Abstract
The chromosomal features that influence retroviral integration site selection are not well understood. Here, we report the mapping of 226 avian sarcoma virus (ASV) integration sites in the human genome. The results show that the sites are distributed over all chromosomes, and no global bias for integration site selection was detected. However, RNA polymerase II transcription units (protein-encoding genes) appear to be favored targets of ASV integration. The integration frequency within genes is similar to that previously described for murine leukemia virus but distinct from the higher frequency observed with human immunodeficiency virus type 1. We found no evidence for preferred ASV integration sites over the length of genes and immediate flanking regions. Microarray analysis of uninfected HeLa cells revealed that the expression levels of ASV target genes were similar to the median level for all genes represented in the array. Although expressed genes were targets for integration, we found no preference for integration into highly expressed genes. Our results provide a more detailed description of the chromosomal features that may influence ASV integration and support the idea that distinct, virus-specific mechanisms mediate integration site selection. Such differences may be relevant to viral pathogenesis and provide utility in retroviral vector design.
Retroviral DNA integration is catalyzed by a viral enzyme, integrase (IN), which nicks the two ends of linear viral DNA and splices them into a site in the host DNA (9, 10). This highly orchestrated reaction produces DNA sequence signatures at the virus-host junctions: the loss of usually 2 bp at the ends of the linear viral DNA and duplication of several base pairs of host DNA at the integration site. However, no gross rearrangements or deletions of either the viral or host DNAs are incurred. The integration reaction can be reproduced in vitro using purified IN and viral and target DNA model substrates. Despite the precision of the integration reaction, there are no strict host DNA sequence requirements. Nevertheless, numerous studies indicate that integration site selection is not likely to be entirely random. For example, various features of host chromosomes have been implicated in influencing integration site selection, including primary DNA sequence (7, 14, 18), DNA structure (16, 20, 24), nucleosome structure (27-31), chromatin structure (32, 36, 37, 40), and transcriptional activity (23, 33, 34, 41, 43). In addition to the passive influences of chromosomal structure, it has been suggested that retroviral integration could be actively targeted by tethering to specific, chromatin-bound host factors (5). Lastly, the IN proteins from different retroviruses produce unique in vitro integration patterns in naked DNA targets (18). These intriguing but disparate observations have not yet led to a unifying model, and the mechanisms that govern integration site selection in vivo remain obscure.
In an infected cell, retroviral DNA is organized in a preintegration complex that includes IN, as well as other viral and host components (2-4, 22). Such complexes can be isolated from infected cells, and their integration activity can be measured on naked DNA targets in vitro. However, in vivo, interactions between host chromatin and the preintegration complex likely influence site selection. The basic unit of chromatin, the nucleosome, is further assembled into chromatin fibers and more highly organized regions (“open” euchromatin and “closed” constitutive or facultative heterochromatin). Such regions are subject to dynamic structural changes during chromosome condensation, transcription, and DNA replication.
A series of studies have addressed the fundamental question of how nucleosomes might influence the retroviral integration reaction. Rather than blocking this process, wrapping of the host DNA into nucleosomes appears to distort the DNA in a way that promotes periodic integration events within the nucleosome, both in vitro and in vivo (27-32). Recognition of distorted DNA may be a fundamental feature of the IN catalytic mechanism (15, 35). Although wrapping of DNA around individual nucleosomes may enhance integration, it is unknown if the preintegration complex has free access to host DNA within higher-order chromatin or if such access depends on other factors.
Although the precise mechanisms of site selection remain elusive, recent discoveries in genomics and transcriptional profiling have provided a wealth of both technical and informational resources for studying integration site selection on a genomic scale. Use of such resources is beginning to reveal how the diverse physical and functional features of host cell chromosomes may influence integration site selection (34, 43). The human genome includes approximately 25,000 genes, comprising about 33% of all DNA sequences, and is organized into gene-dense and gene-sparse regions. A large subset of human genes (>20,000) have been more precisely defined by comparing the genomic DNA sequence with expressed RNAs (the RefSeq genes) (25, 26). Further analysis of the human transcriptome has revealed highly and weakly expressed clusters of genes, so-called RIDGEs and anti-RIDGEs, respectively (6, 39). Thus, identification of nucleotide positions of integration sites in human DNA allows their immediate characterization with respect to gene organization and provides information about possible transcriptional activity of the target site at the time of integration. In some cases, the results of such analyses can also provide information concerning the likely biochemical status of the integration site in terms of euchromatic versus heterochromatic structure (7).
Knowledge of the determinants of retroviral integration site selection is critical to understanding the early steps in retroviral replication and viral pathogenesis and may also be relevant to the design of safer retroviral vectors. The advantage of this class of vectors is that integration into the host cell genome is a normal step in the replication cycle. However, a major limitation is the possibility that integration may disrupt normal cellular functions. For example, a well-recognized mechanism for retroviral oncogenesis is the activation of a cellular proto-oncogene by a nearby integrated retroviral long terminal repeat (LTR) (promoter insertion oncogenesis) (13). This limitation was made apparent recently when 2 of 11 patients being treated for X-linked severe combined immunodeficiency disease with murine leukemia virus (MLV)-based gene therapy vectors developed leukemia induced by promoter insertion near the growth-promoting gene LMO2 (8, 11). Although these adverse events may be driven mainly by accumulation of sufficient integrations for subsequent selection, an understanding of the mechanism of integration site selection might prove useful for future vector design.
