Abstract
The pneumovirus respiratory syncytial virus (RSV) is a leading cause of epidemic respiratory tract infection. Upon entry, RSV replicates in the epithelial cytoplasm, initiating compensatory changes in cellular gene expression. In this study, we have investigated RSV-induced changes in the nuclear proteome of A549 alveolar type II-like epithelial cells by high-resolution two-dimensional gel electrophoresis (2DE). Replicate 2D gels from uninfected and RSV-infected nuclei were compared for changes in protein expression. We identified 24 different proteins by peptide mass fingerprinting after matrix-assisted laser desorption ionization-time of flight mass spectrometry (MS), whose average normalized spot intensity was statistically significant and differed by ±2-fold. Notable among the proteins identified were the cytoskeletal cytokeratins, RNA helicases, oxidant-antioxidant enzymes, the TAR DNA binding protein (a protein that associates with nuclear domain 10 [ND10] structures), and heat shock protein 70- and 60-kDa isoforms (Hsp70 and Hsp60, respectively). The identification of Hsp70 was also validated by liquid chromatography quadropole-TOF tandem MS (LC-MS/MS). Separate experiments using immunofluorescence microscopy revealed that RSV induced cytoplasmic Hsp70 aggregation and nuclear accumulation. Data mining of a genomic database showed that RSV replication induced coordinate changes in Hsp family proteins, including the 70, 70-2, 90, 40, and 40-3 isoforms. Because the TAR DNA binding protein associates with ND10s, we examined the effect of RSV infection on ND10 organization. RSV induced a striking dissolution of ND10 structures with redistribution of the component promyelocytic leukemia (PML) and speckled 100-kDa (Sp100) proteins into the cytoplasm, as well as inducing their synthesis. Our findings suggest that cytoplasmic RSV replication induces a nuclear heat shock response, causes ND10 disruption, and redistributes PML and Sp100 to the cytoplasm. Thus, a high-resolution proteomics approach, combined with immunofluorescence localization and coupled with genomic response data, yielded unexpected novel insights into compensatory nuclear responses to RSV infection.
The paramyxovirus respiratory syncytial virus (RSV) is the primary etiologic agent of epidemic lower respiratory tract infections in infants and young children (29). In the United States, almost all children by the age of 3 years will have been infected by RSV (25), manifesting a clinical spectrum ranging from otitis media (34) to upper respiratory tract infections and severe lower respiratory tract (LRT) infection, including pneumonitis and bronchiolitis (29, 30). Although numerically LRT infection is the least common clinical manifestation of RSV infection, RSV-induced LRT infections are an increasing cause of morbidity, as the proportion of hospitalizations associated with bronchiolitis in infants rose from 22 to 47% from 1980 to 1996 (65). Strikingly, >70% of children with RSV LRT infections have impaired pulmonary function for up to 10 years afterward (46). In view of its ability to produce LRT infections in otherwise-healthy infants, induce recurrent wheezing in children with established atopy, and produce mortality in children with underlying pulmonary or cardiac diseases, RSV remains a significant health problem worldwide (28, 48, 66).
The molecular biology of RSV replication has been intensively investigated (reviewed in reference 20). After its adsorption to the cell surface, 10 major viral proteins are transcribed in the cytoplasm by the viral RNA-dependent RNA polymerase, using the antisense genome as a template through a sequential stop-start mechanism guided by short template signals (17). Subsequently, the viral transcriptase switches to a replicative mode, replicating the full-length antigenome that serves as a template for producing progeny virions that subsequently bud from the apical surface (16, 72). In natural infections, RSV replication produces epithelial damage (2) and perivascular mononuclear infiltration (21). Because the epithelium is an important site for initiation and coordination of pulmonary inflammation (1), the ability of RSV to induce epithelial signaling has been intensively studied (6, 10, 11, 22, 23).
Although the RSV life cycle is entirely cytoplasmic, RSV replication induces profound nuclear responses in the host epithelial cell. For example, previous studies employing high-density oligonucleotide arrays have provided evidence that RSV replication alters global gene expression pattern responses in infected alveolar type II-like A549 epithelial cells (68, 74). As a mechanism for its ability to induce mononuclear infiltration into the infected airway, we found that RSV induced time-dependent expression of 17 distinct chemokines, including those of the CC (I-309, Exodus-1, TARC, RANTES, MCP-1, MDC, and MIP-1α/β), CXC (GROα/β/γ, ENA-78, interleukin-8 [IL-8], and I-TAC), and CX3C (fractalkine) subclasses (74). The mechanism by which RSV induces chemokine expression in response to cytoplasmic viral replication is, at least in part, mediated through the induction of nuclear-to-cytoplasmic translocation of low-abundance regulatory proteins, including the transcription factors NF-κB (6, 23), interferon response factor (10), and signal transducers and activators of transcription (45), as well as enhanced synthesis of the bZIP transcription factors, nuclear factor-IL-6 and activator protein 1 (11, 38). This combination of inducible nuclear proteins together form nucleoprotein complexes (enhanceosomes) on target chemokine genes, stimulating their transcription (7, 11).
These earlier studies indicated that significant alterations in the abundance and spectrum of nuclear regulatory proteins occurs in response to cytoplasmic RSV replication. Furthermore, these studies also raised questions regarding whether other important nuclear-protein alterations may occur as a result of RSV replication that were not detected by genomic approaches. To investigate this, we applied a proteomics approach using high-resolution two-dimensional gel electrophoresis (2DE) and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) to evaluate relative changes in nuclear-protein abundance in response to RSV infection. Our 2DE analysis focused on highly purified sucrose step gradient-purified nuclear extracts from control and RSV-infected A549 cells. Separate 2DE analyses were conducted using subnuclear fractions of soluble proteins prepared by high-salt extraction and the remaining insoluble postnuclear pellet. Proteins whose average abundance was changed by twofold (or greater) in six replicate gels for each treatment were then identified by peptide mass fingerprinting by MALDI-TOF-MS. These analyses identified 24 distinct proteins whose expression was significantly altered, including components of the nuclear cytoskeleton, heat shock proteins, and nuclear domain 10 (ND10) structures. Strikingly, immunofluorescence microscopic analysis demonstrated that RSV infection induced cytoplasmic Hsp70 aggregation and redistribution to punctate nuclear structures. In addition, RSV infection produced dissolution of the ND10 structures with redistribution of their major components, promyelocytic leukemia (PML) and Sp100 proteins, into the perinuclear space and cytoplasm. Taken together, these results provide new insights into specific changes in nuclear-protein abundance after cytoplasmic RSV replication and show that RSV induces global nuclear structural rearrangements and heat shock response as parts of its coordinated cellular response to viral replication.
MATERIALS AND METHODS
Cell culture and infection.
Human A549 pulmonary type II epithelial cells (American Type Culture Collection) were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C in a 5% CO2 incubator (23). The human RSV A2 strain was grown in Hep-2 cells and purified by centrifugation on discontinuous sucrose gradients (69). The virus titer of the purified RSV pools was 7.5 to 8.5 log PFU/ml, determined by methylcellulose plaque assay. No contaminating cytokines, including IL-1, tumor necrosis factor alpha, IL-6, IL-8, granulocyte-macrophage colony-stimulating factor, and interferon (IFN), were found in these sucrose-purified viral preparations (23). Lipopolysaccharide, assayed using the Limulus hemocyanin agglutination assay, was also not detected. Virus pools were aliquoted, quick frozen on dry ice-ethanol, and stored at −70°C until they were used. For viral adsorption, cells were transferred into Dulbecco's modified Eagle's medium containing 2% (vol/vol) fetal bovine serum and infected with sucrose step gradient-purified RSV at a multiplicity of infection (MOI) of 1 for 24 h prior to harvest and assay.
Preparation of nuclear extracts.
Control or RSV-infected A549 cells were incubated in hypotonic buffer, buffer A (50 mM HEPES, pH 7.4, 10 mM KCl, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 0.1 μg of phenylmethylsulfonyl fluoride/ml, 1 μg of pepstatin A/ml, 1 μg of leupeptin/ml, 10 μg of soybean trypsin inhibitor/ml, 10 μg of aprotinin/ml, and 0.1% IGEPAL CA-630). After 10 min on ice, the lysates were centrifuged at 4,000 × g for 10 min at 4°C. The pellet (containing the nuclei) was resuspended in buffer B (buffer A containing 1.7 M sucrose) and centrifuged at 15,000 × g for 30 min at 4°C (8). For whole nuclear extracts, the purified nuclei were lysed in DeStreak Rehydration buffer (Amersham). For the high-salt soluble fraction, purified nuclei were incubated in buffer C (10% glycerol, 50 mM HEPES, pH 7.4, 400 mM KCl, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 0.1 μg of phenylmethylsulfonyl fluoride/ml, 1 μg of pepstatin A/ml, 1 μg of leupeptin/ml, 10 μg of soybean trypsin inhibitor/ml, 10 μg of aprotinin/ml) with vortexing for 30 min at 4°C. After centrifugation at 15,000 × g for 5 min at 4°C, the supernatant was acetone precipitated and the dried pellet was resuspended in DeStreak Rehydration buffer. Nucleic acid was removed by the addition of 300 U of endonuclease (catalog no. E 8263; Sigma)/ml and incubated for 30 min at 22°C. All extracts were normalized for protein amounts determined by Coomassie G-250 staining (Bio-Rad, Hercules, Calif.).
2DE.
Isoelectric focusing (IEF) was performed with 11-cm-long precast immobilized pH gradient (IPG) strips (pH 3 to 10 or 5 to 8 as appropriate; Bio-Rad). Two hundred microliters of 1-mg/ml protein aliquots were loaded onto an IPG strip and rehydrated overnight. IEF was performed at 20°C with the following parameters: 50 V for 11 h, 250 V for 1 h, 500 V for 1 h, 1,000 V for 1 h, 8,000 V for 2 h, and 8,000 V for 6 h. After IEF, the IPG strips were stored at −80°C until 2D sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed. For the 2D SDS-PAGE, the IPG strips were incubated in 4 ml of equilibration buffer (6 M urea, 2% SDS, 50 mM Tris-HCl, pH 8.8, 20% glycerol) containing 10 μl of tri-2(2-carboxyethyl) phosphine (Geno Technology, Inc., St. Louis, Mo.)/ml for 15 min at 22°C with shaking. The samples were then incubated in another 4 ml of equilibration buffer with 25 mg of iodoacetamide/ml for 15 min at 22°C with shaking. Electrophoresis was performed at 150 V for 2.25 h at 4°C with precast 8 to 16% polyacrylamide gels in Tris-glycine buffer (25 mM Tris-HCl, 192 mM glycine, 0.1% SDS, pH 8.3). After 2DE, the gels were fixed in fix buffer (10% methanol, 7% acetic acid in double-distilled H2O), stained with SYPRO-Ruby (Bio-Rad), and destained in fix buffer.
