Abstract
Reactivation in Epstein-Barr virus (EBV) is closely associated with a G0/G1 cell cycle arrest which can be induced either by lytic cycle-inducing agents or by the immediate-early gene product Zta. Accumulating evidence shows that in epithelial cells, downregulation of the proto-oncogene, c-myc, plays an important role in lytic cycle-associated cell growth arrest. Here, we provide evidence that c-Myc provides a gatekeeper function to ensure that certain cell cycle inhibitory events have been capitulated prior to full progression into the lytic cycle. Specifically, we show that reconstitution of c-Myc expression during the lytic cycle to levels observed in cycling uninduced cells inhibits the transactivation function of Zta. Nuclear localization studies show that c-Myc does not grossly alter the nuclear localization of Zta or its association with the insoluble nuclear fraction. Enforced expression of another transcription factor that promotes cell cycle progression, E2F1, also inhibits Zta transactivation. Analysis of c-Myc- and E2F1-mediated inhibition of a panel of Zta mutants shows parallel genetics and inhibition maps to a small bipartite sequence located between amino acids 29 and 53 of Zta, containing homology to the proline-rich domain of the tumor suppressor protein p53. Mutation of a conserved tryptophan residue located at amino acid 49 of Zta largely prevents inhibition by both c-Myc and E2F1. These studies identify a negative regulatory element within the Zta activation domain that is regulated by the cell cycle-promoting factors c-Myc and E2F1.
The Epstein-Barr virus (EBV) is a DNA tumor virus that is associated with a number of epithelial and lymphoid malignancies in humans, including nasopharyngeal carcinoma, Burkitt's lymphoma, Hodgkin's disease, and a small percentage of gastric carcinomas and T-cell malignancies (14, 20, 29). EBV likely contributes to tumor development and maintenance through the expression of viral genes that play a role in cell cycle promotion, inhibition of apoptotic pathways, and inhibition of inflammatory responses. Like other herpesviruses, EBV can exist in either a latent or a lytic replicative state. A number of EBV genes expressed during latency are involved in promoting cell cycle progression, and expression of the complete repertoire of latency-associated viral genes is fully capable of driving primary B lymphocytes into the cell cycle (28). In contrast, accumulating evidence indicates that EBV lytic replication occurs in growth-arrested cells, a property that appears to be conserved with other herpesviruses, including herpes simplex virus and cytomegalovirus (CMV) (3, 4, 8, 15, 18, 21-23, 27). Previous studies have shown that a number of agents that can induce the switch from latency to the lytic replication cycle in EBV-infected cells can induce growth arrest (11-13, 22). In addition, expression of the immediate-early BZLF1 gene in EBV-negative cells can induce cell growth arrest, indicating that the virus itself encodes the capacity to establish growth arrest during entry into the lytic replication cycle (3, 4, 8, 15, 21, 23, 27).
The mechanisms through which growth arrest is achieved during reactivation appear to differ, depending on the lytic cycle-inducing agent (11-13, 22). In addition, although the BZLF1 gene product, Zta, can induce growth arrest in both B lymphocytes and epithelial cells, it is clear that the mechanism through which Zta inhibits the cell cycle is distinct in these two cell types (3, 4, 15, 23, 27). In epithelial cell systems, we have found that downregulation of the proto-oncogene, c-myc, is a key event in inducing cell cycle arrest during reactivation (3, 23). Expression of Zta in EBV-negative epithelial cells causes downregulation of c-Myc, and this downregulation is required for Zta-mediated growth arrest (3). In addition, induction of the lytic cycle in the EBV-positive epithelial cell line, NPC-KT, by treatment with iododeoxyuridine (IDU) results in downregulation of c-Myc expression (23).
Although the reason why lytic replication occurs in cell cycle-arrested cells has not been established, it has previously been suggested that inhibition of cellular DNA replication may prevent competition for resources required for viral DNA replication (3, 8). Since it may be advantageous for EBV to replicate in a growth-arrested environment, we have postulated that there may be checkpoints to ensure that cell growth arrest has occurred prior to engagement in the full lytic replication cycle. Here, we show that in epithelial cells, one such checkpoint is downregulation of c-Myc. Enforced expression of c-Myc by transfection of a c-Myc expression vector inhibits reactivation. Further, we show that inhibition of reactivation by c-Myc occurs through inhibition of the transactivation function of the immediate-early factor, Zta.
MATERIALS AND METHODS
Cell culture and transfection.
All cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM; Life Technologies), supplemented with 10% fetal bovine serum (FBS; Life Technologies), penicillin, streptomycin, and glutamine. Cells were grown at 37°C in a humidified 5% CO2-containing atmosphere.
Transient transfection experiments were performed by using a modified version of the calcium phosphate precipitation procedure (a detailed protocol is available at www.flemingtonlab.com). Briefly, 106 cells were plated onto 100-mm-diameter tissue culture dishes. The following day, the medium was replaced with 8 ml of fresh supplemented DMEM; 4 h later, DNA precipitates were generated by mixing 0.5 ml of 1× HEPES-buffered saline (0.5% HEPES, 0.8% NaCl, 0.1% dextrose, 0.01% anhydrous Na2HPO4, 0.37% KCl [pH 7.10]) with a total of 30 μg of plasmid DNA (effector and reporter plasmids were added in the amounts indicated in the figure legends, and carrier DNA [pUHD10] was added to make a total of 30 μg). A total of 30 μl of 2.5 M CaCl2 was added, and samples were mixed immediately. Precipitates were allowed to form at room temperature for 20 min before being added dropwise to cells. Cells were incubated at 37°C with 5% CO2 for 16 h before the medium was replaced with 10 ml of fresh DMEM (plus 10% FBS).
Plasmid construction.
