Abstract
To investigate interactions between hepatitis C virus (HCV) RNA replication complexes, a system was developed to simultaneously select different HCV subgenomic replicons within the same cell. Transcomplementation of defective replicons was not observed, suggesting an isolated and independent nature of the HCV RNA replication complex. In contrast, a high level of competition between replicons was observed, such that the presence and increased fitness of one replicon reduced the capacity of a second one to stably replicate. These results suggest that at least one factor in Huh7 cells required for HCV RNA replication is limiting and saturable.
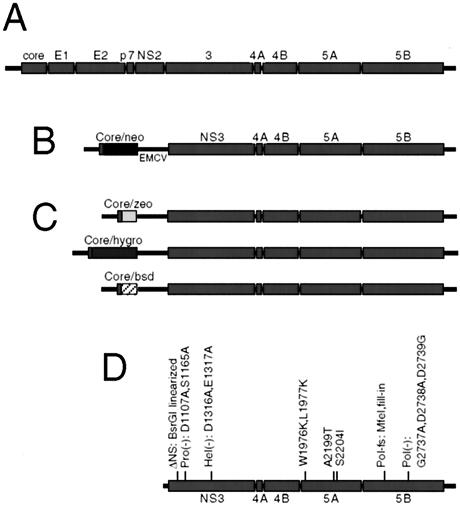
Hepatitis C virus (HCV), the sole member of the Hepacivirus genus, possesses a single-stranded, sense RNA genome approximately 9,600 nucleotides in length (Fig. 1A) (reviewed in references 14, 19, and 20). The HCV RNA replication complex, likely composed of the viral nonstructural (NS) proteins and yet-unidentified cellular proteins, replicates the viral genome through a negative-stranded RNA intermediate. The development of HCV subgenomic replicons has allowed examination of viral RNA replication in cell culture (18). HCV replicons were originally developed as subgenomic bicistronic RNAs encoding the neomycin phosphotransferase (neo)-selectable marker in place of the viral structural proteins (Fig. 1B). Stable replication can be scored by the frequency of G418 (a neomycin analog)-resistant colonies following replicon RNA transfection.
FIG. 1.
HCV genome and replicon organization. (A) Schematic representation of HCV genomic RNA. The amino-terminal one third of the polyprotein, translated through the action of an IRES, encodes the HCV structural proteins, including the capsid protein, core, and the envelope glycoproteins E1 and E2. The remainder of the polyprotein encodes the viral nonstructural proteins NS2, -3, -4A, -4B, -5A, and -5B. (B) Organization of the prototype HCV subgenomic replicon. The majority of the structural region of the Con1 HCV polyprotein was replaced with the neomycin phosphotransferase gene (neo), cloned as a fusion with the first 16 amino acids of the core protein, such that translation is driven by the HCV IRES. Translation of the remaining NS proteins is mediated by the encephalomyocarditis virus IRES element (EMCV). (C) Organization of alternatively selectable subgenomic replicons tested in this study. Each drug resistance gene was cloned as fusions to the core protein, as in the neo replicon. (D) Location and description of replicon mutations utilized in this study.
Most HCV genomes require specific mutations in the NS proteins, termed adaptive mutations, to replicate efficiently (1, 2, 6, 13, 16, 17). In this study, two NS5A adaptive mutations were utilized: S2204I, the strongest NS5A adaptive mutation, and A2199T, which enhances replication to 80% of the efficiency of the S2204I mutation (1). All replicons presented here were derived from the HCV Con1 isolate, were based on the original bicistronic construct (Fig. 1B) (1, 18), and will be referred to by the adaptive mutation, other pertinent mutations, and the selectable marker (i.e., S2204I neo).
Development of an alternative selectable replicon.