Measurement of relative spot intensities.
The destained gels were scanned at a 100-μm resolution using the Perkin-Elmer (Boston, Mass.) ProXPRESS Proteomic Imaging System with 480-nm excitation and 620-nm emission filters. The exposure time was adjusted to achieve a value of ∼55,000- to 63,000-pixel intensity on the most intense protein spots on the gel. The 2D gel images were subsequently analyzed using Progenesis Discovery software version 2003.03 (Nonlinear Dynamics, Ltd., Newcastle Upon Tyne, United Kingdom). An average gel was created from gels run on nuclear extracts from six separate samples from mock-infected cells (controls) and six separate samples from RSV-infected cells (24 h). The software automatically selected 1 of the 12 gels as the base image of the reference gel. The gel with the highest number of spots was set as the reference gel. Unmatched spots present in 8 of the 12 other gels were subsequently added to the reference gel image by the software to give a comprehensive reference gel. Subsequent to automatic spot detection, spot filtering was manually performed; spots with an area of <270 pixels were filtered out, and spots with a volume (intensity)/area ratio of <375 pixels (whose abundance was insufficient for MS identification) were also filtered. Typically, some manual spot editing was required to correct for spots that were not split correctly, not detected, or split unnecessarily during the automated detection process. The matching of spots between the gels was manually reviewed and adjusted as necessary. Consistent with previous work (47), the SYPRO Ruby staining was linear with respect to input protein over a large dynamic range spanning the spot intensities of the analyzed proteins (data not shown). Moreover, the log-transformed normalized spot volumes were normally distributed, indicating that nonparametric statistical comparisons, such as t tests, could be applied to identify those proteins whose expression was significantly changed by infection. The spot volumes were normalized based on the total spot volume for each gel, and the control and RSV-infected nuclei were compared. A ±2-fold change in the normalized spot volume was considered significantly changed, and these spots were subsequently robotically picked and trypsin digested, and peptide masses were identified by MALDI-TOF. Protein identification was performed using a Bayesian algorithm (73), in which high-probability matches are indicated by an expectation score, an estimate of the number of matches that would be expected in that database if the matches were completely random. Hierarchical clustering was performed by the hclust algorithm in the Splus 6 statistical package (Insightful Inc.), using Euclidian distance.
Protein identification.
Protein gel spots were excised and prepared for MALDI-TOF-MS analysis using Genomic Solutions' ProPic and ProPrep robotic instruments following the manufacturer's protocols. Briefly, gel pieces were incubated with trypsin (20 μg/ml in 25 mM ammonium bicarbonate, pH 8.0; Promega Corp.) at 37°C for 4 h. MALDI-TOF-MS was performed using an Applied Biosystems Voyager model DE STR for peptide mass fingerprinting. Tryptic digests of some gel spots selected for validation were subsequently subjected to analysis by liquid chromatography ion-trap microspray tandem MS (LC-MS/MS) using a Micromass quadrupole time of flight MS. LC was performed on a C18 New Objective PicoFrit column that allowed the eluate to be sprayed directly into the source of the mass spectrometer. MS/MS was performed in a data-dependent mode, and the data were processed using a Proteometrics Sonar MS/MS search engine (Genomic Solutions).
Western immunoblot analysis.
For Western blots, nuclear proteins were fractionated on SDS-10% PAGE and transferred to polyvinylidene difluoride membranes (Millipore, Bedford, Mass.) (39). The membranes were treated with 5% milk-Tris-buffered saline (100 mM Tris-HCl, 150 mM NaCl, pH 7.5) containing 0.1% (vol/vol) Tween for 1 h and then incubated with an appropriate antibody overnight at 4°C. Anti-lamin-B, -PML, and -Sp100 antibodies were affinity-purified rabbit polyclonal antibodies (Santa Cruz Biotechnology, Santa Cruz, Calif.); anti-β-tubulin (Santa Cruz Biotechnology) and -β-actin (Santa Cruz Biotechnology) were mouse monoclonal antibodies. The membranes were washed four times in Tris-buffered saline with 0.1% (vol/vol) Tween 20 and then incubated with horseradish peroxidase-conjugated secondary antibody. After being washed, immune complexes were detected by reaction in the enhanced-chemiluminescence assay (Amersham) according to the manufacturer's recommendations.
Immunofluorescence microscopy.
A549 cells (105) plated on coverslips were mock or RSV infected (MOI, 5). The mock- and RSV-infected cells were washed, air dried, and fixed for 10 min with 4% paraformaldehyde in phosphate-buffered saline (PBS), pH 7.4. Lipids were removed by extraction with acetone-methanol (1:1). To remove the cytoplasm and expose nuclear epitopes, where indicated, the cells were washed with PBS, fixed in 4% paraformaldehyde at 4°C for 15 min, and placed into 0.1 N HCl containing 100 μg of pepsin (Sigma Biochemicals, Inc.)/ml for 30 min at 37°C. The cells were then incubated for 60 min at 37°C with the appropriate primary antibody (diluted 1:200 in PBS-Tween 20) to Hsp70 (StressGen Biotechnologies), anti-PML or anti-Sp100 (both from Santa Cruz Biotechnology). After being washed three times for 15 min each time with PBS-Tween 20, the cells were incubated for 1 h at room temperature with fluorescein isothiocyanate-conjugated secondary antibodies diluted 1:200 (Santa Cruz Biotechnology). Nuclei of cells were stained for 15 min with DAPI (4′6-diamidino-2-phenylindole dihydrochloride; 10 ng/ml). The cells were then mounted in antifade medium (Dako Inc., Carpinteria, Calif.) on a microscope slide and photographed. Confocal microscopy was performed on a Zeiss LSM510 META system. Images were captured at ×60 to ×192 magnification.
RESULTS
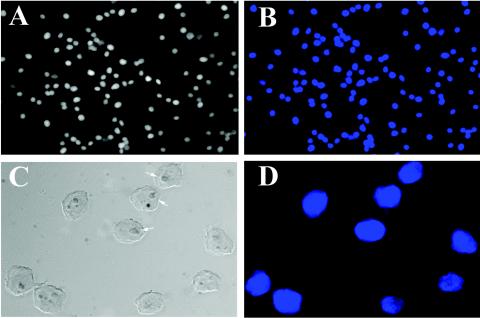
A549 cells, a human lung carcinoma cell line that retains features of alveolar cells, including surfactant secretion, have been widely used to investigate the effects of RSV replication in vitro (5, 11, 23, 38). Under our standardized conditions for in vitro infection, where sucrose-purified RSV (MOI, 1.0) are adsorbed to A549 cells, transcribed viral RNAs are first detectable 6 h later. Cell surface expression of the RSV glycoproteins (F and G) and the first round of virion release follow, both occurring ∼12 h after adsorption (74). Although viral cytopathic effect is present 12 to 24 h later, infected cells retain viability for 48 to 72 h, when syncytium formation and apoptotic cell death predominate. In this study, we isolated highly purified nuclei from uninfected and infected A549 cells prepared 24 h after viral adsorption to ensure cells were uniformly infected. Nuclei purified by sucrose step gradient centrifugation served as the source of protein for the 2DE analysis (8, 32). To demonstrate the quality of the nuclear preparations, the extracts were stained with DAPI and visualized by phase-contrast microscopy (Fig. 1). As seen in Fig. 1A to D, nuclear preparations consisted of homogeneous nuclear forms that uniformly stained with DAPI. The nuclear structures were well maintained, as intact nucleoli could be identified by phase-contrast microscopy under higher magnification (Fig. 1C and D). As biochemical markers for enrichment of nuclear protein, Western immunoblot analysis was performed on A549 nuclear and cytoplasmic fractions that stained either with an antibody to the nuclear matrix protein lamin B or with an antibody to the cytoplasmic marker tubulin. As seen in Fig. 1E, a strong 80-kDa lamin B band was detected specifically in the nuclear fractions, whereas a 60-kDa β-tubulin band was strongly detected in the cytoplasmic, but not the nuclear, fraction. Together, these observations gave strong evidence that the nuclear fractions were homogeneous and enriched in nuclear proteins.
FIG. 1.
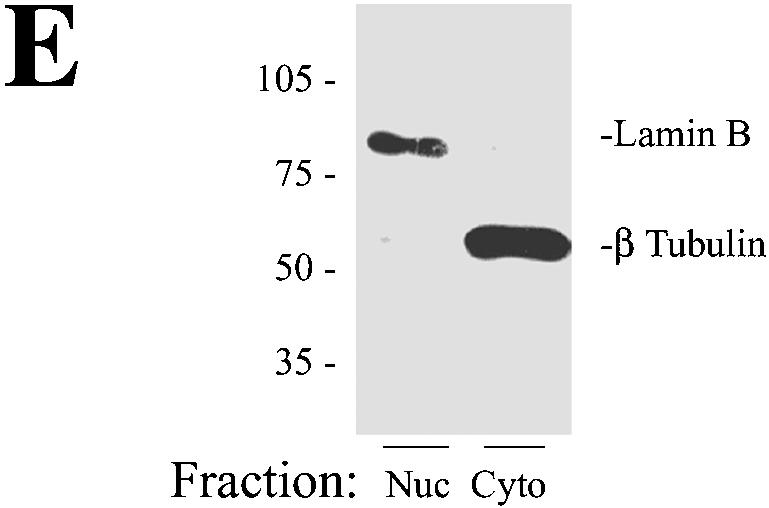
Characterization of A549 cell nuclear preparations. (A to D) Microscopic analysis of sucrose step gradient-purified nuclei. Purified nuclei were diluted in PBS, plated on a microscope coverslip, and stained with DAPI (see Materials and Methods). Panels A and C, phase-contrast microscopy; panels B and D, DAPI staining. Panels A and B, low-resolution images. Panels C and D, high-resolution images. In panel C, nucleoli are visible (arrows). (E) Western immunoblot for nuclear and cytoplasmic markers. Duplicate 75-μg samples of nuclear and cytoplasmic extracts were fractionated by one-dimensional SDS-PAGE, transferred to polyvinylidene difluoride, and stained with lamin B and β-tubulin antibodies. The locations of molecular weight markers (10−3) are indicated on the left. The lamin B stain is localized to the nuclear fraction (Nuc), whereas the β-tubulin is localized to the cytoplasmic lysates (Cyto).
2DE analysis of nuclear proteins.