To generate the reporter plasmids 2X(ZIIIB)BG-Luc, 2X(BMRF1-AP1)BG-Luc, and 2X(ORI-Lyt)BG-Luc, the double-stranded oligonucleotides 2X(ZIIIB) (5′-CTAGCAGGCATTGCTAATGTACCAGGCATTGCTAATGTACA-3′ and 5′-GATCTGTACATTAGCAATGCCTGGTACATTAGCAATGCCTG-3′), 2X(BMRF1.AP1) (5′-CTAGCGACCTTTGAGTCAGGTGGCTGACCTTTGAGTCAGGTGGCA-3′ and 5′-GATCTGCCACCTGACTCAAAGGTCAGCCACCTGACTCAAAGGTCG-3′), and 2X(Ori.Lyt) (5′-CTAGCGGTCTCTGTGTAATACTTTTGGTCTCTGTGTAATACTTTA-3′ and 5′-GATCTAAAGTATTACACAGAGACCAAAAGTATTACACAGAGACCG-3′), respectively, were cloned into plasmid BG-Luc (2). The reporter plasmid −221ZpLuc was generated by transferring the Zp promoter region from the −221ZpCAT plasmid (10) into the vector pGL3Basic. Deletion mutations and point mutation of the BZLF1 transactivation domain were introduced into pBS(SVp/e)-Zta (9) with either the Bio-Rad Muta-Gene kit or the Stratagene QuickChange site-directed mutagenesis kit according to the manufacturer's instructions. All mutants were screened and verified by sequencing. The plasmid pcDNA3-c-myc contains CMV promoter-driven c-Myc cDNA (23). The plasmid encoding HA-E2F1 was a generous gift from Wilhelm Krek (Friedrich Miescher Institute, Basel, Switzerland).
Cell fractionation.
A total of 106 cells was plated and transfected as described above. Forty-eight hours posttransfection, cells were harvested. Cell pellets were suspended in 750 μl of buffer A (10 mM HEPES, 15 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 0.6% NP-40, 1 mM dithiothreitol [DTT] [pH 7.60]), incubated for 10 min on ice, and vortexed vigorously for 1 min. Samples were then centrifuged in an Eppendorf microfuge at 4°C (3,000 rpm) for 2 min. The supernatant (cytoplasm extract) was collected and stored at −80°C. The pellet (nuclear fraction) was washed three times with 750 μl of buffer B (10 mM HEPES, 15 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 0.2% NP-40, 1 mM DTT [pH 7.60]), and suspended in 75 μl of buffer C (25 mM HEPES, 50 mM KCl, 0.1 mM EDTA, 10% glycerol, 0.4 M NaCl, 1.0 mM DTT [pH 8.0]). Samples were incubated at 4°C for 1 h, during which time the samples were vortexed vigorously for 1 min at 10-min intervals. Samples were centrifuged at 4°C at 14,000 × g for 15 min. The supernatant (soluble nuclear extract) and pellet (insoluble nuclear extract) were collected and stored at −80C.
Immunofluorescence assays.
NPC-KT cells were seeded on coverslips in 10-cm plates at 106 cells per plate. Cells were transfected with a total of 30 μg of DNA by the modified calcium phosphate method as described above. Sixteen hours posttransfection, the media were changed; 48 h later, cells were washed two times with 1× phosphate-buffered saline (PBS). Cells were fixed with 3.7% formaldehyde in PBS for 15 min, neutralized with 50 mM Tris in PBS (pH 7.6) for 10 min, permeabilized with 0.1% Triton X-100 in PBS for 10 min, and blocked with 20% FBS in PBS for 15 min at room temperature. Cells were then incubated with the primary antibody (anti-Zta monoclonal antibody [MAb], 1:50; Argene) for 60 min, and the secondary antibody (fluorescein isothiocyanate [FITC]-conjugated goat anti-mouse immunoglobulin, 1:200; Biosource) for 45 min at room temperature. Coverslips were mounted with Vectashield mounting medium with DAPI (4′,6-diamidino-2-phenylindole). PBS washes were carried out consecutively after each step. Representative images were collected with either a Leica DM IRB microscope for epifluorescence or a Leica DM RXA microscope for deconvolution microscopy.
Reporter gene assays.
A total of 106 cells was plated and transfected as described above. Forty-eight hours posttransfection, cells were harvested. A fraction of collected cells was washed once with 1× PBS and then suspended in extraction buffer (90 mM K2HPO4, 10 mM KH2PO4, 1 mM DTT [pH 7.8]). Cells were lysed by four freeze-thaw vortex cycles (the freezing cycle consisted of 15 min at −80°C, and the thawing cycle consisted of 2 min at 37°C, followed by vortexing to mix well). Cellular debris was removed by centrifugation at 4°C (14,000 rpm) for 15 min, and supernatants were subjected to the firefly luciferase reporter assay according to the manufacturer's protocol (Promega).
Western blot analysis.
After a single 1× PBS wash, a fraction of harvested cells was separated for Western blot analysis. Cells were immediately suspended in 300 μl of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis loading buffer (125 mM Tris [pH 6.80], 10% glycerol, 2% SDS, 5% 2-mercaptoethanol, 0.05% bromphenol blue) and boiled for 30 min to shear the genomic DNA. Whole-cell extracts were measured with the Bio-Rad protein assay kit according to the manufacturer's instructions. An equal weight of cell lysates was subjected to SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. The blots were blocked for 30 min in Tris-buffered saline containing 5% low-fat powdered milk and 1% FBS and then incubated with the primary antibody (in blocking buffer) overnight at 4°C. The blots were washed three times with 1× TBST (140 mM NaCl, 3mM KCl, 25 mM Tris-Hcl [pH 7.4], 0.1% Tween 20) (each wash was carried out for approximately 10 min). The blots were then incubated with horseradish peroxidase-conjugated secondary antibody (Bio-Rad) in blocking buffer for 1 h at room temperature. Blots were washed as described above and analyzed with an enhanced chemiluminescence detection system (Perkin-Elmer) according to the manufacturer's recommendations, and filters were exposed to Fuji Super RX film. The following primary antibodies were used for Western blot analysis: anti-c-Myc MAb (sc-42; Santa Cruz), anti-E2F1 MAb (sc-251; Santa Cruz), and anti-BMRF1 MAb (EBV 12900; Capricorn). The anti-Zta polyclonal antibody N5 was generated with the bZIP domain of Zta and affinity purified with a glutathione S-transferase Zta affinity column.
RESULTS
Inhibition of reactivation by enforced c-Myc expression.