To develop a system in which two distinct subgenomic replicons could be maintained in a single cell, we replaced the neo gene with other drug resistance genes in the S2204I replicon (Fig. 1C) (sequences are available upon request). RNAs transcribed in vitro from these constructs were tested for the ability to replicate and provide drug resistance following transfection of Huh7 cells, as previously described (1). Unless otherwise stated, 6 × 106 cells were electroporated with 1 μg of each RNA for each transfection. Transfections were performed a minimum of three independent times, with representative experiments shown in figures. Although not all platings were precisely quantified, those that were showed minimal variation between experiments.
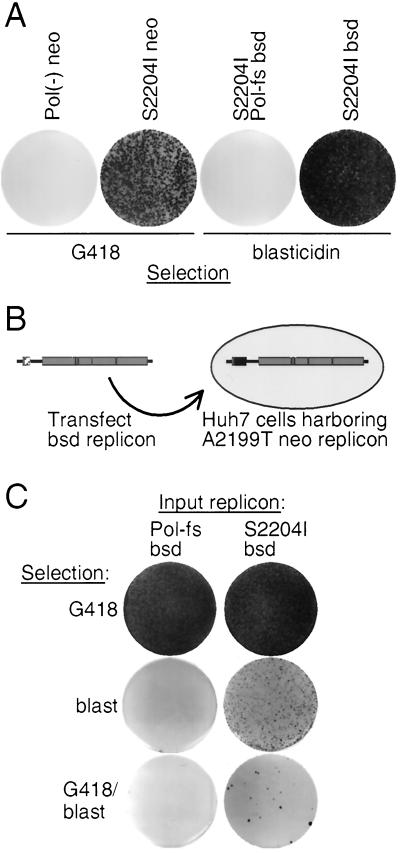
While replicons containing zeocin (zeo) or hygromycin (hygro) resistance genes were not selectable (data not shown), those containing the blasticidin S deaminase (bsd) gene (S2204I bsd) efficiently induced blasticidin-resistant colonies (Fig. 2A). No drug-resistant colonies were obtained after transfection of a replicon with a frameshift mutation in the NS5B RNA polymerase open reading frame (S2204I Pol-fs bsd), confirming that drug selection required RNA replication. At the applied drug concentrations (1 mg of G418/ml and 3 μg of blasticidin/ml), S2204I bsd appeared 5- to 10-fold more efficient at inducing drug-resistant colonies than S2204I neo (Fig. 2A).
FIG. 2.
(A) Comparison of neo- and bsd-resistant replicon efficiencies. Huh7 cells were electroporated with 1 μg of indicated neo- or bsd-resistant replicon RNA. Equal numbers of cells (106) were plated and selected in media containing the pertinent drug. Shown are Coomassie-stained plates following 2 to 3 weeks of selection. (B) Schematic representation of sequential replicon transfections. Huh7 cells previously transfected and selected for maintenance of A2199T neo replicons were transfected with 1 μg of S2204I bsd or, as a negative control, the replication-incompetent Con1 S2204I Pol-fs. (C) Following transfection, 106 cells were plated and selected with the indicated drugs. Shown are Coomassie-stained plates following 2 to 3 weeks of selection.
Blasticidin and neomycin replicons can be independently selected in the same cell.
Transfection of a clonal population of Huh7 cells already harboring A2199T neo with S2204I bsd RNA (Fig. 2B) produced colonies resistant to selection with blasticidin or the combination of blasticidin plus G418 (Fig. 2C, bottom two rows). These cells maintained high G418 resistance following transfection of the secondary RNAs (Fig. 2C, top row). As expected, cells transfected with S2204I Pol-fs bsd failed to produce blasticidin-resistant colonies or combined G418- and blasticidin-resistant colonies. This result confirmed that the neo and bsd replicons were capable of replicating in the same cell and providing resistance to both drugs simultaneously.
S2204I bsd introduced into cells already harboring A2199T neo yielded fewer colonies under dual selection than under blasticidin selection alone (a mean of 108 ± 16 CFU/μg of RNA compared to 2,650 ± 290 CFU/μg, respectively) (Fig. 2C, column 3). In addition, transfection of bsd replicons encoding other less efficient adaptive mutations into these cells did not result in colonies resistant to both drugs (data not shown). This low efficiency of dual selection was even more apparent when the inverse experiment was carried out, as no dual-drug-resistant colonies were obtained when S2204I neo replicons were transfected into a population of Huh7 cells already carrying S2204I bsd (data not shown). These results suggested that Huh7 cells already maintaining one replicon might be refractory to the establishment of another.
