Abstract
A subpopulation of hepatitis C virus (HCV) core protein in cells harboring full-length HCV replicons is biochemically associated with detergent-resistant membranes (DRMs) in a manner similar to that of markers of classical lipid rafts. Core protein does not, however, colocalize in immunofluorescence studies with classical plasma membrane raft markers, such as caveolin-1 and the B subunit of cholera toxin, suggesting that core protein is bound to cytoplasmic raft microdomains distinct from caveolin-based rafts. Furthermore, while both the structural core protein and the nonstructural protein NS5A associate with membranes, they do not colocalize in the DRMs. Finally, the ability of core protein to localize to the DRMs did not require other elements of the HCV polyprotein. These results may have broad implications for the HCV life cycle and suggest that the HCV core may be a valuable probe for host cell biology.
Hepatitis C virus (HCV) is a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. HCV has a positive-sense single-stranded RNA genome that encodes a polyprotein of ∼3,000 amino acids. The polyprotein can be separated into two functional regions: the structural components of the virion (which include core protein and two envelope proteins, E1 and E2) and the nonstructural proteins (NS2 to NS5B), which participate in viral replication but are not believed to be contained in virus particles (4).
HCV core protein is synthesized as a 191-amino-acid precursor (p23). Subsequent proteolytic processing by signal peptidase and signal peptide peptidase generates a truncated mature form of core protein (p21) consisting of 179 amino acids (13, 20). This maturation process is important to release core protein from anchorage to endoplasmic reticulum (ER) membranes and for trafficking to lipid droplets (20). The mature protein predominates both in transfected tissue culture cells and in virus particles isolated from infected sera (40).
In addition to its presumed role in virus particle assembly and budding, HCV core protein interacts with a variety of host cell signaling pathways (14, 19, 26, 30, 41). Most of the core protein expressed in transfected cells is localized in the cytoplasm, associated either with what appears to be intracellular membrane organelles or with the surfaces of lipid bodies (11, 20).
Detergent-resistant membranes (DRMs), or rafts, are specialized and heterogeneous cellular membrane subdomains defined by their resistance to solubilization with cold nonionic detergents, e.g., Triton X-100 (2, 25, 32, 39). Classical lipid rafts are located predominantly on the plasma membrane, and the proteins associated with these rafts are key mediators of many biological events, such as trafficking (37) and signal transduction pathways (36). DRMs also play important roles in the replication cycles of several viruses. Previous reports have shown that viruses like simian virus 40, human immunodeficiency virus, influenza virus, rotavirus, and Ebola virus use lipid rafts as a portal for viral entry, as a platform for the assembly of viral components, or for the budding of viruses from their host cells (8-10, 17, 34). Although recent data suggest that NS5A interacts with DRMs (33), the interactions of other HCV components (e.g., the structural proteins) with DRMs have not yet been investigated. Based on core protein's ability to participate in host cell signaling pathways and the fact that other viruses exploit DRMs for critical aspects of their propagation, we hypothesized that the HCV core protein might also associate with lipid rafts. Here we report for the first time the appearance of a significant proportion of core protein in DRMs. Interestingly, core protein DRMs have properties that distinguish them from classical plasmalemal lipid rafts. These results have important implications with respect to the function of core protein in the HCV life cycle.
HCV replicon-expressed core protein associates with detergent-resistant membranes.
