Abstract
The nonstructural 5A (NS5A) protein of the hepatitis C virus (HCV) is a multifunctional phosphoprotein that is implicated in viral replication and HCV-mediated pathogenesis. We report here that the NS5A protein from the HCV genotype 1a is processed into shorter distinct forms when expressed in mammalian cells (Vero, HepG2, HuH-7, and WRL68) infected with an NS5A-expressing HSV-1-based amplicon vector or when transiently transfected with NS5A-expressing plasmids in the absence of exogenous apoptotic stimuli. Inhibitor studies combined with cell-free cleavage assays suggest that calcium-dependent calpain proteases, in addition to caspase-like proteases, are involved in NS5A processing. Interestingly, His-tagging experiments indicated that all the detectable NS5A-cleaved products are N-terminal forms of the protein. Additionally, immunofluorescence studies showed that, despite proteolytic cleavage, the NS5A protein exhibits a cytoplasm-perinuclear localization similar to that of the full-length protein. Thus, our results are consistent with recent data that demonstrated that NS5A is capable of perturbing intracellular calcium homeostasis and suggest that NS5A is both an inducer and a substrate of the calcium-dependent calpain protease(s). This may imply that cleavage of NS5A by calpain(s) could play a role in the modulation of NS5A function.
The hepatitis C virus (HCV) is the major etiological agent worldwide of chronic hepatitis, which often leads to liver cirrhosis and hepatocellular carcinoma (8, 11, 50). HCV is a small hepatotropic virus classified within the Flaviviridae family (40). The viral genome consists of a 9.6-kb single-stranded positive-sense RNA that encodes a precursor polyprotein of about 3,000 amino acid residues. The polyprotein is processed by host and viral proteases to produce at least 10 mature proteins. These proteins include at least three structural proteins, the nucleocapsid and two envelope glycoproteins (E1 and E2), the p7 protein, and six nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) with various enzymatic activities (47). An additional protein known as ARFP (for alternative reading frame protein), F (for frameshift), or core+1 (to describe the localization of the protein) has recently been identified. This protein is encoded by an alternative reading frame within the core coding region, but its function remains unknown (63, 64, 69).
Among the HCV nonstructural proteins, NS5A has received a lot of attention due to its proposed implication in the interferon response and its apparent key role in controlling the host antiviral properties (17, 26, 44, 45, 57). The HCV NS5A protein is a multifunctional serine phosphoprotein with a mass of 56 to 58 kDa (28, 48, 59) that displays a wide range of activities related to viral replication and HCV-mediated pathogenesis (2, 3, 33, 60). NS5A modulates viral replication by its direct association to the viral replicase complex formed at the cytoplasmic side of the endoplasmic reticulum (ER) (54, 62). Additionally, NS5A interacts with a number of cellular proteins, thereby affecting numerous host functions, including modulation of signal transduction pathways (19, 27, 58, 60), suppression of apoptosis (18, 20, 35, 39, 43), perturbation of cell growth and differentiation (1, 22), disruption of lipid metabolism (53), and modulation of transcription (10, 21, 32, 39, 46, 55). In addition, NS5A was recently shown to perturb calcium homeostasis, both in transfected cells and in the replicon system, leading to oxidative stress and activation of STAT3 and NF-κB transcription factors as well as the calcium-dependent calpain protease(s) (25, 65, 66).
The native NS5A protein is predominantly localized to the cytoplasm and the perinuclear area despite the presence of a functional nuclear localization signal (NLS) in its C-terminal region (29). A small amphipathic α-helix in the N terminus of NS5A was recently shown to acts as an ER membrane retention signal (5). On the other hand, N-terminal deletion mutants of the protein have been almost exclusively localized in the nucleus and reported to function as potent transcriptional activators (32, 55). These findings suggested that cleavage of the NS5A protein could operate as a posttranslational modification aiming to unmask its NLS, thus leading the C-terminal fragments of the protein to the nucleus where they could act as transcriptional factors (24, 52, 60). In this regard, subsequent studies demonstrated that NS5A from genotype 1b is cleaved at a few sites by caspase-like proteases in the presence of apoptotic stimuli generating NS5A products with N-terminal deletions, which could enter the nucleus and function as transcriptional activators (24, 52). On the basis of these data, caspase-mediated proteolytic processing of NS5A was proposed to activate the cryptic NLS and allow the protein to function as a transcription factor (52). However, the physiological relevance of these observations remains unclear. Furthermore, only a small amount of NS5A, if any, was found in the nucleus of cells expressing native HCV-1b NS5A protein under apoptotic conditions (52).
In this study we show that the HCV NS5A protein, in addition to caspases, is a substrate for the Ca2+-dependent calpain proteases. Furthermore, despite cleavage, we were unable to detect nuclear localization of the NS5A protein, and the detection of C-terminal truncated NS5A protein products remained elusive.
MATERIALS AND METHODS
Chemicals.
The following inhibitors were purchased from Affiniti Research products (Mamhead, Exeter, United Kingdom): MG132 (Nα-benzyloxycarbonyl-l-leucyl-l-leucl-l-leucinal); lactacystin; N-acetyl-Leu-Leu-Met-H (ALLM) or calpain II inhibitor; N-acetyl-Leu-Leu-Nle-H (ALLN) or calpain I inhibitor, and Z-Val-Ala-Asp(OMe)-fluoromethylketone (Z-VAD-FMK). All products were used within the indicated times. The purified 80-kDa subunit of rabbit m-calpain (calpain II) was obtained from Sigma.
Plasmids.
All plasmids were constructed by using standard technology, and a summary of all plasmids is shown in Table 1. The entire NS5A sequence (nucleotides [nt] 6258 to 7601) from HCV 1a strain H77 was PCR amplified from plasmid p90-FL (kindly provided by C. Rice). The following primers were used: sense, 5′-AGATATCATGAGCTCCGGTTCCTG-3′ (EcoRV and SacI restriction sites are underlined; the translation initiation codon is indicated in bold); antisense, 5′-CTCGAGAAGCTTAGCAGCACACGA-3′ (XhoI and HindIII restriction sites are underlined; the complementary sequence of a stop codon is shown in bold). The PCR conditions were as follows: 94°C for 1 min followed by 35 cycles of 94°C for 1 min, 65°C for 30 s, and 72°C for 2 min, with a final extension at 72°C for 10 min. The PCR product was digested with EcoRV and XhoI-Klenow and inserted into the SmaI site of pGEM-3zf(+) (Promega), yielding plasmid pHPI 691. The sequence of the NS5A coding region was verified by dideoxy sequencing analysis. For expression in mammalian cells, the HindIII-blunt-ended fragment, containing the entire NS5A coding sequence from plasmid pHPI 691, was inserted into the XbaI-blunt-ended site of vector pCI (Promega), generating plasmid pHPI 728. The NS5A open reading frame is under the control of the human cytomegalovirus (HCMV) immediate-early promoter. To generate the herpes simplex virus type 1 (HSV-1)-based amplicon vectors, the HindIII-blunt-ended fragment, encoding the entire NS5A protein from plasmid pHPI 691, was inserted into the NotI-blunt-ended site of vector pA-SKlacZ, generating plasmid pHPI 757. The NS5A open reading frame is under the control of the HSV-1 IE4 (α22/α47) promoter. The vector pA-SKlacZ is a pBluescript II derivative (Stratagene) that contains the amplicon module, ori-S, and α sequences from the HSV-1 genome and the LacZ β-galactosidase expression cassette [based on the HCMV promoter and the simian virus 40 poly(A) sequences] (61). For the construction of the C-terminal six-His-tagged NS5A protein, the SalI-blunt-ended fragment of the NS5A coding sequence from plasmid pHPI 691 was cloned into the XhoI-blunt-ended site of the pIND-V5 His A plasmid (Invitrogen), giving rise to plasmid pHPI 758. For expression from the amplicon plasmid, the PmeI fragment encoding the C-terminal six-His-tagged NS5A from plasmid pHPI 758 was cloned into the NotI-blunt-ended site of the pA-SKlacZ amplicon plasmid, giving rise to pHPI 782 (NS5A/His-C). For the construction of the N-terminal six-His-tagged NS5A protein, the (EcoRI-XbaI)-blunt-ended fragment derived from pHPI 765, a plasmid containing the entire NS5A coding sequences from pHPI 691 (nt 6258 to 7601) but lacking the initiation codon was cloned into the BamHI-blunt-ended site of vector pEBV-His C (Invitrogen), giving rise to plasmid pHPI 1402. For expression from the amplicon plasmid, the EcoRI-blunt-ended fragment encoding the N-terminal six-His-tagged NS5A protein from pHPI 1402 was cloned into the XbaI-blunt-ended site of the pA-EUA2 amplicon plasmid, yielding plasmid pHPI 1403 (NS5A/His-N), such that the NS5A coding sequence was under the control of the HCMV promoter and the simian virus 40 poly(A) sequence (see Fig. 5). The pA-EUA2 vector has been derived from the pA-SK amplicon plasmid and carries the green fluorescent protein-coding sequence and the BGH poly(A)-taken from pIRES-EGFP (Clonetech)-under the HSV-1 IE4 (α22/α47) promoter (A. L. Epstein and V. Revol-Guyot, unpublished data). Plasmid pHPI 1406 encodes an N-terminal deletion (−162 amino acids) fragment of NS5A and was constructed following amplification of the corresponding sequence from pHPI 611 (31). The sense primer was 5′-CCAAGCTTGCCATGGCGCCCCCTTGC-3′ (HindIII and NcoI restrictions sites are underlined; the translation initiation codon is indicated in bold), and the antisense primer was as described above. PCR conditions were as described above. For expression in mammalian cells the HindIII-blunt-ended fragment of the PCR product was inserted into the XbaI-blunt-ended site of pCI, generating plasmid pHPI 1406. The nucleotide sequence of this C-terminal fragment of NS5A was verified by sequencing analysis (MWG-Biotech Co.). pHPI 662, expressing the MBP-NS5A.1 protein, was used to generate the rabbit polyclonal antibody against the NS5A protein (31). pGEM-Luc, which was used for the in vitro cleavage assays, was purchased from Promega. A list of all plasmids is provided in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Vector | Description of insert |
|---|---|---|
| pHPI 691 | pGEM-3zf(+) | The entire NS5A coding sequence (nt 6258-7601) from HCV-1a |
| pHPI 728 | pCI | The HindIII fragment encoding the entire NS5A from pHPI 691 |
| pHPI 757 | pA-SKlacZ | The HindIII fragment encoding the entire NS5A from pHPI 691 |
| pHPI 758 | pIND-V5 His A | The Sal-I fragment encoding most of the NS5A from pHPI 691 |
| pHPI 782 | pA-SKlacZ | PmeI fragment containing the C-terminal six-His-tagged NS5A sequences from pHPI 758 |
| pHPI 730 | pGEM-3zf(+) | PvuII-HindIII C-terminal fragment of NS5A from pHPI 691 |
| pHPI 765 | pHPI 730 | SacI N-terminal fragment of NS5A from pHPI 691 |
| pHPI 1402 | pEBV-His C | EcoRI-XbaI fragment containing the entire NS5A from pHPI 765 |
| pHPI 1403 | pA-EUA2 | EcoRI fragment containing the N-terminal six-His-tagged NS5A from pHPI 1402 |
| pHPI 1406 | pCI | PCR product of N-terminal deleted (−162 amino acids) NS5A from pHPI 611 (reference 31) |
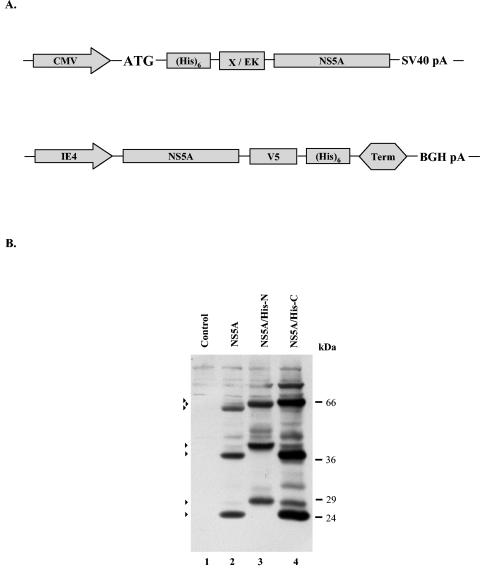
FIG. 5.