We speculate that host cell machinery required for replicase assembly and/or function may be limiting in Huh7 cells. Factors required for replicon maintenance may have already been monopolized by viral proteins from the first RNA, thus prohibiting their use by the second RNA. In cells selected with blasticidin alone, there would be no pressure to maintain the neo replicon, allowing for its replacement with the bsd replicon. It is unlikely that the colony numbers were influenced by interference between the G418 and blasticidin mechanisms of cell killing, because these drugs have been reported to function independently (8). As confirmation, Huh7 cells carrying an integrated DNA expressing bsd, pcDNA6/TR (Invitrogen), did not show a dramatic difference in neo replicon permissivity, regardless of whether blasticidin was included during replicon selection (data not shown).
Cotransfection enhances replicon dual-selection efficiency.
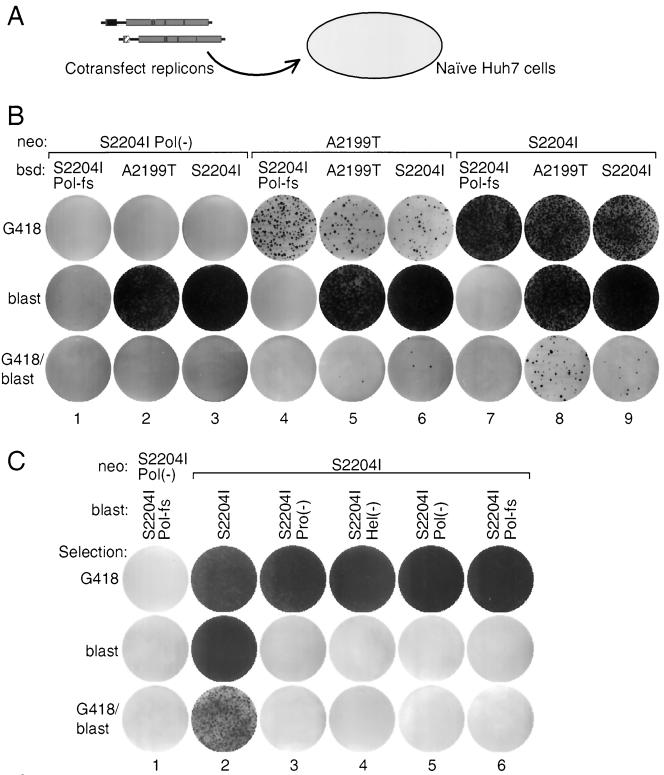
Simultaneous cotransfection of cells with two replicons may provide each RNA equal opportunity to establish productive replication and thus increase the efficiency of dual selection. To test this, equal amounts (1 μg each) of bsd and neo replicons, carrying either the A2199T or S2204I mutation, were mixed, transfected into naïve Huh7 cells, and plated under single or dual selection (Fig. 3A). This markedly enhanced the recovery of dual-drug-resistant colonies (Fig. 3B). Even a pair of moderately adapted A2199T replicons could be coselected, albeit at a low frequency (18 ± 3 CFU/μg of each RNA) (Fig. 3B, column 5, bottom row). Replication-incompetent replicons with either the Pol-fs, described above, or an NS5B active-site mutation (Pol−) were used as negative controls and were unselectable in these experiments. To further increase replicon dual-selection efficiency, a more highly replicon-permissive Huh7 subpopulation, termed Huh7.5 (3), was used. Cotransfection of these cells by any adapted replicon pair produced approximately 100-fold more dual-selected colonies than did the parental Huh7 cells (Fig. 3C, column 2).
FIG. 3.
