Abstract
The tripartite ATP-independent periplasmic (TRAP) transporters are a widespread class of membrane transporters in bacteria and archaea. Typical substrates for TRAP transporters are organic acids including the sialic acid N-acetylneuraminic acid. The substrate binding proteins (SBP) of TRAP transporters are the best studied component and are responsible for initial high-affinity substrate binding. To better understand the dynamics of the ligand binding process, pulsed electron-electron double resonance (PELDOR, also known as DEER) spectroscopy was applied to study the conformational changes in the N-acetylneuraminic acid-specific SBP VcSiaP. The protein is the SBP of VcSiaPQM, a sialic acid TRAP transporter from Vibrio cholerae. Spin-labeled double-cysteine mutants of VcSiaP were analyzed in the substrate-bound and -free state and the measured distances were compared to available crystal structures. The data were compatible with two clear states only, which are consistent with the open and closed forms seen in TRAP SBP crystal structures. Substrate titration experiments demonstrated the transition of the population from one state to the other with no other observed forms. Mutants of key residues involved in ligand binding and/or proposed to be involved in domain closure were produced and the corresponding PELDOR experiments reveal important insights into the open-closed transition. The results are in excellent agreement with previous in vivo sialylation experiments. The structure of the spin-labeled Q54R1/L173R1 R125A mutant was solved at 2.1 Å resolution, revealing no significant changes in the protein structure. Thus, the loss of domain closure appears to be solely due to loss of binding. In conclusion, these data are consistent with TRAP SBPs undergoing a simple two-state transition from an open-unliganded to closed-liganded state during the transport cycle.
Introduction
All bacteria enclose themselves from their environment with at least one membrane. To survive in a given environment, they use membrane transporters to actively import any available nutrients. Although bacteria possess a large variety of substrate-specific active transporters, they can be grouped into a small number of major classes: ABC transporters (1), secondary active transporters (2), the phosphotransferase system (3), and tripartite ATP-independent periplasmic (TRAP) transporters (4). TRAP transporters are currently the least well studied class. They are absent in eukaryotic organisms but widespread in bacteria and are also found in archaea. A typical TRAP transporter consists of three structural domains: a high affinity substrate binding protein (SBP) and two transmembrane domains (TMDs) with four and twelve predicted transmembrane helices (4). The domains are commonly referred to as P domain (substrate binding protein), Q domain (smaller TMD), and M domain (larger TMD). The Q- and M domains are either fused into one protein or expressed as separate proteins that form a tight complex (4). As indicated by their name, TRAP transporters are independent of ATP hydrolysis and some representatives have been shown to rely on a Na+ gradient and membrane potential to power the transport mechanism (5, 6). This is considered a reason why TRAPs are especially widespread in marine microorganisms (7). Molecules known to be transported by TRAP transporters range from small organic acids including C4-dicarboxylates, larger sugar acids like N-acetylneuraminic acid (Neu5Ac), to amino acids (4, 8). Most TRAP transporter substrates contain a carboxylic acid group, which is specifically recognized by the P domain of the transporter (9).
High-resolution structural information about TRAP transporters is currently only available for the soluble P domains. The first crystal structure of such a domain was solved in 2006 (10) and several more structures either with or without substrate followed (reviewed in (11)). All P domain structures can be characterized by two αβ-domains that are connected by an extended hinge helix and a substrate-binding cleft between the two αβ-domains (Fig. 1). In the substrate-bound state, the two αβ-domains close around the substrate reminiscent of a Venus flytrap and the reverse motion is thought to occur when the substrate is channeled into the transporter (4), likely by an allosteric mechanism through conformational changes in the membrane domains (12). The overwhelming majority of P domains have a conserved arginine in the substrate binding cleft (position 147 in HiSiaP). This residue is crucial for the substrate interaction by recognizing the aforementioned carboxylic acid group in the substrate. It thereby acts as a selectivity filter for the transporter, allowing the SBP to recognize organic acids with high affinity and specificity (9). Thus, P domains are structurally well characterized in their two resting states, namely “open ligand-free” and “closed ligand-bound”, and the interactions between substrate and protein are well studied. However, as with all dynamic systems, it is of high interest to analyze how well the crystal structures reflect the solution state. An important question with implications for the mechanism of the whole transporter is, whether in solution, the P domain is present in equilibrium between open- and closed forms or if the conformational change is strictly substrate induced. Also, it is possible that there are additional stable intermediate states of the protein that have not yet been discovered by crystallography. Here, pulsed electron-electron double resonance (PELDOR) spectroscopy (also known as double electron-electron resonance (DEER) spectroscopy) was applied to analyze the structure of the P domain of VcSiaPQM, a Neu5Ac transporter from Vibrio cholerae, in solution (13, 14). Site-directed spin labeling (15, 16) was used to introduce nitroxide spin labels at positions that allow to readily distinguish the open- and closed states of the protein. These labeled forms were then used to study the structure of the protein in solution. Further, residues that have been proposed as crucial for the function of the P domain have been mutated and the effects were analyzed. The crystal structure of one of these spin-labeled VcSiaP mutants (R125A) was solved at 2.1 Å. The structure verifies that neither the R125A mutation nor the spin-labeling process disturbed the overall structure of the protein and it validates the PELDOR distances. Taken together, the results demonstrate, for the first time to our knowledge, that a TRAP SBP has two clear states in solution—an open unliganded- and a closed ligand-bound form. This supports current models of an allosteric mechanism for ligand release that is catalyzed by conformational changes in the membrane domains.
Figure 1.
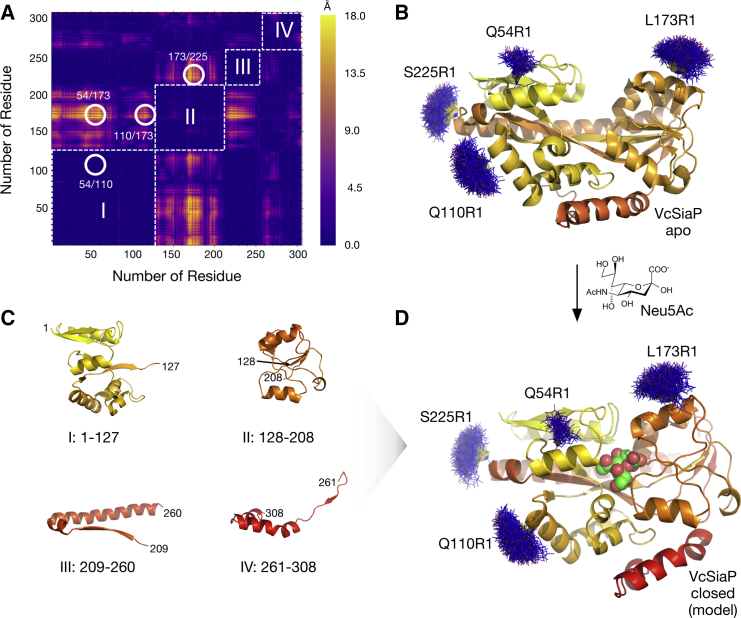
Structural changes of P domains. (A) Difference distance matrix (diffDM) for the substrate-bound and -free forms of HiSiaP (PDB: 3B50 (37), PDB: 2CEY (10)). (Dark violet regions) Pairs of residues, where the Cβ-Cβ distance does not change between both conformations. (Yellow peaks) Large distance changes of up to 18 Å. (White circles) Pairs of residues that were selected as spin labeling sites. The violet squares along the diagonal of the matrix (dashed, white lines) can be interpreted as rigid domains (I–IV) of the P domain. Note that the matrix is symmetric along its diagonal. (B) The substrate-free structure of VcSiaP (PDB: 4MAG (23)). The protein is shown as cartoon model. A color gradient is running from yellow (N-terminus) to red (C-terminus) to indicate the trace of the polypeptide chain. Models of spin labels at positions highlighted in (A) were attached with MtsslWizard (blue lines). (C) Cartoon models of the individual structures of the rigid domains I-IV of substrate-free VcSiaP. (D) Model of the closed form of VcSiaP. The model was produced by superposing the rigid domains I-IV in (C) onto the structure of closed HiSiaP (PDB: 3B50 (37)). The model of the bound Neu5Ac is shown as spheres.