To test the hypothesis that HCV core protein can associate with DRMs, FLRP1 cells (a Huh7 clonal cell line harboring a full-length genotype 1b replicon [5]) were washed in cold TNE buffer (25 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA) and lysed with a ball-bearing homogenizer, and aliquots were incubated for 30 min on ice with or without 1% Triton X-100. The lysates were overlaid with a 5 to 40% OptiPrep (Sigma) gradient and centrifuged at 40,000 rpm for 4 h at 4°C in a SW60 ultracentrifuge rotor, as described previously (12). Fractions were collected from the top of the tube, and proteins were precipitated with chloroform-methanol. Precipitated proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted for the presence of HCV core protein (using monoclonal antibodies [MAbs] to HCV core protein kindly provided by Harry Greenberg). Without detergent treatment, all core protein is found essentially in the membrane-containing fraction (fraction 2 from the top) (Fig. 1A, upper panel). When treated with Triton X-100, a considerable amount of core protein remained associated with DRMs and the rest was found in the bottom fractions of the gradient, which contain detergent-soluble proteins (Fig. 1A, bottom panel). To further characterize the detergent-resistant core protein fraction, we studied its distribution relative to that of raft and nonraft markers on Nycodenz (Sigma) gradients, which have a wider range of steps and better separation of fractions containing detergent-resistant membranes from the detergent-soluble fractions than those of the previous format. For this, FLRP1 and Huh7 cells were lysed in TNE buffer containing 1% Triton X-100. Following incubation for 30 min on ice, cell lysates were overlaid with a 8 to 35% Nycodenz gradient and subjected to ultracentrifugation at 40,000 rpm for 14 h at 4°C with a SW60 rotor (for a detailed description of the protocol, see reference 21). Eleven equal fractions were collected from the top of the centrifuge tube, and proteins were precipitated and analyzed by Western blotting as described above. In addition to anti-core protein, the blots were probed with antibodies to the caveolar raft marker caveolin-1 (1) (pAb; Transduction Laboratories) and the ER membrane-associated protein calnexin (pAb; StressGen Biotechnologies Corporation). To detect glycosylphosphatidylinositol (GPI)-linked proteins, which are also markers for classical lipid rafts (7), membranes were incubated for 1 h with a 1-μg/ml concentration of proaerolysin—a toxin displaying high affinity for GPI-linked proteins (38), and subsequently incubated with antiaerolysin MAbs coupled to horseradish peroxidase according to the manufacturer's instructions (Protox Biotech).
FIG. 1.
HCV core protein associates with a detergent-resistant membrane biochemically similar to classical lipid rafts. (A) FLRP1 cells (Huh7 cells harboring HCV replicons) were lysed, and aliquots were incubated at 4°C in the absence or presence of 1% Triton X-100 (Tx-100). The lysates were overlaid with a 5 to 40% OptiPrep gradient. Following ultracentrifugation, fractions were collected from the top of the tube and the proteins were precipitated and analyzed by Western blotting using an antibody against HCV core protein. The locations of fractions containing membranes, DRMs, and soluble proteins are indicated. (B) FLRP1 and Huh7 cells were lysed in cold 1% Triton X-100 and loaded in an 8 to 35% Nycodenz gradient. Following ultracentrifugation, fractions were collected from the top of the tube and the proteins were precipitated and analyzed by Western blotting using antibodies against core protein, caveolin-1, aerolysin (to detect GPI-linked proteins), and calnexin. (C) FLRP1 cells were treated with 10 mM mβCD or αCD for 30 min prior to lysis and analyzed as described above for HCV core protein. Memb., membrane.
As shown in Fig. 1B, a significant fraction (about 20%) of core protein floated to the top of the gradient, concentrating predominantly in fraction 2, where it cofractionated along with the raft markers, caveolin-1 and GPI-linked proteins. The remainder of the core protein was found in the bottom fractions, which contain detergent-soluble proteins. Calnexin, an ER-associated protein that is not resistant to Triton X-100, distributed to the bottom of the gradient and was not detected in the DRM fractions. From this data we conclude that a subpopulation of HCV core protein associates with DRMs.
mβCD disrupts the association of HCV core protein with DRMs.
Lipid rafts are enriched in cholesterol, and the biophysical properties of cholesterol help to determine a key structural feature of rafts (6). Cholesterol, along with other lipids and proteins that preferentially partition into liquid-ordered phases in the exoplasmic and cytoplasmic leaflets of the membrane, generates constrained raft microdomains with decreased lateral mobility (23, 35). Indeed, sensitivity of the raft domains to cholesterol depletion by agents such as methyl-β-cyclodextrin (mβCD) has been widely used as a functional biochemical criterion of classical lipid rafts. We therefore next sought to test the hypothesis that the core protein DRMs were sensitive to cholesterol chelation by mβCD. FLRP1 cells were incubated in the presence of mβCD or control α-cyclodextrin (an inactive analogue of mβCD) for 30 min at 37°C before Triton X-100 solubilization and gradient formation. As shown in Fig. 1C, the subpopulation of core protein that was associated with the DRM fraction in the gradient disappeared upon mβCD, but not αCD, treatment. Thus, in this respect, the core protein-containing DRM behaves in a manner biochemically similar to that of classical lipid rafts.