Construction and production of the His-tagged NS5A proteins. (A) The NS5A protein tagged at the N terminus with the six-His tag was produced under the control of the HCMV immediate-early promoter from the pA-EUA2 amplicon plasmid (Epstein and Revol-Guyot, unpublished). The Xpress peptide and an enterokinase cleavage site were interposed between the six-His tag and the NS5A coding sequence. The NS5A tagged at the C terminus with the six-His tag was produced under the control of the IE4 (α22/α47) promoter of the HSV-1 virus from the pA-SKlacZ amplicon plasmid. The V5 epitope was interposed between the six-His tag and the NS5A coding sequence. (B) Vero cells (2 × 106) seeded in six-well plates were infected with the amplicon vector pA-SKlacZNS5A (lane 2), pA-EUA2NS5A/His-N (lane 3), pA-SKlacZNS5A/His-C (lane 4) (MOI = 1; A/H = 1:10), or with the control virus HSV-1 strain LaL (MOI = 10) (lane 1). Cells were harvested at 10 h p.i. and the proteins were analyzed by Western blotting, as described in the legend of Fig. 1. Arrows indicate the major products that reacted with the NS5A polyclonal antibody. The positions of the molecular mass markers are shown on the right.
Cells and viruses.
Vero (green monkey kidney fibroblasts), HepG2 (epithelium from human hepatocellular carcinoma), and HuH-7 (human hepatome) cells were obtained from the American Type Culture Collection. WRL68 (human liver embryonic hepatoma) cells were kindly provided by A. Budkowska (Institut Pasteur, Paris, France). Vero, WRL68, and HuH-7 cells were maintained in Dulbecco's modified Eagle medium (Biochrom KG) supplemented with 5 (Vero) or 10% (WRL68 and HuH-7) fetal bovine serum (Gibco BRL), penicillin and streptomycin (at concentrations of 5 IU ml−1 and 50 mg ml−1, respectively), 2 mM l-glutamine, and nonessential amino acids (only for HuH-7) (1×; Biochrom KG). HepG2 were grown in minimal essential medium (Gibco BRL) containing 10% fetal bovine serum, penicillin-streptomycin, and l-glutamine, as above.
A high-titer amplicon virus stock was generated as previously described (61). The viability of the cells following the treatment with cellular protease inhibitors was assessed by trypan blue, as specified by the supplier (Biochrom AG). Infections were performed as previously described (61). Briefly, monolayer cell cultures were infected either with the pA-SKlacZNS5A amplicon vector at a multiplicity of infection (MOI) of 1 and an amplicon/helper virus (A/H) ratio of 1:10 or with the HSV-1 strain LaL at an MOI of 10. After 1 h of adsorption, the medium was replaced with fresh 199V medium (Gibco BRL) supplemented with 1% fetal bovine serum. At the indicated times post infection (p.i.), cells were washed twice with ice-cold phosphate-buffered saline (PBS-A), resuspended in PBS-A, pelleted at 8,000 rpm (Eppendorf centrifuge 5417C) for 2 min, and stored at −20°C until use.
Antibodies.
The MBP-NS5A.1 protein was used to generate a polyclonal antibody against NS5A. The protein was produced in Escherichia coli harboring the pHPI 662 plasmid and purified by affinity chromatography, as previously described (31). Rabbits were immunized against the NS5A protein following three injections, each containing approximately 300 μg of the purified fusion protein, at intervals of about 3 weeks. After the third injection the antibody response was analyzed by Western blotting. The antibody dilution was 1:100. The monoclonal antibody against NS5A was purchased from Biotrend Chemikalien GmbH and used at a dilution of 1:500.
The anti-NS5A polyclonal antibody was purified by a slightly modified affinity chromatography method based on CNBr-activated Sepharose 4B beads, as has been previously described (13). For this purpose the glutathione transferase-NS5A.1 protein, produced and purified from E. coli harboring the pHPI 913 plasmid, was used (31). The protein was dialyzed against the coupling buffer (0.2 M NaHCO3, 0.5 M NaCl [pH 8.5]) and coupled to CNBr-activated Sepharose 4B beads (Sigma) (1 to 10 mg of protein per ml of gel incubated overnight at 4°C). Subsequently, beads were spun out at 2,000 rpm (Beckman GPR centrifuge) and the excess protein was removed by several washes with ∼10 volumes of coupling buffer. Remaining active groups were blocked by incubation of the suspension with 0.2 M glycine, pH 8, overnight at 4°C. The noncovalently adsorbed proteins and the excess of blocking reagents were washed away by three cycles of coupling buffer, followed by acetate buffer (0.1 M Na acetate-HCl, 0.5 M NaCl [pH 4]). Beads coupled with the antigen were then washed with PBS-A and incubated with antiserum overnight at 4°C. Subsequently, they were packed in a column of appropriate size and washed with PBS-A until the optical density at 280 nm reached ∼0. The antibodies bound were eluted with 100 mM glycine-HCl, pH 2.8, and neutralized with 33.7 mM Tris-HCl, pH 8.8, and 0.3 M KCl. Purified antibody was divided into small aliquots and stored in −80°C.
Immunoblotting.
Cell monolayers were harvested at the indicated times, rinsed twice with ice-cold PBS-A and lysed (20 min on ice) in triple detergent buffer (50 mM Tris-HCl [pH 8], 150 mM NaCl, 0.1% sodium dodecyl sulfate [SDS], 100 μg of phenylmethylsulfonyl fluoride per ml, 1% NP-40, 0.5% sodium deoxycholate) in the presence of protease inhibitor cocktail (Sigma) or the complete mini protease inhibitor cocktail (Roche), as specified by the suppliers. SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer was added to each sample, and the samples were boiled for 3 min before separation on 12% denaturing polyacrylamide gels and transfer onto nitrocellulose sheets. After blocking in PBS containing 0.02% (vol/vol) Tween 20 (PBST) and 5% (wt/vol) dried milk for 40 min at room temperature, the membranes were incubated with the primary antibodies in PBST-1% dried milk overnight at 4°C. The membranes were then washed in PBST-1% dried milk three times for 30 min in total and incubated at room temperature for 2 h with a horseradish peroxidase-conjugated secondary antibody (DAKO), diluted in PBST-1% dried milk. After three washes with PBST-1% dried milk (total time, 30 min) and three washes with PBS (total time, 10 min), the membranes were soaked in Pierce enhanced chemiluminescence reagent and used to expose a film (Kodak).
Immunofluorescence analysis.
For immunofluorescence analysis, Vero cells were cultured on 10-mm cover glasses (Mikroskopische-Deckgläser) the day before transfection. The transfection procedure based on the Lipofectamine Plus reagent (Invitrogen) was as specified by the supplier. At 36 h posttransfection (p.t.), the cells were fixed with 3.7% paraformaldehyde for 30 min at room temperature and neutralized for 10 min with 100 mM glycine in PBS-A. The cells were then washed once with PBS-A and permeabilized with 0.1% Triton-X in PBS-A in the presence of 2 mg of bovine serum albumin per ml for 30 min at room temperature. Next, the cells were incubated for 2 h at room temperature with the affinity-purified NS5A rabbit polyclonal antibody in the permeabilization buffer. After at least three washes with the permeabilization buffer, the cells were incubated with the anti-rabbit Alexa Fluor 488-conjugated secondary antibody (Molecular Probes), diluted 1:1,000 in the permeabilization buffer, for 1 h at room temperature. After several washes first with the permeabilization buffer and then with PBS-A, coverslips were mounted with Mowiol (Sigma) and examined by laser scanning confocal microscopy (Leica TCS-SP microscope equipped with Leica confocal software).
Transfection.
Vero cells were seeded in six-well culture plates (Nunc) the day before transfection in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. After 18 h, the cells (60 to 70% confluence) were transfected with 1 μg of CsCl-purified DNA per well by using the Lipofectamine Plus reagent (Invitrogen), as specified by the supplier. At the indicated times p.t., cells were treated with the indicated protease inhibitors for 12 h. For the reversion experiments, after the 12-h treatment with the drugs, cells were washed three times with culture medium and further incubated until the time of harvesting. In each case, half of the cell lysates were used for protein analysis.
Purification of the six-His-tagged NS5A protein.
Approximately 108 Vero cells were infected with the pA-SKlacZNS5A/His-C amplicon vector at an MOI of 0.5 and an A/H ratio of 1:10. Cells were collected when a 100% cytopathic effect was observed. The six-His-tagged NS5A protein was purified by using a Ni-nitrilotriacetic acid agarose matrix according to the manufacturer's protocol (QIAGEN).