Materials and Methods
Cloning, protein expression, purification, and spin labeling
The VcSiaP encoding gene (omitting the N-terminal signal sequence) was PCR-amplified from genomic V. cholerae DNA using oligos: 5′-GTT ATT CCA TGG GGG CGA CGA CTT TAA AGA TGG GG-3′ (forward) and 5-TTC TTC GTC GAC TTA CAT TGC TGC CAA TTT CGA CAC AAT CGG-3′ (reverse). The PCR product was cloned into the pBADHisTEV vector (Huanting Liu, University of St. Andrews, St. Andrews, Scotland) via the NcoI and SalI restriction sites. For protein production, the plasmid was transformed into E. coli C43 cells. M9 minimal media supplemented with 5% glycerol, 100 μg/mL ampicillin, 2 mM MgSO4, and 0.1 mM CaCl2 was used to avoid copurification of Neu5Ac from the medium (17). First, an overnight culture was prepared in LB media. On the next day, a second culture was prepared and inoculated with the overnight culture. The cells were grown to an OD600 of 5.0–6.0. The cells were then washed twice by centrifuging at 4000 g for 15 min and resuspending in M9 media. Six liters of M9 media were inoculated with 5 mL of the cells (OD600 = 5.0–6.0) and incubated at 37°C for 14–16 h with shaking until an OD600 of 0.6 was reached. Each culture was then induced with 500 mg/L L(+)-arabinose and grown for further 5 h at 25°C. The cells were harvested by centrifugation at 4000 rcf for 20 min and then flash-frozen in liquid nitrogen for storage.
For purification, the cell pellet was resuspended in buffer containing 50 mM Tris-Cl, pH 8, 50 mM NaCl, and 10 % glycerol (buffer A). A cell disrupter (Constant Systems, Daventry, Northamptonshire, UK) was used to lyse the cells twice at 30 kpsi, followed by centrifugation of the lysate at 48.384 rcf for 20 min. The obtained supernatant was incubated for 1 h with Ni2+ NTA resin (GE Healthcare, Port Washington, NY), which was previously equilibrated with buffer A. The resin was washed with 50 mL of buffer A, then with 25 mL of buffer A supplemented with 1 mM Tris(2-carboxyethyl)phosphine to reduce the cysteines of the protein. After another washing step with buffer A to remove the Tris(2-carboxyethyl)phosphine, the protein was labeled and eluted in one step with 15 ml of buffer A containing 31 μL of 100 mM S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSSL) and 200 mM imidazole. The sample was then loaded onto an ENrich Q 10/100 column (Bio-Rad, Hercules, CA) with buffer A and eluted with a linear gradient from 0.05 to 1 M NaCl. Finally, the protein was subjected to size exclusion chromatography on an equilibrated Superdex 75 16/60 or a Superdex 200 16/60 (both GE Healthcare) with buffer A as running buffer.
Continuous-wave x-band electron paramagnetic resonance spectroscopy
The doubly spin-labeled proteins were concentrated to 50 μM. Electron paramagnetic resonance (EPR) spectra were recorded on an EMXnano X-band EPR Spectrometer from Bruker (Billerica, MA). The samples were measured at room temperature with a microwave power of 2.51 mW, a video amplifier gain of 30 dB, a modulation amplitude of 1 G, a time constant of 20.48 ms, a conversion time of 21.33 ms, and a resolution of 10 points per G.
PELDOR spectroscopy
For PELDOR spectroscopy, the doubly spin-labeled VcSiaP samples (25 μM) were dissolved in PELDOR buffer (100 mM TES pH 7.5, 100 mM NaCl in D2O). If needed, the samples were supplemented with Neu5Ac and incubated for 30 min on ice. The samples were transferred to a 3 mm quartz Q-band EPR tube and flash-cooled in liquid nitrogen. The PELDOR time traces were recorded on an ELEXSYS E580 pulsed Q-band EPR spectrometer (Bruker), with an ER 5106QT-2 Q-band resonator. The instrument was equipped with a continuous flow helium cryostat (CF935) and temperature control system (ITC 502), both from Oxford Instruments (Abingdon, Oxfordshire, UK). The second microwave frequency was coupled into the microwave bridge using a commercially available setup from Bruker. All pulses were amplified via a 150 W pulsed traveling wave tube amplifier. PELDOR experiments were performed with the pulse sequence π/2(νΑ)-τ1-π(νΑ) − (τ1 + t) − π(νΒ)-(τ2 − t)-π(νΑ)-τ2-echo. The detection pulses (νΑ) were set to 12 ns for the π/2 and 24 ns for the π-pulses and applied at a frequency 80 MHz lower than the resonance frequency of the resonator. The pulse amplitudes were chosen to optimize the refocused echo. The π/2-pulse was phase-cycled to eliminate receiver offsets. The pump pulse (νΒ) was set at the resonance frequency of the resonator and its optimal length (typically 16 ns) was determined using a transient nutation experiment for each sample. The field was adjusted such that the pump pulse is applied to the maximum of the nitroxide spectrum. The pulse amplitude was optimized to maximize the inversion of a Hahn-echo at the pump frequency. All PELDOR spectra were recorded at 50 K with an experiment repetition time of 1 ms, a video amplifier bandwidth of 20 MHz, and an amplifier gain of 42 dB. The parameter τ1 was set to 260 ns and the maximum of τ2 was set to values ranging from 4 to 12 μs. Deuterium modulation was suppressed by addition of 8 spectra of variable τ1 with a Δτ1 of 16 ns. The obtained time traces were divided by a monoexponential decay to eliminate intermolecular contributions and renormalized. Distance distributions were obtained from the background-corrected data by using the program DEER Analysis 2016 (http://www.epr.ethz.ch/software.html) developed by Jeschke et al. (18) (the uncorrected time traces are shown in Fig. S1 in the Supporting Material). The influence of different starting points for the background fitting was analyzed with the evaluation feature of DEER Analysis. Linear combination fitting of time traces and integration of distance distributions were performed with Python (www.python.org) scripts using the NumPy (www.numpy.org) and SciPy (www.scipy.org) functions. The PyMOL (www.pymol.org) plugin MtsslWizard (19) and MMM (http://www.epr.ethz.ch/software.html) were used to predict distance distributions.
Crystallography
Purified VcSiaP R125A Q54R1/L173R1 at ∼17 mg/mL was used to setup crystallization trials with the JCSG+ Screen (Molecular Dimensions, Altamonte Springs, FL) and 96 well MRC plates (Molecular Dimensions). For each drop, 0.5 μL of protein was mixed with 0.5 μL of reservoir solution. A single crystal was observed in condition D7. The crystal was allowed to grow for several weeks at room temperature before harvesting. Before flash-cooling in liquid nitrogen, the crystal was cryo-protected with 35% glycerol. Data were collected at beamline BL14.3 of BESSYII (Berlin, Germany), using a MarMOSAIC 225 CCD detector. The data were processed using XDS (20) as implemented in XDSAPP (21). Data collection and processing statistics are listed in Table 1. The structure of VcSiaP was solved using PHASER (22) and PDB: 4MAG (23) as the search model. The PHENIX suite (24) and COOT (25) were used to refine the structure. The geometry of the model was optimized and validated using the software MolProbity (26).