It is interesting that in spite being associated with ER-like intracellular organelles, the core protein-bearing raft domains are sensitive to cholesterol depletion by mβCD from the plasma membrane. This finding suggests the existence of a possible cross talk between elements derived from the plasma membrane and core protein in specialized rafts. Another scenario is that cholesterol depletion by mβCD has more-global effects on the cell as was recently suggested by Kwik et al. (15).
HCV core protein has two subcellular localization patterns: rings and patches.
HCV core protein has been shown to interact with lipid droplets (3). The above data also suggest that there is a subpopulation of core protein in HCV replicon cells that biochemically associates with DRMs. We next asked whether there were two morphologically distinct subpopulations of core protein in replicon cells. We also wished to determine the respective sensitivities of these subpopulations to treatment with Triton X-100 and mβCD. FLRP1 cells, which harbor full-length replicons, were cultured on coverslips and fixed with 4% paraformaldehyde. Following permeabilization with saponin, HCV core protein was detected with a primary anti-core protein MAb and secondary goat anti-mouse antibodies conjugated to either Alexa 488 or Alexa 594. Lipid droplets were stained with Oil Red O. Fluorescence microscopy was performed on a Nikon Eclipse E600 microscope supplemented with a SPOT digital camera (Diagnostic Instruments) and analyzed with the OpenLab (Improvision) software package. We also performed experiments on cells treated with either 10 mM mβCD for 30 min at 37°C or dipped in 1% Triton X-100 (four times for 5 s each time) prior to fixation as described previously (31).
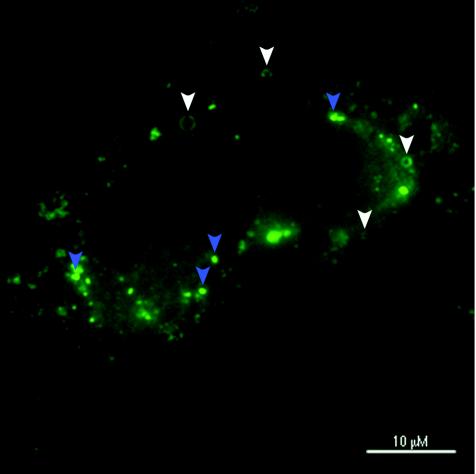
We found two types of core protein staining patterns in Huh7 cells harboring the full-length HCV replicons: dots or patches and ring-like structures (Fig. 2). When the replicon cells were costained for core protein and lipid droplets, the ring-like structures completely colocalized with the surfaces of lipid bodies, whereas the dot-like signals did not (Fig. 3A to C). When treated with 1% Triton X-100 before fixation, the ring-like structures disappeared while the dots or patches remained (Fig. 3D to F). In contrast, when treated with 10 mM mβCD to deplete cholesterol, the rings remained but the patches disappeared (Fig. 3G to I). Taken together, these results suggest that there are indeed two morphologically distinct populations of core protein in replicon cells and that the dot-like core protein signal, but not the lipid droplets, represents the subpopulation that is associated with the DRMs.
FIG. 2.
There are two morphologically distinct populations of core protein in replicon cells. FLRP1 cells stained for HCV core protein. Note the two patterns: rings (white arrowheads) and dots or patches (blue arrowheads).
FIG. 3.