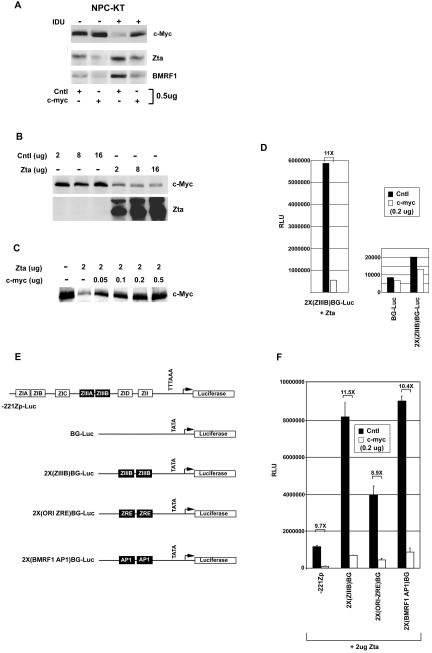
It has previously been shown that treatment of the EBV-positive epithelial cell line, NPC-KT, with the lytic cycle-inducing agent IDU results in downregulation of c-Myc expression and that c-Myc downregulation precedes noticeable induction of the immediate-early transcription factor Zta (22). To test whether inhibition of c-Myc expression is required for maximal induction of reactivation in this system, we transfected NPC-KT cells with a control expression vector or a c-Myc expression vector and treated with IDU. Seventy-two hours later, expression of Zta or the Zta responsive early gene, the BMRF1 gene, was monitored by Western blotting. It should be noted that transfection efficiencies in NPC-KT cells are typically near 80% (see Fig. 3; data not shown). As shown in Fig. 1A, expression of both Zta and BMRF1 is inhibited in cells transfected with the c-Myc expression vector. This indicates that downregulation of c-Myc in this system is required for maximal induction of reactivation.
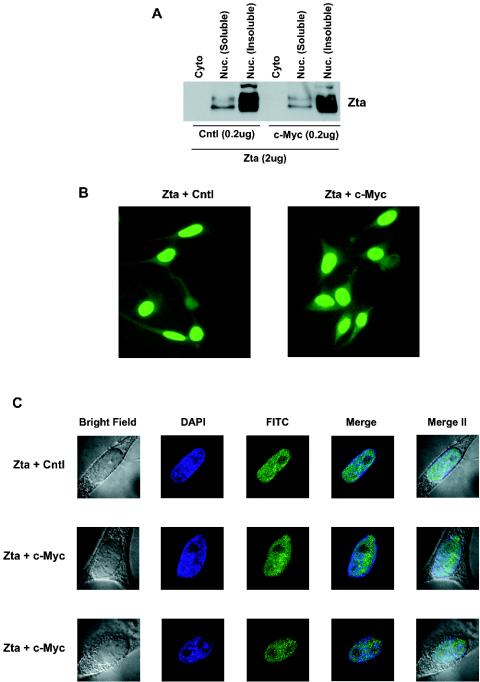
FIG. 3.
c-Myc does not alter the nuclear localization of Zta. (A) NPC-KT cells were transfected with the indicated amounts of expression vectors, and cells were fractionated as described in Materials and Methods. Zta expression was monitored by Western blot analysis. (B) Immunofluorescence assays were performed on NPC-KT cells transfected with either Zta or Zta plus c-Myc expression plasmids. Cells were stained for Zta with a Zta MAb (Argene) and FITC-conjugated goat anti-mouse immunoglobulin (Biosource). (C) Deconvolution images of NPC-KT cells transfected with either Zta or Zta plus c-Myc expression plasmids. Cells were stained for Zta with a Zta MAb and DAPI (DNA). Merge, overlays of FITC and DAPI images; Merge II, overlays of FITC, DAPI, and bright fields. Cells shown are representative examples of multiple cells that were analyzed.
FIG. 1.
Inhibition of reactivation by enforced c-Myc expression. NPC-KT cells were transfected in 100-mm plates by the calcium phosphate precipitation method described in Materials and Methods. (A to D and F) Cells were transfected with the indicated amounts of either a c-Myc expression vector (pcDNA3-c-myc), a c-Myc control plasmid (pcDNA3), the Zta expression vector BS(SVp/e)-Zta, or the Zta control vector BS(SVp/e). Cells were harvested 48 h after transfection, protein levels were assessed by Western blot analysis, and reporter activity was analyzed by a luciferase assay (D) Cells were transfected with 2 μg of BS(SVp/e) (right panel) or BS(SVp/e)-Zta (left panel) and 200 ng of either pcDNA3 (Cntl) or pcDNA3-c-myc (c-myc). The data shown are a representative result of three experiments. RLU, relative light units. (E) Schematic representation of reporter constructs.
c-Myc inhibits the transactivation function of Zta.
Since activation of Zta expression is one of the first viral events in the lytic cascade, the results shown in Fig. 1A indicate that c-Myc can inhibit the initial stages of reactivation. Induction of the Zta promoter, Zp, is thought to occur through a two-step model (9, 10, 24). Initial induction of Zp occurs through the activation of promoter elements that bind cellular transcription factors (10). Once functional levels of Zta are obtained through these initial signaling events, Zta is thought to bind to two Zta binding sites in Zp to establish maximal expression of Zta (9, 16, 17, 25). To begin to address the mechanism of c-Myc-mediated inhibition of Zta induction, we tested whether c-Myc could inhibit either the initial promoter activation stage or the autoactivation step.
To test whether c-Myc can inhibit Zp induction by cellular transcription factors during the initial stage of induction, we transfected the EBV-negative epithelial cell line HeLa with a reporter plasmid containing the regulatable region of Zp (extending to −221) cloned upstream from a luciferase gene. Zp was induced with the phorbol ester tetradecanoyl phorbol acetate, which concomitantly inhibits the expression of c-Myc (data not shown). Reconstitution of c-Myc levels through cotransfection with a c-Myc expression vector consistently showed a moderate two- to threefold inhibition of Zp induction (data not shown), indicating that c-Myc downregulation may in part favor induction of Zta by cellular transcription factors.