Simultaneous replicon transfections into naïve Huh7 and Huh7.5 cells. (A) Schematic representation of simultaneous transfection of naïve Huh7 cells with mixtures of neo and bsd replicons. (B) Naïve Huh7 cells were electroporated with 1 μg each of indicated combinations of neo and bsd replicons, and 106 cells were plated and selected in media containing the indicated drug(s). Shown are Coomassie-stained plates following 2 to 3 weeks of selection. (C) Example of transcomplementation assays evaluated in Huh7.5 cells. Naïve Huh7.5 cells were electroporated with 1 μg each of indicated combinations of neo and bsd replicons. Cells (106) were plated and selected in media containing the indicated drug(s).
No observed transcomplementation of defective replicons.
To identify replicon defects that could be complemented in trans, a variety of mutations were created in the neo and bsd replicons (Fig. 1D). Replication-defective replicons tested included NS3 protease and helicase active-site mutants (Pro− and Hel−, respectively) (12), replicons lacking polymerase activity (Pol-fs and Pol−, a mutation of the enzyme active site) (12), and a replicon containing lysine substitutions of two hydrophobic residues of the NS5A amino-terminal amphipathic alpha-helix (W1976K and L1977K) (M. J. Evans, T. L. Tellinghuisen, C. M. Rice, and S. P. Goff, unpublished data). Extremely inefficient replicons, such as the Con1 replicon containing no adaptive mutations (1, 18) and replicons derived from the H77 genome (2), with or without the S2204I adaptive mutation, were also tested. Transcomplementation assays were carried out by both sequential and simultaneous replicon transfections and were scored by replication of the defective replicon when cotransfected with a viable replicon. We attempted to dual select the defective and replication-competent replicon and, alternatively, merely select for the defective species. In none of these experiments did we observe evidence for transcomplementation (Fig. 3C and data not shown).
These data suggest that HCV replication complexes are strikingly autonomous, with little exchange of factors between them. HCV viral proteins do not appear to stray from their respective complexes or may not be able to access other complexes. Although observed for other members of the Flaviviridae, including bovine diarrhea virus (5) and Kunjin virus (9-11, 15), transcomplementation has been generally inefficient and limited to only a few viral defects. It remains possible that other defective HCV replicons can be transcomplemented in this system. The lack of observed transcomplementation suggests that RNA genomes are strongly linked to the replication complexes they encode and cannot be readily exchanged with others, because defective replicon RNAs were not acquired and replicated by replication-competent complexes. Defective replicons were also not repaired by recombination between genomes, although such an event may be possible based on the identification of naturally occurring recombinant HCV isolates (4, 7).
Competition between replicons.
The neo and bsd S2204I replicons were the most efficient replicons when selected independently (for example, Fig. 3B, row 1, column 7, and row 2, column 3). However, cotransfection of these replicons actually produced fewer dual-drug-resistant colonies (84 ± 6 CFU/μg of each RNA) than the S2204I neo and A2199T bsd combination (390 ± 23 CFU/μg of each RNA) (Fig. 3B, compare row 3, columns 8 and 9). Interestingly, S2204I neo and A2199T bsd had nearly equivalent colony-forming activities under single selection (Fig. 3B, compare middle row, column 2, and top row, column 7). Coselection of two replicons may require that they share limiting host machinery, which may be most efficient with replicons that have similar efficiencies. Another line of evidence for interference between replicons was the observation that cotransfection of one replicon reduced the selectability of a second. For example, the relative G418-resistant colony-forming activity of both neo replicons was inversely proportional to the efficiency of the cotransfected bsd replicon (2,480 ± 120, 580 ± 32, and 450 ± 18 CFU/μg of neo RNA) (Fig. 3B, top row, columns 4 to 6 and 7 to 9, respectively). Replicon competition was also apparent, although the colonies were too dense for quantification, in the reciprocal direction, as cotransfection of progressively more efficient neo replicons reduced the colony-forming activity of a bsd replicon (Fig. 3B, middle row, columns 2, 5, and 8). Because all replicons were transfected in equal amounts regardless of the adaptive mutation context, it is again unlikely that transfection efficiency influenced the number of dual-selectable colonies obtained.