Results of recent studies have suggested that there are differences in site selection by MLV and human immunodeficiency virus type 1 (HIV-1). These findings may have relevance to both virus biology and vector design (5, 34, 43). We recently observed that, in vitro, the efficiency of avian sarcoma virus (ASV) IN-mediated integration is increased when a nucleosome-packaged DNA substrate is compacted. HIV-1 IN-mediated integration is inhibited under the same conditions (37). We also have found that an ASV vector is susceptible to chromatin-mediated silencing in HeLa cells, while a matched HIV-1 vector is highly resistant (R. A. Katz et al., unpublished data). These results support the idea that there are fundamental differences in retrovirus-host genome interactions and have led us to investigate ASV integration site selection in vivo. Here we report results from mapping and analyzing over 200 ASV integration sites in the human genome.
MATERIALS AND METHODS
Sequencing of host-virus DNA junctions.
Human cells (HeLa) were infected with the amphotropic ASV vector (RCASACMVEGFP) (17), which was produced in DF-1 chicken cells. Supernatant from virus-producing cells was passed through a 0.45-μm-pore-size filter and added to HeLa cells with fresh growth medium and DEAE-dextran (final concentration, 10 μg/ml). The avian retroviral DNA can be integrated in mammalian cells, but they cannot support productive replication, thus limiting the infection to one round. Infected-cell DNA was harvested 48 h postinfection. The PCR-based strategy for cloning virus-host junctions was similar to that described previously (43). The concentration of isolated DNA was estimated by UV absorbance, and 2.5 μg was digested with AluI, BstEII, and EcoRI endonucleases. AluI cleaves the human genomic DNA frequently and was used to generate host-virus junction fragments (downstream LTR-host). BstEII cleaves the viral DNA just 3′ of the upstream LTR, thus suppressing internal viral amplifications. EcoRI cleavage suppressed amplification from circular viral DNA forms. Resulting DNA fragments were ligated to the AluI adaptor provided with the Universal Genome Walker kit (lontech) according to the manufacturer's recommendations. PCR was performed with one primer specific to the LTR and another to the adaptor.
The reaction mix (50 μl) contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 250 μM deoxynucleoside triphosphates, and 0.3 μl of AmpliTaq DNA polymerase (Applied Biosystems). Reactions were cycled as follows: 30 cycles of 94°C for 40 s, 68°C for 30 s, and 72°C for 45 s in a MultiCycler PTC 225 Tetrad (MJ Research). PCR products were diluted 50-fold and then subjected to nested PCR performed under the same conditions. Amplification products were isolated after agarose gel electrophoresis and cloned in the pCR4-TOPO plasmid, which was used to transform Escherichia coli TOP10 competent cells with the use of the TOPO TA cloning kit for sequencing (Invitrogen). Inserts from isolated clones were amplified by PCR with M13 primers complementary to the pCR4-TOPO plasmid, and the resultant products were purified and sequenced. Primers used for this study were: ASV 3′ LTR, 5′-ACC TGG GTT GAT GGC CGG ACC GTT GAT T-3′; adaptor primer, 5′-GTA ATA CGA CTC ACT ATA GGG C-3′; ASV 3′ LTR nested primer, 5′-CCT GAC GAC TAC GAG CAC CTG CAT GAA G-3′; adaptor nested primer, 5′-ACT ATA GGG CAC GCG TGG T-3′; M13 forward primer, 5′-AAA CGA CGG CCA G-3′; M13 reverse primer, 5′-CAG GAA ACA GCT ATG AC-3′.
Mapping integration sites.
To map integration site sequences to the human genome, we used the BLAT program (University of California, Santa Cruz, Human Genome Project Working Draft July 2003 Freeze; http://www.genome.ucsc.edu/cgi-bin/hgBlat).
An integration site was considered to be authentic only if it contained both the downstream LTR and adaptor sequence, matched the genomic location after the end of the downstream LTR (…CA), represented ≥95% identity with the genomic sequence, and matched no more than one genomic region with ≥95% identity. Integration was judged to have occurred in a gene only if it was located within the boundaries of one of the RefSeq genes (National Center for Biotechnology Information Reference Sequence Project). These sequences were designated as genes on the basis of human mRNAs and their translation products, rather than gene prediction programs. We will refer to genes that have incurred an integration event as “target genes.”
Preparation of probes for DNA microarrays.
Total HeLa cell RNA was isolated with the Trizol reagent (Invitrogen) and purified with the RNeasy kit (Qiagen). For the preparation of cDNA probes, 25 μg of total RNA was reverse transcribed in the presence of oligo(dT)12-18 and aminoallyl-dUTP. The cDNAs were labeled with N-hydroxysuccinimide ester linked to either Cy3 or Cy5 dye, respectively (Amersham Biosciences). The labeled probes were purified using the QIAquick PCR purification kit (Qiagen).
DNA microarrays.
The DNA microarray chips were prepared at the Fox Chase Cancer Center Microarray Facility with a set of 15,552 human oligodeoxynucleotides (MWG Biotech). The DNA was spotted onto polylysine-coated glass slides by using the Omnigrid arrayer (Analytical Instruments). The processing of the slides and the hybridization reaction were performed essentially as described elsewhere (http://cmgm.stanford.edu/pbrown/mguide/).
Microarray analysis.
To examine the level of relative expression of ASV target genes, we used two data sets obtained with HeLa cells (H55 and H56). Genes with intensities greater than 2 standard deviations above the background were considered expressed. This assignment of threshold values for expressed versus nonexpressed genes is arbitrary. For analyses of these data sets, we set this threshold value low, resulting in scoring a majority of genes as expressed. This approach appeared to be most appropriate for examining the relationship between the level of gene expression and integration site selection. The H55 data set contained 13,750 expressed genes out of the total of 15,552, with a range of expression levels from 81 to 61,000 arbitrary units. The H56 data set contained 11,251 expressed genes, with a range of expression values from 83 to 65,000 arbitrary units. Of the 50 ASV target genes represented in the library, 2 and 11 fell in the nonexpressed category for the H55 and H56 data sets, respectively.