Table 1.
Data Collection and Refinement Statistics
| VcSiaP Q54R1/L173R1 | |
|---|---|
| Wavelength | 0.89429 |
| Resolution range | 45.29–2.101 (2.176–2.101) |
| Space group | P 21 2 21 |
| Unit cell | 72.3 78.1 116.2 90 90 90 |
| Total reflections | 73,484 (6631) |
| Unique reflections | 37,987 (3606) |
| Multiplicity | 1.9 (1.8) |
| Completeness (%) | 97 (94) |
| Mean I/σ(I) | 7.25 (1.18) |
| Wilson B-factor | 40.2 |
| R-merge | 0.043 (0.51) |
| R-meas | 0.061 (0.73) |
| CC1/2 | 0.997 (0.507) |
| CC∗ | 0.999 (0.82) |
| Reflections used in refinement | 37,954 (3605) |
| Reflections used for R-free | 2019 (188) |
| R-work | 0.222 (0.357) |
| R-free | 0.259 (0.359) |
| CC(work) | 0.967 (0.517) |
| CC(free) | 0.957 (0.438) |
| RMS(bonds) | 0.005 |
| RMS(angles) | 0.78 |
| Ramachandran favored (%) | 98 |
| Ramachandran allowed (%) | 1.5 |
| Ramachandran outliers (%) | 0 |
| Rotamer outliers (%) | 0.78 |
| Clashscore | 7.60 |
| PDB-ID | 5LTC |
Values in parentheses correspond to the shell of highest resolution.
Results
Selection of labeling sites for PELDOR spectroscopy
To investigate the structures of substrate-bound and -free VcSiaP in solution, PELDOR spectroscopy was applied (13, 14). This EPR technique can accurately measure distances between paramagnetic centers in a range of 15 up to 170 Å (27, 28) and has frequently been employed to study conformational changes in transporters and channels (29, 30, 31, 32, 33). Like most proteins, VcSiaP is diamagnetic, and therefore invisible for EPR. Thus, site-directed spin labeling was used to attach two spin labels to its molecular surface (15). To find optimal labeling positions for VcSiaP, a difference distance matrix (diffDM) between the substrate-bound and -free crystal structures of the homolog HiSiaP (50% identical amino acids) was calculated (Fig. 1 A) (34, 35). HiSiaP was used, because no structure of substrate-bound VcSiaP is currently available. The residue numbering between HiSiaP and VcSiaP differs by one or two amino acids (depending on the position in the sequence; see sequence alignment in Fig. S2). In the following, the VcSiaP numbering is used. The diffDM reveals the absolute value of the spatial displacement between substrate-bound and -free HiSiaP for each possible pair of Cβ atoms. Consequentially, the distinct yellow peaks in the diffDM (Fig. 1 A) represent pairs of residues, where the conformational changes between the two crystal structures are especially large (up to 18.0 Å). Based on this analysis, we selected the residue pairs Q54/L173, Q110/L173, L173/S225, and Q54/Q110 (control) as labeling sites (Fig. 1 A). The corresponding double cysteine mutants were cloned, expressed, and labeled with the MTSSL spin label (36), creating the VcSiaP mutants Q54R1/L173R1, Q110R1/L173R1, L173R1/S225R1, and Q54R1/Q110R1. Judged by room temperature cw-X-band EPR spectroscopy, an average labeling efficiency of 90% was achieved.
Building a model of substrate-bound VcSiaP
A diffDM can also be used to identify rigid subdomains within protein structures (35). Because rigid subdomains do not, by definition, change their conformation between two different states of the protein, they show up as unicolored squares along the diagonal of the diffDM. Here, four such squares were identified (Fig. 1 A, I–IV). Note that it would be possible to subdivide the squares, if a more fine-grained model was needed and if the coordinate error of the underlying structures was sufficiently small (35). To build a model of substrate-bound VcSiaP, the open structure (PDB: 4MAG (23), Fig. 1 B) was split at the positions indicated by the diffDM to create the rigid subdomains I–IV (I, 1–127; II, 128–208; III, 209–260; IV, 261–308) (Fig. 1 C). The border between III and IV is close to the kink in helix α9, which has previously identified as a hallmark of closed P domains (10). These rigid subdomains of VcSiaP were then superimposed onto the substrate-bound HiSiaP crystal structure (PDB: 3B50 (37)), leading to a coarse model of substrate-bound VcSiaP. The geometry of the cleavage sites was regularized in COOT (25) (Fig. 1 D).
Comparing the solution and crystal structures of VcSiaP with PELDOR spectroscopy
PELDOR experiments on the doubly spin-labeled VcSiaP mutants were conducted. Fig. 2 A shows the Q-band PELDOR time traces of the control mutant, VcSiaP Q54R1/Q110R1, which, according to the crystal structures, should lead to the same distance in the substrate-bound- and -free states (in Fig. 1 A, both residues, Q54 and Q110, are located in the same rigid body, I). After the initial decay, both time traces show several clear oscillations, indicating narrow underlying spin-spin distance distributions. Indeed, as expected for the control sample, the two time traces (± Neu5Ac) were virtually identical (Fig. 2 A). Both time traces were analyzed with the DEER Analysis 2016 software (18), leading to the distance distributions in Fig. 2 B. For both samples, a narrow peak at 27 Å with a shoulder at 30 Å was observed. Models of the open and closed structure with the R1 side chain at positions Q54 and Q110 were produced with MtsslWizard (blue sticks in Fig. 1, B and D) and theoretical distance distributions were calculated with MtsslWizard and MMM (shaded areas in Fig. 2 B) (19). The experimental and expected distributions for VcSiaP Q54R1/Q110R1 agree very well for both experiments (with and without Neu5Ac). A possible explanation for the shoulder at 30 Å is a second conformation of the R1 spin label, which has been frequently observed in available crystal structures of the R1 side chain (38, 39, 40).
Figure 2.
PELDOR measurements on spin-labeled VcSiaP. (A, C, E, and G) Background corrected PELDOR time traces of the indicated VcSiaP double mutant either with or without Neu5Ac, as indicated in the figure. (B, D, F, and H) Distance distributions (solid lines) calculated from time traces on the left using DEER Analysis 2016. Predicted distance distributions (mtsslWizard (mW) and MMM) are shown as translucent shades, as indicated. Hence, darker shades correspond to distances that are predicted by both programs. The error bars were calculated with DEER Analysis 2016. Spin-Spin distances from the crystal structure of the spin labeled 54/173 mutant are shown as vertical lines in (D). To see this figure in color, go online.
The same procedure was applied to the VcSiaP Q54R1/L173R1 mutant. Again, high-quality time traces with clearly visible oscillations were observed (Fig. 2 C). But, in this case, the PELDOR time traces of the two samples (± Neu5Ac) differed strongly. Accordingly, the two corresponding distance distributions show different but well-defined peaks at 27 Å (+ Neu5Ac) or 43 Å (− Neu5Ac) (Fig. 2 D). Also for this mutant, the experimental distributions show a good match to the predictions made with MtsslWizard and MMM, although for both programs, the predicted distributions are broader than the experimentally determined distributions. The x-ray structure of this mutant revealed that the difference between prediction and experiment is simply due to the prediction error (see below). The experiment was repeated for the VcSiaP Q110R1/L173R1 and L173R1/S225R1 mutants. Also here, clear differences between ± Neu5Ac were found. Again, the observed distances fit to the MtsslWizard and MMM predictions (Fig. 2, E–H). Interestingly, for all double mutants, the room-temperature cw-X-band EPR spectra are virtually identical for the apo- or Neu5Ac-bound state, despite the large changes of the distance distributions. This indicates that the mobilities and possibly the conformations of the R1 labels do not significantly change between the two states (Fig. S3).
