta-amino ester)s on gene delivery efficiency and toxicity. Biomaterials. 2012;33(13):3594–603. doi: 10.1016/j.biomaterials.2012.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo S, Huang Y, Wei T, Zhang W, Wang W, Lin D, Zhang X, Kumar A, Du Q, Xing J, Deng L, Liang Z, Wang PC, Dong A, Liang XJ. Amphiphilic and biodegradable methoxy polyethylene glycol-block-(polycaprolactone-graft-poly(2-(dimethylamino)ethyl methacrylate)) as an effective gene carrier. Biomaterials. 2011;32(3):879–89. doi: 10.1016/j.biomaterials.2010.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun X, Zhang N. Cationic polymer optimization for efficient gene delivery. Mini reviews in medicinal chemistry. 2010;10(2):108–25. doi: 10.2174/138955710791185109. [DOI] [PubMed] [Google Scholar]
- 71.Bacalocostantis I, Mane VP, Kang MS, Goodley AS, Muro S, Kofinas P. Effect of thiol pendant conjugates on plasmid DNA binding, release, and stability of polymeric delivery vectors. Biomacromolecules. 2012;13(5):1331–9. doi: 10.1021/bm3004786. [DOI] [PubMed] [Google Scholar]
- 72.Luo XH, Huang FW, Qin SY, Wang HF, Feng J, Zhang XZ, Zhuo RX. A strategy to improve serum-tolerant transfection activity of polycation vectors by surface hydroxylation. Biomaterials. 2011;32(36):9925–39. doi: 10.1016/j.biomaterials.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 73.Jia H-z, Luo X-h, Cheng H, Yang J, Li C, Liu CW, Feng J, Zhang X-z, Zhuo R-x. Extraordinarily enhanced gene transfection and cellular uptake by aromatic hydrophobicization to PEI25K. J Mater Chem. 2012;22(45):24092–24101. [Google Scholar]
- 74.Sanjoh M, Miyata K, Christie RJ, Ishii T, Maeda Y, Pittella F, Hiki S, Nishiyama N, Kataoka K. Dual environment-responsive polyplex carriers for enhanced intracellular delivery of plasmid DNA. Biomacromolecules. 2012;13(11):3641–9. doi: 10.1021/bm301095a. [DOI] [PubMed] [Google Scholar]
- 75.Kim YH, Park JH, Lee M, Park TG, Kim SW. Polyethylenimine with acid-labile linkages as a biodegradable gene carrier. J Control Release. 2005;103(1):209–19. doi: 10.1016/j.jconrel.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 76.Walker GF, Fella C, Pelisek J, Fahrmeir J, Boeckle S, Ogris M, Wagner E. Toward synthetic viruses: endosomal pH-triggered deshielding of targeted polyplexes greatly enhances gene transfer in vitro and in vivo. Mol Ther. 2005;11(3):418–25. doi: 10.1016/j.ymthe.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 77.Shim MS, Kwon YJ. Acid-transforming polypeptide micelles for targeted nonviral gene delivery. Biomaterials. 2010;31(12):3404–13. doi: 10.1016/j.biomaterials.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 78.Lee Y, Miyata K, Oba M, Ishii T, Fukushima S, Han M, Koyama H, Nishiyama N, Kataoka K. Charge-conversion ternary polyplex with endosome disruption moiety: a technique for efficient and safe gene delivery. Angewandte Chemie. 2008;47(28):5163–6. doi: 10.1002/anie.200800963. [DOI] [PubMed] [Google Scholar]
- 79.Knorr V, Russ V, Allmendinger L, Ogris M, Wagner E. Acetal linked oligoethylenimines for use as pH-sensitive gene carriers. Bioconjug Chem. 2008;19(8):1625–34. doi: 10.1021/bc8001858. [DOI] [PubMed] [Google Scholar]
- 80.Hwang do W, Son S, Jang J, Youn H, Lee S, Lee D, Lee YS, Jeong JM, Kim WJ, Lee DS. A brain-targeted rabies virus glycoprotein-disulfide linked PEI nanocarrier for delivery of neurogenic microRNA. Biomaterials. 2011;32(21):4968–75. doi: 10.1016/j.biomaterials.2011.03.047. [DOI] [PubMed] [Google Scholar]