The dots but not the rings are resistant to Triton X-100 extraction. (A-C) FLRP1 cells stained for core protein (green) and for lipid droplets with Oil Red O (red). The rings (white arrowheads) colocalize with lipid droplets, and the dots or patches (blue arrowheads) do not. (D-F) FLRP1 cells were treated with 1% Triton X-100 (Tx-100) before immunofluorescence (31) and stained for core protein (green) and lipid bodies (red). (G-I) FLRP1 cells treated with 10 mM mβCD for 30 min prior to fixation and stained for core protein (green) and lipid bodies (red). Note that the dot pattern of core protein is resistant to Triton X-100 and sensitive to mβCD.
The intracellular localization of the HCV core protein is different from markers of classical rafts.
We next wished to determine whether a subpopulation of core protein colocalizes with classical markers of lipid rafts in intact cells. For this we performed immunofluorescence and confocal analyses of FLRP1 cells stained for core protein and two markers for rafts, the B subunit of cholera toxin and caveolin-1. The B subunit of cholera toxin, which binds as a pentamer to glycosphingolipid GM1 in the plasma membrane, is one of the most widely used markers for a subpopulation of lipid rafts that reside primarily in the exoplasmic leaflet of biological membranes (22).
FLRP1 cells were fixed with 4% formaldehyde-5% sucrose in phosphate-buffered saline (PBS) for 20 min at room temperature and permeabilized with 0.2% Triton X-100 for 10 min at room temperature. Cells were blocked for 1 h at 37°C with PBS-2% bovine serum albumin, and then stained with anti-core protein MAb and a 25-μg/ml concentration of either the biotin-labeled B subunit of cholera toxin (Sigma) or anticaveolin-1 pAb. Cells were then incubated with 1 μg of streptavidin-fluorescein isothiocyanate (Pharmingen) per ml and goat anti-mouse or goat anti-rabbit antibody coupled to either Alexa-488 or Alexa 594 for 30 min at 37°C in PBS-0.2% bovine serum albumin. Cells were visualized with Nikon upright and Bio-Rad confocal microscopes.
HCV core protein did not colocalize with either of these raft markers (Fig. 4). These analyses suggest that the core protein associates with a subtype of raft domains that is morphologically distinct from caveolin-1- and sphingolipid-containing lipid rafts. This conclusion is consistent with the facts that both caveolin-1 and GM1 are typically concentrated at steady state in the plasma membrane and that core protein is likely to be linked to ER or ER-derived endo-membranes.
FIG. 4.
Core protein does not colocalize with markers of classical lipid rafts. (A-C) FLRP1 cells stained for HCV core protein (red) and with the B subunit of cholera toxin (CTX) (green); (D-F) FLRP1 cells stained for HCV core protein (green) and caveolin-1 (Cav-1) (red).
The association of HCV core protein with the DRMs is independent of other members of the HCV polyprotein.
To test the hypothesis that the association of HCV core protein with detergent-resistant membranes does not require the aid of other viral proteins, we inserted the HCV core protein coding sequence into a mammalian expression vector, pEF6, and exogenously expressed core protein in Huh7 cells. The cells were then subjected to membrane flotation analysis as described for Fig. 1. As shown in Fig. 5, core protein expressed by itself in Huh7 cells behaved very similarly to core protein expressed in the context of the full-length replicon, with a significant fraction being found in the detergent-resistant low-density membrane fractions. HCV core protein is thus capable of associating with DRMs in the absence of the other members of the HCV polyprotein. To date, we have not detected any differences (as assessed by size) between the DRM-associated and Triton X-100-soluble forms of core protein.
FIG. 5.
HCV core protein association with the DRMs does not require other viral proteins. The HCV core protein sequence was inserted into the pEF6 mammalian expression vector and transfected into Huh7 cells. Twenty-four hours posttransfection, cells were lysed, in parallel with replicon-harboring FLRP1 cells, in 1% Triton X-100 and subjected to flotation on an 8 to 35% Nycodenz gradient, followed by analysis of the fractions for HCV core protein, as described for Fig. 1.