Following Neu5Ac binding to VcSiaP with PELDOR spectroscopy
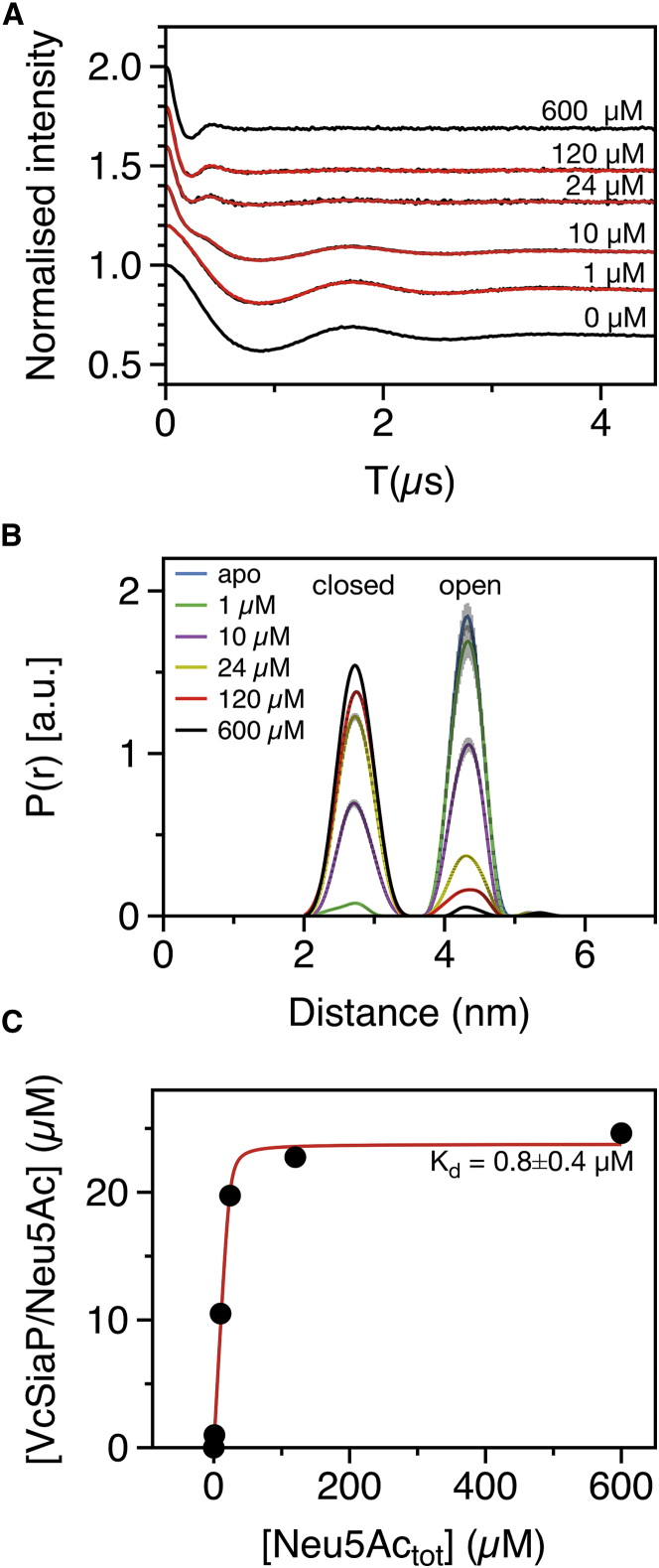
To analyze if any stable intermediate states of the P domain exist, the binding of Neu5Ac to VcSiaP was quantitatively analyzed by PELDOR spectroscopy. For this purpose, samples of VcSiaP Q54R1/L173R1 (25 μM) supplemented with different amounts of Neu5Ac were produced (0–600 μM Neu5Ac). This particular mutant was chosen, because it showed the clearest difference between substrate-bound and -unbound states (Fig. 2). A Q-band PELDOR time trace was recorded for each sample (Fig. 3 A). The corresponding distance distributions were calculated with DEER Analysis and each normalized to an integral value of 1.0 (Fig. 3 B). Assuming that the PELDOR distance distributions quantitatively reflect the state of VcSiaP in solution, the peak area of the substrate-bound (closed) form of each sample (Fig. 3 B) should be directly proportional to concentration of the VcSiaP/Neu5Ac complex. Vice versa, the peak area of the substrate-free (open) form should be proportional to the VcSiaPfree concentration. The two peaks were therefore integrated for each sample and the integral values converted to concentrations (Table S1). Tikhonov regularization as implemented in DEER Analysis 2016 was used to extract the distance distributions from the PELDOR time traces. On principle, the same PELDOR time trace will yield distance distributions of differing width and/or shape, depending on the choice of the Tikhonov regularization parameter α. Although DEER Analysis automatically chooses the optimal α-value based on an L-curve criterion, it cannot be excluded that this procedure influenced the integrals that were calculated above. It was therefore also tried to extract the fractions/concentrations of apo- and Neu5Ac-bound VcSiaP directly from the background-corrected PELDOR time traces (Fig. 3 A). For this purpose, the time traces from the intermediate (1–120 μM) Neu5Ac concentrations were fitted as linear combinations of the apo- and fully substrate-bound time traces. The red lines in Fig. 3 A show that the resulting fits almost perfectly reproduce the experimental data. The resulting fractions/concentrations of apo- and Neu5Ac-bound VcSiaP and their estimated uncertainties are listed in Table S1. Reassuringly, the concentrations from both the linear combination and integration methods matched very well (Table S1). A binding isotherm was plotted using the mean of the calculated concentrations from both methods (Fig. 3 C). Then, a dissociation constant of 0.8 ± 0.4 μM was determined by nonlinear fitting of equation y = ((Ptot + Ligtot + Kd) − sqrt((Ptot + Ligtot + Kd)2 −4 × Ptot × Ligtot))/2 to the data points (41). Note, that for an optimal binding experiment, the concentration of VcSiaP should have been significantly below the expected Kd value to avoid substrate depletion (41). Here, to record PELDOR time traces with good signal/noise, much higher concentrations of 25 μM VcSiaP were used. The consequence was a very sharp transition in the binding isotherm, which makes it difficult to determine the Kd value with high accuracy. Nevertheless, the obtained Kd value is reasonably close to previously published values (0.3 μM (23), and 0.1 μM (5, 6)), suggesting that the PELDOR distance distributions of VcSiaP 54R1/173R1 can be quantitatively analyzed in the described ways.
Figure 3.
Open-close transition of VcSiaP followed by PELDOR spectroscopy. (A) PELDOR time traces of VcSiaP Q54R1/L173R1 titrated with the indicated amounts of Neu5Ac (black traces). (Superposed curves) Fits resulting from linear combinations of the 0 μM (open) and 600 μM (closed) Neu5Ac time traces using the equation y = a × open + (1−a) × closed. Note that small differences in modulation depths were corrected by scaling the time traces to a modulation depth of 100% before the fitting procedure. The fitting results were then back-scaled to the original modulation depth. (B) Distance distributions (DeerAnalysis 2016) corresponding to the time traces shown in (A). The distributions were normalized, so that their integral equals 1.0. The error bars were calculated using the evaluation procedure from DEER Analysis 2016. (C) Binding isotherm of the VcSiaP Q54R1/L173R1 × Neu5Ac interaction. (Black dots) Calculated VcSiaP/Neu5Ac concentrations (see main text). (Solid red line) Fit of the equation y = ((Ptot +Ligtot + Kd) −sqrt((Ptot + Ligtot + Kd)2 −4 × Ptot × Ligtot))/2 (41) to the data points. Ptot is the total concentration of VcSiaP, Ligtot is the total amount of Neu5Ac, and Kd is the dissociation constant. A Kd of 0.8 ± 0.4 μM was determined. To see this figure in color, go online.