High-resolution 2DE was performed on the nuclear proteins from mock- and RSV-infected cells (24 h; MOI, 1), in which the first dimension was fractionated over an IEF range of pH 5 to 8. Here, six control samples and six RSV-infected nuclear samples (each representing a separate isolation) were analyzed to allow meaningful statistical comparison. In this analysis, we resolved a range of 488 to 537 spots on each individual gel. Of these, 527 spots were present in 8 of the 12 gels (representing reference spots) and therefore were used in the comparative analysis. 2DE was also performed on total nuclear proteins from six separate nuclear samples from mock- and RSV-infected cells fractionated over an IEF range of pH 3 to 10. Over this IEF range, 673 reference spots were identified, representing 146 spots in addition to those identified by the narrower-range IEF. Similar comparative 2DE analysis was also performed on soluble (salt-extractable) and insoluble (postnuclear pellet) nuclear extracts to enrich lower-abundance proteins. In these experiments, sucrose step gradient-purified whole nuclei were extracted under high-salt conditions (0.4 M KCl) to enrich soluble nucleoplasmic proteins (including transcription factors [7, 23, 31, 37]), leaving chromatin and structural proteins in the insoluble postnuclear pellet. The soluble and postnuclear fractions were also analyzed by 2DE in six independent replicates for each treatment group. In the soluble fraction, 421 reference spots were identified, and in the postnuclear-pellet fractions, 1,065 reference spots were identified and compared.
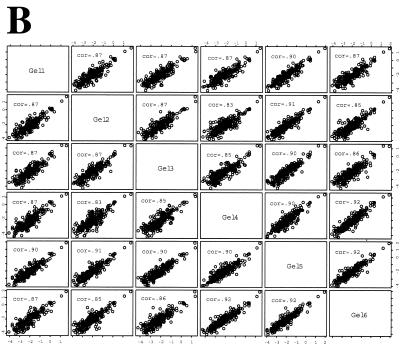
Representative images of a single SYPRO Ruby-stained 2DE gel (IEF range, pH 5 to 8) from high-salt extractions of the control and RSV-infected nuclei are shown in Fig. 2A. Overall, the patterns of proteins were largely similar, indicating that at this time, RSV did not induce global perturbations of the nuclear proteome. To determine whether the spot intensity patterns were similar for the replicates under the same treatment conditions, the normalized spot volumes were compared pairwise across the treatment set. For each spot resolved by 2DE, the normalized spot volume in the base gel was plotted against that value in gels 2 to 5, and the Pearson correlation coefficient (r2) was calculated. As seen in Fig. 2B, for gels 1 to 6, the value of r2 ranged from 0.83 to 0.92. These findings indicated that the 2DE analysis was highly reproducible under the given conditions. Similar correlation coefficients were found for the 2DE analysis of the RSV-infected gels (not shown).
FIG. 2.
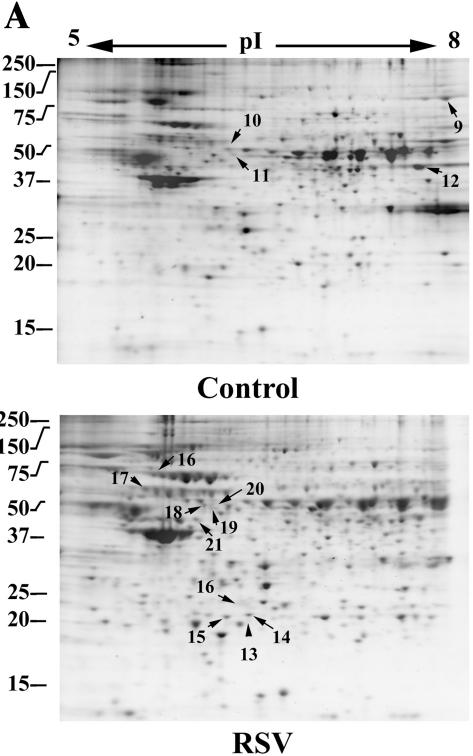
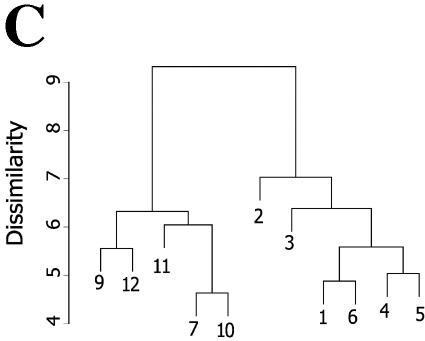
2DE of soluble nuclear proteins. (A) SYPRO-Ruby-stained 2DE. High-salt extracts containing the soluble proteins from control or RSV-infected (24 h; MOI, 1.0) A549 cells. Proteins were fractionated over immobilized pH gradients from pH 5 to 8 in the horizontal dimension, followed by fractionation by SDS-PAGE in the vertical dimension. Left, apparent migration of molecular mass standards (in kilodaltons). The numbers indicate the spots identified by tryptic peptide mass fingerprinting in Table 1. (B) Gel-to-gel correlation of control replicates. Log normalized spot volumes for gel 1 were plotted pairwise versus gels 2 to 5, and the Pearson's correlation coefficient (cor) was calculated. (C) Hierarchical clustering. Normalized spot intensities were subjected to hierarchical clustering by treatment condition, and their relationships were plotted as a dendrogram. The y axis is dissimilarity. Gels 1 to 6, control nuclei; gels 7 to 12, RSV infected (one RSV-infected gel was excluded due to poor spot resolution). Note that the gels from similar treatments cluster in the same node.
To determine whether 2DE spot data extracted from the control and RSV-infected nuclear extracts reflected differences by treatment group, the spots from the control and treated gels were matched, and the normalized spot volumes were subjected to hierarchical clustering (Fig. 2C). In this technique, the protein expression profile for an individual 2DE is grouped with its nearest neighbor based on the mathematical proximity of the two gels. These gels are combined into a single node, and that node is then grouped with the next-nearest gel. This process is repeated iteratively until all gels are contained within the dendrogram. The degree of relatedness is indicated by the height of a common line on the vertical dendrogram that connects the two nodes (the y axis is the dissimilarity metric calculated by Euclidian distance). As can be seen in Fig. 2C, two major groups are identified by the clustering algorithm. On the left side of the dendrogram, the gels from RSV-infected cells (numbered 7 to 12) are contained within a single branch of the dendrogram, indicating that the spot patterns are closely related. In the right half, gels from mock-infected cells (numbered 1 to 6) form another cluster more closely related to one another than to the RSV-treated gels. Together, these findings that the individual gels clustered by treatment condition indicated that reproducible differences in global spot volume patterns were detected, and spot intensities were representative of the effects of viral treatment.
Identification of differentially expressed proteins.
For each 2DE analysis, the change (the ratio of the mean normalized spot volume of treated versus control gels) and a paired t test were calculated (adjusted for multiple-hypothesis testing using the Bonferroni correction) for spots identified in four of the six replicates of each treatment group. We focused on identifying proteins whose RSV-induced change was >2.0-fold and whose corrected t test statistic was <0.05. From this analysis, 12 spots were identified in the whole-nuclear extracts fractionated in the first dimension over the pH range 3 to 10, 7 spots were identified in the whole nuclei fractionated over the pH range 5 to 8, 18 spots were identified in the soluble (high-salt) fraction, and 28 spots were identified in the insoluble (postnuclear-pellet) fraction and subjected to tryptic peptide mass fingerprint analysis by MALDI-TOF. The resulting peptide masses were then used to match proteins in the National Center for Biotechnology Information human database using a Bayesian search algorithm (73). If a protein did not match any human proteins, the search was repeated against a viral-protein database. Proteins whose statistical indicators gave a high-probability identification are shown in Table 1 and are labeled in the representative gels (Fig. 2A). In the intact nuclear preparations fractionated over a pH range from 3 to 10, cytoskeletal proteins (lamin A and C and cytokeratin 18), Mn superoxide dismutase (MnSOD), and 3′-5′ RNA exonuclease were upregulated by RSV. In the whole-nuclear samples fractionated over the pH range 5 to 8, upregulation of the TAR DNA binding protein was identified in two separate spots in the RSV-infected nuclei. The TAR DNA binding protein is an RNA binding protein that is associated with nuclear structures, including GEM and the ND10 structures (71). In the soluble nuclear subfractions, we found downregulation of nuclear RNA helicase and upregulation of thioredoxin peroxidase, Hsp 70- and 60-kDa isoforms, and phospholipase C-α. In the insoluble postnuclear pellet, the abundance of enolase 1 was downregulated, whereas that of cytokeratin 18 was upregulated (in agreement with its upregulation in the whole-nuclear analysis). We did not identify any RSV proteins in the nuclear fractions.
TABLE 1.