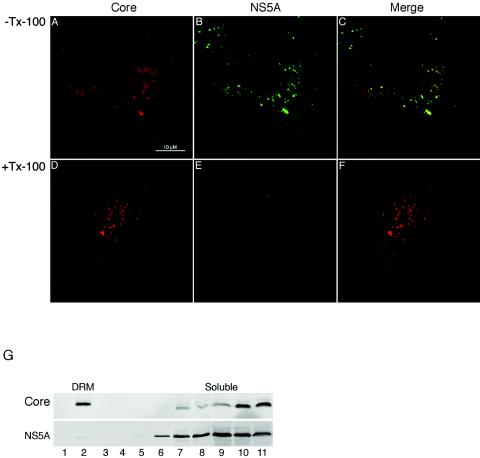
The ability of core protein to target to the DRMs in the absence of the nonstructural (NS) proteins is consistent with the presumed different functions and ultimate fate associated with the latter. In particular, while the NS proteins are believed to play a critical role in the replication of the viral genome, they are not thought to be part of the secreted virus particle. We therefore asked whether these differences might also be reflected at the level of the DRM. For this purpose, we performed immunofluorescence colocalization studies on FLRP1 cells using a polyclonal antibody against HCV core protein (kindly provided by Amy Weiner and Michael Houghton) and a MAb against NS5A (Virostat, Portland, Maine). For the Triton X-100 extraction treatment, the coverslips were treated as detailed for Fig. 3 and visualized by confocal microscopy. We also performed biochemical assays by subjecting FLRP1 cells to DRM gradient flotation and Western blot analyses as described for Fig. 1. As shown in Fig. 6, while core protein and NS5A can be seen to colocalize in the absence of detergent (Fig. 6C), they behave quite differently upon Triton X-100 treatment, with little of NS5A remaining associated with the core protein DRMs (Fig. 6E and bottom panel of G). Thus, while core protein and NS5A are in close proximity in replicon-harboring cells, they appear to indeed be associated with functionally different membrane domains.
FIG. 6.
HCV NS5A colocalization with core protein is lost in the presence of Triton X-100. (A-F) FLRP1 cells stained for core protein (red) and NS5A (green). In panels D-F, FLRP1 cells were treated with 1% Triton X-100 (Tx-100) before immunofluorescence (31). (G) FLRP1 cells were lysed in 1% Triton X-100 and subjected to flotation on a 8 to 35% Nycodenz gradient, followed by analysis of the fractions for HCV core protein (upper panel) and NS5A (bottom panel) as described for Fig. 1B.
Finally, because of the large body of data linking lipid rafts to intracellular signaling events and virus morphogenesis, the association of a subpopulation of HCV core protein with DRMs provides an attractive mechanism of how core protein may mediate its reported signaling activities (16, 18, 24, 27-29) and presumed role in HCV assembly. In the absence of an efficient cell culture system for HCV particle production, it may be difficult to directly test the latter hypothesis. It will be interesting, however, to determine whether the core protein-containing DRMs are induced by HCV or preexist in cells and, in the latter case, what normal and/or pathological host functions may be mediated by these DRMs. HCV core protein may thus prove to be a particularly valuable probe of host cell biology.
Acknowledgments
We thank Allen Cooper and Harry B. Greenberg for helpful discussions and critical reading of the manuscript.
J.S.G. is supported by a Burroughs Welcome Fund Career Award. This work was also supported by PHS grants CA57973 and 1RO1-DK064223-01 and a Veterans Administration Merit Review Award.