We also analyzed two public (GEO) HeLa cDNA array data sets (GSM2145 and GSM2177; http://www.ncbi.nlm.nih.gov/geo/). In these data sets, the background and background standard deviation levels were not available and thus we were unable to analyze these in a way similar to our own data sets. The platform for the GSM2145 and GSM2177 data sets used 9,985 genes, and 34 ASV target genes were represented. The ranges of expression levels for the GSM2145 and GSM2177 data sets were 123 to 50,000 and 104 to 34,000 arbitrary units, respectively.
Median expression values were obtained for target genes versus all genes. For this analysis, all four data sets were used (GSM2145, GSM2177, H55, and H56). The median expression levels of target genes and all genes were expressed as a ratio of target genes to all genes for each pair of data sets.
Nucleotide sequence accession numbers.
The GenBank accession numbers for 226 integration sites are AY653309 through AY653534.
RESULTS
ASV integration sites are distributed broadly in the human genome.
We used an ASV vector pseudotyped with the murine amphotropic envelope protein to allow one-step infection of HeLa cells (1, 17). Cells were isolated 48 h postinfection to minimize selective forces that might bias the analysis of integration site selection. Cellular genomic DNA was isolated, and virus-host DNA junctions were amplified by a linker-mediated PCR method (43). The PCR products were cloned and sequenced, and the locations of the integration sites were determined using the human DNA sequence (19, 38). A total of 226 integration sites were analyzed. Mapping of these sites revealed that integration events were distributed over all 23 human chromosomes (Fig. 1). The number of integration events per chromosome was generally proportional to chromosome size (Fig. 1 legend), validating our methods and suggesting that there were no global restrictions for integration site selection. Furthermore, we found no evidence for integration hotspots as observed for HIV-1, by using the criteria described previously (34).
FIG. 1.
Distribution of ASV integration events in human chromosomes. Results are plotted as the percentage of integration events in each chromosome (n = 226). The parameters used to calculate the relative target size of each chromosome included the estimated size (19, 38) as well as corrections for differential chromosome aneuploidies, minute chromosomes, and translocations. For these corrections we used the HeLa cell karyotype and chromosome composition analyses described by Macville et al. (21). The hypothesis that the number of integration sites is proportional to chromosome length could not be rejected (χ2 = 27.6, 22 df, P = 0.194).
Genes are favored targets for ASV integration: comparison with MLV and HIV-1 results.
We next determined the frequency with which integrations occurred into RNA polymerase II genes (protein-encoding genes), by using human RefSeq genes (25, 26) as a criterion. (We refer to genes that have incurred an integration event as target genes.) The results showed that 95 of 226 (42%) integration events occurred in the defined RefSeq gene set (n = 21,404) (Table 1). The use of the RefSeq gene set allowed us to compare our results with those reported by Wu et al. (43), who also used RefSeq genes to map MLV (and HIV-1) integration sites in HeLa cells. The percentage that we observed (42%) was somewhat higher than the published value for MLV (34.2%) (43). However, the RefSeq gene assembly that we used (July 2003) is slightly larger than the assembly used in the study by Wu et al. (43) (November 2002). We therefore calculated extrapolated values that would normalize the two studies, so that a more accurate comparison could be made (Table 1). With this correction, the percentages were quite similar (42 versus 40.2%), and both values were significantly higher than the value determined for random integration (26.3%) (Table 1). We also recalculated our values using the November 2002 RefSeq gene assembly, yielding a value of 39.8% integrations into genes for ASV. This value can be directly compared to the 34.2% value reported for MLV (P = 0.134) and the 22.4% value for random integration (P = 3.1 × 10−9) (Table 1). By these criteria, we conclude that the observed 42% integration into RefSeq genes with the ASV vector is greater than that predicted for random integration (Table 1) and that the value is similar to that observed with MLV.
TABLE 1.
Percentage of ASV integration events into RefSeq genes: comparison with HIV-1 and MLV studiesa
| Virus | Total no. of integrations | Cell type | % Integration into RefSeq genesb
|
|
|---|---|---|---|---|
| N02 | J03 | |||
| ASV | 226 | HeLa | 39.8e | 42.0e |
| HIV-1c | 524 | SupT1 | 61.0 | (71.7) |
| HIV-1d | 379 | H9/HeLa | 57.8f | (67.9) |
| HIV-1d | 135 | HeLa | 50.0 | (58.8) |
| MLVd | 903 | HeLa | 34.2g | (40.2) |
| Randomd | 10,000 | Simulated | 22.4 | (26.3) |
Results from the present study are shown in boldface.
N02, November 2002 assembly of RefSeq genes (18,214 genes); J03, July 2003 assembly of RefSeq genes (21,404 genes). Values in parentheses indicate extrapolated proportional increase in RefSeq targets by using the July 2003 assembly versus the November 2002 assembly used by Wu et al. (43).
Raw data are from the work of Schröder et al. (34) and were reevaluated by Wu et al. (43) using RefSeq genes. Values were compared with ASV by using the 2 × 2 chi-square test with the Yates correction (P ≤ 1.3 × 10−7).
From the work of Wu et al. (43).
Values are significantly different from random integration (P ≤ 2.13 × 10−7) with the 2 × 2 chi-square test with the Yates correction.
Value was reported to be significantly different from random integration (43).