Although it has previously been shown that treatment of NPC-KT cells with IDU results in downregulation of c-Myc prior to detectable Zta expression, transfection of a Zta expression vector into NPC-KT cells in the absence of IDU similarly inhibits c-Myc expression (Fig. 1B). Transfection of approximately 200 to 500 ng of a c-Myc expression vector can reconstitute c-Myc expression to its normal level (Fig. 1C). To assess whether c-Myc influences Zta transactivation function, we transfected NPC-KT cells with the Zta-responsive reporter 2X(ZIIIB)BG-Luc, 2 μg of a Zta expression vector, and 200 ng of either a control or a c-Myc expression vector. Reconstitution of c-Myc expression strongly suppressed Zta-mediated transactivation. In contrast, c-Myc did not influence expression of the control plasmid, BG-Luc, or 2X(ZIIIB)BG-Luc in the absence of Zta (Fig. 1D), indicating that inhibition is specific to Zta function. Inhibition of Zta transactivation by c-Myc was also observed with other Zta-responsive promoters such as the Zta promoter −221Zp, as well as artificial reporters containing two copies of either a ZRE from the lytic origin of replication or the BMRF1 AP1 site (which Zta binds with high affinity) (Fig. 1F). Inhibition of Zta-mediated transactivation is dose responsive, with 500 ng of the c-Myc expression plasmid resulting in nearly a 25-fold inhibition of Zta transactivation functions (Fig. 2A). Importantly, inhibition of Zta-mediated transactivation is not an artifact, due to c-Myc inhibiting Zta expression from the Zta expression vector (Fig. 2A).
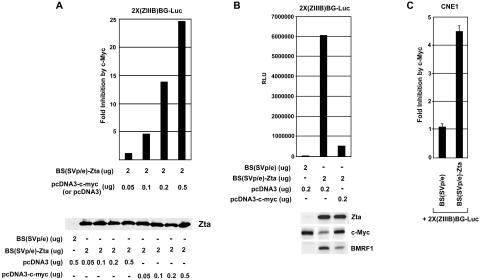
FIG. 2.
Cells were transfected and analyzed as described in the legend to Fig. 1. (A and B) Results of representative experiments are shown. RLU, relative light units.(A) Inhibition of Zta transactivation by c-Myc is dose dependent. NPC-KT cells were transfected with the indicated amounts of Zta expression plasmid BS(SVp/e)-Zta or Zta control expression plasmid BS(SVp/e) plus either the control c-Myc plasmid pcDNA3 or the c-Myc expression plasmid pcDNA3-c-myc. (B) Inhibition of Zta transactivation of the endogenous Zta-responsive BMRF1 gene in NPC-KT cells. (C) Inhibition of Zta transactivation in the EBV-negative nasopharyngeal cell line CNE1. CNE1 cells were transfected with 2 μg of BS(SVp/e) or BS(SVp/e)-Zta plus 200 ng of pcDNA3 or pcDNA3-c-myc.
To assess whether c-Myc inhibits Zta-mediated transactivation of Zta-responsive genes in their natural genomic context, we transfected NPC-KT cells with a Zta expression vector plus either a control or a c-Myc expression vector; expression of the endogenous Zta-responsive BMRF1 promoter was monitored by Western blot analysis of BMRF1 expression. As shown in Fig. 2B, c-Myc inhibits Zta-mediated transactivation of endogenous BMRF1. Lastly, we assessed whether inhibition of Zta function occurs in other epithelial cell lines. As shown in Fig. 2C, c-Myc similarly inhibits Zta-mediated transactivation in the EBV-negative nasopharyngeal carcinoma cell line CNE1.
c-Myc does not alter the gross localization of Zta.
To begin to investigate the mechanism through which c-Myc influences Zta function, we first tested whether c-Myc altered the nuclear localization of Zta. As expected, Zta was localized in the nuclear fraction, and the bulk of Zta was found in the insoluble portion of the nuclear fraction which is typically associated with chromatin (Fig. 3A). Notably, reconstituted c-Myc expression did not alter the gross localization of Zta in the soluble or insoluble nuclear fraction. Analysis of Zta localization by fluorescence microscopy similarly did not detect any notable alteration of Zta nuclear localization in the presence of c-Myc (Fig. 3B and C). This does not preclude the possibility that some subnuclear redistribution cannot be detected by these assays, but it indicates that there is no gross alteration in the localization of Zta to the nucleus or to the insoluble nuclear fraction.
The cell cycle regulatory factor E2F1 inhibits Zta transactivation.
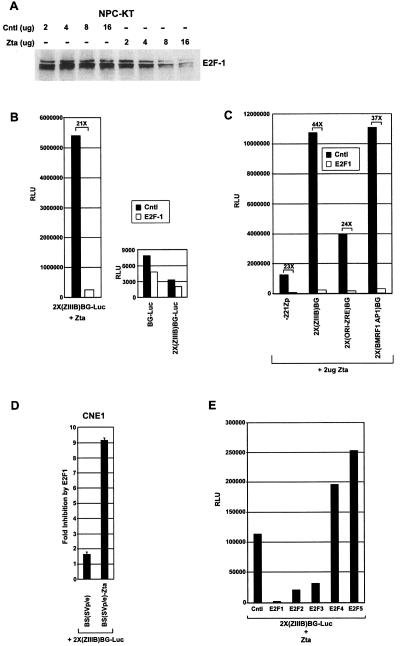
c-Myc and E2F1 are thought to have related functions and are known to signal in part through similar signal transduction pathways. Like c-Myc, the transcription factor E2F1 can induce quiescent cells to enter the cell cycle (5). Moreover, c-Myc has been shown to induce the expression of E2F1, and E2F1 has been shown to induce the expression of c-Myc (1). It had previously been shown that Zta can inhibit expression of E2F1 in HeLa cells (3); as shown in Fig. 4A, Zta similarly suppressed E2F1 expression in NPC-KT cells. We therefore tested whether E2F1 could inhibit Zta transactivation. Figure 4B shows that E2F1 inhibits Zta-mediated transactivation of the reporter plasmid 2X(ZIIIB)BG-Luc, but importantly, E2F1 does not inhibit the control plasmid BG-Luc or the 2X(ZIIIB)BG-Luc plasmid in the absence of Zta. As was observed with c-Myc, E2F1 also inhibits a panel of Zta-responsive promoters including Zp (Fig. 4C), and inhibition is observed in the cell line CNE1 (Fig. 4D).