Mutational analysis of the open-closed transition
Whereas the role of R147 as a selectivity filter in P domains is established (9), it is currently not known exactly how the bound substrate triggers the conformational change of the P domain. In Neu5Ac binding P domains, a group of three conserved, polar amino acids, R125, E184, and H207 were observed to form an intricate network of interactions (23). It has been proposed that these residues play an important role in the open-closed transition of P domains, also because they are close to two hinge regions in the structure (Fig. 4 A) (23).
Figure 4.
Conformational changes upon Neu5Ac binding. (A) Detail of the substrate-bound HiSiaP structure (PDB: 3B50 (37)), showing the Neu5Ac molecule and its interaction with the R125, E184, and H207 triade (VcSiaP numbering). (B) Superposition of the four rigid bodies of substrate-bound HiSiaP (compare Fig. 1) with substrate-free HiSiaP (PDB: 2CEY (10)). (C) Detail of the superposition in (B), showing the R125, E184, H207 triade. (D) Open/closed state (percentage) of VcSiaP mutants as determined by PELDOR spectroscopy. The PELDOR data is shown in Fig. S4. The ++/+/− indicates if 10 mM (++), 1 mM (+), or no (−) Neu5Ac was present in the experiment. The error bars represent ±3 times the SD calculated in the linear combination fitting procedure. To see this figure in color, go online.
To better visualize the conformational changes of these amino acids upon substrate binding, the closed structure of HiSiaP was split into four rigid bodies as indicated by the diffDM (Fig. 1) and the rigid bodies were superposed onto the open structure (Fig. 4 B, root mean square deviation (RMSD) = 0.45 for 308 Cα atoms). In this way, the conformational changes of the side chains are not obstructed by the larger scale rigid body movements of the protein backbone and therefore easier to analyze. Indeed, the superposition revealed that the conformation of R125 changes upon substrate binding, whereas H207 and E184 appear unchanged (Fig. 4 C).
The three residues were systematically mutated in the VcSiaP Q54R1/L173R1 construct to analyze their individual influence on Neu5Ac binding. PELDOR measurements in the presence and absence of 1 mM Neu5Ac were conducted with the purified mutants. According to the titration experiment above (Fig. 3), 1 mM Neu5Ac suffices to induce the closed state in the wild-type protein. The time traces and distance distributions are compiled in Fig. S4. For each experiment, the percentage of open versus closed VcSiaP was determined by linear combination fitting as described above (Figs. 4 D and S4). The error bars in Fig. 4 D represent the estimated uncertainty of the open/closed fractions (Table S2). First, R125 was mutated to alanine. The PELDOR experiments reveal that with 1 mM Neu5Ac, no significant amount of the substrate-bound (closed) conformation could be detected (Fig. 4 D). However, at 10 mM Neu5Ac, the amount of closed VcSiaP increased to 6 ± 2%. To conserve the positive charge of R125, the residue was also mutated to lysine. In this case, a small but significant percentage (15 ± 2%) of VcSiaP/Neu5Ac complex was observed with 1 mM ligand, while most of the protein (85 ± 2%) remained in the open conformation (Fig. 4 D). Also here, increasing the Neu5Ac concentration to 10 mM led to a larger percentage of closed VcSiaP (31 ± 3%). Next, E184 was mutated to alanine. The PELDOR measurement with 1 mM Neu5Ac revealed a ∼1:1 mix of open and closed VcSiaP. In contrast, the charge-conserving E184D mutant behaved almost like the wild-type in our experiments (Fig. 4 D). Interestingly, the charged to polar mutant E184Q behaved similar to the E184A mutant, indicating that the ionic interaction of residue 184 with R125 is important, while small structural changes (E→D) can be tolerated (Fig. 4 D). Finally, H207 was mutated to alanine or glutamine. According to Fig. 4 A, a glutamine at position 207 should still be able to form a polar interaction with E184. Both H207 mutants appeared to be less stable than the wild-type protein. For example, aggregate peaks were observed (and removed) in gel filtration experiments (Fig. S5). However, once purified, both H207A and H207Q behaved almost like the wild-type protein in the PELDOR experiments (Fig. 4 D).
X-ray structure of spin-labeled VcSiaP R125A Q54R1/L173
To check whether the very low binding activity of the R125A Q54R1/L173R1 mutant is caused by a change in its overall structure, the protein was crystallized. A single crystal was obtained in condition D7 of the JCSG+ screen and a 2.1 Å diffraction dataset was collected. The structure was solved by molecular replacement with PHASER (22), using the wild-type structure as search model (PDB: 4MAG (23)).
The asymmetric unit contained two copies of the protein. Refinement with phenix.refine (PHENIX; https://www.phenix-online.org/documentation/reference/refinement.html) led to R/Rfree-factors of 22.2/25.9. MolProbity was used to validate the stereochemistry of the model (26). Data collection and refinement statistics are listed in Table 1. The final, refined VcSiaP Q54R1/L173R1 structure and the search model superpose with an RMSD of 0.3 Å for 260 Cα atoms for chain A and with an RMSD of 0.5 Å for 282 Cα atoms for chain B. Fig. S6 shows that chain A fits almost perfectly to the previously published wild-type structure, while chain B is in a slightly more closed conformation. Such slightly closed states of unliganded P domains were predicted by MD simulations (12). While both spin labels are clearly visible in the electron density, the 54R1 side chain is better defined and therefore apparently less mobile (Fig. 5). This fits to the observation that the conformational ensemble produced by MtsslWizard and MMM is much smaller for Q54R1 than for L173R1 (Fig. 1, B and D). Details about the conformation of the two spin labels including the dihedral angles of the side chains are compiled in Fig. S7. The distance between the Q54R1 and L173R1 spin centers was measured for both chains in the crystal structure and amounts to 42.7/40.9 Å. Both distances fit well to the corresponding PELDOR result (Fig. 2 D, gray lines). The cw-X-band EPR spectra of Q54R1/L173R1 ± Neu5Ac indicated that the mobilities of the spin labels (and therefore likely also their molecular surroundings) did not significantly change upon Neu5Ac binding (Fig. S3). Thus, as explained above, the crystal structure was separated into rigid-bodies I–IV (Fig. 1 C) and superimposed onto the closed-state HiSiaP structure. The distance between the two spin labels was again measured and now fits very well to the measured PELDOR distance of the closed state (Fig. 2 D, red lines).
Figure 5.
X-ray structure of spin-labeled VcSiaP R125A Q54R1/L173R1. (A) The R1 side chain at position 54. The protein backbone is shown as a cartoon/stick model. A neighboring molecule in the crystal is indicated. (Gray mesh) The refined 2mFo-DFc electron density contoured at 1.0 σ. Residual difference electron density (mFo-DFc) contoured at 3.0 σ is indicated. (B) The R1 side chain at position 173. The figure is analogous to (A). (Broken arrow) Distance vector between the two spin centers. Its absolute value is 42.7 Å (The N-N distance was measured). To see this figure in color, go online.
In summary, neither the R125A mutation nor the attachment of the two spin labels significantly disturbed the overall structure of VcSiaP. Further, our structure is another indication that crystal structures of the R1 side chain can be good approximations for the rotameric state of the side chain in frozen solution (i.e., in PELDOR samples) (39, 40).