- 81.Gary DJ, Lee H, Sharma R, Lee JS, Kim Y, Cui ZY, Jia D, Bowman VD, Chipman PR, Wan L, Zou Y, Mao G, Park K, Herbert BS, Konieczny SF, Won YY. Influence of nano-carrier architecture on in vitro siRNA delivery performance and in vivo biodistribution: polyplexes vs micelleplexes. Acs Nano. 2011;5(5):3493–505. doi: 10.1021/nn102540y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qi R, Liu S, Chen J, Xiao H, Yan L, Huang Y, Jing X. Biodegradable copolymers with identical cationic segments and their performance in siRNA delivery. J Control Release. 2012;159(2):251–60. doi: 10.1016/j.jconrel.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 83.Kano A, Moriyama K, Yamano T, Nakamura I, Shimada N, Maruyama A. Grafting of poly(ethylene glycol) to poly-lysine augments its lifetime in blood circulation and accumulation in tumors without loss of the ability to associate with siRNA. J Control Release. 2011;149(1):2–7. doi: 10.1016/j.jconrel.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 84.Maji S, Zheng M, Agarwal S. Functional Degradable Polymers via Radical Ring-Opening Polymerization and Click Chemistry. Macromol Chem Physic. 2011;212(23):2573–2582. [Google Scholar]
- 85.Fujigaya T, Yamamoto Y, Kano A, Maruyama A, Nakashima N. Enhanced cell uptake via non-covalent decollation of a single-walled carbon nanotube-DNA hybrid with polyethylene glycol-grafted poly(l-lysine) labeled with an Alexa-dye and its efficient uptake in a cancer cell. Nanoscale. 2011;3(10):4352–8. doi: 10.1039/c1nr10635j. [DOI] [PubMed] [Google Scholar]
- 86.Miyata K, Nishiyama N, Kataoka K. Rational design of smart supramolecular assemblies for gene delivery: chemical challenges in the creation of artificial viruses. Chem Soc Rev. 2012;41(7):2562–74. doi: 10.1039/c1cs15258k. [DOI] [PubMed] [Google Scholar]
- 87.Yin L, Song Z, Kim KH, Zheng N, Tang H, Lu H, Gabrielson N, Cheng J. Reconfiguring the architectures of cationic helical polypeptides to control non-viral gene delivery. Biomaterials. 2013;34(9):2340–2349. doi: 10.1016/j.biomaterials.2012.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Taori VP, Lu H, Reineke TM. Structure-activity examination of poly(glycoamidoguanidine)s: glycopolycations containing guanidine units for nucleic acid delivery. Biomacromolecules. 2011;12(6):2055–63. doi: 10.1021/bm101537f. [DOI] [PubMed] [Google Scholar]
- 89.Ladd J, Zhang Z, Chen S, Hower JC, Jiang S. Zwitterionic polymers exhibiting high resistance to nonspecific protein adsorption from human serum and plasma. Biomacromolecules. 2008;9(5):1357–61. doi: 10.1021/bm701301s. [DOI] [PubMed] [Google Scholar]
- 90.Kulkarni A, DeFrees K, Schuldt RA, Hyun SH, Wright KJ, Yerneni CK, VerHeul R, Thompson DH. Cationic alpha-cyclodextrin:poly(ethylene glycol) polyrotaxanes for siRNA delivery. Mol Pharm. 2013;10(4):1299–305. doi: 10.1021/mp300449t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang J, Mao HQ, Leong KW. A novel biodegradable gene carrier based on polyphosphoester. J Am Chem Soc. 2001;123(38):9480–1. doi: 10.1021/ja016062m. [DOI] [PubMed] [Google Scholar]
- 92.Williams CK. Synthesis of functionalized biodegradable polyesters. Chem Soc Rev. 2007;36(10):1573–80. doi: 10.1039/b614342n. [DOI] [PubMed] [Google Scholar]
- 93.Son S, Namgung R, Kim J, Singha K, Kim WJ. Bioreducible polymers for gene silencing and delivery. Acc Chem Res. 2012;45(7):1100–12. doi: 10.1021/ar200248u. [DOI] [PubMed] [Google Scholar]