In vitro transcription and translation.
For in vitro transcription, plasmids pHPI 691 and pGEM-Luc were linearized with the EcoRI and XhoI enzymes (Biolabs), respectively. In both cases, in vitro transcription was performed by using the SP6 RNA polymerase according to manufacturer's protocol (Promega). The in vitro translation of the NS5A and Luc RNAs was performed in the presence of [35S]methionine (Amersham) under standard conditions in rabbit reticulocyte lysate (RRL) extracts, as specified by the supplier (Promega). Alternatively, a TNT transcription-translation kit (Promega) was used with the circular pHPI 691 plasmid. Translation was conducted at 30°C.
In vitro cleavage assay and autoradiography.
Cell-free cleavage assays were carried out at 37°C in a final volume of 50 μl. In vitro translated 35S-labeled protein (3 to 5 μl) was mixed with PBS (150 mM NaCl, 10 mM sodium phosphate [pH 7.5]) and CaCl2 (2 mM) and EGTA (4 mM) were added when necessary. The 80-kDa subunit of rabbit m-calpain was used at a concentration of 4 U/ml unless otherwise indicated. When one component was omitted, the volume was adjusted to 50 μl with PBS. To study the effect of protease inhibitors on m-calpain, 4 U of the 80-kDa subunit of rabbit m-calpain per ml was preincubated with ALLM (800 and 100 μΜ), ALLN (800 and 100 μΜ), or their solvent dimethyl sulfoxide (DMSO; equal volumes) in cleavage buffer (25 mM Tris-HCl [pH 7.5], 100 mM NaCl, and 3 mM dithiothreitol) on ice for 30 min. 35S-labeled proteins (3 to 5 μl) and CaCl2 (2 mM) were added when necessary, and all the samples (final volume, 50 μl) were incubated at 37°C for 1 h. Finally, the samples were separated on a 12% denaturing polyacrylamide gel, blotted onto nitrocellulose sheets, and analyzed by autoradiography.
RESULTS
Construction of HSV-1-based amplicon vectors expressing the HCV-1a NS5A protein.
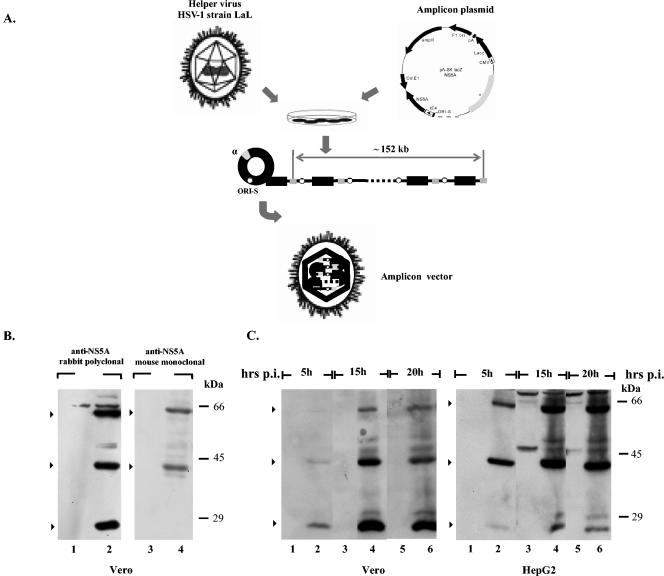
To achieve efficient expression of the HCV NS5A protein, we used HSV-1-based amplicon vectors (Fig. 1A) (14, 16, 38). These viral vectors allow the expression of foreign genes into different cell lines, including primary hepatocytes, and they have been previously used to produce properly modified HCV structural proteins (61). For this purpose, the nucleotide sequence encoding the NS5A polypeptide (nt 6258 to 7601) was cloned into the pA-SKlacZ amplicon plasmid (38), and the amplicon vector was prepared as described in Materials and Methods (Fig. 1A). First, Vero cells were infected either with the pA-SKlacZNS5A amplicon vector or with helper HSV-1 virus strain LaL (38). Cells were collected at 10 h p.i., and the production of the NS5A protein was assessed by Western blot analysis by using either a rabbit polyclonal antibody raised against an MBP-NS5A recombinant protein, as described in Materials and Methods, or a commercially available NS5A monoclonal antibody. As shown in Fig. 1B, both antibodies reacted with a 56-kDa protein, corresponding to the full-length NS5A protein. However, two additional proteins of about 40 and 24 kDa strongly reacted with the NS5A polyclonal antibody (Fig. 1B, lane 2), whereas only the 40-kDa product was detectable with the NS5A monoclonal antibody (Fig. 1B, lane 4). No bands were observed in extracts of Vero cells infected with the control virus (Fig. 1B, lanes 1 and 3). Thus, HSV-1-based amplicon vectors support efficient production of the full-length HCV NS5A protein in Vero cells. The unexpected detection of the 24- and 40-kDa products appeared at first to be in agreement with recent studies, which showed cleavage of the HCV-1b NS5A by caspase-like protease(s) under certain conditions (24, 52). Therefore, both products were assumed to be truncated forms of the NS5A protein.
FIG. 1.
Expression of the NS5A coding sequence from the pA-SKlacZNS5A amplicon vector. (A) Schematic representation depicting the pA-SKlacZNS5A amplicon plasmid used to generate the HSV-1-based amplicon vectors. The NS5A coding sequence is expressed under the control of the HSV-1 IE4 (α22/α47) promoter. The sequences designated α and ori-S represent the packaging signal sequence and the origin of replication of the HSV-1 DNA, respectively. The lacZ gene is under the control of the HCMV immediate-early promoter. For the generation of the HSV-1-based amplicon vectors, mammalian cells were transfected with the amplicon plasmid and superinfected with the helper virus HSV-1 strain LaL. In the presence of the helper virus, the plasmid DNA is amplified (presumably by a rolling-circle mechanism) into a head-to-tail concatemer, which is then packaged into defective HSV-1 particles that are up to the size of one genome (150 kb) (15, 16). (B) Production of the NS5A protein from the HSV-1-based amplicon vectors. Vero cells were infected either with the pA-SKlacZNS5A amplicon vector (lanes 2 and 4) (MOI = 1; A/H = 1:10) or with the virus HSV-1 strain LaL (control; MOI = 10) (lanes 1 and 3). Cells were lysed at 10 h p.i., separated on SDS-12% PAGE gels and analyzed by Western blotting by using the NS5A polyclonal antibody produced in this study (half of the lysates) or with a commercially available mouse monoclonal antibody (half of the lysates) (Biotrend Chemikalien GmbH). Arrows indicate the major products that were detected with the NS5A monoclonal or polyclonal antibodies. The positions of the molecular mass markers are shown on the right. (C) Monolayers of Vero or HepG2 cells seeded in six-well plates were infected either with the pA-SKlacZNS5A amplicon vector (MOI = 1; A/H = 1:10) (lanes with even numbers) or with the HSV-1 strain LaL (control, MOI = 10) (lanes with odd numbers). Cells were harvested at the indicated times p.i., and the proteins were separated on SDS-12% PAGE followed by Western blot analysis by using the NS5A polyclonal antibody. Arrows indicate the major products that reacted with the NS5A polyclonal antibody. The positions of the molecular mass markers are shown on the right.
To further characterize the processing of the NS5A protein, we performed kinetic studies in Vero and HepG2 cells infected with the NS5A-expressing HSV amplicon vector. Cells were infected either with the pA-SKlacZNS5A amplicon vector or with the control virus. Infected cells were harvested at selected times p.i. and proteins were analyzed by immunoblotting. As shown in Fig. 1C, both cell lines supported NS5A cleavage, albeit differences in the kinetics of the NS5A cleavage pattern were observed. Unexpectedly, in Vero cells generally only the shorter forms (the 24- or both the 24- and the 40-kDa proteins) were detected at early times p.i., indicating that efficient cleavage of NS5A occurs prior to protein accumulation in the cells (Fig. 1C and data not shown). All the forms of the NS5A protein (full-length and shorter forms) are easily detected at 15 h p.i. The kinetics of the NS5A cleavage pattern was affected by the physiological state of the Vero cells, such as cell density and passage. On the other hand, HepG2 cells supported almost equivalent levels of production of the full-length and the 40-kDa fragment during early and late times after infection, whereas the amount of the 24-kDa fragment was reduced at late times p.i. (Fig. 1C). Similar results were observed with HuH-7 and WRL68 cells (data not shown). These data suggest that the HCV-1a NS5A is cleaved into shorter forms in cells infected with the NS5A-expressing amplicon vector. Furthermore, the properties of the host cell appear to influence the efficiency of NS5A cleavage.
Caspase inhibitors only partially block the NS5A cleavage.
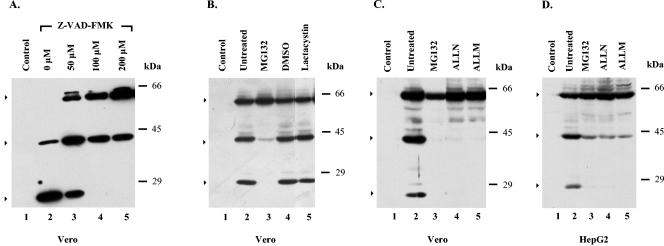
To examine whether caspase-like proteases are responsible for the cleavage of the HCV-1a NS5A protein, we tested the effect of Z-VAD-FMK, a pancaspase inhibitor, on NS5A processing in cells infected with the NS5A-expressing amplicon vector. Initial experiments were performed in Vero cells, as they appear to support the most efficient production of both the 40- and 24-kDa proteins. Cells were infected either with the pA-SKlacZNS5A amplicon vector or with the control virus in the absence or presence of Z-VAD-FMK, and cell lysates were analyzed by immunoblotting by using the NS5A polyclonal antibody. To ensure effective inhibition, a wide range of drug concentrations (50 to 200 μΜ) was used. Representative results are shown in Fig. 2A. Consistent with a caspase-like-mediated proteolytic cleavage of NS5A, the abundance of the full-length NS5A protein increased, whereas the levels of the 24-kDa protein band were markedly decreased in a concentration-dependent manner in the presence of Z-VAD-FMK (Fig. 2A, lanes 3 to 5). In the presence of 100 to 200 μM concentrations of the inhibitor, the 24-kDa protein band was undetectable (Fig. 2A, lanes 4 and 5). Interestingly, however, the level of the 40-kDa band was slightly increased at a 50 μM concentration of inhibitor (Fig. 2A, lane 3) and remained unaffected at higher concentrations (Fig. 2A, lanes 4 and 5). In the absence of Z-VAD-FMK only the truncated forms of the NS5A protein were detected (Fig. 2A, lane 2). No evidence for toxic effects of the inhibitor was observed (data not shown). Therefore, consistent with previous studies (24, 52), the cleavage of the HCV-1a NS5A protein appears to be affected by caspase-like protease(s). However, Z-VAD-FMK had only a limited effect on the stabilization of the protein, as the production of the 40-kDa fragment was totally unaffected by the pancaspase inhibitor, suggesting that additional proteases are involved in the processing of the HCV-1a NS5A protein.