FIG. 4.
Regulation of Zta transactivation by E2F1. (A) Cells were transfected as described in the legend to Fig. 1 with the indicated amounts of either a control [Cntl; BS(SVp/e)] or Zta [BS(SVp/e)-Zta] expression vectors, cells were harvested 48 h later, and E2F1 expression was monitored by Western blot analysis. (B) NPC-KT cells were transfected with 2 μg of either a control vector (right panel) or a Zta expression vector (left panel) plus 200 ng of either a control vector (pRC-CMV) or a E2F1 (pRC-CMV-HA-E2F1) expression vector, and reporter activity was monitored by a luciferase assay. (C) NPC-KT cells were transfected and analyzed as described above. (D) CNE1 cells were transfected and analyzed as above. (E) NPC-KT cells were transfected with 200 ng of the control or the indicated E2F expression vector plus 2 μg of BS(SVp/e)-Zta expression vector. The data shown in panels B, C, and E are representative results of three experiments. RLU, relative light units.
Like E2F1, the E2F family members E2F2 and E2F3 have also been shown to be able to drive quiescent cells into the cell cycle, while the E2F family members E2F4 and E2F5 cannot (5, 6). Although not as potent as E2F1, E2F2 and E2F3 inhibit Zta-mediated transactivation, whereas E2F4 and E2F5 failed to inhibit Zta-mediated transactivation (Fig. 4E). These data suggest a possible connection between the ability of c-Myc and E2F1 to drive cell proliferation and their ability to inhibit Zta.
Requirement for the Zta activation domain for c-Myc- and E2F1-mediated inhibition of Zta transactivation.
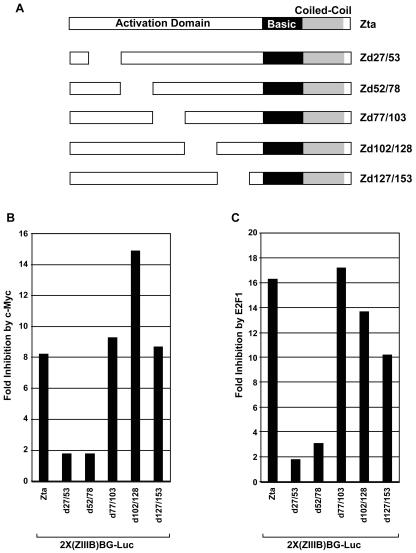
As discussed above, localization studies suggest the possibility that c-Myc does not influence the ability of Zta to localize to the nucleus or the nuclear insoluble fraction. This suggested that c-Myc may instead specifically influence the transactivation function of Zta. We therefore carried out a genetic analysis of the Zta activation domain to determine whether c-Myc inhibits Zta function through this region. Analysis of the 27-amino-acid scanning mutants shown in Fig. 5 showed a dependence on sequences between amino acids 27 and 78. Moreover, E2F1-mediated inhibition of Zta transactivation showed a similar genetic profile, suggesting that the mechanism of action of c-Myc and E2F1 is similar.
FIG. 5.
Genetic analysis of c-Myc- and E2F1-mediated inhibition of Zta transactivation. (A) Schematic of Zta deletions. (B) NPC-KT cells were transfected with 200 ng of either a control (pcDNA3) or a c-Myc (pcDNA3-c-myc) expression vector plus the reporter plasmid 2X(ZIIIB)BG-Luc and 2 μg of the indicated Zta or Zta mutant expression plasmid. (C) Cells were transfected with 200 ng of either a control (pRC-CMV) or E2F1 (pRC-CMV-HA-E2F1) expression plasmid plus the reporter plasmid 2X(ZIIIB)BG-Luc and 2 μg of the indicated Zta or Zta mutant expression plasmid.
c-Myc- and E2F1-mediated inhibition of Zta transactivation maps to a crucial tryptophan residue at position 49 of the Zta activation domain.
To further refine sequences in the Zta activation domain that are required for c-Myc- and E2F1-mediated inhibition, a series of five-amino-acid deletion mutants were generated spanning amino acids 9 to 53 of Zta; these mutants were tested for inhibition by c-Myc and E2F1. As was observed with the 27-amino-acid deletions shown in Fig. 5, the genetics of c-Myc- and E2F1-mediated inhibition with this five-amino-acid deletion series are similar. Further, this analysis revealed a bipartite motif spanning amino acids 29 to 33 and amino acids 44 to 53 that is required for maximal inhibition by both c-Myc and E2F1 (Fig. 6B). Notably, a tryptophan residue is located within the second domain (i.e., the region spanning amino acids 44 to 53). Since tryptophan residues are rare amino acids and since they are commonly involved in protein-protein interactions, we generated a single-amino-acid Zta mutant in which this residue was changed to an alanine. This mutation severely mitigates the ability of either c-Myc or E2F1 to inhibit Zta transactivation function. This indicates that c-Myc- and E2F1-mediated inhibition of Zta transactivation is mediated through a common pathway, likely involving protein-protein interactions with tryptophan 49 of Zta. As shown in Fig. 6C, mutation of tryptophan 49 to alanine also prevents c-Myc-mediated inhibition of the endogenous Zta-responsive gene BMRF1, indicating that these results are not specific to reporter assays and reflect the true mechanism through which c-Myc inhibits Zta's transactivation function. It should also be noted that the loss of c-Myc responsiveness observed with the deletion mutants and the tryptophan 49 mutant is not due to a loss of Zta function (or a decrease in baseline activity), since all of these mutants retain significant transactivation potential with >106 relative light units in the absence of c-Myc (Fig. 6B, right panels, and data not shown). In addition, there was no loss in the ability of the Zta mutants ZW49A, Zd44/48, and Zd27/53 to activate the endogenous BMRF1 gene (Fig. 6C). This argues strongly that these regions of the Zta activation domain play an important role in mediating inhibition by c-Myc and E2F1.
FIG. 6.