Differentially expressed proteins in nuclei of RSV-infected cellsa
| Fraction | Common name | Acc. number | Expectation score | Cov | FC | P value |
|---|---|---|---|---|---|---|
| WN, pH 3-8 | ||||||
| 1 | MnSOD | AAP34410.1 | 1.20E-02 | 15 | 4.7 | 0.003 |
| 2 | Lamin A/C | NP_733822.1 | 3.10E-04 | 20 | NA (I) | 0.000 |
| 3 | Cytokeratin 18 | AAH09754.1 | 4.00E-03 | 15 | 4.7 | 0.013 |
| 4 | 3′-5′ RNA exonuclease | NP_149100 | 2.30E-03 | 12 | NA (I) | <0.001 |
| 5 | G6PD | 7546523 | 1.10E-02 | 18 | NA (I) | 0.001 |
| WN, pH 5-8 | ||||||
| 6 | Unknown | AAH36000.1 | 3.00E-09 | 18 | 2.3 | 0.005 |
| 7 | TAR DNA binding protein | NP<031401.1> | 8.90E-08 | 14 | NA (I) | 0.006 |
| 8 | TAR DNA binding | NP_031401.1 | 6.80E-03 | 14 | NA (I) | 0.006 |
| Hi salt, pH 5-8 | ||||||
| 9 | Transl. elongation 2 | NP_001952.1 | 3.70E-06 | 15 | NA (D) | 0.025 |
| 10 | Cytokeratin 8 | AAA35748.1 | 2.00E-04 | 28 | NA (D) | <0.001 |
| 11 | Nuclear RNA helicase | AAB50231.1 | 3.30E-02 | 16 | ||
| 12 | Hypothetical | CAD97642.1 | 6.00E-03 | 14 | −3.3 | 0.002 |
| 13 | Thioredoxin peroxidase | NP_006397.1 | 2.20E-16 | 14 | 3 | 0.002 |
| 14 | MHC I ligand | 1EFX | 2.20E-05 | 9 | ||
| 15 | HLA-Cw304 | BAA19535.1 | 3.00E-05 | 8 | ||
| 16 | Hsp 70 | NP_694881.1 | 2.70E-10 | 32 | 3 | 0.007 |
| 17 | Hsp 60 | AAA36022.1 | 6.90E-05 | 19 | 2.6 | 0.001 |
| 18 | PLC alpha | BAA03759.1 | 9.10E-08 | 21 | 2.6 | <0.001 |
| 19 | ERp57 | 2201353A | 7.50E-04 | 12 | ||
| 20 | ATPase | NP_001684.2 | 4.60E-03 | 14 | ||
| 21 | TBP interacting protein | NP_006657.1 | 7.60E-07 | 32 | 3 | 0.020 |
| PNP, pH 5-8 | ||||||
| 22 | TCP1 ring complex | 2136253 | 3.30E-03 | 16 | −2.3 | 0.021 |
| 23 | Proliferation-associated | 13632817 | 1.50E-03 | 19 | NA (D) | <0.001 |
| 24 | Enolase 1 | NP_001419.1 | 1.20E-04 | 32 | −3 | 0.006 |
| 25 | Cytokeratin 18 | 30311 | 1.00E-03 | 17 | 7.5 | <0.001 |
Shown are high-probability identifications from peptide mass fingerprinting in MALDI-TOF (Expectation score) grouped by subcellular fractionation and 2DE resolution method. WN, whole nuclei; Hi salt (0.4 M KCl extract), PNP, postnuclear pellet. The pH range is the first-dimension isoelectric focusing gradient. For each protein, the common name, GenBank accession number, expectation score (statistical significance of the match), percent coverage of the protein by peptides measured by MALDI-TOF (Cov), fold change in normalized spot volume (FC; signed ratio of treated to control), and P value of the two-tailed t test comparing control versus RSV-infected normalized spot volumes (after Bonferroni correction) are indicated. NA, not applicable (the spot volume for either the control or RSV-treated sample is undetectable, precluding a reliable FC calculation). Each spot identified as NA is designated as increased (I), indicating RSV increases its expression, or decreased (D), indicating RSV decreases its expression.
Validation of selected differentially expressed proteins.
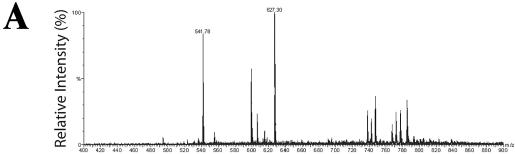
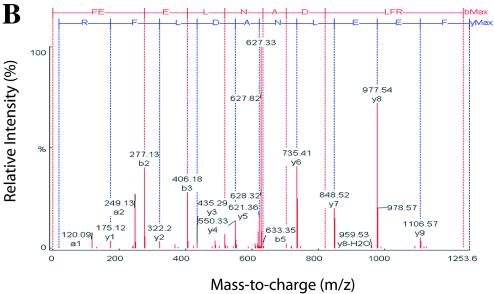
To partially validate our 2DE data set, we confirmed our identification of cytokeratin 18, Hsp70, and the 3′-5′ RNA exonuclease by sequencing some of their tryptic peptides using LC-MS/MS. Here, the relevant spot identified in the difference comparison was picked from the gel and digested with trypsin and peptides fractionated by reverse-phase high-pressure liquid chromatography. Individual peptides were then fragmented and sequenced by mass spectrometry. In the case of Hsp70, six peptides were sequenced that matched inducible Hsp70. For the parent ion identified in the total-ion chromatogram in Fig. 3A with mass-to-charge (m/z) values of 627.30, tandem MS showed the peptide sequence to be FEELNADLFR (Fig. 3B), corresponding exactly to amino acids 305 to 314, respectively, of Hsp70. In a similar manner, we used LC-MS/MS to validate the identification of cytokeratin and 3′-5′ exonuclease (data not shown).
FIG. 3.
Validation of nuclear Hsp70 expression. (A) Total ion chromatogram of tryptic digest corresponding to spot 16 (Fig. 1A and Table 1). Both parent ions, with m/z of 541.78 and 627.30, were selected for MS/MS analysis. (B) MS/MS spectrum of parent ion 627.30. Fragment ions produced by collision induced dissociation in tandem MS. The deduced sequence of the peptide, from the NH2 terminus, is shown in single-letter amino acid code (red lettering, top), and that from the COOH-terminus is shown beneath (blue). The NH2-terminal sequence is FEELNADLFR, matching the coding sequence of Hsp70 (amino acids 305 to 314). MS/MS analysis of parent ion 541.78 also exactly matched Hsp70.
Nuclear stress response in RSV-infected A549 cells.
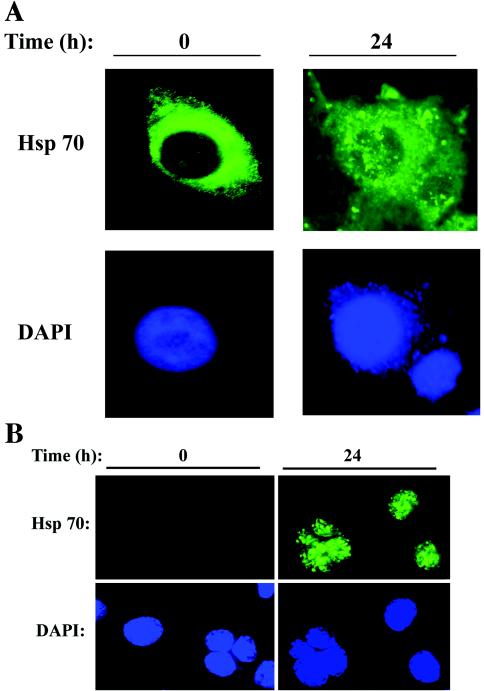
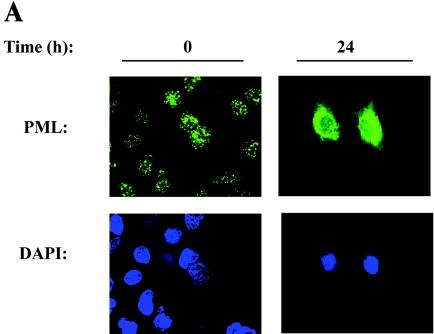
We further examined the significance of our findings, first focusing on the inducible Hsp70 protein by indirect immunofluorescence using confocal microscopy. In uninfected A549 cells, Hsp70 expression was uniformly distributed in a granular pattern throughout the cytoplasm (Fig. 4A). However, this pattern was markedly changed in response to RSV replication, where cytoplasmic and nuclear aggregates of Hsp70 immunostaining were seen. This signal was not due to overlying cytoplasmic Hsp70, because the image shown was a confocal slice through the nuclear compartment. To further demonstrate nuclear accumulation of Hsp70, control and RSV-infected cells were fixed, permeabilized, and digested with pepsin to remove the surrounding cytoplasm and increase the availability of nuclear epitopes. As shown in Fig. 4B, almost no nuclear Hsp70 was seen in the control cells, indicating that the nucleus-associated Hsp70 in uninfected cells was only weakly associated and was probably washed out from the fixation and digestion procedure. Under these conditions, the nuclear staining of Hsp70 in the RSV-infected cells was clearly demonstrated when we observed Hsp70 distributed in a punctate pattern throughout the nucleus.
FIG.4.
RSV-induced Hsp70 redistribution. (A) Immunofluorescence microscopy. Control (mock-infected; 0 h) or RSV-infected (24 h; MOI, 1.0) cells were fixed and stained with anti-Hsp70 (top), or nuclei were stained with DAPI (bottom). Shown is a single confocal slice of the stained cells. Hsp70 was detected in a finely granular distribution throughout the cytoplasm in uninfected cells. Conversely, in RSV-infected cells, the distribution of cytoplasmic Hsp70 was more punctate, and some apparent nuclear redistribution could be observed. (B) Nuclear accumulation of Hsp70. Control or RSV-infected cells were fixed, and the cytoplasm was permeabilized and digested with pepsin prior to being stained with Hsp70 (top) or DAPI (bottom). Hsp70 was strongly associated with the nucleus after RSV infection.
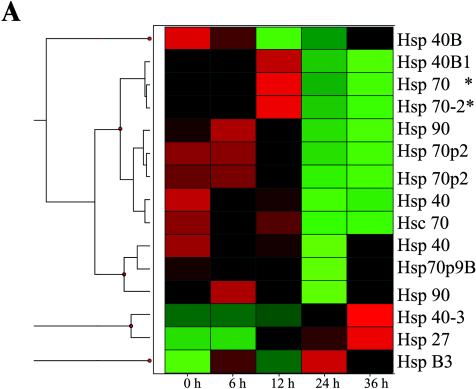
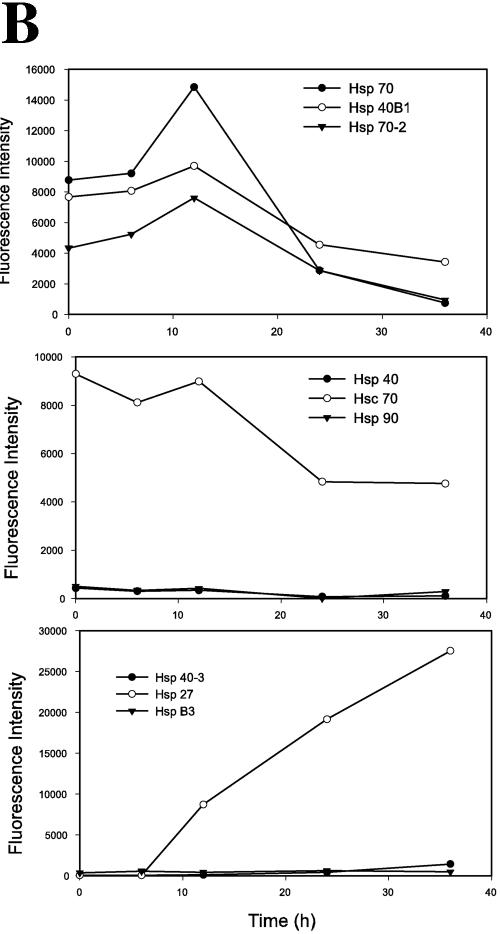
The activities associated with inducible Hsp70, including prevention of denatured protein aggregation, modulation of signal transduction, and inhibition of apoptosis (4, 24, 59, 62), are dependent on Hsp70 association with cochaperones, which themselves are members of the Hsp family (33, 57). For example, Hsp70-catalyzed folding of nascent polypeptide chains occurs with the Hsp40 (DnaJ) cochaperone prior to release of the target protein for Hsp90 binding (33). To determine whether RSV replication affected the expression of Hsp gene family members, an established database of RSV-inducible genes in A549 cells (74) was mined for changes in Hsp gene expression. These data are visually represented by hierarchical clustering on a heat map, where the Z score-normalized signal intensity determined in three independent time course experiments is shown (Fig. 5A). The Z score normalization expresses each gene expression profile as a deviation from the mean in standard-deviation units and allows the comparison of gene expression patterns whose absolute expression levels may differ by orders of magnitude. The individual genes are grouped with other genes that share expression profiles and are represented by a dendrogram (essentially as described above for Fig. 2C). From these data, it can be seen that RSV alters the expression of multiple members of the Hsp gene family. Two major clusters are seen. The first major group is composed of 12 probe sets corresponding to genes expressed in mock-infected cells (time zero) that are variably induced by RSV and whose expression later falls (representing Hsp 40B to Hsp90 isoforms as indicated). The second major group is composed of three probe sets corresponding to genes that are not expressed in uninfected cells and whose expression is induced 24 and 36 h after RSV infection (Hsp40-3, -27, and -B3). In the first group, a subgroup of closely related expression profiles is composed of Hsp40B1, -70, and -70-2. These genes are expressed in uninfected cells, are transiently induced 12 h after RSV adsorption, and fall to very low levels 24 h (and more) after infection. In contrast, the subgroup containing Hsp40 and the constitutive Hsc70 isoforms are regulated slightly differently, being highly expressed in uninfected cells and downregulated 6 to 12 h after viral adsorption, and falling to low levels 24 h after infection. To more clearly demonstrate the changes in Hsp subgroup expression, the absolute signal intensity is plotted in Fig. 5B for the inducible Hsp40B1, -70, and -70-2 subgroup (top), the Hsp40 and Hsc70 subgroup (middle), and the RSV-induced Hsp40-3, -27, and -B3 subgroup (bottom). Note that the average signal intensity of Hsp70 mRNA is induced 12 h after RSV infection and falls after 24 h. Consistent with its grouping by hierarchical clustering, a similar qualitative pattern is seen for Hsp40B1 and -70-2. Of the Hsp family members in the middle graph of Fig. 5B, Hsc70 is most highly expressed and shows a gradual decline in expression. Of those in the lower graph, Hsp27 is most highly induced within 12 h of RSV infection and monotonically increases throughout the experiment. Together, these data suggest that RSV induces transient coordinate changes in Hsp gene expression prior to the nuclear accumulation of Hsp70 detected by 2DE, constituting a heat shock-like response.