Discussion
The PELDOR-based binding study on the VcSiaP Neu5Ac interaction (Fig. 3) led to a Kd value, which is close to previously determined values using isothermal titration calorimetry (ITC) or Trp fluorescence quenching (5, 6, 23). This suggests that the PELDOR data can be quantitatively analyzed and, judged by the error calculated from the linear combination fitting of VcSiaP Q54R1/L173R1 time traces, it appears that as little as 3% of the closed conformation can be detected (Table S2). This knowledge is vital for the discussion of the mutational analysis below. It should be noted that for mutants that do not produce an equally drastic change of the PELDOR time traces upon addition of Neu5Ac (Fig. 2 C) and for lower signal/noise, this detection limit will be higher. Such PELDOR-based binding experiments are time-consuming but offer the opportunity to measure the concentration of both the ligand-bound and -free states of the protein and at the same time to gain information about the structural state of the protein. This is usually not possible with simpler biochemical binding assays. Nevertheless, it will be difficult to precisely determine low Kd values (≪1 μM) with PELDOR using the described procedure, because it is currently experimentally not feasible to use the necessary nanomolar concentrations of spin-labeled protein, while still measuring time traces with sufficiently high signal/noise. Of course, it has to be determined for each particular case, which signal/noise is really needed to accurately distinguish the two states. However, the method might be advantageous for large Kd values: for the R125A and R125K mutants, no Neu5Ac binding had previously been detected via ITC (23); in contract, the PELDOR experiment indicates weak binding. It should be noted that the PELDOR samples were flash-frozen. Therefore, if a Kd can be determined, it might differ from a Kd that was determined at room temperature, depending on the kon and koff rates compared to the time needed for the freezing process. The recently developed trityl spin labels, which can be used at room temperature might be a possibility to avoid this problem (42, 43, 44). However, these labels are considerably larger than the MTSSL label and are currently not used on a routine basis. Förster resonance energy transfer spectroscopy is another possible alternative and can even determine the kon and koff rates of the interaction. However, because Förster resonance energy transfer labels are usually quite large compared to spin labels such as the R1 side chain, the structural information might be less informative than what can be gained from PELDOR experiments.
Several high-resolution crystal structures of TRAP transporter P domains in both the substrate-bound and -free states have been solved in the last decade (reviewed in Fischer et al. (11) and Berntsson et al. (45)), providing a detailed picture of the overall structures of P domains and their interaction with their particular substrate. The PELDOR results from this study indicate that the crystal structures of Neu5Ac binding P domains are very good models for the solution state of the proteins. The distance distributions that were predicted from the crystal structures using MtsslWizard and MMM fit nicely to the experimental data. An even better fit was obtained for the spin-labeled crystal structure (Fig. 2), indicating that the small differences between experiment and prediction can be explained by the known error of the prediction algorithms (±3 Å (19, 46, 47)). Within the detection limit of the PELDOR experiments, the P domains of Neu5Ac TRAP transporters appear to almost exclusively adopt the open state in the absence of ligand. For ABC transporters, the SBPs of the GlnPQ amino acid ABC transporter from Lactococcus lactis and of the maltose ABC transporter were shown to fluctuate between the closed- and open state even in the absence of ligand (48, 49), while the SBP of a glutamine ABC transporter (GlnBP) appears to remain in the open state without ligand (50). The data above further demonstrate that within the detection limit of the PELDOR experiments there is no trace of any stable intermediate states of the P domain in solution (Fig. 3). These results are in agreement with MD simulations, were no stable intermediate states were predicted for the P domain of the ectoine TRAP transporter TeaABC (12). The slightly different open conformations that were present in our crystal structure (Fig. S6) fit to the relatively broad energetic minimum for the open structure that was observed in those calculations (12). Considering the current hypothesis for the transport cycle of TRAP transporters (4), large concentrations of closed ligand-free P domains would trigger unproductive closing and opening of the transporter. One might speculate that VcSiaP was evolutionarily optimized to only close when the substrate is bound, thereby increasing the efficiency of the transporter. It has to be noted again that the PELDOR experiments are conducted using flash-frozen VcSiaP solutions. It cannot be ruled out that during the freezing process any transition states or closed ligand-free molecules have snapped back to the open state, and thus were not observed. Also, the PELDOR data only show the steady state of the sample. Transient states that were indicated by stopped-flow fluorescence spectroscopy analysis of other TRAP SBPs (51, 52, 53) might be present at low concentrations (≲3%). Time-resolved PELDOR experiments using freeze-quench instrumentation (54, 55) might be a possible (but experimentally demanding) way to investigate the structure of such transient states.
As mentioned above, the R125-E184-H207 triad (henceforth referred to as “triad”) is located in the hinge I and II regions of Neu5Ac transporter P domains, implicating a role in triggering the conformational change between substrate-free- (open) and -bound (closed) states of the protein (23). In a previous study, isothermal titration calorimetry was used to determine the Neu5Ac binding characteristics of triad mutants (23). The PELDOR data from this work now gives the opportunity to correlate the mere ability to bind Neu5Ac with the ability of the particular mutant to perform an open-close transition. Firstly, all triad mutants were able to adopt the open state with wild-type-like, sharp distance distributions. Also, if the closed state was observed for a particular mutant, the same average distance as for the wild-type protein was observed (Fig. S4). This strongly suggests that the triad is not necessary for the P domain to adopt its native open or closed conformation. The R125A mutant was structurally intact (Fig. S4), but at 1 mM Neu5Ac, only a very low percentage of the closed state was observed (Fig. 4 D). To verify that this very small fraction of closed state was not an artifact, the PELDOR experiment was repeated with 10 mM Neu5Ac, resulting in a 4 ± 2% closed state (Fig. 4 D). Thus, although the R125A mutant binds Neu5Ac very weakly (23), it can still correctly adopt the closed state. The same weak-binding phenotype had also been observed for the R125K mutant (23), but, using PELDOR, a small but significant percentage (15 ± 2%) of the protein was clearly observed in the closed state (Fig. 4 D). Also here, increasing the Neu5Ac concentration to 10 mM led to an increase of the closed-state percentage to 36 ± 3% (Fig. 4 D). So, similar to R125A, the R125K mutant binds Neu5Ac very weakly, but has clearly not lost its ability to reach the closed state. According to the crystal structure of substrate-bound HiSiaP (Fig. 4 A), R125K should still be able to form an ionic interaction with E184 (2.8 Å), but its amino group will be too far from the Neu5Ac binding site to strongly interact with the substrate (>4.8 Å). This explains the very weak Neu5Ac binding of both mutants and why R125K binds stronger than R125A. The mutational data on E184 reveal that the residue is important, but not crucial for the function of the P domain. While the E→A mutant is still 55 ± 1% closed at 1 mM Neu5Ac, the charge-conserving E→D mutant was almost indiscernible from the wild-type protein. Also in this case, the PELDOR data agree well with available binding data (23). E184 thus seems to simply stabilize R125 by an ionic interaction, keeping the latter in an optimal state to interact with the substrate. H207 does not seem to play any important role in the substrate-induced closing mechanism of the P domain, because even the H→A mutant was 96 ± 1% closed in the presence of 1 mM Neu5Ac. However, as mentioned above, the protein was less stable when this mutation was introduced. This might be the reason for the reduced binding affinity that was previously observed (23). The triad residues have also been mutated in an earlier study on nontypable H. influenzae and the effects of the mutations on LPS sialylation were analyzed in vivo by complementation assays (37). Strikingly, the in vivo effects fit perfectly to the PELDOR results in Fig. 4 D: R125A showed no sialylation, E184Q and R125K showed partial sialylation, and H207A showed full sialylation (residue numbers are given in VcSiaP numbering).