- 94.Shi J, Johnson RN, Schellinger JG, Carlson PM, Pun SH. Reducible HPMA-co-oligolysine copolymers for nucleic acid delivery. Int J Pharm. 2012;427(1):113–22. doi: 10.1016/j.ijpharm.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DW, Stebbing D, Crosley EJ, Yaworski E, Hafez IM, Dorkin JR, Qin J, Lam K, Rajeev KG, Wong KF, Jeffs LB, Nechev L, Eisenhardt ML, Jayaraman M, Kazem M, Maier MA, Srinivasulu M, Weinstein MJ, Chen Q, Alvarez R, Barros SA, De S, Klimuk SK, Borland T, Kosovrasti V, Cantley WL, Tam YK, Manoharan M, Ciufolini MA, Tracy MA, de Fougerolles A, MacLachlan I, Cullis PR, Madden TD, Hope MJ. Rational design of cationic lipids for siRNA delivery. Nature biotechnology. 2010;28(2):172–6. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 96.Rehman Z, Zuhorn IS, Hoekstra D. How cationic lipids transfer nucleic acids into cells and across cellular membranes: recent advances. J Control Release. 2013;166(1):46–56. doi: 10.1016/j.jconrel.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 97.Chen Y, Wu JJ, Huang L. Nanoparticles targeted with NGR motif deliver c-myc siRNA and doxorubicin for anticancer therapy. Mol Ther. 2010;18(4):828–34. doi: 10.1038/mt.2009.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Manjappa AS, Chaudhari KR, Venkataraju MP, Dantuluri P, Nanda B, Sidda C, Sawant KK, Murthy RS. Antibody derivatization and conjugation strategies: application in preparation of stealth immunoliposome to target chemotherapeutics to tumor. J Control Release. 2011;150(1):2–22. doi: 10.1016/j.jconrel.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 99.Sawant R, Torchilin V. Intracellular transduction using cell-penetrating peptides. Molecular bioSystems. 2010;6(4):628–40. doi: 10.1039/b916297f. [DOI] [PubMed] [Google Scholar]
- 100.Yu SS, Lau CM, Barham WJ, Onishko HM, Nelson CE, Li H, Smith CA, Yull FE, Duvall CL, Giorgio TD. Macrophage-specific RNA interference targeting via “click”, mannosylated polymeric micelles. Mol Pharm. 2013;10(3):975–87. doi: 10.1021/mp300434e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen Y, Bathula SR, Yang Q, Huang L. Targeted nanoparticles deliver siRNA to melanoma. The Journal of investigative dermatology. 2010;130(12):2790–8. doi: 10.1038/jid.2010.222. [DOI] [PubMed] [Google Scholar]
- 102.Tseng YC, Mozumdar S, Huang L. Lipid-based systemic delivery of siRNA. Adv Drug Deliv Rev. 2009;61(9):721–31. doi: 10.1016/j.addr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wolff JA, Rozema DB. Breaking the bonds: non-viral vectors become chemically dynamic. Mol Ther. 2008;16(1):8–15. doi: 10.1038/sj.mt.6300326. [DOI] [PubMed] [Google Scholar]
- 104.Zhou HS, Liu DP, Liang CC. Challenges and strategies: the immune responses in gene therapy. Medicinal research reviews. 2004;24(6):748–61. doi: 10.1002/med.20009. [DOI] [PubMed] [Google Scholar]
- 105.El-Sayed A, Harashima H. Endocytosis of Gene Delivery Vectors: From Clathrin-dependent to Lipid Raft-mediated Endocytosis. Mol Ther. 2013;21(6):1118–30. doi: 10.1038/mt.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stoter M, Epstein-Barash H, Zhang L, Koteliansky V, Fitzgerald K, Fava E, Bickle M, Kalaidzidis Y, Akinc A, Maier M, Zerial M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nature biotechnology. 2013 doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 107.Sahay G, Querbes W, Alabi C, Eltoukhy A, Sarkar S, Zurenko C, Karagiannis E, Love K, Chen D, Zoncu R, Buganim Y, Schroeder A, Langer R, Anderson DG. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nature biotechnology. 