FIG. 5.
RSV induces Hsp family gene expression in distinct profiles. (A) Hierarchical clustering of mRNA profiles. A previously reported database of RSV-inducible gene expression in A549 cells was mined for RSV-induced expression changes in all Hsp genes (74). The following Hsp family members were represented on the chip, and their profiles were extracted: Hsp40B1 (GenBank accession no. D85429), -70 (GenBank accession no, M11717), -70-2 (GenBank accession no. M59830), -90 (GenBank accession no. J04988), -70p2 (GenBank accession no. L26336), and -40 (GenBank accession no. U40992); Hsc70 (GenBank accession no. L12723), -40 (DnaJ homolog; GenBank accession no. U40992), -70p9B (GenBank accession no. L15189), -40-3 (GenBank accession no. AF088982), -27 (IFN inducible; GenBank accession no. X67325), and -B3 (GenBank accession no. U15590). Because of the variable level of expression of the individual genes, the average signal intensity (SI) was normalized by the Z score, where deviation from the mean is measured in standard-deviation units. The Z score is determined for any cell i by the formula Z = (SIi − SIrow)/SD, where SIrow is the average signal intensity for the gene (across the row) and SD is the standard deviation. The data are represented as a heat map, where each value is the colored representation of the calculated Z score for each time point. The scale is represented by red (Z > +1.2), green (Z < −1.2), and black (Z = 0). At the left is a dendrogram indicating the mathematical dissimilarity of the expression profiles. Genes with similar expression profiles are grouped together and are connected by a short line that connects the two nodes. Two major clusters are seen; the first group are genes expressed at time zero, transiently induced by RSV 6 to 12 h after infection and later falling (representing Hsp40B to Hsp90, indicated at the right), and the second group are genes not expressed at time zero and induced 24 and 36 h after RSV infection (Hsp 40-3, Hsp 27, and Hsp B3). (B) Profile of responses for Hsp subgroups. The average signal intensity changes from three independent microarrays are plotted as a function of time. Top, the profile of the 12-h induced Hsp genes, including Hsp70. Hsp70 mRNA abundance is significantly influenced by RSV infection [analysis of variance with replicates, Pr(F) = 0.0143]. Middle, the profile of Hsc70 and coclustering genes. Bottom, induction profile of Hsp27 and associated genes.
RSV induces ND10 reorganization.
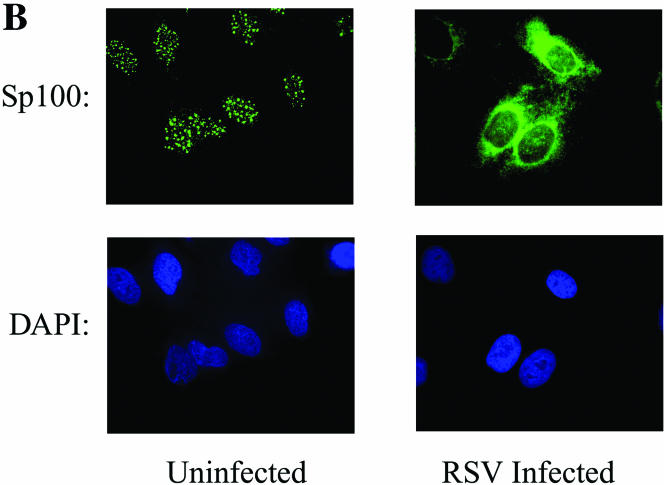
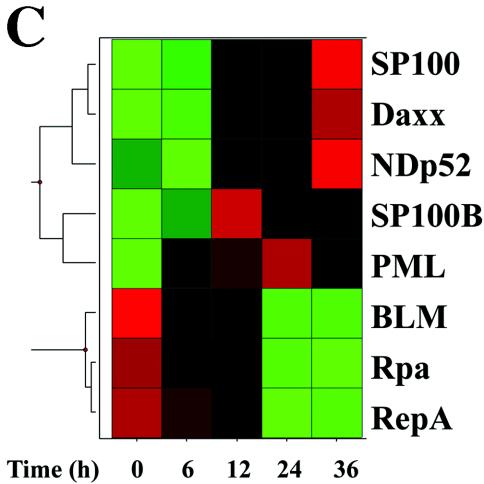
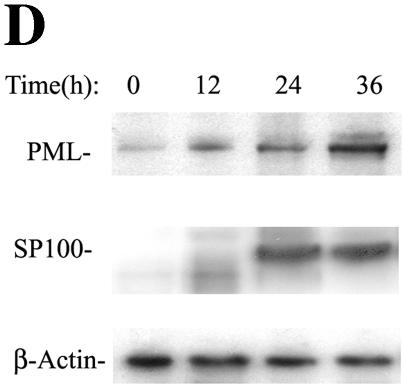
ND10s are dynamic nuclear structures, composed of PML, death domain-associated protein (Daxx), Sp100, and the Blooms' syndrome helicase, that function as depots of nuclear transcription factors (51). Our findings by 2DE suggested that RSV replication induced nuclear appearance of the TAR DNA binding protein, an ND10-associated protein (71). Because an antibody to the TAR DNA binding protein was unavailable, we evaluated the effects of RSV on ND10 abundance and composition by indirect immunofluorescence microscopy using antibodies available to the major structural proteins, PML (Fig. 6A) and Sp100 (Fig. 6B). In uninfected A549 cells, consistent with earlier work, PML staining appeared as discrete nuclear dots that localized to nuclei (identified by DAPI staining). In marked contrast, the PML staining intensity was increased and the protein was redistributed into the perinuclear space and cytoplasm of RSV-infected cells (Fig. 6A). A similar staining pattern was seen for Sp100 (Fig. 6B). The expression patterns of the ND10-associated structural proteins in response to viral infection were queried in the microarray database, where two general patterns of gene expression were observed (Fig. 6C). Here, Sp100B, PML, Daxx, ND10 52-kDa protein (NDp52), and Sp100 were expressed at low abundance in uninfected cells, and their expression was induced 12 to 36 h after viral adsorption. In contrast, expression of the Bloom's syndrome helicase and the 70- and 14-kDa replication A proteins were initially high in uninfected cells and fell over 6 to 12 h after viral infection. The patterns of PML and Sp100 protein expression were confirmed by Western blotting, which showed that, relative to β-actin, the abundances of PML and Sp100 were both strongly induced (Fig. 6D). The altered mobility of the PML and SP100 isoforms may be due to their posttranslational modifications, such as their well-described conjugation to the small ubiquitin modifier, SUMO-1 (reviewed in reference 51). Together, these data indicated that cytoplasmic RSV replication induces significant changes in the ND10 structures through transcriptional and posttranscriptional mechanisms.
FIG. 6.
ND10 redistribution as a function of RSV infection. (A) Redistribution of PML. Immunofluorescence microscopy was performed using anti-PML antibody (top). The bottom images are DAPI-stained nuclei. Left, control cells; right, RSV infected. In control cellular nuclei, PML is present in distinct ND10 structures. In RSV-infected cells, strong PML immunofluorescence is redistributed into the cytoplasm. (B) Redistribution of Sp100. Immunofluorescence using anti-Sp100. The bottom images are as in Fig. 6A. (C) Effect of RSV in expression of ND10 major structural proteins. Hierarchical clustering and heat map of ND10 structural proteins represented in the A549 genomic database, including Sp100 (GenBank accession no. M60618), Daxx (GenBank accession no. AB015051), NDp52 (GenBank accession no. U22897), Sp100B (GenBank accession no. U36501), PML (GenBank accession no. M79463), Blooms' helicase (BLM; GenBank accession no. U39817), replication protein A 14 kDa (Rpa; GenBank accession no. L07493), and replication protein A 70 kDa (RepA; GenBank accession no. M63488). The data are calculated and presented as described for Fig. 5A. RSV induces the coordinate expression of PML, Sp100B, NDp52, Daxx, and Sp100. (D) Western immunoblot analysis of PML and Sp100 protein expression in A549 cells. Cells infected with RSV for 0 (uninfected), 12, 24, and 36 h were fractionated by one-dimensional SDS-PAGE and probed with the indicated antibody. β-Actin was used as a loading control. ND10 redistribution occurs in the setting of increased expression of PML and Sp100.