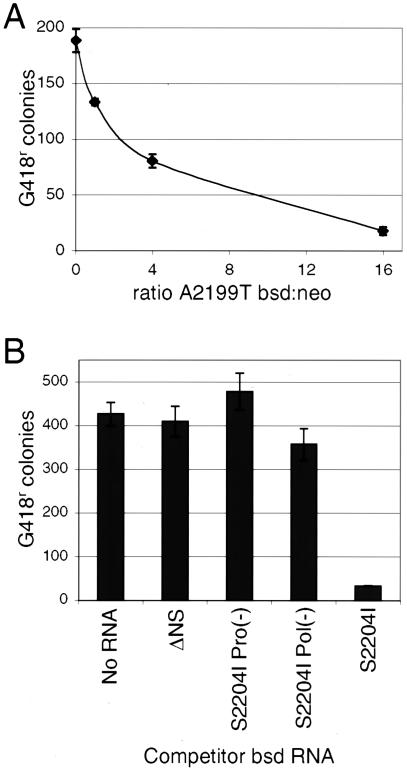
To further investigate replicon competition, naïve Huh7 cells were transfected with a constant amount of A2199T neo and increasing amounts of A2199T bsd (Fig. 4A). The total amount of RNA was held constant by addition of S2204I Pol− bsd. Transfections were selected with G418 to score the replication potential of the neo replicon. As shown in Fig. 4A, even a one-to-one ratio of neo-to-bsd replicon RNA resulted in a 29% reduction in G418-resistant colonies. At the highest ratio tested, a 16-fold excess of bsd RNA, selection of the neo replicon was reduced approximately 20-fold.
FIG. 4.
Replicon competition experiments. (A) G418-resistant colonies derived following cotransfection of naïve Huh7 cells with a fixed amount (0.5 μg) of the Con1 A2199T neo replicon and, as competitor, increasing amounts of the Con1 A2199I bsd replicon. To maintain a constant amount of RNA (total of 8.5 μg) per transfection, the replication-incompetent Con1 S2204I Pol− bsd RNA was added as filler. Transfections were plated (106 cells) and selected in media containing G418. Following selection, plates were stained and counted. Error bars represent the standard deviations across three separate transfections. (B) Various bsd replicons were cotransfected in 10-fold excess as competitor against a fixed amount (0.5 μg) of A2199T neo replicon RNA. G418-resistant colony numbers derived from these transfections are reported. Error bars represent the standard deviation across three separate transfections.
In another experiment, cells were transfected by a fixed amount of A2199T neo and a 10-fold excess of different competing bsd replicons (Fig. 4B). A truncated bsd replicon (ΔNS), containing just the HCV internal ribosome entry site (IRES), bsd gene, encephalomyocarditis virus IRES, and only a short segment of the NS3 gene, which can neither replicate nor translate any functional viral proteins, had no effect on the efficiency of establishment of A2199T neo (Fig. 4B). Neither a protease active-site mutant bsd replicon (S2204I Pro− bsd) nor a polymerase active-site mutant bsd replicon (S2204I Pol− bsd) significantly affected recovery of G418-resistant colonies (Fig. 4B). In contrast, S2204I bsd reduced the number of G418-resistant colonies by approximately 13-fold (Fig. 4B), indicating that replication of the competitor species was necessary to observe appreciable interference in this setting. Transfection efficiency likely did not influence the results of the above experiments, because in each experiment a fixed quantity of neo replicon was selected and the overall quantity of competitor bsd replicon was held constant. In addition, the vast excess of bsd replicon competitor utilized in these experiments (Fig. 4A and B) suggests that cytotoxicity resulting from transfection of replicon RNA did not contribute to the competition phenomenon.