Refined genetic analysis of c-Myc- and E2F1-mediated inhibition of Zta transactivation. (A and B) Transfections were carried out as described in the legend to Fig. 5, and BMRF1 and Zta expression levels were assayed (C) by Western blot analysis. The right panels are representative raw data from results shown in the middle panels to illustrate that loss of responsiveness for the Zta mutants is not due to low levels of Zta transactivation activity.
c-Myc inhibition of Zta transactivation is mediated through a limiting regulatory factor.
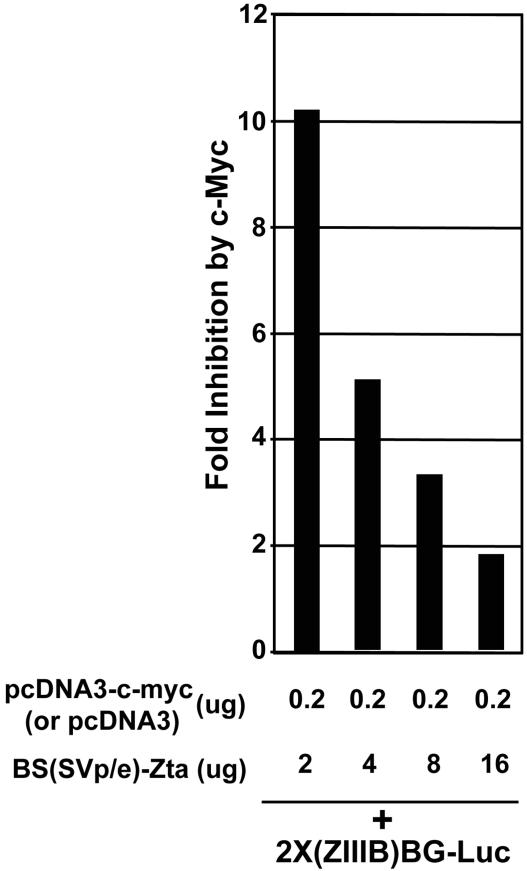
To further assess the nature of the c-Myc-mediated inhibitory factor(s), we tested whether excess Zta expression can overcome c-Myc inhibition. As shown in Fig. 7, higher levels of Zta expression can effectively titrate away the inhibitory factor, suggesting that the inhibitory factor is limiting. This also suggests that once the virus has committed to a high level of Zta expression, c-Myc-mediated inhibition no longer plays a regulatory role in reactivation. This could be a mechanism to ensure that once the virus has committed to reactivation (i.e., through high-level induction of Zta), c-Myc can no longer interfere with Zta function and therefore cannot impede high-level progression through the lytic cycle.
FIG. 7.
c-Myc-mediated inhibition of Zta transactivation is overcome by high levels of Zta expression.
DISCUSSION
Numerous studies have established that reactivation of EBV is associated with cell cycle arrest (3, 4, 15, 18, 21-23, 27), and it is assumed that this supports an environment for more-robust viral replication. The studies presented here provide evidence that in epithelial cells, EBV has evolved with a gatekeeper mechanism to control reactivation until cell growth arrest has been achieved. A number of latency-associated EBV genes have been shown to promote cell cycle progression, in part through the induction of c-Myc (28). As such, in latently infected cells, high-level c-Myc expression may provide a mechanism to help suppress reactivation (Fig. 8). It has previously been shown that treatment of NPC-KT cells with IDU results in downregulation of c-Myc expression prior to the induction of detectable Zta levels (22). It has also been found that Zta can inhibit c-Myc expression, suggesting that in cases where the inducing agent cannot specifically inhibit c-Myc expression, Zta itself can accomplish this task (3, 23). Notably, even low levels of Zta can efficiently induce growth arrest (reference 3 and data not shown), indicating that Zta may be able to downregulate c-Myc expression during the early stages of reactivation. In addition, previous studies have shown that suppression of c-Myc expression is independent of Zta's transactivation function (23). Therefore, during the early stages of Zta expression, Zta should be fully capable of carrying out its c-Myc downregulatory function. At the same time, its ability to activate transcription of downstream viral promoters is suppressed until it has successfully inhibited c-Myc expression.
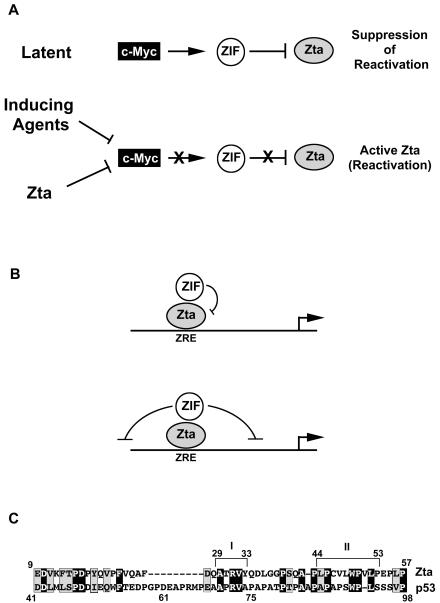
FIG. 8.
(A) Model depicting role of c-Myc-mediated inhibition of Zta in the EBV life cycle. (B) Model for mechanism of action of c-Myc- and E2F1-regulated ZIF. (C) Illustration of bipartite c-Myc-E2F1 inhibitory sequences in Zta (domains I and II) and alignment with the proline-rich domain of p53.