2013 doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Benjaminsen RV, Mattebjerg MA, Henriksen JR, Moghimi SM, Andresen TL. The Possible “Proton Sponge” Effect of Polyethylenimine (PEI) Does Not Include Change in Lysosomal pH. Mol Ther. 2013;21(1):149–57. doi: 10.1038/mt.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Funhoff AM, van Nostrum CF, Koning GA, Schuurmans-Nieuwenbroek NM, Crommelin DJ, Hennink WE. Endosomal escape of polymeric gene delivery complexes is not always enhanced by polymers buffering at low pH. Biomacromolecules. 2004;5(1):32–9. doi: 10.1021/bm034041+. [DOI] [PubMed] [Google Scholar]
- 110.Rehman ZU, Hoekstra D, Zuhorn IS. Mechanism of Polyplex- and Lipoplex-Mediated Delivery of Nucleic Acids: Real-Time Visualization of Transient Membrane Destabilization without Endosomal Lysis. Acs Nano. 2013 doi: 10.1021/nn3049494. [DOI] [PubMed] [Google Scholar]
- 111.Parsa S, Wang Y, Rines K, Pfeifer BA. A high-throughput comparison of recombinant gene expression parameters for E. coli-mediated gene transfer to P388D1 macrophage cells. J Biotechnol. 2008;137(1–4):59–64. doi: 10.1016/j.jbiotec.2008.07.1815. [DOI] [PubMed] [Google Scholar]
- 112.Parsa S, Wang Y, Fuller J, Langer R, Pfeifer BA. A Comparison Between Polymeric Microsphere and Bacterial Vectors for Macrophage P388D1 Gene Delivery. Pharm Res. 2008;25:1202–1208. doi: 10.1007/s11095-008-9563-x. [DOI] [PubMed] [Google Scholar]
- 113.Li J, Chen YC, Tseng YC, Mozumdar S, Huang L. Biodegradable calcium phosphate nanoparticle with lipid coating for systemic siRNA delivery. J Control Release. 2010;142(3):416–21. doi: 10.1016/j.jconrel.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lukacs GL, Haggie P, Seksek O, Lechardeur D, Freedman N, Verkman AS. Size-dependent DNA mobility in cytoplasm and nucleus. J Biol Chem. 2000;275(3):1625–9. doi: 10.1074/jbc.275.3.1625. [DOI] [PubMed] [Google Scholar]
- 115.Lechardeur D, Sohn KJ, Haardt M, Joshi PB, Monck M, Graham RW, Beatty B, Squire J, O’Brodovich H, Lukacs GL. Metabolic instability of plasmid DNA in the cytosol: a potential barrier to gene transfer. Gene Ther. 1999;6(4):482–97. doi: 10.1038/sj.gt.3300867. [DOI] [PubMed] [Google Scholar]
- 116.Ward CM, Read ML, Seymour LW. Systemic circulation of poly(L-lysine)/DNA vectors is influenced by polycation molecular weight and type of DNA: differential circulation in mice and rats and the implications for human gene therapy. Blood. 2001;97(8):2221–9. doi: 10.1182/blood.v97.8.2221. [DOI] [PubMed] [Google Scholar]
- 117.van der Aa MA, Mastrobattista E, Oosting RS, Hennink WE, Koning GA, Crommelin DJ. The nuclear pore complex: the gateway to successful nonviral gene delivery. Pharm Res. 2006;23(3):447–59. doi: 10.1007/s11095-005-9445-4. [DOI] [PubMed] [Google Scholar]
- 118.Wente SR. Gatekeepers of the nucleus. Science. 2000;288(5470):1374–7. doi: 10.1126/science.288.5470.1374. [DOI] [PubMed] [Google Scholar]
- 119.Gorlich D, Mattaj IW. Nucleocytoplasmic transport. Science. 1996;271(5255):1513–8. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 120.Zanta MA, Belguise-Valladier P, Behr JP. Gene delivery: a single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleus. Proc Natl Acad Sci U S A. 1999;96(1):91–6. doi: 10.1073/pnas.96.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cartier R, Reszka R. Utilization of synthetic peptides containing nuclear localization signals for nonviral gene transfer systems. Gene Ther. 2002;9(3):157–67. doi: 10.1038/sj.gt.3301635. [DOI] [PubMed] [Google Scholar]