FIG. 2.
Effect of protease inhibitors on the NS5A cleavage in pA-SKlacZNS5A-infected cells. (A) Vero cells (2 × 106) were infected either with the amplicon vector pA-SKlacZNS5A (MOI = 1; A/H = 1:10) (lanes 2 to 5) or with the control virus HSV-1 strain LaL (MOI = 10) (lane 1). Cells were treated with different concentrations of Z-VAD-FMK (50 to 200 μM) (lanes 3 to 5), beginning at the time of adsorption. Cell lysates, harvested at 10 h p.i., were analyzed by Western blotting, as described in the legend of Fig. 1. The positions of the molecular mass markers are shown on the right. (B) Vero cells were infected either with the amplicon vector pA-SKlacZNS5A (lanes 2 to 5) or with the control virus HSV-1 strain LaL (lane 1), as above. Cells were either left untreated (lanes 1 and 2) or were treated with MG132 (5 μM; lane 3), DMSO (the solvent of MG132; lane 4) or lactacystin (25 μM; lane 5), beginning at the time of adsorption. Cells were harvested at 10 h p.i. and proteins were visualized by Western blotting, as above. The positions of the molecular mass markers are shown on the right. (C) Vero cells were infected either with the amplicon vector pA-SKlacZ NS5A (lanes 2 to 5) or with the control virus HSV-1 strain LaL (lane 1), as above. Cells were either left untreated (lanes 1 and 2) or treated with MG132 (5 μΜ; lane 3), ALLN (10 μΜ; lane 4), or ALLM (100 μΜ; lane 5), beginning at the time of adsorption. Cells were harvested at 10 h p.i., and the proteins were visualized by Western blot analysis, as above. The positions of the molecular mass markers are shown on the right. (D) Monolayers of HepG2 cells, seeded in six-well plates, were infected either with the amplicon vector pA-SKlacZNS5A (lanes 2 to 5) or with the control virus HSV-1 strain LaL (lane 1), as above. Cells were either left untreated (lanes 1 and 2) or were treated with MG132 (5 μΜ; lane 3), ALLN (10 μΜ; lane 4), or ALLM (100 μΜ; lane 5), beginning at the time of adsorption. Cells were collected 10 h p.i., and the proteins were visualized by Western blot analysis, as above. The positions of the molecular mass markers are shown on the right. Arrows indicate the intact NS5A and the major proteolytic products.
Effect of proteasomal inhibitors on NS5A processing.
Proteasome is the most abundant cellular protease, and many viral proteins are substrates for the ubiquitin-proteasome pathway (30). Furthermore, examples of limited cleavage by proteasomes have been reported (42). To determine whether the cleavage of the NS5A protein is controlled by the proteasome-mediated pathway, the cell-permeable, pharmacological proteasomal inhibitors MG132 and lactacystin were tested. MG132, a tripeptide aldehyde that reversibly inhibits the chymotrypsin- and trypsin-like activities of the proteasome, is the most widely used proteasomal inhibitor, even though it can also inhibit a number of cathepsins and cysteine proteases (4, 37). Lactacystin, a Streptomyces metabolite, is more specific than MG132, as it only blocks proteasome and cathepsin A activities (37). In contrast to MG132, lactacystin inhibits proteasomal activity irreversibly (4, 37).
As shown in Fig. 2, MG132 efficiently inhibited the production of both the 40- and 24-kDa truncated forms of the NS5A protein (Fig. 2B, lane 3), whereas in the presence of DMSO, the solvent of MG132, the levels of both the 40- and 24-kDa proteins remained the same as in untreated cells (Fig. 2B, lanes 2 and 4). In contrast, treatment with lactacystin had virtually no effect on NS5Α processing (Fig. 2B, lane 5). Similar data with lactacystin were obtained after treatment with epoxomycin (data not shown). To ensure that the proteasomal inhibitors were active under our experimental conditions, the accumulation of total ubiquitinated proteins was analyzed by Western blotting. As expected, ubiquitinated proteins were accumulated only in the presence of the proteasomal inhibitors but not in the presence of Z-VAD-FMK or in untreated cells (data not shown).
Based on the lack of an effect of the most specific proteasome inhibitors (lactacystin and epoxomycin) on NS5A cleavage, we conclude that NS5A processing does not depend on the proteolytic activity of the proteasomes. The effect of MG132 on NS5A processing is most likely due to the inhibition of a proteasome-independent proteolytic activity.
Calpain inhibitors totally block NS5A cleavage.
While these studies were in progress, it was reported that the expression of NS5A in transfected cells perturbs intracellular Ca2+, suggesting that Ca2+-dependent proteases might also be activated in cells that produce the NS5A protein (25, 66). Furthermore, calcium-dependent proteases are also inhibited by MG132 (4, 37). Therefore, we examined the effect of calpains, Ca2+-dependent cysteine proteases, on NS5A processing by using two cell-permeable pharmacological inhibitors, ALLM (a calpain II inhibitor) and ALLN (a calpain I inhibitor), which affect the isoenzymes m-calpain and μ-calpain, respectively. For this purpose, Vero (Fig. 2C) or HepG2 (Fig. 2D) cells infected either with the pA-SKlacZNS5A amplicon vector (lanes 2 to 5) or with the control virus (lane 1) were treated separately with ALLN, ALLM, or MG132, and cell lysates were analyzed by Western blotting by using the NS5A polyclonal antibody. As before, MG132 suppressed the cleavage of the full-length NS5A protein (Fig. 2C and D, lanes 3). Additionally, both calpain inhibitors effectively inhibited the production of both the 40- and 24-kDa products (Fig. 2C and D, lanes 4 and 5). The effect of the inhibitors was concentration dependent, and 0.1 μΜ MG132 or 5 μΜ ALLM was enough to block NS5A cleavage (data not shown). Overall, these results are in agreement with the involvement of calpain protease(s) in the posttranslational processing of the HCV-1a NS5A protein.
Enzyme m-calpain is involved in Ca2+-dependent NS5A cleavage in vitro.
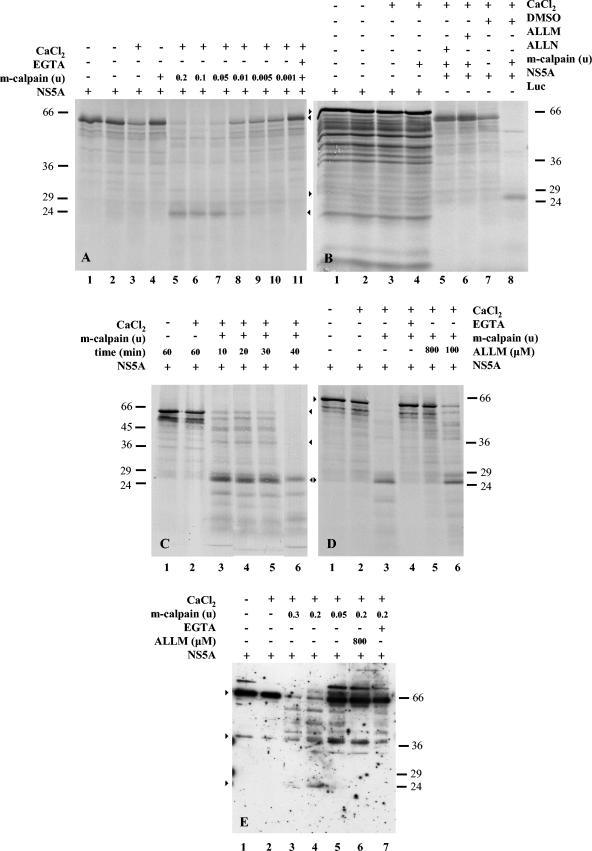
Next, we sought to determine whether the NS5A protein acts as a substrate for purified calpain in vitro. The 35S-labeled NS5A protein translated in Flexi (Fig. 3A and B) or TNT (Fig. 3C and D) RRLs was incubated with the purified 80-kDa subunit of rabbit m-calpain under different conditions and analyzed by SDS-PAGE. A titration (0.2 to 0.001 units) of purified m-calpain (Fig. 3A, lanes 5 to 10) shows that calpain concentrations ranging from 0.2 to 0.05 units (lanes 5 to 7) efficiently cleaved the full-length NS5A protein in the presence of 2 mM CaCl2 (34). Under these conditions only a 24-kDa fragment was detectable. Furthermore, consistent with the Ca2+ requirement for calpain activation, cleavage of NS5A was completely blocked when the lysates were incubated with the calcium chelator EGTA (Fig. 3A, lane 11). The 35S-labeled full-length NS5A protein remained intact in the absence of Ca2+, with or without m-calpain (Fig. 3A, lanes 4 and 1, respectively). However, in the presence of Ca2+, there was a decrease in the amount of the protein, suggesting the existence of Ca2+-dependent proteolytic activity in Flexi lysates (Fig. 3A, lane 3). These data prompted us to use the TNT in vitro transcription-translation system as described below (Fig. 3C and D), in which the NS5A protein remained intact under the previously described conditions (in fact, the protein remained stable in the presence of higher Ca2+ concentrations, up to 5 mM).
FIG. 3.