We have shown that c-Myc and E2F1 can both inhibit Zta-mediated transactivation, and we have provided a genetic correlation suggesting that both of these factors inhibit Zta through a common pathway. Although c-Myc and E2F1 likely have functions that are unique, they can also clearly signal through a common pathway, as evidenced in part by the observation that E2F1 can activate c-Myc expression and that c-Myc can signal E2F1 expression (1, 5). In addition, both of these factors have previously been shown to induce quiescent cells to enter the cell cycle (1, 5, 6). We have also tested whether other cell cycle-promoting genes such as cyclin E, which is similarly able to drive cells to enter the cell cycle, can inhibit Zta transactivation; however, enforced expression of these factors had minimal effects on Zta function (Z. Lin and E. Flemington, unpublished data). This suggests that the ability of c-Myc and E2F1 to drive cells into the cell cycle alone may not be sufficient to account for their ability to inhibit Zta transactivation and that there is something unique to the c-Myc/E2F pathway that mediates inhibition of Zta. Notably, Zta has also been found to induce expression of the tumor suppressor protein p53 in epithelial cells (3, 19). The coordinate induction of p53 along with the downregulation of c-Myc and E2F1 has been correlated with induction of growth arrest, while in some settings, induction of p53 along with enforced c-Myc or E2F1 expression has been shown to induce apoptosis (1, 5, 6), although we did not observe c-Myc or E2F1 inducing apoptosis in our experiments with NPC-KT or CNE1 cells (data not shown). Therefore, in some cases, downregulation of c-Myc and E2F1 during EBV reactivation may serve not only to inhibit cell cycle progression but also to protect against p53-mediated apoptosis. As such, it makes sense that there may be a selective use of these cell cycle effectors in a gatekeeper function.
It has previously been postulated that although EBV, like other members of the herpesvirus family, induces growth arrest, it may also induce certain cyclin and/or cyclin-dependent kinase functions to promote a pseudo-S-phase environment, which in turn would help support robust viral DNA replication (8). A recent report has indeed shown that in the B-lymphoblastoid cell line B95-8, expression of Zta leads to induction of cyclins A and E (15). Therefore, the selective ability of c-Myc and E2F1, but not cyclins, to inhibit Zta transactivation may also allow the concomitant development of a pseudo-S-phase environment.
We have shown that inhibition of Zta by c-Myc is not due to changes in its ability to localize to soluble or insoluble nuclear fractions. Instead, our studies suggest that inhibition is mediated through protein-protein interactions with the Zta activation domain. We have tested whether E2F1 or c-Myc bind Zta in vivo, using coimmunoprecipitation assays (data not shown), but we have thus far failed to detect any interaction between these factors. We cannot rule out the possibility that the inability to detect an interaction between these factors is due to technical issues. However, we favor a model whereby c-Myc and E2F1 are in a common pathway that leads to the activation of a Zta inhibitory factor(s) (ZIF) (Fig. 8) that associates with the activation domain of Zta.
There are two general means by which c-Myc and E2F1 might inhibit Zta transactivation function. First, it is possible that c-Myc and E2F1 simply inhibit the function of a coactivator that binds to Zta. Importantly, however, the Zta mutants that are nonresponsive to inhibition consistently have a slightly higher level of activation potential than wild-type Zta in the presence of c-Myc or E2F1 in reporter assays (Fig. 6 and data not shown). This issue is even more dramatic when assessing transactivation of the endogenous BMRF1 promoter, where the mutants show a much higher induction of BMRF1 in the presence of c-Myc than of wild-type Zta (Fig. 6C). These data argue against a mechanism involving the inhibition of a Zta coactivator and suggest that c-Myc and E2F1 instead specifically elicit a configuration that actively suppresses Zta transactivation or suppresses the function of promoters bound by Zta (Fig. 8B). Such an inhibitory factor could either alter Zta function (for example, through covalent modification) or act on local chromatin structure or factors in the basal transcriptional apparatus (Fig. 8B).
Interestingly, we have found that both sequences in the bipartite c-Myc inhibitory domain (amino acids 29 to 33 and 44 to 53) of Zta have homology to the proline-rich domain of the tumor suppressor protein p53 (Fig. 8C). The proline-rich domain of p53 has been shown to be crucial for its apoptotic activity and has been shown to be required for p53-mediated inhibition of a certain class of promoters (26). In another study, a short region of p53 spanning amino acids 61 to 75 (which contains the homology to the domain of Zta spanning amino acids 30 to 34), was shown to be critical for binding to the transcriptional repressor SIN3A (30). These data suggest the possibility that inhibition of promoters by p53 could be mediated by a factor that plays a role in modulating Zta function by c-Myc and E2F1. Another potential implication of the homology between Zta and p53 is the possibility that like Zta, p53 can be downregulated by c-Myc and E2F1 through the binding of a factor to its proline-rich domain. Like Zta, p53 induces growth arrest, and suppression of p53 by ZIF would represent a novel mechanism by which c-Myc and E2F1 could counter the growth arrest activity of p53 in proliferating cells.
The studies presented here have been carried out with epithelial cell systems. We have begun addressing by transient reporter assays whether Zta is similarly regulated by c-Myc in several B-cell lines, but thus far we have observed either minimal or no inhibition by c-Myc (data not shown). At this time, we do not know whether these differences are due to technical issues related to distinct means of introducing DNA into B cells or whether there is a tissue-specific difference in the way that reactivation is regulated by cell cycle regulatory factors. Nevertheless, Fais et al. (7) have reported that stable transfection of a c-Myc expression vector into lymphoblastoid cell lines resulted in a decrease in spontaneous reactivation and expression of endogenous early genes. Therefore, it is possible that Zta function can be inhibited by c-Myc in B lymphocytes and that this can be observed by a different technical approach (i.e., stable transfection and/or analysis of endogenous Zta-responsive promoters). Alternatively, it is possible that inhibition of reactivation by c-Myc is mediated through a different mechanism, such as the inhibition of cellular transcription factors that bind to Zp. Further investigations will be required to explore these issues and to address the relationship between cell cycle control factors and reactivation in B lymphocytes.
Acknowledgments
We thank Gilbert Morris for critically reading the manuscript.
This work was supported by National Institutes of Health research grant R01 GM48045 (E.F.).