DISCUSSION
RSV is an RNA virus encoded by a negative-sense genome that induces significant nuclear changes upon its infection of a host epithelial cell. Studies have shown that RSV replication influences significant genomic perturbations (68, 74), as well as translocation of low-abundance regulatory proteins (11). In this study, we have exploited this well-characterized model of viral-epithelial interaction to begin to determine the effects of viral replication on the abundance of nuclear proteins by high-resolution 2DE. These proteins included members of the nuclear cytoskeleton (lamin A and C and cytokeratin 18), proteins involved in maintenance of the intracellular redox state (MnSOD and thioredoxin peroxidase) and protein folding and antiapoptosis (Hsp70 and -60), and structural members of ND10s. We have validated the identification of Hsp70, cytokeratin, and 3′-5′ exonuclease and have extended our understanding of the coordinated response to viral replication using microscopic imaging assays coupled with focused data mining of a gene expression database of the same system. Together, these studies have provided new insights into the compensatory nuclear response to cytoplasmic Paramyxovirus infection.
2DE using immobilized pH gradients coupled with MS is now able to spatially resolve and identify thousands of proteins and has become an effective tool for global proteomics analyses. Although the use of IPG strips has improved the gel-to-gel reproducibility of this technique (27), few studies have undertaken a rigorous statistical treatment because of the labor-intensive nature of 2DE gel analysis. Using a newly available automated spot detection algorithm, our experimental approach was designed, in part, to evaluate this semiquantitative approach to differential protein expression. Rather than using a typical single pairwise comparison for which statistical confidence could not be determined, we ran six independent gels for each treatment condition or extraction method and identified proteins by a Bonferroni-adjusted t test. In our analysis, we found that, even for identically treated samples, in six replicates, the coefficient of variance of each spot averaged 0.32 and, surprisingly, was independent of the normalized spot intensity. We have evaluated multiple methods for gel-to-gel comparison to determine which method best minimized the coefficient of variation (data not shown). These methods included scaling each gel to the total spot intensity, normalizing spot intensities to a group of spots on the same gel whose expression did not change as a result of treatment, normalizing spot intensities to externally spiked (reference) proteins, or normalizing spot intensities to the total signal of the individual gel. Surprisingly, none of these methods was consistently superior to analyzing the gels using the raw (unnormalized or unscaled) spot intensity. Because normalizing spot intensities to the total signal of the individual gel is an accepted practice in the field (56), we chose that method of analysis. Our data, therefore, may be limited, in that proteins whose levels significantly changed may not have been identified by our filtering strategy because of the noise in the determination of spot intensity. This failure to detect true positives would represent an unknown type II error. It is important to note that although the overall spot distributions between control and RSV-infected gels were distinct (hierarchical clustering in Fig. 2C), only 24 differentially regulated proteins were identified. Better approaches in sample preparation and methods for gel-to-gel comparison and statistical adjustments for multiple-hypothesis testing are needed. In addition, 2DE analysis is skewed toward identification of higher-abundance proteins, which proved to be the case in this study as well. Equally important regulatory proteins, such as transcription factors and kinases, are likely below the limit of detection of current 2DE systems using crude cellular proteins. Identification of these lower-abundance proteins will require sample prefractionation or other enrichment techniques. Thus, questions regarding the global changes in regulatory proteins are still unresolved and will require further investigation.
In spite of these limitations, our findings provide intriguing new insight into homeostatic nuclear changes in response to RSV infection. In both the whole-nuclear and insoluble-protein fractions, we identified alterations in cytokeratin expression. Cytokeratins are important components of intermediate filaments of the cytoskeleton and are expressed in cell-type-restricted patterns in the respiratory tract. In fact, patterns of cytokeratin expression have been used as markers of cellular differentiation. Previous work has shown that cytokeratin isoforms 7, 18, and 19 are normally expressed in bronchial epithelial cells (64). Although it has been reported that toxicant exposure can induce alterations in cytokeratin isoforms in the respiratory tree (67), the effects of viral replication on cytokeratin expression have not been investigated. Using differential-display analysis of RSV-infected Hep2 cells, cytokeratin 17 expression was found to be induced late in the course of RSV replication (after 4 days [19]). Here, the protein localized to the cell membrane in regions of syncytium formation, perhaps suggesting its participation in virus-mediated cell fusion. Although we observed downregulation of cytokeratin 8 and upregulation of cytokeratin 18 isoforms, our analysis was at an earlier time point, and we therefore would not have detected changes in cytokeratin 17 expression. The cellular consequences of altered cytokeratin expression have not been fully evaluated. One study showed that expression of cytokeratins 8 and 18 induces enhanced cellular migration (15). Also, we note earlier studies which indicated that components of the actin cytoskeleton stimulate RSV transcription. Although it is interesting to speculate that induction of cytokeratin expression may be adaptive for viral transcription (36), the consequences of differential cytokeratin isoform expression for RSV spread or pathogenesis are unknown.
The heat shock proteins are encoded by a family of highly specialized genes with pleiotropic actions. In this study, we observed the nuclear accumulation of Hsp 70- and 60-kDa isoforms in response to RSV replication. Under noninfectious conditions, Hsp70 is known to inhibit the aggregation of nascent or misfolded proteins (33), control nuclear import of transcription factors, and prevent apoptosis by blocking formation of the apoptosome or inhibiting the apoptosis-inducing factor (61, 63). Viral infection with RSV likely produces a misfolded-protein stress response, and therefore, coordinate Hsp expression may be highly adaptive for the virus to prevent the aggregation of viral structural proteins during replication. In fact, Hsp70 expression is required for optimal adenovirus replication (3) through mechanisms that have not yet been fully elucidated. In the separate example of canine distemper virus, cytoplasmic Hsp70 complexes with viral nucleocapsid, leading to enhanced viral transcription. In parallel, Hsp70 association with nucleocapsid (N) protein leads to its trafficking to complex intranuclear inclusion bodies. As a result, Hsp70 plays a role in viral transcription (58) and induction of cytopathic effect (70). To our knowledge, Hsp70 induction has not been reported in RSV infection; interestingly, our data suggest that nuclear Hsp70 is not homogeneously distributed throughout the nucleus, perhaps suggesting that it is associated with other cellular or (yet to be identified) viral proteins.
In addition to its role in preventing protein aggregation, Hsp70 has been reported to be part of the poly(A) binding protein-AU factor 1 binding complex, a complex that controls the turnover of unstable RNAs containing 3′-untranslated-region AU-rich elements (44). In this capacity, nuclear Hsp70 may be important for sequestering this complex and playing a part in cytokine and chemokine mRNA induction in RSV-infected cells. It was observed earlier that IFN stimulation induced changes in RANTES mRNA stability (12); it should be noted that beta interferon secretion is strongly induced in this model of RSV replication (40). Thus, it is intriguing to speculate that nuclear Hsp70 may be responsible for sequestering poly(A) binding protein-AU factor 1, thereby playing a role in the posttranscriptional component of RANTES gene expression in RSV-infected cells. This hypothesis requires additional study.
Hsp70 expression is highly inducible by a variety of cellular stressors, including heat, abnormal amino acids, intracellular oxidation, and viral infections. For the DNA viruses, herpes simplex virus (HSV), cytomegalovirus (CMV), and adenovirus, the heat shock response is induced in infected cells as a consequence of specific virally encoded proteins. Adenovirus protein E1A (55), the avian adenovirus Gam1 antiapoptotic protein (26), the CMV immediate-early proteins 1 and 2 (13), and HSV ICP0 (42) have all been observed to induce Hsp70 expression. Although the proteins responsible have not yet been identified, enhanced Hsp70 expression has been shown in response to Newcastle disease virus (18) and the morbillivirus canine distemper virus (58). The proteins involved in Hsp70 expression in response to RSV will require further investigation.
The TAR DNA binding protein was previously identified as an RNA-splicing factor that associates with ND10 structures (9, 71). Our identification of increased nuclear TAR DNA binding protein by 2DE analysis led to a surprising finding that ND10 structural proteins are induced by RSV, and the structures themselves apparently dissolve, allowing PML and Sp100 to translocate into the cytoplasm. ND10s are dynamic macromolecular complexes of helicases and transcription factors that are important sites for transcriptional control (51, 54). ND10s are known to be sites for genomic replication of several DNA viruses, including herpesviruses, adenoviruses, and simian virus 40 (reviewed in reference 49). Here, specific proteins encoded by the DNA viruses disrupt ND10 by modifying interactions between the constituent proteins; these viral protein disruptors include the HSV type 1 ICP0 protein (50) and the human CMV immediate-early 1 gene product IE72 (43). In these examples, ND10 disruption is apparently accomplished by interfering with the modification of PML by the small ubiquitin-like modifier, SUMO-1, a posttranslational modification important in forming ND10s (53). Other agents have been observed to disrupt ND10 structures as well, including heat shock and cadmium (54). Heat shock induces a transient, selective loss of Sp100 from ND10s but preserves the association of PML, whereas cadmium induces selective PML dissociation from ND10s (54). In contrast to our findings for RSV infection, where ND10 disruption is associated with cytoplasmic redistribution of PML and Sp100, in these previous studies, the PML and Sp100 released from ND10 structures apparently remain in the nucleus (52, 54).
Our findings are the first to indicate that Paramyxovirus infection affects ND10 structure, even though viral transcription and replication are entirely cytoplasmic. Interestingly, IFN treatment has been found to induce the expression of PML, Sp100, and NDp52, doubling the number and size of ND10s and preventing their subsequent disruption by virus (reviewed in reference 49). Because these proteins are IFN inducible, some have suggested that they may also play a role in antiviral defense. For example, it has been observed that PML expression inhibits vesicular stomatitis virus and influenza virus replication and cytopathic effect (14). It may be possible, therefore, that cytoplasmic translocation of PML and Sp100 to the site of RSV replication is antiviral, serving to limit RSV replication. This question merits further study. Moreover, it is surprising that, despite the induction of many structural ND10 proteins, including the TAR DNA binding protein, Daxx, Sp100, and PML, the ND10s dissolve during the course of RSV infection. Recently, it has been shown that heavy metals induce intranuclear redistribution of ND10s through a p38MAP kinase pathway (54). It has been shown that MAP kinases are rapidly activated by RSV replication (60); perhaps this signaling pathway is also partly responsible for ND10 redistribution in RSV infection.