Value was reported to be significantly different from random integration and HIV-1 value of 57.8 (43). Value is indistinguishable from ASV value (39.8) (P = 0.134), by the 2 × 2 chi-square test with the Yates correction.
A concern in determining the absolute frequencies of integration events in genes is that the percentage of genes in HeLa cells may not correspond to the percentage represented in the human genome sequence, due to gene amplifications or deletions. To address this issue, we have calculated the number of genes and total size of the HeLa cell genome size by using the detailed karyotype (21) and the NCBI Map Viewer. We found that the ratio between the genome size and the gene number in HeLa cells is equivalent to that of the human genome.
Our conclusion that ASV integration favors genes is also highly dependent on the frequency measured for random integration into RefSeq genes. As mentioned previously, this frequency was experimentally determined using computer-simulated integration events (42). In an attempt to confirm this value, we used a second computational method for determining the frequency of random integration. We calculated the percentage of RefSeq gene sequences in the total human genome sequence; however, this calculation yielded a higher value for random RefSeq integration than was reported. We note that our calculation is subject to biases including, but not limited to, an underestimate of the total human genome size (e.g., due to estimates of unsequenced gap sizes) and the relatively high percentage of overlapping gene sequences that were not taken into account. We therefore rejected this calculation, but note that our results regarding the apparent preference for ASV integration into genes should be viewed with caution, as we have not calculated a random integration value independently. Despite these uncertainties with respect to random integration, we can clearly identify statistically distinct similarities and differences with MLV integration site selection (Table 1) and HIV-1 selection (as described below).
Two previous studies have indicated that genes are highly favored for HIV-1 integration. In the first high-throughput study of HIV-1 integration site selection, Schröder et al. (34) reported that 69% of HIV-1 integration events occurred within genes (in SupT1 cells), compared to an experimental in vitro control for random integration (35%). In this study, the “gene” designation was not limited to RefSeq sequences, and therefore we cannot directly compare these findings with our ASV results. However, Wu et al. (43) reevaluated the results of Schröder et al. (34) and determined that 61% of the HIV-1 integrations were located in RefSeq genes (Table 1). These authors also surveyed integration sites in HeLa cells, independently, by using an HIV-1 vector and found that 50% of integrations were in genes (43) (Table 1). Taken together, these earlier studies and our own suggest that, although all three viruses favor integration into genes, HIV-1 integration displays a much stronger preference for such sequences than does either ASV or MLV.
Potential functional ramifications of integration into genes.
Previous cell sorting experiments indicated that expression of the human cytomegalovirus immediate-early promoter-driven green fluorescent protein (GFP) reporter in our ASV vector was subject to rapid chromatin-based, epigenetic silencing in a large subset of infected HeLa cells (Katz et al., unpublished). One interpretation of these observations is that integration within genes might promote GFP expression, as such regions (e.g., euchromatin) are accessible to transcription factors. Correspondingly, integration into heterochromatic, nongenic regions might promote silencing. We therefore prepared a second integration site library from cells that had been sorted for high expression of GFP (data not shown). Analysis of a small data set of 22 integration events showed that 36% were within genes in this population; this value is comparable to the 42% observed with unsorted cells. Although limited, this analysis suggests that high ASV reporter gene expression is not correlated with integration into gene sequences.
Distribution of integration sites with respect to gene structure.
The results reported previously with MLV indicated that transcriptional start sites were favored for integration by this virus, with 20.2% occurring within a window of ±5 kb. With ASV, we also found a statistically significant bias for integrations in this window (8.4%) compared to the calculated random events (2.1%) (P = 0.0044) (Table 2). A similar value (10.8%) was also reported for HIV-1 (Table 2) (43). However, when we compared the distribution of ASV integration sites along the length of genes, including the 5-kb upstream and downstream windows, we observed no bias for transcriptional start sites (Fig. 2). As ASV does not appear to favor transcriptional start regions, we conclude therefore that ASV and MLV integration site selection preferences are distinct in HeLa cells.
TABLE 2.
Percentage of ASV integration events with respect to RefSeq gene structure
| Location | % of integrationsa
|
|||
|---|---|---|---|---|
| ASV | MLVb | HIV-1b | Randomb | |
| 5 kb upstream of genes | 4.4c | 11.2 | 2.9 | 2.1 (2.5)d |
| ±5 kb from transcription start | 8.4c | 20.2e | 10.8e | 4.3 (5.1)d |
| ±1 kb from CpG islands | 3.1f | 16.8e | 2.1 | 2.1 |
Values for ASV and MLV were found to be significantly different (P ≤ 0.0034).
Data from the work of Wu et al. (43).
Distinguishable from random integration (P ≤ 0.02) by using one-sided test of the binomial proportion.
Values in parentheses indicate expected proportional increase in RefSeq targets by using the July 2003 assembly versus the November 2002 assembly used by Wu et al. (43).
Distinguishable from random integration, as described by Wu et al. (43).
Value is indistinguishable from random value (P = 0.869) by one-sided test of the binomial proportion.
FIG. 2.
Analysis of ASV integration sites with respect to target transcription unit (RefSeq gene) length and flanking regions. Target gene lengths were divided into eight equal bins, and the percentage of integration events in each bin was determined. For upstream (us) and downstream (ds) regions, fixed 5-kb windows were used. These criteria were as described by Wu et al. (43) for MLV, allowing direct comparisons of the results. The hypothesis that all designated regions were equally likely to contain integration events could not be rejected (P ≥ 0.498) by the chi-square test.