REFERENCES
- 1.Anderson, R. G. 1998. The caveolae membrane system. Annu. Rev. Biochem. 67:199-225. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, R. G., and K. Jacobson. 2002. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science 296:1821-1825. [DOI] [PubMed] [Google Scholar]
- 3.Barba, G., F. Harper, T. Harada, M. Kohara, S. Goulinet, Y. Matsuura, G. Eder, Z. Schaff, M. J. Chapman, T. Miyamura, and C. Brechot. 1997. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc. Natl. Acad. Sci. USA 94:1200-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., and V. Lohmann. 2000. Replication of the hepatitis C virus. Bailliere's Best Pract. Res. Clin. Gastroenterol. 14:241-254. [DOI] [PubMed] [Google Scholar]
- 5.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 6.Brown, D. A., and E. London. 2000. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275:17221-17224. [DOI] [PubMed] [Google Scholar]
- 7.Brown, D. A., and J. K. Rose. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68:533-544. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, S. M., S. M. Crowe, and J. Mak. 2001. Lipid rafts and HIV-1: from viral entry to assembly of progeny virions. J. Clin. Virol. 22:217-227. [DOI] [PubMed] [Google Scholar]
- 9.Chazal, N., and D. Gerlier. 2003. Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 67:226-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuadras, M. A., and H. B. Greenberg. 2003. Rotavirus infectious particles use lipid rafts during replication for transport to the cell surface in vitro and in vivo. Virology 313:308-321. [DOI] [PubMed] [Google Scholar]
- 11.Dubuisson, J., F. Penin, and D. Moradpour. 2002. Interaction of hepatitis C virus proteins with host cell membranes and lipids. Trends Cell Biol. 12:517-523. [DOI] [PubMed] [Google Scholar]
- 12.Elazar, M., K. H. Cheong, P. Liu, H. B. Greenberg, C. M. Rice, and J. S. Glenn. 2003. Amphipathic helix-dependent localization of NS5A mediates hepatitis C virus RNA replication. J. Virol. 77:6055-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hope, R. G., and J. McLauchlan. 2000. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J. Gen. Virol. 81:1913-1925. [DOI] [PubMed] [Google Scholar]
- 14.Hosui, A., K. Ohkawa, H. Ishida, A. Sato, F. Nakanishi, K. Ueda, T. Takehara, A. Kasahara, Y. Sasaki, M. Hori, and N. Hayashi. 2003. Hepatitis C virus core protein differently regulates the JAK-STAT signaling pathway under interleukin-6 and interferon-gamma stimuli. J. Biol. Chem. 278:28562-28571. [DOI] [PubMed] [Google Scholar]
- 15.Kwik, J., S. Boyle, D. Fooksman, L. Margolis, M. P. Sheetz, and M. Edidin. 2003. Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin. Proc. Natl. Acad. Sci. USA 100:13964-13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu, W., S. Y. Lo, M. Chen, K. Wu, Y. K. Fung, and J. H. Ou. 1999. Activation of p53 tumor suppressor by hepatitis C virus core protein. Virology 264:134-141. [DOI] [PubMed] [Google Scholar]
- 17.Manes, S., G. del Real, and A. C. Martinez. 2003. Pathogens: raft hijackers. Nat. Rev. Immunol. 3:557-568. [DOI] [PubMed] [Google Scholar]
- 18.Marusawa, H., M. Hijikata, T. Chiba, and K. Shimotohno. 1999. Hepatitis C virus core protein inhibits Fas- and tumor necrosis factor alpha-mediated apoptosis via NF-κB activation. J. Virol. 73:4713-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLauchlan, J. 2000. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. J. Viral Hepat. 7:2-14. [DOI] [PubMed] [Google Scholar]
- 20.McLauchlan, J., M. K. Lemberg, G. Hope, and B. Martoglio. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21:3980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naslavsky, N., R. Stein, A. Yanai, G. Friedlander, and A. Taraboulos. 1997. Characterization of detergent-insoluble complexes containing the cellular prion protein and its scrapie isoform. J. Biol. Chem. 272:6324-6331. [DOI] [PubMed] [Google Scholar]
- 22.Nichols, B. J. 2003. GM1-containing lipid rafts are depleted within clathrin-coated pits. Curr. Biol. 13:686-690. [DOI] [PubMed] [Google Scholar]