Finally, it is noteworthy that RSV induces a number of distinct enzymes involved in the maintenance of the redox state. RSV replication induces the formation of reactive oxygen species that play important roles in inflammatory-chemokine expression in A549 cells (10). Our 2DE analysis has identified MnSOD, ERp57/Grp58, and thioredoxin peroxidase as being upregulated by RSV infection. ER57/Grp58 is a mitogen-inducible (thiol-dependent) reductase (35). Thioredoxin peroxidase is an antioxidant enzyme controlling the NF-κB pathway (41), a transcription factor induced by RSV replication (23). These enzymes may play important roles in antioxidant defense in response to RSV-induced reactive oxygen species.
In summary, we have applied both 2DE-MS and microarray approaches to investigate the nuclear responses to cytoplasmic RSV replication in A549 cells. Our findings indicate that cytoplasmic RSV replication induces a coordinated expression of heat shock protein family members and produces a nuclear heat shock response involving the accumulation of the Hsp 70- and 60-kDa isoforms, whose effects on viral replication require further investigation. Although RSV induces expression of PML and Sp100, structural proteins forming the ND10, surprisingly, RSV disrupts the ND10 structures and produces their nuclear-to-cytoplasmic redistribution. Here, cytoplasmic PML-Sp100 may confer an antiviral response. Thus, a high-resolution proteomics discovery approach, combined with cellular imaging and targeted genomic data mining, has led to novel insights into compensatory cellular responses to RSV infection.
Acknowledgments
We thank Michael Oglesbee (Ohio State University) for helpful discussions.
This work was supported by NIAID grant AI40218 (A.R.B.); NHLBI contract N01-HV-28184, Proteomic Technologies in Airway Inflammation (A.K.); NCI grant 1R24CA88317 (A.K.); and NIEHS P30 ES06676, Center in Environmental Toxicology (J. Halpert, University of Texas Medical Branch).
REFERENCES
- 1.Adler, K. B., B. M. Fischer, D. T. Wright, L. A. Cohn, and S. Becker. 1994. Interactions between respiratory epithelial cells and cytokines: relationships to lung inflammation. Ann. N. Y. Acad. Sci. 725:28-145. [DOI] [PubMed] [Google Scholar]
- 2.Aherne, W. T., T. Bird, S. D. B. Court, P. S. Gardner, and J. McQuillin. 1970. Pathological changes in virus infections of the lower respiratory tract in children. J. Clin. Pathol. 23:7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anest, V., J. L. Hanson, P. C. Cogswell, K. A. Steinbrecher, B. D. Strahl, and A. S. Baldwin. 2003. A nucleosomal function for IκB kinase-alpha in NF-κB-dependent gene expression. Nature 423:659-663. [DOI] [PubMed] [Google Scholar]
- 4.Beere, H. M., B. B. Wolf, K. Cain, D. D. Mosser, A. Mahboubi, T. Kuwana, P. Tailor, R. I. Morimoto, G. M. Cohen, and D. R. Green. 2000. Heat shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat. Cell Biol. 2:469-475. [DOI] [PubMed] [Google Scholar]
- 5.Bennett, C. R., Jr., and D. Hamre. 1962. Growth and serological characteristics of respiratory syncytial virus. J. Infect. Dis. 110:8-16. [DOI] [PubMed] [Google Scholar]
- 6.Bitko, V., A. Velazquez, L. Yank, Y.-C. Yang, and S. Barik. 1997. Transcriptional induction of multiple cytokines by human respiratory syncytial virus requires activation of NF-κB and is inhibited by sodium salicylate and aspirin. Virology 232:369-378. [DOI] [PubMed] [Google Scholar]
- 7.Brasier, A. R., M. Jamaluddin, A. Casola, W. Duan, Q. Shen, and R. Garofalo. 1998. A promoter recruitment mechanism for TNFα-induced IL-8 transcription in type II pulmonary epithelial cells: dependence on nuclear abundance of Rel A, NF-κB1 and c-Rel transcription factors. J. Biol. Chem. 273:3551-3561. [DOI] [PubMed] [Google Scholar]
- 8.Brasier, A. R., M. Lu, T. Hai, Y. Lu, and I. Boldogh. 2001. NF-κB inducible BCL-3 expression is an autoregulatory loop controlling nuclear p50/NF-κB1 residence. J. Biol. Chem. 276:32080-32093. [DOI] [PubMed] [Google Scholar]
- 9.Buratti, E., and F. E. Baralle. 2001. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem. 276:36337-36341. [DOI] [PubMed] [Google Scholar]
- 10.Casola, A., N. Burger, T. Liu, M. Jamaluddin, A. R. Brasier, and R. P. Garofalo. 2001. Oxidant tone regulates RANTES gene expression in airway epithelial cells infected with RSV. J. Biol. Chem. 276:19715-19722. [DOI] [PubMed] [Google Scholar]
- 11.Casola, A., R. P. Garofalo, H. Haeberle, T. Elliott, R. Lin, M. Jamaluddin, and A. R. Brasier. 2000. Multiple inducible cis elements control RANTES promoter activation in alveolar epithelial cells infected with RSV. J. Virol. 75:6428-6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casola, A., A. Henderson, T. Liu, R. P. Garofalo, and A. R. Brasier. 2002. Regulation of RANTES promoter activation in alveolar epithelial cells after cytokine stimulation. Am. J. Physiol. 283:L1280-L1290. [DOI] [PubMed] [Google Scholar]
- 13.Caswell, R., L. Bryant, and J. Sinclair. 1996. Human cytomegalovirus immediate-early 2 (IE2) protein can transactivate the human hsp70 promoter by alleviation of Dr1-mediated repression. J. Virol. 70:4028-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chelbi-Alix, M. K., F. Quignon, L. Pelicano, M. H. Koken, and H. de The. 1998. Resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J. Virol. 72:1043-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu, Y. W., R. B. Runyan, R. G. Oshima, and M. J. Hendrix. 1993. Expression of complete keratin filaments in mouse L cells augments cell migration and invasion. Proc. Natl. Acad. Sci. USA 90:4261-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins, P. L. 1991. The molecular biology of the human respiratory syncytial virus (RSV) of the genus Pneumovirus, p. 103-162. In D. W. Kingsbury (ed.), The paramyxoviruses. Plenum, New York, N.Y.
- 17.Collins, P. L., L. E. Dickens, A. Buckler-White, R. A. Olmsted, M. K. Spriggs, E. Camargo, and K. V. W. Coelingh. 1986. Nucleotide sequences for the gene junctions of human respiratory syncytial virus reveal distinctive features of intergenic structure and gene order. Proc. Natl. Acad. Sci. USA 83:4594-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins, P. L., and L. E. Hightower. 1982. Newcastle disease virus stimulates the cellular accumulation of stress (heat shock) mRNAs and proteins. J. Virol. 44:703-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domachowske, J. B., C. A. Bonville, and H. F. Rosenberg. 2000. Cytokeratin 17 is expressed in cells infected with respiratory syncytial virus via NF-κB activation and is associated with the formation of cytopathic syncytia. J. Infect. Dis. 182:1022-1028. [DOI] [PubMed] [Google Scholar]
- 20.Easton, A. J., J. B. Domachowske, and H. F. Rosenberg. 2004. Animal pneumoviruses: molecular genetics and pathogenesis. Clin. Microbiol. Rev. 17:390-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferris, J. A., W. A. Aherne, and W. S. Locke. 1973. Sudden and unexpected deaths to infants: histology and virology. Br. Med. J. 2:439-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiedler, M. A., and K. Wernke-Dollries. 1999. Incomplete regulation of NF-κB by IκBa during respiratory syncytial virus infection in A549 cells. J. Virol. 73:4502-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garofalo, R., M. Sabry, M. Jamaluddin, R. K. Yu, A. Casola, P. L. Ogra, and A. R. Brasier. 1996. Transcriptional activation of the interleukin-8 gene by RSV infection in alveolar epithelial cells: nuclear translocation of the RelA transcription factor as a mechanism producing airway mucosal inflammation. J. Virol. 70:8773-8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrido, C., S. Gurbuxani, L. Ravagnan, and G. Kroemer. 2001. Heat shock proteins: endogenous modulators of apoptotic cell death. Biochem. Biophys. Res. Commun. 286:433-442. [DOI] [PubMed] [Google Scholar]
- 25.Glezen, W. P., L. H. Taber, and A. L. Frank. 1986. Risk of primary infection and reinfection with respiratory syncytial virus. Am. J. Dis. Child. 140:543-546. [DOI] [PubMed] [Google Scholar]
- 26.Glotzer, J. B., M. Saltik, S. Chiocca, A.-I. Michou, P. Moseley, and M. Cotten. 2000. Activation of heat-shock response by an adenovirus is essential for virus replication. Nature 407:207-211. [DOI] [PubMed] [Google Scholar]
- 27.Gorg, A., C. Obermaier, G. Boguth, A. Harder, B. Scheibe, R. Wildgruber, and W. Weiss. 2000. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21:1037-1053. [DOI] [PubMed] [Google Scholar]
- 28.Groothuis, J. R., K. M. Gutierre, and B. A. Lauer. 1988. Respiratory syncytial virus infection in children with bronchopulmonary dysplasia. Pediatrics 82:199-203. [PubMed] [Google Scholar]
- 29.Hall, C. B. 2001. Respiratory syncytial virus and parainfluenza virus. N. Engl. J. Med. 344:1917-1928. [DOI] [PubMed] [Google Scholar]
- 30.Hall, C. B., and C. A. McCarthy. 1995. Respiratory syncytial virus, p. 1501. In G. L. Mandel, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases. Churchill Livingston, New York, N.Y.