As CpG islands are frequently associated with transcription start sites, we determined the percentage of integrations within 1 kb of these elements. In contrast to the 16.8% observed with MLV, only 3.1% of the integration events occurred in this window, and this value is close to that predicted for random integration (Table 2) (43). However, our sample size may be too small to distinguish the difference from a random value (P = 0.869). We also determined the frequency of ASV integrations into various features of human chromosomes, including satellite repeat elements, retrotransposons, and endogenous retroviruses (Table 3). The results showed that the relative number of integrations generally paralleled the target size of each element. We found that satellite DNA was a less favored target, as was observed for HIV-1 (7, 34).
TABLE 3.
Percentage of ASV integration events into sequence features of human DNA
| DNA featured | % of ASV integrationa | % in human genomeb |
|---|---|---|
| SINE | ||
| Alu | 11.5 | 10.6 |
| MIR | 5.3 | 2.2 |
| LINE | 16.4 | 20.0 |
| LTR | 8.4 | 8.3 |
| DNA repeat element | 4.9 | 2.8 |
| tRNA | 0.4 | NDc |
| Satellite | None | ND |
All values (except MIR) are indistinguishable from the percentage in human genome (P ≥ 0.0551) by two-sided test of the binomial proportion.
From Schröder et al. (34).
ND, not determined.
Abbreviations: SINE, short interspersed element; LINE, long interspersed element; MIR, mammalian interspersed repeat.
Relationship between integration site selection and transcriptional activity of the target genes.
Because the open chromatin state of active genes (during access by transcription factors or engagement with actively transcribing RNA polymerases) may also allow access by the viral preintegration complexes, a simple hypothesis is that transcriptionally active genes may be preferred target sites for integration. Having observed that ASV DNA integration occurred with a higher-than-expected frequency within genes, we asked if integration could be correlated with transcriptional activity at the time of infection. The ASV target genes that we identified included housekeeping-type genes, as well as tissue-specific genes not expected to be expressed preferentially in HeLa cells.
To examine the expression of target genes in detail, we analyzed the transcriptional activity of uninfected HeLa cells by microarray analysis. Of the 95 target genes, 50 were represented in the test array. HeLa gene expression levels were collected into groups based on increments of 500 arbitrary units, and the 50 target genes fell into six bins (closed triangles, Fig. 3). As shown in Fig. 3, analysis of these data showed that most target genes were expressed at levels above background, but no bias for integration into highly expressed genes was observed. The ratio of the median expression values of ASV target genes to all genes was 0.99 (an average of duplicate data sets). This comparison indicates that target genes are expressed at average, rather than high, levels. In an independent test of these results we also analyzed two HeLa cell microarray data sets from a public database that was also used for the previous MLV study (43). Of the 95 genes that were targets for integration, 34 were available in this database (some common to our platform). In this analysis, the ratio of median expression values of target genes to total genes was ca. 0.97, confirming that there was no preference for integration into highly expressed genes. We applied the Wilcoxon test (testing the same hypothesis as a t test) to all four data sets and found that we could not distinguish the median expression levels of target genes and all genes (P ≥ 0.497).
FIG. 3.
Transcriptional activity of target genes. Microarray gene expression profiling was carried out for the HeLa cells as described in Materials and Methods (data set H55). Only expressed genes were used for this analysis (see Materials and Methods). HeLa gene expression levels were collected into groups based on increments of 500 arbitrary units. Each data point corresponds to the number of genes in the group (y axis, log scale) having the arbitrary units indicated on the x axis. The values for all expressed genes are indicated by filled diamonds, and ASV target gene values are indicated by filled triangles. The median value for all genes is shown as a dotted vertical line. The x axis is truncated at ca. 35,000 array units, eliminating irrelevant data points for expressed genes, for which no integrations were observed.
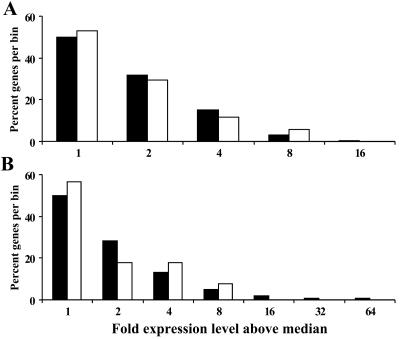
To further assess the relationship between transcriptional activity and integration site selection, we reanalyzed two data sets by sorting the genes into bins based on their expression levels and then determined the number of integration events in each bin. As shown in Fig. 4, the number of integrations was generally proportional to the number of genes per bin. Based on our sampled genes, we conclude that there is no strong bias for ASV integration into highly active genes. Instead, integration appears to be random with respect to transcriptional activity of genes.
FIG. 4.
Relationship between transcriptional activity of target genes and frequency of ASV integration events. Expression levels of total genes and target genes were collected in bins based on their fold value above the median. Fold values above the median are indicated as 2 (one through twofold), 4 (two- through fourfold), etc., for each bin. Percentages of total genes (filled bars) or target genes (open bars) in each bin are plotted. Target site values on the x axis are truncated, eliminating higher irrelevant values for which no integrations were observed. (A) Analysis applied to the GEO GSM2145 data set. (B) Analysis applied to the H56 data set. Using Spearman's test, we determined a rank correlation between total genes and target genes (for panel A, P = 0.0083, R = 1.0; for panel B, P = 0.05, R = 0.9; R = Spearman's correlation).
We also compared the number of integration events that occurred in chromosomes that contain highly expressed gene clusters (6, 39) (RIDGEs on chromosomes 1, 11, 16, and 19) with the number that occurred in those with poorly expressed gene clusters (anti-RIDGEs on chromosomes 4, 8, 13, and 18) (39). The collective target sizes for these two chromosome sets were similar, and we found that the total number of integration events in each chromosome set was nearly equivalent (37 versus 36 events, respectively). Strikingly, only 4 of 226 (1.8%) integration events occurred into chromosome 19 (Fig. 1), which is rich in highly expressed genes; this is in contrast to the much greater percentage observed for HIV-1 (34).