A recent report also described competition between replicons (16). However, in contrast to our results, competition was found to be dependent on translation of the viral proteins but not on replication, as a polymerase-defective RNA was still capable of interfering. Differences in assay techniques may explain this discrepancy with our findings. We assayed stable replication efficiency (colony-forming activity) over several weeks, while Lohmann et al. (16) assayed transient replication at 24 h posttransfection, which may be less sensitive to the stability of the competitor RNA. In a stable assay such as ours, amplification and maintenance of the effector through many rounds of replication is likely required to observe competition.
Conclusions.
In this report, we describe a system to study the interplay between HCV subgenomic replicons in the same cell. This report provides a new view of the relationship between HCV replication complexes. We suggest that HCV replication takes place in distinct units within the host cell, with little exchange of viral proteins between them. Competition between replicons indicates a limited capacity of Huh7 cells to support subgenomic replicon function. Identification of the titratable cellular factor(s) required for HCV RNA replication will provide new insight into the composition and functions of the HCV replication complex and possibly allow for the development of more efficient replicon-permissive cell environments.
Acknowledgments
We thank Daniel Shaye, Carina Storrs, and Brett Lindenbach for reading the manuscript and for their helpful comments and Keril J. Blight for reagents.
C.M.R. is supported by grants from the Public Health Service (CA57973 and AI40034) and the Greenberg Medical Research Institute. S.P.G. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 2.Blight, K. J., J. A. McKeating, J. Marcotrigiano, and C. M. Rice. 2003. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J. Virol. 77:3181-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colina, R., D. Casane, S. Vasquez, L. Garcia-Aguirre, A. Chunga, H. Romero, B. Khan, and J. Cristina. 2004. Evidence of intratypic recombination in natural populations of hepatitis C virus. J. Gen. Virol. 85:31-37. [DOI] [PubMed] [Google Scholar]
- 5.Grassmann, C. W., O. Isken, N. Tautz, and S. E. Behrens. 2001. Genetic analysis of the pestivirus nonstructural coding region: defects in the NS5A unit can be complemented in trans. J. Virol. 75:7791-7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu, B., A. T. Gates, O. Isken, S. E. Behrens, and R. T. Sarisky. 2003. Replication studies using genotype 1a subgenomic hepatitis C virus replicons. J. Virol. 77:5352-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalinina, O., H. Norder, S. Mukomolov, and L. O. Magnius. 2002. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 76:4034-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karreman, C. 1998. A new set of positive/negative selectable markers for mammalian cells. Gene 218:57-61. [DOI] [PubMed] [Google Scholar]
- 9.Khromykh, A. A., P. L. Sedlak, K. J. Guyatt, R. A. Hall, and E. G. Westaway. 1999. Efficient trans-complementation of the flavivirus Kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. J. Virol. 73:10272-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khromykh, A. A., P. L. Sedlak, and E. G. Westaway. 2000. Cis- and trans-acting elements in flavivirus RNA replication. J. Virol. 74:3253-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khromykh, A. A., P. L. Sedlak, and E. G. Westaway. 1999. Trans-complementation analysis of the flavivirus Kunjin ns5 gene reveals an essential role for translation of its N-terminal half in RNA replication. J. Virol. 73:9247-9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knight and P. M. Howley (ed.), Fields virology. Lippincott-Raven, Philadelphia, Pa.
- 15.Liu, W. J., P. L. Sedlak, N. Kondratieva, and A. A. Khromykh. 2002. Complementation analysis of the flavivirus Kunjin NS3 and NS5 proteins defines the minimal regions essential for formation of a replication complex and shows a requirement of NS3 in cis for virus assembly. J. Virol. 76:10766-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 19.Moradpour, D., V. Brass, R. Gosert, B. Wolk, and H. E. Blum. 2002. Hepatitis C: molecular virology and antiviral targets. Trends Mol. Med. 8:476-482. [DOI] [PubMed] [Google Scholar]
- 20.Rosenberg, S. 2001. Recent advances in the molecular biology of hepatitis C virus. J. Mol. Biol. 313:451-464. [DOI] [PubMed] [Google Scholar]