Processing of the NS5A protein by m-calpain in vitro. (A) NS5A was produced in vitro in Flexi RLL extracts (lane 1). NS5A was incubated at 37°C for 1 h in the absence (lane 2) or in the presence (lane 3) of 2 mM CaCl2. NS5A was treated with 0.2 U of m-calpain in the absence of CaCl2 (lane 4). The NS5A protein was incubated with 0.2 to 0.001 U of m-calpain (lanes 5 to 10) in the presence of 2 mM CaCl2. The effect of EGTA (4 mM) is shown in a sample prepared as in lane 5 (lane 11). The positions of the molecular mass markers are shown on the left. (B) Production of Renilla luciferase (Luc) in Flexi RRLs (lane 1). Luc was incubated at 37°C for 1 h in the absence (lane 2) or in the presence (lane 3) of 2 mM CaCl2. Luc was treated with 0.2 U of m-calpain and 2 mM CaCl2 (lane 4). The NS5A protein produced in vitro (panel A) was incubated at 37°C for 1 h with 0.2 U of m-calpain and 2 mM CaCl2 in the presence of ALLN (800 μM; lane 5), ALLM (800 μM; lane 6), or DMSO (the solvent of inhibitors; lane 8). NS5A was treated with DMSO and 2 mM CaCl2 in the absence of m-calpain (lane 7). The positions of the molecular mass markers are shown on the right. Starting from the top, arrows indicate the luciferase, full-length NS5A, and the 24-kDa NS5A bands. (C) The NS5A protein produced in RRLs by using the TNT transcription-translation kit was incubated at 37°C for 1 h in the absence (lane 1) or in the presence (lane 2) of 2 mM CaCl2. The protein was treated with 0.2 U of m-calpain in the presence of 2 mM CaCl2 at 37°C for 10 (lane 3), 20 (lane 4), 30 (lane 5), or 40 min (lane 6). The positions of the molecular mass markers are shown on the left. (D) NS5A produced by the TNT transcription-translation kit was incubated at 37°C for 1 h in the absence (lane 1) or in the presence (lane 2) of 2 mM CaCl2. The protein was treated with 0.2 U of m-calpain in the presence of 2 mM CaCl2 at 37°C for 60 min (lane 3). Effects of EGTA (4 mM) and ALLM (800 μM) are shown in samples prepared as in lane 3 (lanes 4 and 5, respectively). Activity of m-calpain was partially inhibited by 100 μΜ ALLM (lane 6). The positions of the molecular mass markers are shown on the right. (E) Partially purified NS5A/His-C (9 μg) was incubated at 37°C for 60 min in the absence (lane 1) or in the presence (lane 2) of 2 mM CaCl2. The protein was treated with 0.3 to 0.05 U of m-calpain (lanes 3 to 5) in the presence of 2 mM CaCl2. Samples prepared as in lane 4 were treated with ALLM (800 μM) (lane 6) or EGTA (4 mM) (lane 7). The positions of the molecular mass markers are shown on the right. Arrows indicate the intact NS5A protein and the two calpain-dependent 40- and 24-kDa products.
In parallel, we assessed the specificity of the cleavage assay by testing the cleavage of 35S-labeled Renilla luciferase produced in Flexi reticulocyte lysates (Fig. 3B). Purified m-calpain (0.2 U) had no effect on the integrity of the 35S-labeled luciferase protein in the presence of 2 mM CaCl2 (Fig. 3B, lane 4). The protein was also equally as stable as the untreated protein (Fig. 3B, lane 1) when incubated in the presence of Ca2+ (Fig. 3B, lane 3) or at 37°C for 1 h (Fig. 3B, lane 2). Furthermore, to verify that calpain was responsible for the cleavage of the NS5A protein, calpain inhibitors were used in the in vitro assay (Fig. 3B). Both ALLN and ALLM completely inhibited the cleavage of NS5A produced in vitro by the calcium-activated m-calpain (Fig. 3B, lanes 5 and 6), whereas DMSO, the solvent of the inhibitors, had no effect on the enzyme activity (Fig. 3B, lanes 7 and 8).
We also studied the effect of m-calpain on the NS5A protein produced from the TNT transcription-translation kit, as no endogenous Ca2+-dependent activity was observed. Calpain digested NS5A in a time-dependent manner (Fig. 3C, lanes 3 to 6) in the presence of 2 mM CaCl2, and complete cleavage was observed after at least a 40-min incubation (Fig. 3C and D, lanes 6 and 3, respectively). The protein was stable in the presence or absence of Ca2+ even 1 h after at 37°C (Fig. 3C and D, lanes 1 and 2 of both panels). NS5A cleavage was blocked by EGTA (Fig. 3D, lane 4) and by ALLM (Fig. 3D, lanes 5 and 6).
To further verify these results, recombinant His-tagged NS5A protein produced in Vero cells infected by the NS5A/His-C amplicon vector was affinity purified and used as a substrate for m-calpain in an in vitro cleavage assay. For this purpose, partially purified six-His-tagged NS5A was treated by different concentrations (0.3 to 0.05 U) of purified m-calpain in the presence of 2 mM CaCl2. Samples were incubated for 1 h at 37°C, and protein(s) was separated by SDS-PAGE, followed by immunoblot analysis with the NS5A polyclonal antibody. Consistent with the previous in vitro assays, in the presence of Ca2+ m-calpain effectively cleaved the full-length NS5A protein in a concentration-dependent manner (Fig. 3E, lanes 3 to 5), whereas the full-length NS5A protein remained intact in the presence of Ca2+ alone (Fig. 3E, compare lanes 1 and 2). The cleavage of NS5A was inhibited in the presence of EGTA (Fig. 3E, lane 7) or ALLM (Fig. 3E, lane 6). As before, the 40-kDa product was hardly detected. Overall, these data suggest that the NS5A protein is a substrate for calpain protease(s).
Intramolecular cleavage of the HCV NS5A protein in transfected cells.
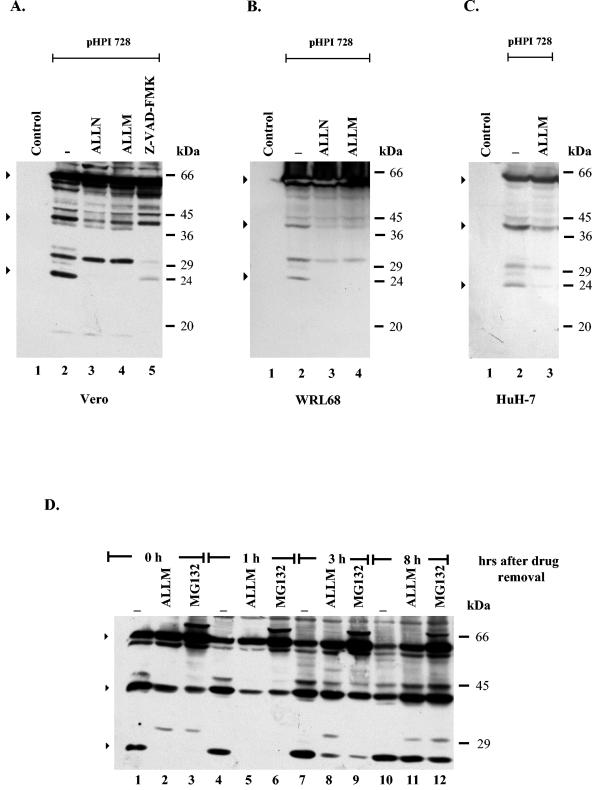
To assess the role of the HSV viral vector on NS5A processing, the production of the NS5A protein was analyzed in transiently transfected cells. In the first series of experiments, the effects of Z-VAD-FMK and calpain inhibitors on the cleavage of the NS5A protein were compared in transfected Vero and liver cells. First, Vero cells were transfected either with plasmid pHPI 728, which encodes the entire NS5A protein, under the control of the HCMV promoter, or with the pCI empty vector (control). At 36 h p.t., the cells were treated with the calpain inhibitors ALLN or ALLM (Fig. 4A, lanes 3 and 4) or Z-VAD-FMK (Fig. 4A, lane 5) and harvested at 48 h p.t. The NS5A protein was visualized by immunoblot analysis with the NS5A polyclonal antibody. When NS5A was transiently produced in Vero cells, the full-length NS5A protein, plus the 40- and 24-kDa forms, was detected, indicating that the cleavage of NS5A is independent of HSV-1 infection (Fig. 4A, lane 2). Consistent with the infection experiments in the presence of the calpain inhibitors (ALLN or ALLM), the levels of the full-length NS5A protein were increased and the levels of the 40- and 24-kDa proteins decreased (Fig. 4A, lanes 3 and 4). Furthermore, as noted previously, the cleavage of the NS5A protein was only partially inhibited by Z-VAD-FMK, as the production of the 24-kDa but not of the 40-kDa fragment was inhibited (Fig. 4A, lane 5). These data are consistent with the involvement of both caspases and calpains in the cleavage of the NS5A protein. Most importantly, as calpain activity is normally low in nonstimulated cells, these data suggest that the NS5A protein by itself is responsible for the activation of calpains (25, 66). Interestingly, a novel 31-kDa fragment as well as a 17-kDa fragment was reproducibly present only in transfected cells. The production of these fragments was unaffected by the presence of calpain inhibitors, but it was completely inhibited by Z-VAD-FMK (Fig. 4A, lanes 3 to 5). Next, these findings were confirmed in HuH-7 and WRL68 cells. As shown in Fig. 4B and C, consistent with the above data, both HuH-7 and WRL68 cells transfected with the NS5A-expressing plasmid (pHPI 728) produced the full-length and the shorter forms of the NS5A protein. Furthermore, calpain inhibitors blocked the production of both the 40- and 24-kDa fragments (Fig. 4B and C).
FIG. 4.
Effect of protease inhibitors on NS5A cleavage in transiently transfected cells. (A) Vero cells, seeded in six-well plates (60 to 70% confluence) were transfected either with plasmid pHPI 728 (lanes 2 to 5) or with the empty plasmid pCI (control; lane 1) (1 μg of DNA/well). At 36 h p.t., cells were treated for 12 h with ALLN (10 μΜ; lane 3), ALLM (100 μM; lane 4), or Z-VAD-FMK (50 μM; lane 5) or left untreated (lanes 1 and 2). Cells were harvested at 48 h p.t., and half of the cell lysates were analyzed by Western blotting, as described in the legend of Fig. 1. (B) WRL68 cells were transfected either with plasmid pHPI 728 (lanes 2 to 4) or with the pCI (control; lane 1), as above. At 36 h p.t. the cells were treated for 12 h with ALLN (10 μM; lane 3) or ALLM (100 μM; lane 4) or left untreated (lanes 1 and 2). Cells were harvested at 48 h p.t. and proteins were visualized by Western blot analysis, as above. (C) HuH-7 cells were transfected either with plasmid pHPI 728 (lanes 2 to 3) or with the pCI (control; lane 1), as above. At 36 h p.t. cells were treated for 12 h with ALLM (100 μM; lane 3) or left untreated (lanes 1 and 2). Cells were harvested at 48 h p.t. and proteins were visualized by Western blot analysis, as above. (D) For the reversion experiments, Vero cells were transfected with plasmid pHPI 728 (lanes 1 to 12). At 36 h p.t. cells were treated for 12 h with MG132 (5 μM; lanes 3, 6, 9, and 12) or ALLM (100 μM; lanes 2, 5, 8, and 11) or left untreated (lanes 1, 4, 7, and 10). Subsequently, the drugs were removed by washing several times with culture medium. Cells were harvested at 0, 1, 3, and 8 h after drug removal and proteins were analyzed by Western blotting, as described above. Arrows indicate the intact NS5A protein and the major products. The positions of the molecular mass markers are shown on the right.