- 122.Branden LJ, Mohamed AJ, Smith CI. A peptide nucleic acid-nuclear localization signal fusion that mediates nuclear transport of DNA. Nature biotechnology. 1999;17(8):784–7. doi: 10.1038/11726. [DOI] [PubMed] [Google Scholar]
- 123.Baker A, Cotten M. Delivery of bacterial artificial chromosomes into mammalian cells with psoralen-inactivated adenovirus carrier. Nucleic acids research. 1997;25(10):1950–6. doi: 10.1093/nar/25.10.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kazuki Y, Oshimura M. Human artificial chromosomes for gene delivery and the development of animal models. Mol Ther. 2011;19(9):1591–601. doi: 10.1038/mt.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dias N, Stein CA. Antisense oligonucleotides: basic concepts and mechanisms. Molecular cancer therapeutics. 2002;1(5):347–55. [PubMed] [Google Scholar]
- 126.Asoh S, Lee-Kwon W, Mouradian MM, Nirenberg M. Selection of DNA clones with enhancer sequences. Proc Natl Acad Sci U S A. 1994;91(15):6982–6. doi: 10.1073/pnas.91.15.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Noss KR, Wolfe SA, Grimes SR. Upregulation of prostate specific membrane antigen/folate hydrolase transcription by an enhancer. Gene. 2002;285(1–2):247–56. doi: 10.1016/s0378-1119(02)00397-9. [DOI] [PubMed] [Google Scholar]
- 128.Lu J, Zhang F, Kay MA. A Mini-intronic Plasmid (MIP): A Novel Robust Transgene Expression Vector In Vivo and In Vitro. Mol Ther. 2013;21(5):954–63. doi: 10.1038/mt.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lu J, Zhang F, Xu S, Fire AZ, Kay MA. The extragenic spacer length between the 5′ and 3′ ends of the transgene expression cassette affects transgene silencing from plasmid-based vectors. Mol Ther. 2012;20(11):2111–9. doi: 10.1038/mt.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen ZY, He CY, Ehrhardt A, Kay MA. Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol Ther. 2003;8(3):495–500. doi: 10.1016/s1525-0016(03)00168-0. [DOI] [PubMed] [Google Scholar]
- 131.Liu X, Yang Z, Feng T. A minicircle DNA vector-mediated siRNA to stably suppress hepatitis B virus replication and expression. Wei sheng wu xue bao = Acta microbiologica Sinica. 2012;52(2):191–7. [PubMed] [Google Scholar]
- 132.Kay MA, He CY, Chen ZY. A robust system for production of minicircle DNA vectors. Nature biotechnology. 2010;28(12):1287–9. doi: 10.1038/nbt.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Morcos PA. Achieving targeted and quantifiable alteration of mRNA splicing with Morpholino oligos. Biochem Biophys Res Commun. 2007;358(2):521–7. doi: 10.1016/j.bbrc.2007.04.172. [DOI] [PubMed] [Google Scholar]
- 134.Huang L, Jin J, Deighan P, Kiner E, McReynolds L, Lieberman J. Efficient and specific gene knockdown by small interfering RNAs produced in bacteria. Nature biotechnology. 2013;31(4):350–6. doi: 10.1038/nbt.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123(4):607–20. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 136.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–40. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 137.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nature structural & molecular biology. 2005;12(4):340–9. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 138.Xiang S, Fruehauf J, Li CJ. Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nature biotechnology. 2006;24(6):697–702. doi: 10.1038/nbt1211. [DOI] [PubMed] [Google Scholar]