- 23.Niv, H., O. Gutman, Y. Kloog, and Y. I. Henis. 2002. Activated K-Ras and H-Ras display different interactions with saturable nonraft sites at the surface of live cells. J. Cell Biol. 157:865-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohkawa, K., H. Ishida, F. Nakanishi, A. Hosui, K. Ueda, T. Takehara, M. Hori, and N. Hayashi. 2004. Hepatitis C virus core functions as a suppressor of cyclin-dependent kinase-activating kinase and impairs cell cycle progression. J. Biol. Chem. 279:11719-11726. [DOI] [PubMed] [Google Scholar]
- 25.Pike, L. J. 2004. Lipid rafts: heterogeneity on the high seas. Biochem J. 378:281-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ray, R. B., K. Meyer, and R. Ray. 1996. Suppression of apoptotic cell death by hepatitis C virus core protein. Virology 226:176-182. [DOI] [PubMed] [Google Scholar]
- 27.Ray, R. B., K. Meyer, R. Steele, A. Shrivastava, B. B. Aggarwal, and R. Ray. 1998. Inhibition of tumor necrosis factor (TNF-alpha)-mediated apoptosis by hepatitis C virus core protein. J. Biol. Chem. 273:2256-2259. [DOI] [PubMed] [Google Scholar]
- 28.Ray, R. B., R. Steele, A. Basu, K. Meyer, M. Majumder, A. K. Ghosh, and R. Ray. 2002. Distinct functional role of hepatitis C virus core protein on NF-kappaB regulation is linked to genomic variation. Virus Res. 87:21-29. [DOI] [PubMed] [Google Scholar]
- 29.Ray, R. B., R. Steele, K. Meyer, and R. Ray. 1998. Hepatitis C virus core protein represses p21WAF1/Cip1/Sid1 promoter activity. Gene 208:331-336. [DOI] [PubMed] [Google Scholar]
- 30.Ray, R. B., R. Steele, K. Meyer, and R. Ray. 1997. Transcriptional repression of p53 promoter by hepatitis C virus core protein. J. Biol. Chem. 272:10983-10986. [DOI] [PubMed] [Google Scholar]
- 31.Roper, K., D. Corbeil, and W. B. Huttner. 2000. Retention of prominin in microvilli reveals distinct cholesterol-based lipid micro-domains in the apical plasma membrane. Nat. Cell Biol. 2:582-592. [DOI] [PubMed] [Google Scholar]
- 32.Schuck, S., M. Honsho, K. Ekroos, A. Shevchenko, and K. Simons. 2003. Resistance of cell membranes to different detergents. Proc. Natl. Acad. Sci. USA 100:5795-5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi, S. T., K. J. Lee, H. Aizaki, S. B. Hwang, and M. M. Lai. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 77:4160-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simons, K., and R. Ehehalt. 2002. Cholesterol, lipid rafts, and disease. J. Clin. Investig. 110:597-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simons, K., and E. Ikonen. 2000. How cells handle cholesterol. Science 290:1721-1726. [DOI] [PubMed] [Google Scholar]
- 36.Simons, K., and D. Toomre. 2000. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell. Biol. 1:31-39. [DOI] [PubMed] [Google Scholar]
- 37.Slimane, T. A., G. Trugnan, I. S. C. Van, and D. Hoekstra. 2003. Raft-mediated trafficking of apical resident proteins occurs in both direct and transcytotic pathways in polarized hepatic cells: role of distinct lipid microdomains. Mol. Biol. Cell 14:611-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sotgia, F., B. Razani, G. Bonuccelli, W. Schubert, M. Battista, H. Lee, F. Capozza, A. L. Schubert, C. Minetti, J. T. Buckley, and M. P. Lisanti. 2002. Intracellular retention of glycosylphosphatidyl inositol-linked proteins in caveolin-deficient cells. Mol. Cell. Biol. 22:3905-3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Goot, F. G., and T. Harder. 2001. Raft membrane domains: from a liquid-ordered membrane phase to a site of pathogen attack. Semin. Immunol. 13:89-97. [DOI] [PubMed] [Google Scholar]
- 40.Yasui, K., T. Wakita, K. Tsukiyama-Kohara, S. I. Funahashi, M. Ichikawa, T. Kajita, D. Moradpour, J. R. Wands, and M. Kohara. 1998. The native form and maturation process of hepatitis C virus core protein. J. Virol. 72:6048-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu, N., A. Khoshnan, R. Schneider, M. Matsumoto, G. Dennert, C. Ware, and M. M. Lai. 1998. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J. Virol. 72:3691-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]