In a second series of experiments, we investigated whether the cleavage pattern of NS5A was restored after drug removal, as the action of both MG132 and ALLM inhibitors is reversible (4, 37). This would provide further evidence that inhibition of the NS5A cleavage is not mediated by a general toxic effect of the drugs but is specific for calpain proteases(s). Vero cells were transfected with plasmid pHPI 728 (Fig. 4D, lanes 1 to 12) and 36 h later were treated with MG132 (Fig. 4D, lanes 3, 6, 9, and 12) and ALLM (Fig. 4D, lanes 2, 5, 8, and 11) for an additional 12 h or were left untreated (Fig. 4D, lanes 1, 4, 7, and 10). The abundance of the 40- and 24-kDa products was determined at selected intervals following the removal of the drugs. Both ALLM and MG132 reversibly inhibited the cleavage of the NS5A protein, as 8 h after their removal the cleavage pattern of NS5A was similar to that of untreated cells (Fig. 4D, lanes 10 to 12). The reversibility of the process is consistent with the known biochemical mechanisms of the two inhibitors and provides further evidence that their ability to block NS5A cleavage is calpain specific. Overall, these data suggest that except from caspase(s), calpain activity is implicated in the cleavage of NS5A.
Characterization of the NS5A cleavage products.
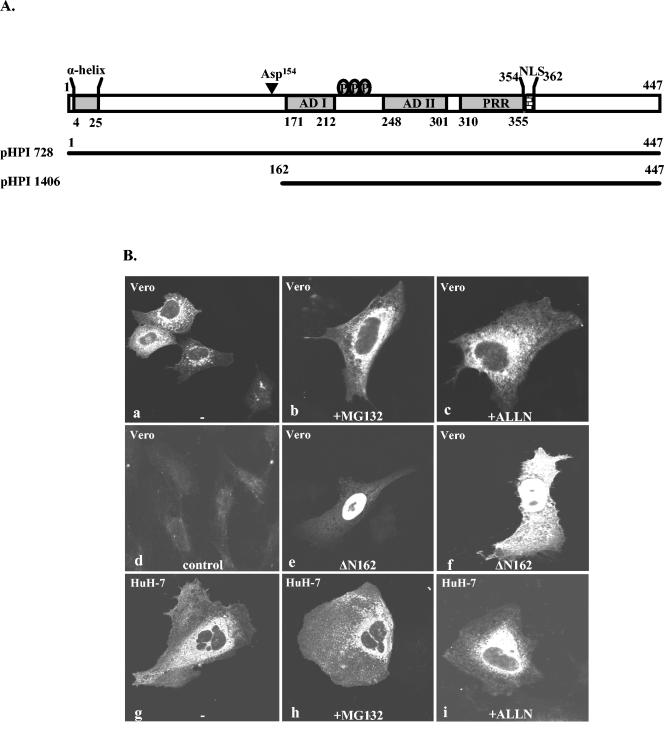
To determine the nature of the proteolytic products of the NS5A protein, two His-tagged forms of NS5A were constructed: one containing a six-His tag at its amino (NS5A/His-N) terminus and one containing this tag at its carboxy (NS5A/His-C) terminus (Fig. 5A). Both His-tagged forms were introduced into amplicon plasmids, and their expression was assessed in Vero cells infected with the tagged or untagged NS5A-expressing amplicon vectors or with the control virus. Cells were harvested at 10 h p.i., and the proteins were visualized by immunoblot analysis with the NS5A polyclonal antibody (Fig. 5B). When expressed in Vero cells, the N-terminal tagged NS5A protein exhibited 59-, 43-, and 27-kDa products, consistent with the predicted size increase (3-kDa) (Fig. 5B, lane 3). In contrast, the C-terminal tagged NS5A protein exhibited an analogous increase only in the size of the full-length product (Fig. 5B, lane 4). Similar results were obtained by immunoblot analysis with the anti-Xpress antibody raised against the X epitope of the N-terminal six-His-tagged NS5A protein (data not shown). These data indicate that both the 40- and 24-kDa polypeptides originate from the N-terminal part of the NS5A protein.
To further verify these results, we sought to compare the subcellular localization of the NS5A protein by immunofluorescence analysis in the presence or absence of calpain and proteasome inhibitors. This experiment was based on previous studies indicating that the entire NS5A protein is anchored to the ER membranes (5) and that recombinant forms of NS5A lacking the N-terminal ER anchoring sequence translocate into the nucleus of transfected cells (32, 55). To examine this, Vero or HuH-7 cells were transfected with the plasmid pHPI 728. At 24 h p.t. cells were treated for 12 h with MG132 (5 μΜ) or ALLN (10 μΜ) for 12 h, and at 36 h p.t. cells were fixed and stained with the affinity-purified NS5A rabbit polyclonal antibody described in the present study. As determined by immunofluorescence analysis, the wild-type NS5A protein, which is proteolytically processed, exhibits a cytoplasmic, ER-like perinuclear staining (Fig. 6B, frames a and g), indicating that the produced NS5A fragments do not translocate to the nucleus. In the presence of MG132 or ALLN (Fig. 6B, frames b, c, h, and i), the staining pattern of the NS5A could not be differentiated from that of the untreated cells (Fig. 6B, frames a and g), expressing the protein. As a control plasmid pHPI 1406 was constructed. This plasmid was designed to lack 162 amino acids from the N-terminal part of the protein, as shown in Fig. 6A. This form of NS5A translocates to the nucleus or exhibits both nuclear and cytoplasmic localization (Fig. 6B, frames e and f). Thus, only the in vitro synthesized recombinant N-terminally truncated forms of NS5A are detectable by Western blotting or by immunofluorescence assays and are found to localize to the nucleus of transfected cells. In contrast the N-terminally truncated forms, which are predicted to be produced by proteolysis of the wild-type NS5A, could not be detected under the experimental conditions used in our study.
FIG. 6.
Subcellular localization of the NS5A protein in transiently transfected cells. (A) Schematic representation of the entire NS5A protein and its major structural components. Representation of the amino acid composition of the NS5A fragments used in the subcellular localization studies. Plasmid pHPI 1406 encodes the NS5A protein that lacks the first 162 amino acids from the N terminus. Abbreviations: AD I, acidic domain I; ADII, acidic domain II; PRR, proline-rich region. Asp154 is a previously identified caspase recognition site (52). (B) Vero or HuH-7 cells, seeded in 10-mm cover glasses, were transfected with either plasmid pHPI 728 (a, b, c, g, h, and i), pHPI 1406 (e and f), or with the empty plasmid pCI (d) and either left untreated (a, d, e, f, and g) or treated with MG132 (5 μΜ; b and h) or ALLN (10 μΜ; c and i), for the last 12 h. Cells were fixed at 36 h p.t. with 3.7% paraformaldehyde, and immunofluorescence analysis was performed by using the affinity-purified NS5A polyclonal antibody. Samples were examined by laser scanning confocal microscopy.
DISCUSSION
By infection studies of HSV-based-amplicon vectors expressing the NS5A protein or transient transfection assays, we show that NS5A when expressed in Vero, HepG2, HuH-7, or WRL68 cells is proteolytically processed into distinctly sized fragments. Most importantly, we provide evidence implicating the calcium-dependent calpain proteases in addition to caspases in the cleavage of the NS5A protein. Interestingly, despite cleavage, no changes could be observed in the subcellular localization of the NS5A protein, which remained in the cytoplasmic-perinuclear region, a finding that correlates with the detection of only the amino-terminal forms of the NS5A protein.
One of the most intriguing findings of our studies is the involvement of the calcium-activated calpain proteases in HCV NS5A cleavage. This is based on the finding that the calpain inhibitors ALLN and ALLM, but not proteosomal-specific inhibitors, completely blocked the production of both the 40- and the 24-kDa N-terminal NS5A cleavage products in infected and transfected NS5A-expressing cells including Vero, HepG2, WRL68, and HuH-7 cells. Although the specificity of calpain inhibitors has been challenged, their primary targets are calpains, and higher concentrations of both inhibitors are required to achieve nonspecific inhibition of other proteases (36). Furthermore, cell-free cleavage assays showed that in vitro synthesized radiolabeled NS5A protein or recombinant His-tagged full-length NS5A protein produced in mammalian cells appears to be a substrate for the 80-kDa subunit of rabbit m-calpain in the presence of calcium (Fig. 3). Differences in the cleavage pattern of NS5A between the in vivo and in vitro experiments may be due to an increased susceptibility of NS5A protein to the calpain protease in the cell-free system that is related to differences in the structure of the protein and/or exposure of amino acid sequences that otherwise are hidden due to the anchorage of the protein to the ER membranes. Notably, unlike caspases that require specific amino acid sequences at their cleavage sites (12), calpains show no apparent sequence preferences, and it is believed that the calpain cleavage site is conformation dependent (6).
Interestingly, while the production of the 40-kDa fragment is affected only by the calpain inhibitors, the production of the 24-kDa fragment is blocked by both a pancaspase (Z-VAD-FMK) and calpain inhibitors, suggesting a different mechanism for the cleavage of the 24-kDa fragment. We speculate that this may be due to the interplay between caspase and calpain proteases (36, 67, 68). Indeed, recent studies have shown that calpain proteases are capable of activating or inhibiting caspase activity (36, 41, 49).
It should be noted that previous studies have shown that caspase-like proteases could cleave HCV-1b NS5A in the presence of exogenous apoptotic stimuli and have mapped the predominant caspase recognition sites at Asp154 and Asp389 (52). However, none of those sites appears to be involved in the production of the 24-kDa form inasmuch as Asp389 is unique for HCV-1b and conversion of Asp154 to Glu154 did not abolish the production of the 24-kDa NS5A cleaved form (data not shown). The identity of caspases and their possible interplay with calpains in the NS5A-expressing cells remains to be elucidated in light of the fact that NS5A has a generally antiapoptotic function (27, 36).