In summary, the impact of mutating the individual three residues of the R125, E184, H207 triad varies strongly. R125 is clearly of high importance, presumably because of its interaction with the substrate. However, based on the available data, the network of interactions between the triad side chains does not seem to act as a substrate sensor, which triggers the conformational changes between substrate-free and -bound states of P domains.
Conclusions
The solution structure and open-close transition of VcSiaP was analyzed with PELDOR spectroscopy, revealing that the crystal structures of both the open- and closed states are good models for the solution structure of the P domain in either state. In the absence of substrate and within the detection limit of the PELDOR experiments, the P domain is exclusively found in the opened state. No indications of stable intermediate states were found in PELDOR-based titration experiments. A mutational analysis of the R125, E184, H207 triad was conducted. R125 is primarily involved in substrate binding and is stabilized by its interaction with E184. H207 does not appear to play a vital role in the open-close transition, but mutating this position leads to a less stable VcSiaP protein. In future experiments, we aim to analyze the structure and function of VcSiaP in the context of the transmembrane domains VcSiaQM.
Author Contributions
J.G., M.F.P., and G.H. performed experiments and analyzed data; G.H. and G.H.T. designed experiments and wrote the article; and G.H. conceived the study.
Acknowledgments
Beamtime and support at BESSY II, BL14.3 is gratefully acknowledged. The pBADHisTEV vector was a gift from Huanting Liu (University of St Andrews). G.H. thanks Olav Schiemann for continued support and access to his spectrometers.
G.H. acknowledges funding by the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG), project No. HA 6805/4-1; and G.H.T. thanks the Biotechnology and Biological Sciences Research Council (BBSRC) for funding, specifically projects No. BBC5098071 and No. BBF0147591.
Editor: David Cafiso.
Footnotes
Seven figures and two tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)34285-0.
Supporting Material
References
- 1.Locher K.P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 2016;23:487–493. doi: 10.1038/nsmb.3216. [DOI] [PubMed] [Google Scholar]
- 2.Law C.J., Maloney P.C., Wang D.-N. Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 2008;62:289–305. doi: 10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siebold C., Flükiger K., Erni B. Carbohydrate transporters of the bacterial phosphoenolpyruvate: sugar phosphotransferase system (PTS) FEBS Lett. 2001;504:104–111. doi: 10.1016/s0014-5793(01)02705-3. [DOI] [PubMed] [Google Scholar]
- 4.Mulligan C., Fischer M., Thomas G.H. Tripartite ATP-independent periplasmic (TRAP) transporters in bacteria and archaea. FEMS Microbiol. Rev. 2011;35:68–86. doi: 10.1111/j.1574-6976.2010.00236.x. [DOI] [PubMed] [Google Scholar]
- 5.Mulligan C., Geertsma E.R., Thomas G.H. The substrate-binding protein imposes directionality on an electrochemical sodium gradient-driven TRAP transporter. Proc. Natl. Acad. Sci. USA. 2009;106:1778–1783. doi: 10.1073/pnas.0809979106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulligan C., Leech A.P., Thomas G.H. The membrane proteins SiaQ and SiaM form an essential stoichiometric complex in the sialic acid tripartite ATP-independent periplasmic (TRAP) transporter SiaPQM (VC1777-1779) from Vibrio cholerae. J. Biol. Chem. 2012;287:3598–3608. doi: 10.1074/jbc.M111.281030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulligan C., Kelly D.J., Thomas G.H. Tripartite ATP-independent periplasmic transporters: application of a relational database for genome-wide analysis of transporter gene frequency and organization. J. Mol. Microbiol. Biotechnol. 2007;12:218–226. doi: 10.1159/000099643. [DOI] [PubMed] [Google Scholar]
- 8.Vetting M.W., Al-Obaidi N., Almo S.C. Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes. Biochemistry. 2015;54:909–931. doi: 10.1021/bi501388y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer M., Hopkins A.P., Thomas G.H. Tripartite ATP-independent periplasmic (TRAP) transporters use an arginine-mediated selectivity filter for high affinity substrate binding. J. Biol. Chem. 2015;290:27113–27123. doi: 10.1074/jbc.M115.656603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Müller A., Severi E., Thomas G.H. Conservation of structure and mechanism in primary and secondary transporters exemplified by SiaP, a sialic acid binding virulence factor from Haemophilus influenzae. J. Biol. Chem. 2006;281:22212–22222. doi: 10.1074/jbc.M603463200. [DOI] [PubMed] [Google Scholar]
- 11.Fischer M., Zhang Q.Y., Thomas G.H. Caught in a TRAP: substrate-binding proteins in secondary transport. Trends Microbiol. 2010;18:471–478. doi: 10.1016/j.tim.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Marinelli F., Kuhlmann S.I., Faraldo-Gómez J.D. Evidence for an allosteric mechanism of substrate release from membrane-transporter accessory binding proteins. Proc. Natl. Acad. Sci. USA. 2011;108:E1285–E1292. doi: 10.1073/pnas.1112534108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schiemann O., Prisner T.F. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Q. Rev. Biophys. 2007;40:1–53. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 14.Jeschke G. DEER distance measurements on proteins. Annu. Rev. Phys. Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 15.Altenbach C., Marti T., Hubbell W.L. Transmembrane protein structure: spin labeling of bacteriorhodopsin mutants. Science. 1990;248:1088–1092. doi: 10.1126/science.2160734. [DOI] [PubMed] [Google Scholar]
- 16.Klare J.P., Steinhoff H.J. Spin labeling EPR. Photosynth. Res. 2009;102:377–390. doi: 10.1007/s11120-009-9490-7. [DOI] [PubMed] [Google Scholar]
- 17.Severi E., Randle G., Thomas G.H. Sialic acid transport in Haemophilus influenzae is essential for lipopolysaccharide sialylation and serum resistance and is dependent on a novel tripartite ATP-independent periplasmic transporter. Mol. Microbiol. 2005;58:1173–1185. doi: 10.1111/j.1365-2958.2005.04901.x. [DOI] [PubMed] [Google Scholar]
- 18.Jeschke G., Chechik V., Godt A. DEER Analysis 2006—a comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 2006;30:473–498. [Google Scholar]
- 19.Hagelueken G., Ward R., Schiemann O. MtsslWizard: in silico spin-labeling and generation of distance distributions in PyMOL. Appl. Magn. Reson. 2012;42:377–391. doi: 10.1007/s00723-012-0314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kabsch W. Automatic-indexing of rotation diffraction patterns. J. Appl. Cryst. 1988;21:67–71. [Google Scholar]
- 21.Krug M., Weiss M.S., Mueller U. XDSAPP: a graphical user interface for the convenient processing of diffraction data using XDS. J. Appl. Cryst. 2012;45:568–572. [Google Scholar]
- 22.McCoy A.J., Grosse-Kunstleve R.W., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gangi Setty T., Cho C., Ramaswamy S. Bacterial periplasmic sialic acid-binding proteins exhibit a conserved binding site. Acta Crystallogr. D Biol. Crystallogr. 2014;70:1801–1811. doi: 10.1107/S139900471400830X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams P.D., Grosse-Kunstleve R.W., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P., Cowtan K. COOT: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Chen V.B., Arendall W.B., 3rd, Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El Mkami H., Norman D.G. Electron Paramagnetic Resonance Investigations of Biological Systems by Using Spin Labels, Spin Probes, and Intrinsic Metal Ions, Part B. Elsevier; Dordrecht, the Netherlands: 2015. EPR distance measurements in deuterated proteins; pp. 125–152. [Google Scholar]