DISCUSSION
In this study, we investigated integration site selection after infection with an ASV-based retroviral vector. We used a vector system that was engineered for infection of human cells (1, 17), which allowed identification of target sites in the human genome and determination of the transcriptional states of the target genes prior to integration. The general experimental design and the mapping methods were as described previously and shown to be free from significant biases (34, 43). For ASV, we found that, in general, integration sites were not restricted to any particular regions or known chromosomal features, as measured by several parameters. One limitation of our study is the use of heterologous human host cells, which was dictated by the availability of human genome sequence and the ability to perform transcriptional profiling. If site selection is influenced by specific interactions between the avian retroviral preintegration complex and chicken-specific host factors, such targeting would be missed in our system. However, the fact that authentic and apparently efficient ASV integration occurs in mammalian cells argues against the existence of critical species-specific interactions. We note that our conclusions regarding global accessibility of integration sites are generally consistent with an earlier study that used a primer extension method to survey ASV integration sites in turkey cells (42). This study detected local integration hotspots for ASV, likely due to the ability to measure a larger number of events. With the noted caveats, our study provides new information regarding how broad chromosomal features and transcriptional activity affect ASV integration site selection and provides a useful comparison with MLV and HIV-1.
Our results suggest that ASV integration favors genes (e.g., 39.8 versus 22.4% expected for random integration) (Table 1). Schröder et al. (34) and Wu et al. (43) reported that genes are favored targets for both HIV-1 and MLV integration, with HIV-1 showing a stronger preference than both ASV and MLV (Table 1). The bias for HIV-1 integration into genes was especially pronounced in genes activated in response to HIV-1 infection (34). The preference for ASV (and MLV) integration into genes is modest and is dependent on the measurement of random integration, as discussed above. Another method for determining the random integration frequency is to perform in vitro integration reactions on naked cellular target DNA (34); however, this method could produce other biases. The measured preferences for integration into genes by ASV and MLV might decrease or increase with future analyses, based on more accurate values for random integration. However, in a comparative sense, we can more definitively conclude that ASV and MLV show similar frequencies of integration into genes, as the same cell type, gene definition (RefSeq), and value for random integration were used in the two studies.
The previous MLV studies revealed a unique bias compared with HIV-1; gene promoter regions were five times more likely than random to be targets for MLV integration, and it is possible that this tendency may be relevant to activation of the LMO2 gene in human patients during retroviral gene therapy (43). With respect to targeting over the length of genes, we found that, in contrast to MLV, ASV does not show a bias for transcriptional start sites (Fig. 2).
Transcriptional analysis of ASV target genes revealed that their expression values fell very close to the median for all analyzed genes. Further analyses showed that integration site selection is essentially random with respect to the intensity of transcriptional activity of target genes (Fig. 3 and 4). It is possible that the preference for ASV integration into genes may reflect increased accessibility associated with some baseline transcriptional activity. Our results show that, although there is a bias for ASV integration into genes, these genes are expressed at average levels; highly active genes (greater than twofold above the median expression level) are not favored (Fig. 4). Recently, it was reported that highly active transcription may inhibit ASV integration (41), and our results are not inconsistent with this proposal. In contrast to our findings with ASV, a positive correlation between high transcriptional activity and integration site selection was seen with HIV-1 (34).
Results from our study together with previous reports (34, 43) suggest that the measurable differences in integration preferences of HIV, MLV, and ASV vectors may result from differences in the integration site selection mechanisms of these retroviruses. Determinants of selection might include virus-specific properties of IN proteins (18, 37), preintegration complexes, or virus-specific interactions with cellular cofactors (4, 14). The yeast retrotransposons Ty3 and Ty5 have evolved mechanisms of site selection that depend on physical interaction between components of the integration complex and different cellular factors bound to DNA, resulting in different targeting specificities (reviewed in reference 5). Although retroviral integration appears to lack specificity with respect to target DNA sequence, similar host factor-based targeting mechanisms may play a role in site selection (5). If such targeting mechanisms are indeed operative, they may be tissue or cell type specific. For example, putative chromatin-bound targeting proteins might be expected to be expressed differentially in various cell types. Furthermore, it is possible that the lack of targeting to transcription start sites that we observed could be due to interspecies infection.
Elaboration of the determinants of site selection will be necessary to completely describe the early events in retroviral replication; with such knowledge, virus-specific differences may be incorporated into retroviral vector design. The predominant retroviral vectors currently in use are based on MLV and HIV-1. It is possible that virus-specific features of integration site selection (such as an apparent lack of preference for transcriptional start sites, highlighted here for ASV) may provide practical advantages (43). We (17) and others (12) recently demonstrated that transduction by ASV vectors is not limited to dividing cells, further highlighting the prospect of exploiting a broader array of retroviral vectors for gene therapy in differentiated cells. Thus, the choice of retroviral vectors might be tailored for specific needs. It has also been suggested that preferences for gene targeting might be exploited for insertional mutagenesis (34).
Beyond providing a more detailed description of the early events of replication, and the possible implications for vector design, our results contribute to an understanding of how genomes are shaped in evolution, as a large percentage of the human genome comprises integrated retroviral sequences (19, 38). Technical refinements, which should eventually allow higher-throughput analysis of integration sites, may allow use of retroviral integration as an in vivo probe of chromatin structure and genome organization, providing an experimental model for genome shaping.