REFERENCES
- 1.Bartek, J., and J. Lukas. 2001. Pathways governing G1/S transition and their response to DNA damage. FEBS Lett. 490:117-122. [DOI] [PubMed] [Google Scholar]
- 2.Campanero, M., M. Armstrong, and E. Flemington. 1999. Distinct cellular factors regulate the c-myb promoter through its E2F element. Mol. Cell. Biol. 19:8442-8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cayrol, C., and E. Flemington. 1996. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 15:2748-2759. [PMC free article] [PubMed] [Google Scholar]
- 4.Cayrol, C., and E. K. Flemington. 1996. G0/G1 growth arrest mediated by a region encompassing the bZIP domain of the Epstein-Barr virus transactivator Zta. J. Biol. Chem. 271:31799-31802. [DOI] [PubMed] [Google Scholar]
- 5.DeGregori, J. 2002. The genetics of E2F family of transcription factors: shared functions and unique roles. Biochim. Biophys. Acta 1602:131-150. [DOI] [PubMed] [Google Scholar]
- 6.DeGregori, J., G. Leone, A. Miron, L. Jakoi, and J. R. Nevins. 1997. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. USA 94:7245-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fais, F., G. Cutrona, M. Ulivi, S. Roncella, M. C. Gagliardi, P. Cornaglia-Ferraris, M. Rowe, V. Barnaba, and M. Ferrarini. 1996. Lymphoblastoid cells transfected with c-myc: downregulation of EBV-lytic antigens and impaired response of autologous CD4+ T cells in vitro. Int. J. Cancer 68:810-816. [DOI] [PubMed] [Google Scholar]
- 8.Flemington, E. K. 2001. Herpesvirus lytic replication and the cell cycle: arresting new developments. J. Virol. 75:4475-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flemington, E., and S. Speck. 1990. Autoregulation of the Epstein-Barr virus putative lytic switch gene BZLF1. J. Virol. 64:1227-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flemington, E., and S. Speck. 1990. Identification of phorbol ester response elements in the promoter of the Epstein-Barr virus putative lytic switch gene BZLF1. J. Virol. 64:1217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuda, M., K. Ikuta, K. Yanagihara, M. Tajima, H. Kuratsune, T. Kurata, and T. Sairenji. 2001. Effect of transforming growth factor-beta1 on the cell growth and Epstein-Barr virus reactivation in EBV-infected epithelial cell lines. Virology 288:109-118. [DOI] [PubMed] [Google Scholar]
- 12.Fukuda, M., T. Satoh, M. Takanashi, K. Hirai, E. Ohnishi, and T. Sairenji. 2000. Inhibition of cell growth and Epstein-Barr virus reactivation by CD40 stimulation in Epstein-Barr virus-transformed B cells. Viral Immunol. 13:215-229. [DOI] [PubMed] [Google Scholar]
- 13.Inman, G. J., U. K. Binné, G. A. Parker, P. J. Farrell, and M. J. Allday. 2001. Activators of the Epstein-Barr virus lytic program concomitantly induce apoptosis, but lytic gene expression protects from cell death. J. Virol. 75:2400-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieff, E. 1996. Epstein-Barr virus and its replication, p. 2343-2396. In B. C. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 15.Kudoh, A., M. Fujita, T. Kiyono, K. Kuzushima, Y. Sugaya, S. Izuta, Y. Nishiyama, and T. Tsurumi. 2003. Reactivation of lytic replication from B cells latently infected with Epstein-Barr virus occurs with high S-phase cyclin-dependent kinase activity while inhibiting cellular DNA replication. J. Virol. 77:851-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieberman, P., and A. Berk. 1990. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J. Virol. 64:2560-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieberman, P. M., J. M. Hardwick, J. Sample, G. S. Hayward, and D. S. Hayward. 1990. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRA sites in target promoter and enhancer regions. J. Virol. 64:1143-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mauser, A., E. Holley-Guthrie, D. Simpson, W. Kaufmann, and S. Kenney. 2002. The Epstein-Barr virus immediate early protein BZLF1 induces both G2 and a mitotic block. J. Virol. 76:10030-10037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mauser, A., S. Saito, E. Appella, C. Anderson, W. Seaman, and S. Kenney. 2002. The Epstein-Barr virus immediate-early protein BZLF1 regulates p53 function through multiple mechanisms. J. Virol. 76:12503-12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, G. 1990. Epstein-Barr virus, p. 1921-1958. In B. N. Fields and D. M. Knipe (ed.), Virology. Raven Press, New York, N.Y.
- 21.Rodriguez, A., M. Armstrong, D. Dwyer, and E. Flemington. 1999. Genetic dissection of cell growth arrest functions mediated by the Epstein-Barr virus lytic gene product, Zta. J. Virol. 73:9029-9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez, A., E. J. Jung, and E. K. Flemington. 2001. Cell cycle analysis of Epstein-Barr virus-infected cells following treatment with lytic cycle-inducing agents. J. Virol. 75:4482-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez, A., E. Jung, Q. Yin, C. Cayrol, and E. Flemington. 2001. Role of c-myc regulation in Zta mediated induction of the cyclin dependent kinase inhibitors, p21 and p27, and cell growth arrest. Virology 284:159-169. [DOI] [PubMed] [Google Scholar]
- 24.Speck, S. H., T. Chatila, and E. K. Flemington. 1997. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 5:399-405. [DOI] [PubMed] [Google Scholar]
- 25.Urier, G., M. Buisson, P. Chambard, and A. Sergeant. 1989. The Epstein-Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J. 8:1447-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venot, C., M. Maratrat, C. Dureuil, E. Conseiller, L. Bracco, and L. Debussche. 1998. The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J. 17:4668-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu, F. Y., H. Chen, S. E. Wang, C. M. J. apRys, G. Liau, M. Fujimuro, C. J. Farrell, J. Huang, S. D. Hayward, and G. S. Hayward. 2003. CCAAT/enhancer binding protein α interacts with ZTA and mediates ZTA-induced p21CIP-1 accumulation and G1 cell cycle arrest during the Epstein-Barr virus lytic cycle. J. Virol. 77:1481-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young, L. S., C. W. Dawson, and A. G. Eliopoulos. 2000. The expression and function of Epstein-Barr virus encoded latent genes. Mol. Pathol. 53:238-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young, L. S., and M. Rowe. 1992. Epstein-Barr virus, lymphomas and Hodgkin's disease. Semin. Cancer Biol. 3:273-284. [PubMed] [Google Scholar]
- 30.Zilfou, J. T., W. H. Hoffman, M. Sank, D. L. George, and M. Murphy. 2001. The corepressor mSin3a interacts with the proline-rich domain of p53 and protects p53 from proteasome-mediated degradation. Mol. Cell. Biol. 21:3974-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]