It should be mentioned that an effort has been made to detect the cleavage of NS5A in the replicon system by using our NS5A polyclonal antibody. However, the low expression levels of the NS5A protein have prevented the evaluation of NS5A cleavage in this system. On the other hand, we were able to detect the production of both the 40- and 24-kDa forms in the presence of all HCV proteins in stable transfected cells expressing the entire HCV polyprotein (data not shown). Alternatively, NS5A cleavage could be negatively associated with genomic replication.
An unexpected finding of this study was that all the detectable cleaved NS5A products, such as the 40- and the 24-kDa polypeptides, are N-terminal forms of the protein. This finding is based on the His-tagged experiments indicating that the size of these fragments was increased only when a six-His tag was placed at the N but not at the C terminus of the NS5A protein (Fig. 5). Therefore, all the detectable cleaved NS5A forms contain the ER anchorage signal and are predicted to follow the same cytoplasmic-perinuclear subcellular distribution as the entire NS5A protein. For reasons that remain unclear at the moment, detection of the remaining C-terminal NS5A products is elusive. In accordance with this finding and with previous studies (52), we were unable to detect nuclear localization for the NS5A protein in the cellular environment that supports cleavage. Conversely, artificially synthesized C-terminal NS5A fragments of a similar amino acid composition are easily detectable by Western blot analysis and are almost exclusively localized in the nucleus. It is likely that extensive cleavage within the C-terminal part of NS5A prevents production of large C-terminal NS5A fragments. Alternatively, differences in the posttranslational modifications between the C-terminal fragments that are artificially synthesized and the same fragments produced following cleavage of the entire NS5A protein may affect the stability of the protein. Last, but not least, limited proteolysis of the protein by calpain(s) and/or caspase(s) could render some of the products susceptible to another proteolytic pathway. This hypothesis is supported by previous studies demonstrating that the calpain-dependent cleavage of p53 is required for subsequent ubiquitin-dependent degradation (56).
Thus, in light of our data the role of a functional NLS in the NS5A protein remains an unsolved puzzle, inasmuch as our data indicate that NS5A cleavage favors the generation of C-terminally truncated NS5A forms which lack these sequences. Consequently, under the conditions used in the present study, NS5A cleavage appears to emphasize the anchorage of NS5A on the ER membranes rather than its transport to the nucleus. On the other hand, failure to detect C-terminal forms of NS5A protein by Western blotting or even by immunofluorescence analysis does not exclude the possibility that small amounts of these proteins are produced that are sufficient to operate in vivo as transcription modulators, as has been suggested (32, 55). It should be noted that recent studies have shown the clustering of important functional domains within the amino-terminal half of NS5A (60). The amphipathic α-helix, which mediates the anchorage of the NS5A protein to the ER membranes, may be related to the disturbance of calcium homeostasis (5, 25, 66). In addition, the N-terminal part of the protein interacts with ApoA1, which is basic component of lipid droplets and is related to lipid metabolism and steatosis (53, 66). Also, it interacts with karyopherin b3, which is related to trafficking of proteins into the nucleus (9), and with NS5B, which is a component of the viral replisome (54). NS5A also interacts with hVAP-33, a SNARE-like molecule, which is associated with the ER and Golgi membranes (62). In contrast, the functional domains that allow the protein to interact with cellular signaling pathways appear to cluster mainly in the C-terminal part of the protein (10, 29, 55).
Calpains are calcium-dependent intracellular cysteine proteases produced either in a ubiquitous and/or tissue-specific manner in a wide range of higher organisms (23, 51). Two major calpain isoforms have been identified: μ-calpain (calpain I) and m-calpain (calpain II), which are distinguished by the optimal Ca2+ concentration for maximal activity (23). Calpain activity is normally low in unstimulated cells because it is strictly regulated by a number of factors including intracellular Ca2+ concentration, the presence of the endogenous calpain inhibitor calpastatin (23), the phosphorylation state of the molecule, and its cellular distribution (23). Activation of calpains is initiated by the elevation of calcium levels, followed by autocleavage (23). Viral infections usually activate a number of cellular proteases including calpains (7). However, since calpain-mediated NS5A cleavage was observed both in infected and transfected cells, we speculate that NS5A protein is directly responsible for calpain activation.
In conclusion, our study establishes a novel cleavage mechanism for HCV NS5A protein through the calpain proteases. Since recent studies have shown that NS5A perturbs Ca2+ homeostasis and induces calpain activation (25, 65, 66), it is intriguing to speculate that there may be a linkage between the NS5A-mediated activation of calpains and calpain-mediated NS5A cleavage. As protein cleavage may represent a posttranslation mechanism required to unmask functional domains of a protein (24, 52), we propose that NS5A cleavage by calpains may provide a previously unrecognized level of control that may determine the biological function of NS5A in response to particular stimuli.
Acknowledgments
We are grateful to C. Rice for kindly providing the plasmid p90-FL, to D. Moradpour for kindly providing extracts from the UHCVcon-57.3 cells, and to A. L. Epstein for providing the amplicon system. We also thank N. Michaelidou and E. Aslanoglou for excellent technical assistance, H. Boleti and U. Georgopoulou for helpful discussions, and H. Boleti for her assistance in editing of the manuscript.
This work was supported by the European Commission grants Quality of Life QLK2-CT-1999-00055 and INCO-IC15-CT98-0304.
REFERENCES
- 1.Arima, N., C. Y. Kao, T. Licht, R. Padmanabhan, and Y. Sasaguri. 2001. Modulation of cell growth by the hepatitis C virus nonstructural protein NS5A. J. Biol. Chem. 276:12675-12684. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 3.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 4.Bogyo, M., and E. W. Wang. 2002. Proteasome inhibitors: complex tools for a complex enzyme. Curr. Top. Microbiol. Immunol. 268:185-208. [DOI] [PubMed] [Google Scholar]
- 5.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. [DOI] [PubMed] [Google Scholar]
- 6.Carillo, S., M. Pariat, A. Steff, I. Jariel-Encontre, F. Poulat, P. Berta, and M. Piechaczyk. 1996. PEST motifs are not required for rapid calpain-mediated proteolysis of c-fos protein. Biochem. J. 313:245-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Z., E. Knutson, A. Kurosky, and T. Albrecht. 2001. Degradation of p21cip1 in cells productively infected with human cytomegalovirus. J. Virol. 75:3613-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 9.Chung, K. M., J. Lee, J. E. Kim, O. K. Song, S. Cho, J. Lim, M. Seedorf, B. Hahm, and S. K. Jang. 2000. Nonstructural protein 5A of hepatitis C virus inhibits the function of karyopherin beta3. J. Virol. 74:5233-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung, K. M., O. K. Song, and S. K. Jang. 1997. Hepatitis C virus nonstructural protein 5A contains potential transcriptional activator domains. Mol. Cells 7:661-667. [PubMed] [Google Scholar]
- 11.Di Bisceglie, A. M. 2000. Natural history of hepatitis C: its impact on clinical management. Hepatology 31:1014-1018. [DOI] [PubMed] [Google Scholar]
- 12.Earnshaw, W. C., L. M. Martins, and S. H. Kaufmann. 1999. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 68:383-424. [DOI] [PubMed] [Google Scholar]
- 13.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Epstein, A. L. 1995. HSV-1 amplicons. Advantages and disadvantages of a versatile vector system. Restor. Neurol. Neurosci. 8:41-43. [DOI] [PubMed] [Google Scholar]
- 15.Fraefel, C., D. R. Jacoby, and X. O. Breakefield. 2000. Herpes simplex virus type 1-based amplicon vector systems. Adv. Virus Res. 55:425-451. [DOI] [PubMed] [Google Scholar]
- 16.Frenkel, N., O. Singer, and A. D. Kwong. 1994. Minireview: the herpes simplex virus amplicon-a versatile defective virus vector. Gene Ther. 1(Suppl. 1):S40-S46. [PubMed] [Google Scholar]
- 17.Gale, M. J., C. M. Blakely, B. Kwieciszewski, S. L. Tan, M. Dossett, N. M. Tang, M. J. Korth, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1998. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol. Cell. Biol. 18:5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gale, M. J., B. Kwieciszewski, M. Dossett, H. Nakao, and M. G. Katze. 1999. Antiapoptotic and oncogenic potentials of hepatitis C virus are linked to interferon resistance by viral repression of the PKR protein kinase. J. Virol. 73:6506-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Georgopoulou, U., K. Caravokiri, and P. Mavromara. 2003. Suppression of the ERK1/2 signaling pathway from HCV NS5A protein expressed by herpes simplex recombinant viruses. Arch. Virol. 148:237-251. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh, A. K., M. Majumder, R. Steele, K. Meyer, R. Ray, and R. B. Ray. 2000. Hepatitis C virus NS5A protein protects against TNF-alpha mediated apoptotic cell death. Virus Res. 67:173-178. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh, A. K., M. Majumder, R. Steele, P. Yaciuk, J. Chrivia, R. Ray, and R. B. Ray. 2000. Hepatitis C virus NS5A protein modulates transcription through a novel cellular transcription factor SRCAP. J. Biol. Chem. 275:7184-7188. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh, A. K., R. Steele, K. Meyer, R. Ray, and R. B. Ray. 1999. Hepatitis C virus NS5A protein modulates cell cycle regulatory genes and promotes cell growth. J. Gen. Virol. 80:1179-1183. [DOI] [PubMed] [Google Scholar]