ADDENDUM IN PROOF
After the acceptance of our manuscript, similar findings on ASV integration site selection were reported by Mitchell et al. (R. S. Mitchell, B. F. Beitzel, A. R. Schröder, P. Shinn, H. Chen, C. C. Berry, J. R. Ecker, and F. D. Bushman. PLoS Biol. 2:E234, 2004).
Acknowledgments
We are grateful to Peter Adams, Glenn Rall, and Ken Zaret for critical comments on the manuscript. We acknowledge the following Fox Chase Cancer Center Core Facilities and individuals for excellent technical assistance: Automated DNA Sequencing Facility (Anita Cywinski), Biostatistics Facility, DNA Microarray Facility (Ketaki Datta), and The Fannie E. Rippel Biochemistry and Biotechnology Facility. We also thank Marie Estes for excellent assistance in preparing the manuscript.
This work was supported by National Institutes of Health grants AI40385, CA71515, CA06927, AI30544, and AI48046 and also by an appropriation from the Commonwealth of Pennsylvania. Partial support for R.A.K. was provided by a grant from the American Cancer Society (IRG-92-027-05).
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute or any other sponsoring organization.
REFERENCES
- 1.Barsov, E. V., and S. H. Hughes. 1996. Gene transfer into mammalian cells by a Rous sarcoma virus-based retroviral vector with the host range of the amphotropic murine leukemia virus. J. Virol. 70:3922-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, P. O., B. Bowerman, H. E. Varmus, and J. M. Bishop. 1987. Correct integration of retroviral DNA in vitro. Cell 49:347-356. [DOI] [PubMed] [Google Scholar]
- 3.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 90:6125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bushman, F. D. 1999. Host proteins in retroviral cDNA integration. Adv. Virus Res. 52:301-317. [DOI] [PubMed] [Google Scholar]
- 5.Bushman, F. D. 2003. Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell 115:135-138. [DOI] [PubMed] [Google Scholar]
- 6.Caron, H., B. van Schaik, M. van der Mee, F. Baas, G. Riggins, P. van Sluis, M. C. Hermus, R. van Asperen, K. Boon, P. A. Voute, S. Heisterkamp, A. van Kampen, and R. Versteeg. 2001. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science 291:1289-1292. [DOI] [PubMed] [Google Scholar]
- 7.Carteau, S., C. Hoffmann, and F. Bushman. 1998. Chromosome structure and human immunodeficiency virus type 1 cDNA integration: centromeric alphoid repeats are a disfavored target. J. Virol. 72:4005-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Check, E. 2003. Second cancer case halts gene-therapy trials. Nature 421:305. [DOI] [PubMed] [Google Scholar]
- 9.Coffin, J. M., S. H. Hughes, and H. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 10.Flint, S. J., L. W. Enquist, V. R. Racaniello, and A. M. Skalka. 2004. Principles of virology. Molecular biology, pathogenesis, and control of animal viruses, 2nd ed. ASM Press, Washington, D.C.
- 11.Hacein-Bey-Abina, S., C. Von Kalle, M. Schmidt, M. P. McCormack, N. Wulffraat, P. Leboulch, A. Lim, C. S. Osborne, R. Pawliuk, E. Morillon, R. Sorensen, A. Forster, P. Fraser, J. I. Cohen, G. de Saint Basile, I. Alexander, U. Wintergerst, T. Frebourg, A. Aurias, D. Stoppa-Lyonnet, S. Romana, I. Radford-Weiss, F. Gross, F. Valensi, E. Delabesse, E. Macintyre, F. Sigaux, J. Soulier, L. E. Leiva, M. Wissler, C. Prinz, T. H. Rabbitts, F. Le Deist, A. Fischer, and M. Cavazzana-Calvo. 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302:415-419. [DOI] [PubMed] [Google Scholar]
- 12.Hatziioannou, T., and S. P. Goff. 2001. Infection of nondividing cells by Rous sarcoma virus. J. Virol. 75:9526-9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayward, W. S., B. G. Neel, and S. M. Astrin. 1981. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature 290:475-480. [DOI] [PubMed] [Google Scholar]
- 14.Holmes-Son, M. L., R. S. Appa, and S. A. Chow. 2001. Molecular genetics and target site specificity of retroviral integration. Adv. Genet. 43:33-69. [DOI] [PubMed] [Google Scholar]
- 15.Katz, R. A., P. DiCandeloro, G. Kukolj, and A. M. Skalka. 2001. Role of DNA end distortion in catalysis by avian sarcoma virus integrase. J. Biol. Chem. 276:34213-34220. [DOI] [PubMed] [Google Scholar]
- 16.Katz, R. A., K. Gravuer, and A. M. Skalka. 1998. A preferred target DNA structure for retroviral integrase in vitro. J. Biol. Chem. 273:24190-24195. [DOI] [PubMed] [Google Scholar]
- 17.Katz, R. A., J. G. Greger, K. Darby, P. Boimel, G. F. Rall, and A. M. Skalka. 2002. Transduction of interphase cells by avian sarcoma virus. J. Virol. 76:5422-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katzman, M., and R. A. Katz. 1999. Substrate recognition by retroviral integrases. Adv. Virus Res. 52:371-395. [DOI] [PubMed] [Google Scholar]
- 19.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 20.Leclercq, I., F. Mortreux, A. S. Gabet, C. B. Jonsson, and E. Wattel. 2000. Basis of HTLV type 1 target site selection. AIDS Res. Hum. Retrovir. 16:1653-1659. [DOI] [PubMed] [Google Scholar]
- 21.Macville, M., E. Schrock, H. Padilla-Nash, C. Keck, B. M. Ghadimi, D. Zimonjic, N. Popescu, and T. Ried. 1999. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res. 59:141-150. [PubMed] [Google Scholar]