- 31.Han, Y., and A. R. Brasier. 1997. Mechanism for biphasic Rel A:NF-κB1 nuclear translocation in tumor necrosis factor α-stimulated hepatocytes. J. Biol. Chem. 272:9823-9830. [DOI] [PubMed] [Google Scholar]
- 32.Han, Y., S. A. Weinman, S. Boldogh, and A. R. Brasier. 1999. TNFα-inducible IκBα proteolysis and NF-κB activation mediated by cytosolic m-Calpain. J. Biol. Chem. 274:787-794. [DOI] [PubMed] [Google Scholar]
- 33.Hartl, F.-U., and M. Hayer-Hartl. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852-1858. [DOI] [PubMed] [Google Scholar]
- 34.Heikknen, T., M. Thint, and T. Chonmaitree. 1999. Prevalence of various respiratory viruses in the middle ear during acute otitis media. N. Engl. J. Med. 340:260-264. [DOI] [PubMed] [Google Scholar]
- 35.Hirano, N., F. Shibasaki, R. Sakai, T. Tanaka, J. Nishida, Y. Yazaki, T. Takenawa, and H. Hirai. 1995. Molecular cloning of the human glucose-regulated protein ERp57/GRP58, a thiol-dependent reductase. Identification of its secretory form and inducible expression by the oncogenic transformation. Eur. J. Biochem. 234:336-342. [DOI] [PubMed] [Google Scholar]
- 36.Huang, Y. T., R. R. Romito, B. P. De, and A. K. Banerjee. 1993. Characterization of the in vitro system for the synthesis of mRNA from human respiratory syncytial virus. Virology 193:862-867. [DOI] [PubMed] [Google Scholar]
- 37.Jamaluddin, M., A. Casola, R. P. Garofalo, Y. Han, T. Elliott, P. L. Ogra, and A. R. Brasier. 1998. The major component of IκBa proteolysis occurs independently of the proteasome pathway in respiratory syncytial virus-infected pulmonary epithelial cells. J. Virol. 72:4849-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jamaluddin, M., R. Garofalo, P. L. Ogra, and A. R. Brasier. 1996. Inducible translational regulation of the NF-IL6 transcription factor by respiratory syncytial virus infection in pulmonary epithelial cells. J. Virol. 70:1554-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jamaluddin, M., T. Meng, J. Sun, I. Boldogh, Y. Han, and A. R. Brasier. 2000. Angiotensin II induces nuclear factor (NF)-κB1 isoforms to bind the angiotensinogen gene acute-phase response element: a stimulus-specific pathway for NF-κB activation. Mol. Endocrinol. 14:99-113. [DOI] [PubMed] [Google Scholar]
- 40.Jamaluddin, M., S. Wang, R. Garofalo, T. Elliott, A. Casola, S. Baron, and A. R. Brasier. 2001. INFβ mediates coordinate expression of antigen-processing genes in RSV infected pulmonary epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 280:L248-L257. [DOI] [PubMed] [Google Scholar]
- 41.Jin, D.-Y., H. Z. Chae, S. G. Rhee, and K.-T. Jeang. 1997. Regulatory role for a novel human thioredoxin peroxidase in NF-κB activation. J. Biol. Chem. 272:30952-30961. [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi, K., E. Ohgitani, Y. Tanaka, M. Kita, and J. Imanishi. 1994. Herpes simplex virus-induced expression of 70 kDa heat shock protein (HSP70) requires early protein synthesis but not viral DNA replication. Microbiol. Immunol. 38:321-325. [DOI] [PubMed] [Google Scholar]
- 43.Korioth, F., G. G. Maul, B. Plachter, T. Stamminger, and J. Frey. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 229:155-158. [DOI] [PubMed] [Google Scholar]
- 44.Laroia, G., R. Cuesta, G. Brewer, and R. J. Schneider. 1999. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284:499-502. [DOI] [PubMed] [Google Scholar]
- 45.Liu, T., S. Castro, A. R. Brasier, M. Jamaluddin, R. P. Garofalo, and A. Casola. 2004. Reactive oxygen species mediate virus-induced STAT activation: role of tyrosine phosphatases. J. Biol. Chem. 279:2461-2469. [DOI] [PubMed] [Google Scholar]
- 46.Long, C. E., J. T. McBride, and C. B. Hall. 1995. Sequelae of respiratory syncytial virus infections: a role for intervention studies. Am. J. Respir. Crit. Care Med. 151:1678-1681. [DOI] [PubMed] [Google Scholar]
- 47.Lopez, M. F., K. Berggren, E. Chernokalskaya, A. Lazarev, M. Robinson, and W. F. Patton. 2000. A comparison of silver stain and SYPRO Ruby protein gel stain with respect to protein detection in two dimensional gels and identification by peptide mass profiling. Electrophoresis 21:3673-3683. [DOI] [PubMed] [Google Scholar]
- 48.MacDonald, N. E., C. B. Hall, and S. C. Suffin. 1982. Respiratory syncytial viral infection in infants with congenital heart disease. N. Engl. J. Med. 307:397-400. [DOI] [PubMed] [Google Scholar]
- 49.Maul, G. G. 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20:660-667. [DOI] [PubMed] [Google Scholar]
- 50.Maul, G. G., H. H. Guldner, and J. G. Spivak. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J. Gen. Virol. 74:2679-2690. [DOI] [PubMed] [Google Scholar]
- 51.Maul, G. G., D. Negorev, P. Bell, and A. M. Ishov. 2000. Properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol. 129:278-287. [DOI] [PubMed] [Google Scholar]
- 52.Maul, G. G., E. Yu, A. M. Ishaov, and A. L. Epstein. 1995. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J. Cell. Biochem. 59:498-513. [DOI] [PubMed] [Google Scholar]
- 53.Muller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nefkens, I., D. G. Negorev, A. M. Ishov, J. S. Michaelson, E. T. H. Yeh, R. M. Tanguay, W. E. G. Muller, and G. G. Maul. 2003. Heat shock and Cd exposure regulate PML and Daxx release from ND10 by independent mechanisms that modify the induction of heat shock proteins 70 and 25 differently. J. Cell Sci. 116:513-524. [DOI] [PubMed] [Google Scholar]
- 55.Nevins, J. R. 1982. Induction of the synthesis of a 70 kDa mammalian heat shock protein by the adenovirus E1A gene product. Cell 29:913-919. [DOI] [PubMed] [Google Scholar]
- 56.Nishihara, J. C., and K. M. Champion. 2002. Quantitative evaluation of proteins in one- and two-dimensional polyacrylamide gels using a fluorescent stain. Electrophoresis 23:2203-2215. [DOI] [PubMed] [Google Scholar]
- 57.Nollen, A. A. A., and R. Morimoto. 2002. Chaperoning signaling pathways: molecular chaperones as stress-sensing “heat shock” proteins. J. Cell Sci. 115:2809-2816. [DOI] [PubMed] [Google Scholar]
- 58.Oglesbee, M., and S. Krakowka. 1993. Cellular stress response induces selective intranuclear trafficking and accumulation of morbillivirus major core protein. Lab. Investig. 68:109-117. [PubMed] [Google Scholar]
- 59.Park, H. S., S. G. Cho, C. K. Kim, H. S. Hwang, K. T. Noh, M. S. Kim, S. H. Huh, M. J. Kim, K. Ryoo, E. K. Kim, W. J. Kang, J. S. Lee, J. S. Seo, Y. G. Ko, S. Kim, and E. J. Choi. 2002. Heat shock protein Hsp72 is a negative regulator of apoptosis signal-regulating kinase 1. Mol. Cell. Biol. 22:7721-7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pazdrak, K., B. Olszewska-Pazdrak, T. Liu, R. Takizawa, A. R. Brasier, R. P. Garofalo, and A. Casola. 2002. MAP kinase activation is involvement in post-transcriptional regulation of RSV-induced RANTES gene expression. Am. J. Physiol. Lung Cell Mol. Physiol. 283:L364-L372. [DOI] [PubMed] [Google Scholar]
- 61.Ravagnan, L., S. Gurbuxani, S. A. Susin, C. Maisse, E. Daugas, N. Zamzami, T. Mak, M. Jaattela, J. M. Penninger, C. Garrido, and G. Kroemer. 2001. Heat-shock protein 70 antagonizes apoptosis-inducing factor. Nat. Cell Biol. 3:839-843. [DOI] [PubMed] [Google Scholar]
- 62.Ray, S., Y. Lu, W. C. Gustafsen, A. P. Fields, and A. R. Brasier. 2004. Genomic mechanisms of p210BCR-ABL signaling: transcriptional induction of heat shock protein 70 through the GATA response element. J. Biol. Chem. 279:35604-35615. [Online.] [DOI] [PubMed] [Google Scholar]
- 63.Saleh, A., S. M. Srinivasula, L. Balkir, P. D. Robbinds, and E. S. Alnemri. 2000. Negative regulation of the Apaf-1 apoptosome by Hsp70. Nat. Cell Biol. 2:476-483. [DOI] [PubMed] [Google Scholar]
- 64.Schlage, W. K., H. Bulles, D. Friedrichs, M. Kuhn, and A. Teredesai. 1998. Cytokeratin expression patterns in the rat respiratory tract as markers of epithelial differentiation in inhalational toxicology. I. Determination of normal cytokeratin expression patterns in the nose, larynx, trachea, and lung. Toxicol. Pathol. 26:324-343. [DOI] [PubMed] [Google Scholar]
- 65.Shay, D. K., R. C. Holman, R. D. Newman, L. L. Liu, J. W. Stout, and L. J. Anderson. 1999. Bronchiolitis-associated hospitalizations among US children, 1980-1996. J. Am. Med. Assoc. 282:1440-1446. [DOI] [PubMed] [Google Scholar]
- 66.Shay, D. K., R. C. Holman, G. E. Roosevelt, M. J. Clarke, and L. J. Anderson. 2001. Bronchiolitis-associated mortality and estimates of respiratory syncytial virus-associated deaths among US children, 1979-1997. J. Infect. Dis. 183:16-22. [DOI] [PubMed] [Google Scholar]
- 67.Stosiek, P., M. Kasper, and R. Moll. 1992. Changes in cytokeratin expression accompany squamous metaplasia of the human respiratory epithelium. Virchows Archiv. 421:133-141. [DOI] [PubMed] [Google Scholar]
- 68.Tian, B., Y. Zhang, B. A. Luxon, R. P. Garofalo, A. Casola, M. Sinha, and A. R. Brasier. 2002. Identification Of NF-κB-dependent gene networks in respiratory syncytial virus-infected cells. J. Virol. 76:6800-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ueba, O. 1978. Respiratory syncytial virus. I. concentration and purification of the infectious virus. Acta Med. Okayama 32:265-272. [PubMed] [Google Scholar]
- 70.Vasconcelos, D., E. Norrby, and M. Oglesbee. 1998. The cellular stress response increases measles virus-induced cytopathic effect. J. Gen. Virol. 79:1769-1773. [DOI] [PubMed] [Google Scholar]
- 71.Wang, I.-F., N. M. Reddy, and C. K. J. Shen. 2002. Higher order arrangement of the eukaryotic nuclear bodies. Proc. Natl. Acad. Sci. USA 99:13583-13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, L., M. E. Peeples, R. C. Boucher, P. L. Collins, and R. J. Pickles. 2002. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J. Virol. 76:5654-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang, W., and B. T. Chait. 2000. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal. Chem. 72:2482-2489. [DOI] [PubMed] [Google Scholar]
- 74.Zhang, Y., B. A. Luxon, A. Casola, R. P. Garofalo, M. Jamaluddin, and A. R. Brasier. 2001. Expression of RSV-induced chemokine gene networks in lower airway epithelial cells revealed by cDNA microarrays. J. Virol. 75:9044-9058. [DOI] [PMC free article] [PubMed] [Google Scholar]