- 28.Schmidt, T., M. A. Wälti, …, G. M. Clore. 2016. Long distance measurements up to 160 Å in the GroEL tetradecamer using Q-band DEER EPR spectroscopy. Angew. Chem. Int. Edit. Nov 17. doi: http://dx.doi.org/10.1002/anie.201609617. [Epub ahead of print] PubMed PMID: 27860003. [DOI] [PMC free article] [PubMed]
- 29.Hänelt I., Wunnicke D., Slotboom D.-J. Conformational heterogeneity of the aspartate transporter Glt(Ph) Nat. Struct. Mol. Biol. 2013;20:210–214. doi: 10.1038/nsmb.2471. [DOI] [PubMed] [Google Scholar]
- 30.Böhm S., Licht A., Bordignon E. Conformational plasticity of the type I maltose ABC importer. Proc. Natl. Acad. Sci. USA. 2013;110:5492–5497. doi: 10.1073/pnas.1217745110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou P., Bortolus M., McHaourab H.S. Conformational cycle of the ABC transporter MsbA in liposomes: detailed analysis using double electron-electron resonance spectroscopy. J. Mol. Biol. 2009;393:586–597. doi: 10.1016/j.jmb.2009.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grote M., Polyhach Y., Bordignon E. Transmembrane signaling in the maltose ABC transporter MalFGK2-E: periplasmic MalF-P2 loop communicates substrate availability to the ATP-bound MalK dimer. J. Biol. Chem. 2009;284:17521–17526. doi: 10.1074/jbc.M109.006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pliotas C., Ward R., Naismith J.H. Conformational state of the MscS mechanosensitive channel in solution revealed by pulsed electron-electron double resonance (PELDOR) spectroscopy. Proc. Natl. Acad. Sci. USA. 2012;109:E2675–E2682. doi: 10.1073/pnas.1202286109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hagelueken G., Abdullin D., Schiemann O. MtsslSuite: probing biomolecular conformation by spin-labeling studies. Methods Enzymol. 2015;563:595–622. doi: 10.1016/bs.mie.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Schneider T.R. Domain identification by iterative analysis of error-scaled difference distance matrices. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2269–2275. doi: 10.1107/S0907444904023492. [DOI] [PubMed] [Google Scholar]
- 36.Berliner L.J., Grunwald J., Hideg K. A novel reversible thiol-specific spin label: papain active site labeling and inhibition. Anal. Biochem. 1982;119:450–455. doi: 10.1016/0003-2697(82)90612-1. [DOI] [PubMed] [Google Scholar]
- 37.Johnston J.W., Coussens N.P., Apicella M.A. Characterization of the N-acetyl-5-neuraminic acid-binding site of the extracytoplasmic solute receptor (SiaP) of nontypeable Haemophilus influenzae strain 2019. J. Biol. Chem. 2008;283:855–865. doi: 10.1074/jbc.M706603200. [DOI] [PubMed] [Google Scholar]
- 38.Hagelueken G., Ingledew W.J., Naismith J.H. PELDOR spectroscopy distance fingerprinting of the octameric outer-membrane protein Wza from Escherichia coli. Angew. Chem. Int. Ed. Engl. 2009;48:2904–2906. doi: 10.1002/anie.200805758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdullin D., Hagelueken G., Schiemann O. Determination of nitroxide spin label conformations via PELDOR and x-ray crystallography. Phys. Chem. Chem. Phys. 2016;18:10428–10437. doi: 10.1039/c6cp01307d. [DOI] [PubMed] [Google Scholar]
- 40.Florin N., Schiemann O., Hagelueken G. High-resolution crystal structure of spin labelled (T21R1) azurin from Pseudomonas aeruginosa: a challenging structural benchmark for in silico spin labelling algorithms. BMC Struct. Biol. 2014;14:16. doi: 10.1186/1472-6807-14-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hulme E.C., Trevethick M.A. Ligand binding assays at equilibrium: validation and interpretation. Br. J. Pharmacol. 2010;161:1219–1237. doi: 10.1111/j.1476-5381.2009.00604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reginsson G.W., Kunjir N.C., Schiemann O. Trityl radicals: spin labels for nanometer-distance measurements. Chemistry. 2012;18:13580–13584. doi: 10.1002/chem.201203014. [DOI] [PubMed] [Google Scholar]
- 43.Yang Z., Bridges M.D., Hubbell W.L. A triarylmethyl spin label for long-range distance measurement at physiological temperatures using T1 relaxation enhancement. J. Magn. Reson. 2016;269:50–54. doi: 10.1016/j.jmr.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddy T.J., Iwama T., Rawal V.H. General synthesis of persistent trityl radicals for EPR imaging of biological systems. J. Org. Chem. 2002;67:4635–4639. doi: 10.1021/jo011068f. [DOI] [PubMed] [Google Scholar]
- 45.Berntsson R.P.A., Smits S.H.J., Poolman B. A structural classification of substrate-binding proteins. FEBS Lett. 2010;584:2606–2617. doi: 10.1016/j.febslet.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 46.Jeschke G. Conformational dynamics and distribution of nitroxide spin labels. Prog. Nucl. Magn. Reson. Spectrosc. 2013;72:42–60. doi: 10.1016/j.pnmrs.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 47.Alexander N.S., Stein R.A., Meiler J. RosettaEPR: rotamer library for spin label structure and dynamics. PLoS One. 2013;8:e72851. doi: 10.1371/journal.pone.0072851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gouridis G., Schuurman-Wolters G.K., Poolman B. Conformational dynamics in substrate-binding domains influences transport in the ABC importer GlnPQ. Nat. Struct. Mol. Biol. 2015;22:57–64. doi: 10.1038/nsmb.2929. [DOI] [PubMed] [Google Scholar]
- 49.Tang C., Schwieters C.D., Clore G.M. Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature. 2007;449:1078–1082. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]
- 50.Bermejo G.A., Strub M.-P., Tjandra N. Ligand-free open-closed transitions of periplasmic binding proteins: the case of glutamine-binding protein. Biochemistry. 2010;49:1893–1902. doi: 10.1021/bi902045p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walmsley A.R., Shaw J.G., Kelly D.J. Perturbation of the equilibrium between open and closed conformations of the periplasmic C4-dicarboxylate binding protein from Rhodobacter capsulatus. Biochemistry. 1992;31:11175–11181. doi: 10.1021/bi00160a031. [DOI] [PubMed] [Google Scholar]
- 52.Walmsley A.R., Shaw J.G., Kelly D.J. The mechanism of ligand binding to the periplasmic C4-dicarboxylate binding protein (DctP) from Rhodobacter capsulatus. J. Biol. Chem. 1992;267:8064–8072. [PubMed] [Google Scholar]
- 53.Thomas G.H., Southworth T., Kelly D.J. Novel ligands for the extracellular solute receptors of two bacterial TRAP transporters. Microbiology. 2006;152:187–198. doi: 10.1099/mic.0.28334-0. [DOI] [PubMed] [Google Scholar]
- 54.Pievo R., Angerstein B., Bennati M. A rapid freeze-quench setup for multi-frequency EPR spectroscopy of enzymatic reactions. ChemPhysChem. 2013;14:4094–4101. doi: 10.1002/cphc.201300714. [DOI] [PubMed] [Google Scholar]
- 55.Kaufmann R., Yadid I., Goldfarb D. A novel microfluidic rapid freeze-quench device for trapping reactions intermediates for high field EPR analysis. J. Magn. Reson. 2013;230:220–226. doi: 10.1016/j.jmr.2013.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.