- 23.Glading, A., D. A. Lauffenburger, and A. Wells. 2002. Cutting to the chase: calpain proteases in cell motility. Trends. Cell Biol. 12:46-54. [DOI] [PubMed] [Google Scholar]
- 24.Goh, P. Y., Y. J. Tan, S. P. Lim, S. G. Lim, Y. H. Tan, and W. J. Hong. 2001. The hepatitis C virus core protein interacts with NS5A and activates its caspase-mediated proteolytic cleavage. Virology 290:224-236. [DOI] [PubMed] [Google Scholar]
- 25.Gong, G., G. Waris, R. Tanveer, and A. Siddiqui. 2001. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-kappa B. Proc. Natl. Acad. Sci. USA 98:9599-9604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He, Y., and M. G. Katze. 2002. To interfere and to anti-interfere: the interplay between hepatitis C virus and interferon. Viral Immunol. 15:95-119. [DOI] [PubMed] [Google Scholar]
- 27.He, Y., H. Nakao, S. L. Tan, S. J. Polyak, P. Neddermann, S. Vijaysri, B. L. Jacobs, and M. G. Katze. 2002. Subversion of cell signaling pathways by hepatitis C virus nonstructural 5A protein via interaction with Grb2 and P85 phosphatidylinositol 3-kinase. J. Virol. 76:9207-9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirota, M., S. Satoh, S. Asabe, M. Kohara, K. Tsukiyama-Kohara, N. Kato, M. Hijikata, and K. Shimotohno. 1999. Phosphorylation of nonstructural 5A protein of hepatitis C virus: HCV group-specific hyperphosphorylation. Virology 257:130-137. [DOI] [PubMed] [Google Scholar]
- 29.Ide, Y., L. Zhang, M. Chen, G. Inchauspe, C. Bahl, Y. Sasaguri, and R. Padmanabhan. 1996. Characterization of the nuclear localization signal and subcellular distribution of hepatitis C virus nonstructural protein NS5A. Gene 182:203-211. [DOI] [PubMed] [Google Scholar]
- 30.Jarrousse, A. S., K. Gautier, S. Apcher, S. Badaoui, G. Boissonnet, M. H. Dadet, L. Henry, J. P. Bureau, H. P. Schmid, and F. Petit. 1999. Relationships between proteasomes and viral gene products. Mol. Biol. Rep. 26:113-117. [DOI] [PubMed] [Google Scholar]
- 31.Kalamvoki, M., V. Miriagou, A. Hadziyannis, U. Georgopoulou, A. Varaklioti, S. Hadziyannis, and P. Mavromara. 2002. Expression of immunoreactive forms of the hepatitis C NS5A protein in E. coli and their use for diagnostic assays. Arch. Virol. 147:1733-1745. [DOI] [PubMed] [Google Scholar]
- 32.Kato, N., K. H. Lan, S. K. Ono-Nita, Y. Shiratori, and M. Omata. 1997. Hepatitis C virus nonstructural region 5A protein is a potent transcriptional activator. J. Virol. 71:8856-8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kubbutat, M. H., and K. H. Vousden. 1997. Proteolytic cleavage of human p53 by calpain: a potential regulator of protein stability. Mol. Cell. Biol. 17:460-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lan, K. H., M. L. Sheu, S. J. Hwang, S. H. Yen, S. Y. Chen, J. C. Wu, Y. J. Wang, N. Kato, M. Omata, F. Y. Chang, and S. D. Lee. 2002. HCV NS5A interacts with p53 and inhibits p53-mediated apoptosis. Oncogene 21:4801-4811. [DOI] [PubMed] [Google Scholar]
- 36.LeBlanc, A. C. 2003. Natural cellular inhibitors of caspases. Prog. Neuro-psychopharmacol. Biol. Psychiatry 27:215-229. [DOI] [PubMed] [Google Scholar]
- 37.Lee, D. H., and A. L. Goldberg. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends. Cell Biol. 8:397-403. [DOI] [PubMed] [Google Scholar]
- 38.Logvinoff, C., and A. L. Epstein. 2000. Intracellular Cre-mediated deletion of the unique packaging signal carried by a herpes simplex virus type 1 recombinant and its relationship to the cleavage-packaging process. J. Virol. 74:8402-8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Majumder, M., A. K. Ghosh, R. Steele, R. Ray, and R. B. Ray. 2001. Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. J. Virol. 75:1401-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy, F. A., C. M. Fauquet, D. H. L. Bishop, S. A. Ghabrial, A. W. Jarvis, G. P. Martelli, M. A. Mayo, and M. D. Summers. 1995. Virus taxonomy: classification and nomenclature of viruses. Sixth report of the International Committee on Taxonomy of Viruses. Springer-Verlag, Vienna, Austria.
- 41.Nakagawa, T., and J. Yuan. 2000. Cross-talk between two cysteine protease families. Activation of caspase-12 by calpain in apoptosis. J. Cell Biol. 150:887-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palombella, V. J., O. J. Rando, A. L. Goldberg, and T. Maniatis. 1994. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell 78:773-785. [DOI] [PubMed] [Google Scholar]
- 43.Park, K. J., S. H. Choi, S. Y. Lee, S. B. Hwang, and M. M. Lai. 2002. Nonstructural 5A protein of hepatitis C virus modulates tumor necrosis factor alpha-stimulated nuclear factor kappa B activation. J. Biol. Chem. 277:13122-13128. [DOI] [PubMed] [Google Scholar]
- 44.Polyak, S. J., K. S. Khabar, D. M. Paschal, H. J. Ezelle, G. Duverlie, G. N. Barber, D. E. Levy, N. Mukaida, and D. R. Gretch. 2001. Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. J. Virol. 75:6095-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polyak, S. J., K. S. Khabar, M. Rezeiq, and D. R. Gretch. 2001. Elevated levels of interleukin-8 in serum are associated with hepatitis C virus infection and resistance to interferon therapy. J. Virol. 75:6209-6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qadri, I., M. Iwahashi, and F. Simon. 2002. Hepatitis C virus NS5A protein binds TBP and p53, inhibiting their DNA binding and p53 interactions with TBP and ERCC3. Biochim. Biophys. Acta 1592:193. [DOI] [PubMed] [Google Scholar]
- 47.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 48.Reed, K. E., J. Xu, and C. M. Rice. 1997. Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: properties of the NS5A-associated kinase. J. Virol. 71:7187-7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reimertz, C., D. Kogel, S. Lankiewicz, M. Poppe, and J. H. Prehn. 2001. Ca(2+)-induced inhibition of apoptosis in human SH-SY5Y neuroblastoma cells: degradation of apoptotic protease activating factor-1 (APAF-1). J. Neurochem. 78:1256-1266. [DOI] [PubMed] [Google Scholar]
- 50.Saito, I., T. Miyamura, A. Ohbayashi, H. Harada, T. Katayama, S. Kikuchi, Y. Watanabe, S. Koi, M. Onji, and Y. Ohta. 1990. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 87:6547-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sato, K., and S. Kawashima. 2001. Calpain function in the modulation of signal transduction molecules. Biol. Chem. 382:743-751. [DOI] [PubMed] [Google Scholar]
- 52.Satoh, S., M. Hirota, T. Noguchi, M. Hijikata, H. Handa, and K. Shimotohno. 2000. Cleavage of hepatitis C virus nonstructural protein 5A by a caspase-like protease(s) in mammalian cells. Virology 270:476-487. [DOI] [PubMed] [Google Scholar]
- 53.Shi, S. T., S. J. Polyak, H. Tu, D. R. Taylor, D. R. Gretch, and M. M. Lai. 2002. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology 292:198-210. [DOI] [PubMed] [Google Scholar]
- 54.Shirota, Y., H. Luo, W. Qin, S. Kaneko, T. Yamashita, K. Kobayashi, and S. Murakami. 2002. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRP) NS5B and modulates RNA-dependent RNA polymerase activity. J. Biol. Chem. 277:11149-11155. [DOI] [PubMed] [Google Scholar]
- 55.Song, J., M. Nagano-Fujii, F. Wang, R. Florese, T. Fujita, S. Ishido, and H. Hotta. 2000. Nuclear localization and intramolecular cleavage of N-terminally deleted NS5A protein of hepatitis C virus. Virus Res. 69:109-117. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki, K., S. Imajoh, Y. Emori, H. Kawasaki, Y. Minami, and S. Ohno. 1988. Regulation of activity of calcium activated neutral protease. Adv. Enzyme Regul. 27:153-169. [DOI] [PubMed] [Google Scholar]
- 57.Tan, S. L., and M. G. Katze. 2001. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology 284:1-12. [DOI] [PubMed] [Google Scholar]
- 58.Tan, S. L., H. Nakao, Y. He, S. Vijaysri, P. Neddermann, B. L. Jacobs, B. J. Mayer, and M. G. Katze. 1999. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc. Natl. Acad. Sci. USA 96:5533-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanji, Y., T. Kaneko, S. Satoh, and K. Shimotohno. 1995. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J. Virol. 69:3980-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tellinghuisen, T. L., and C. M. Rice. 2002. Interaction between hepatitis C virus proteins and host cell factors. Curr. Opin. Microbiol. 5:419-427. [DOI] [PubMed] [Google Scholar]
- 61.Tsitoura, E., M. Lucas, V. Revol-Guyot, A. L. Epstein, R. Manservigi, and P. Mavromara. 2002. Expression of hepatitis C virus envelope glycoproteins by herpes simplex virus type 1-based amplicon vectors. J. Gen. Virol. 83:561-566. [DOI] [PubMed] [Google Scholar]
- 62.Tu, H., L. Gao, S. T. Shi, D. R. Taylor, T. Yang, A. K. Mircheff, Y. Wen, A. E. Gorbalenya, S. B. Hwang, and M. M. Lai. 1999. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 263:30-41. [DOI] [PubMed] [Google Scholar]
- 63.Varaklioti, A., N. Vassilaki, U. Georgopoulou, and P. Mavromara. 2002. Alternate translation occurs within the core coding region of the hepatitis C viral genome. J. Biol. Chem. 277:17713-17721. [DOI] [PubMed] [Google Scholar]
- 64.Walewski, J. L., T. R. Keller, D. D. Stump, and A. D. Branch. 2001. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA 7:710-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waris, G., and A. Siddiqui. 2003. Regulatory mechanisms of viral hepatitis B and C. J. Biosci. 28:311-321. [DOI] [PubMed] [Google Scholar]
- 66.Waris, G., K. D. Tardif, and A. Siddiqui. 2002. Endoplasmic reticulum (ER) stress: hepatitis C virus induces an ER-nucleus signal transduction pathway and activates NF-kappaB and STAT-3. Biochem. Pharmacol. 64:1425-1430. [DOI] [PubMed] [Google Scholar]
- 67.Waterhouse, N. J., D. M. Finucane, D. R. Green, J. S. Elce, S. Kumar, E. S. Alnemri, G. Litwack, K. Khanna, M. F. Lavin, and D. J. Watters. 1998. Calpain activation is upstream of caspases in radiation-induced apoptosis. Cell Death Differ. 5:1051-1061. [DOI] [PubMed] [Google Scholar]
- 68.Wood, D. E., and E. W. Newcomb. 1999. Caspase-dependent activation of calpain during drug-induced apoptosis. J. Biol. Chem. 274:8309-8315. [DOI] [PubMed] [Google Scholar]
- 69.Xu, Z., J. Choi, T. S. Yen, W. Lu, A. Strohecker, S. Govindarajan, D. Chien, M. J. Selby, and J.Ou. 2001. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 20:3840-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]