- 22.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mooslehner, K., U. Karls, and K. Harbers. 1990. Retroviral integration sites in transgenic Mov mice frequently map in the vicinity of transcribed DNA regions. J. Virol. 64:3056-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller, H. P., and H. E. Varmus. 1994. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 13:4704-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pruitt, K. D., K. S. Katz, H. Sicotte, and D. R. Maglott. 2000. Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Trends Genet. 16:44-47. [DOI] [PubMed] [Google Scholar]
- 26.Pruitt, K. D., and D. R. Maglott. 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29:137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pruss, D., F. D. Bushman, and A. P. Wolffe. 1994. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl. Acad. Sci. USA 91:5913-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruss, D., R. Reeves, F. D. Bushman, and A. P. Wolffe. 1994. The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J. Biol. Chem. 269:25031-25041. [PubMed] [Google Scholar]
- 29.Pryciak, P. M., H. P. Muller, and H. E. Varmus. 1992. Simian virus 40 minichromosomes as targets for retroviral integration in vivo. Proc. Natl. Acad. Sci. USA 89:9237-9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pryciak, P. M., A. Sil, and H. E. Varmus. 1992. Retroviral integration into minichromosomes in vitro. EMBO J. 11:291-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pryciak, P. M., and H. E. Varmus. 1992. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell 69:769-780. [DOI] [PubMed] [Google Scholar]
- 32.Rohdewohld, H., H. Weiher, W. Reik, R. Jaenisch, and M. Breindl. 1987. Retrovirus integration and chromatin structure: Moloney murine leukemia proviral integration sites map near DNase I-hypersensitive sites. J. Virol. 61:336-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scherdin, U., K. Rhodes, and M. Breindl. 1990. Transcriptionally active genome regions are preferred targets for retrovirus integration. J. Virol. 64:907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schröder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 35.Scottoline, B. P., S. Chow, V. Ellison, and P. O. Brown. 1997. Disruption of the terminal base pairs of retroviral DNA during integration. Genes Dev. 11:371-382. [DOI] [PubMed] [Google Scholar]
- 36.Stevens, S. W., and J. D. Griffith. 1994. Human immunodeficiency virus type 1 may preferentially integrate into chromatin occupied by L1Hs repetitive elements. Proc. Natl. Acad. Sci. USA 91:5557-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taganov, K. D., I. Cuesta, R. Daniel, L. A. Cirillo, R. A. Katz, K. S. Zaret, and A. M. Skalka. 2004. Integrase-specific enhancement and suppression of retroviral DNA integration by compacted chromatin structure in vitro. J. Virol. 78:5848-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venter, J. C., M. D. Adams, E. W. Myers, P. W. Li, R. J. Mural, G. G. Sutton, H. O. Smith, M. Yandell, C. A. Evans, R. A. Holt, J. D. Gocayne, P. Amanatides, R. M. Ballew, D. H. Huson, J. R. Wortman, Q. Zhang, C. D. Kodira, X. H. Zheng, L. Chen, M. Skupski, G. Subramanian, P. D. Thomas, J. Zhang, G. L. Gabor Miklos, C. Nelson, S. Broder, A. G. Clark, J. Nadeau, V. A. McKusick, N. Zinder, A. J. Levine, R. J. Roberts, M. Simon, C. Slayman, M. Hunkapiller, R. Bolanos, A. Delcher, I. Dew, D. Fasulo, M. Flanigan, L. Florea, A. Halpern, S. Hannenhalli, S. Kravitz, S. Levy, C. Mobarry, K. Reinert, K. Remington, J. Abu-Threideh, E. Beasley, K. Biddick, V. Bonazzi, R. Brandon, M. Cargill, I. Chandramouliswaran, R. Charlab, K. Chaturvedi, Z. Deng, V. Di Francesco, P. Dunn, K. Eilbeck, C. Evangelista, A. E. Gabrielian, W. Gan, W. Ge, F. Gong, Z. Gu, P. Guan, T. J. Heiman, M. E. Higgins, R. R. Ji, Z. Ke, K. A. Ketchum, Z. Lai, Y. Lei, Z. Li, J. Li, Y. Liang, X. Lin, F. Lu, G. V. Merkulov, N. Milshina, H. M. Moore, A. K. Naik, V. A. Narayan, B. Neelam, D. Nusskern, D. B. Rusch, S. Salzberg, W. Shao, B. Shue, J. Sun, Z. Wang, A. Wang, X. Wang, J. Wang, M. Wei, R. Wides, C. Xiao, C. Yan, et al. 2001. The sequence of the human genome. Science 291:1304-1351. [DOI] [PubMed] [Google Scholar]
- 39.Versteeg, R., B. D. van Schaik, M. F. van Batenburg, M. Roos, R. Monajemi, H. Caron, H. J. Bussemaker, and A. H. van Kampen. 2003. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 13:1998-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vijaya, S., D. L. Steffen, and H. L. Robinson. 1986. Acceptor sites for retroviral integrations map near DNase I-hypersensitive sites in chromatin. J. Virol. 60:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weidhaas, J. B., E. L. Angelichio, S. Fenner, and J. M. Coffin. 2000. Relationship between retroviral DNA integration and gene expression. J. Virol. 74:8382-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Withers-Ward, E. S., Y. Kitamura, J. P. Barnes, and J. M. Coffin. 1994. Distribution of targets for avian retrovirus DNA integration in vivo. Genes Dev. 8:1473-1487. [DOI] [PubMed] [Google Scholar]
- 43.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749-1751. [DOI] [PubMed] [Google Scholar]