Abstract
The large tegument protein encoded by the UL36 gene of pseudorabies virus (PrV) physically interacts with the product of the adjacent UL37 gene (B. G. Klupp, W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter, J. Virol. 76:3065-3071, 2002). To analyze UL36 function, two PrV recombinants were generated by mutagenesis of an infectious PrV full-length clone in Escherichia coli: PrV-ΔUL36F exhibited a deletion of virtually the complete UL36 coding region, whereas PrV-UL36BSF contained two in-frame deletions of 238 codons spanning the predicted UL37 binding domain. Coimmunoprecipitation experiments confirmed that the mutated gene product of PrV-UL36BSF did not interact with the UL37 protein. Like the previously described PrV-ΔUL37 (B. G. Klupp, H. Granzow, and T. C. Mettenleiter, J. Virol. 75:8927-8936, 2001) but in contrast to PrV-ΔUL36F, PrV-UL36BSF was able to replicate in rabbit kidney (RK13) cells, although maximum virus titers were reduced ca. 50-fold and plaque diameters were reduced by ca. 45% compared to wild-type PrV. PrV-ΔUL36F was able to productively replicate after repair of the deleted gene or in a trans-complementing cell line. Electron microscopy of infected RK13 cells revealed that PrV-UL36BSF and phenotypically complemented PrV-ΔUL36F were capable of nucleocapsid formation and egress from the nucleus by primary envelopment and deenvelopment at the nuclear membrane. However, reenvelopment of nucleocapsids in the cytoplasm was blocked. Only virus-like particles without capsids were released efficiently from cells. Interestingly, cytoplasmic nucleocapsids of PrV-UL36BSF but not of PrV-ΔUL36F were found in large ordered structures similar to those which had previously been observed with PrV-ΔUL37. In summary, our results demonstrate that the interaction between the UL36 and UL37 proteins is important but not strictly essential for the formation of secondary enveloped, infectious PrV particles. Furthermore, UL36 possesses an essential function during virus replication which is independent of its ability to bind the UL37 protein.
Pseudorabies virus (PrV; suid herpesvirus 1), the causative agent of Aujeszy's disease of swine, has been assigned to the Varicellovirus genus of the Alphaherpesvirinae subfamily of the Herpesviridae (41, 48). Recently, the complete DNA sequence of the 143,461-bp genome of PrV has been determined and shown to contain at least 70 open reading frames (ORFs) which have been demonstrated or suggested to encode viral proteins (31). Since homologues of these ORFs were also found in other alphaherpesvirus genomes, the gene designations originally introduced for herpes simplex virus type 1 (HSV-1) (39) were widely adopted. Approximately half of the conserved alphaherpesvirus gene products are incorporated into mature virions, which consist of an inner nucleoprotein core comprising the linear DNA genome, an icosahedral nucleocapsid, a tegument layer, and an outer envelope containing viral (glyco)proteins (47, 48). The most complex structure within herpesvirus particles is the tegument, which consists of more than 15 different proteins, including the UL36 and UL37 gene products (reviewed in reference 42). However, the precise architecture and the biological functions of the tegument are still only incompletely understood.
Several major tegument proteins contained in alphaherpesvirus virions possess important functions during early steps of the viral life cycle. For example, the UL48 gene products of HSV-1 (VP16, αTIF) and other viruses, including PrV, induce transcription of viral immediate-early genes (1, 7, 20). The UL41 gene product of HSV-1 (vhs) mediates shutoff of host cell gene expression by mRNA destabilization (35). On the other hand, during the late phase of infection, many tegument proteins are relevant for virion morphogenesis (reviewed in reference 42). Herpesvirus nucleocapsids are formed in the host cell nucleus and leave this compartment by consecutive envelopment and deenvelopment at the inner and outer leaflets of the nuclear membrane. This process involves the UL31 and UL34 gene products, which are conserved not only in alpha- but also in beta- and gammaherpesviruses (19, 28, 44, 46). Final tegumentation of herpesvirus nucleocapsids occurs in the cytoplasm, followed by budding into vesicles of the trans-Golgi network and release of mature virions by exocytosis (23, 25, 45, 49, 52).
Electron microscopic studies revealed that secondary envelopment of PrV is affected by mutations in different proteins, including deletions of envelope glycoproteins gE and gM (5, 6), of the UL3.5 protein (18), and of tegument proteins encoded by UL11 (33), UL37 (29), UL47 (32), and UL48 (20). Surprisingly, none of these mutations led to complete inhibition of productive virus replication, and even simultaneous deletion of the four major tegument protein genes, UL46, UL47, UL48, and UL49, of PrV did not abolish virus propagation in noncomplementing cells (22). However, simultaneous deletion of gE and gM or of gM and UL11 led to more than additive replication defects (5, 6, 34), indicating that these proteins possess redundant functions or participate in alternative assembly pathways.
In other alphaherpesviruses, several tegument proteins have been shown or suggested to be essential for virus replication, e.g., the products of the UL36, UL37, and UL48 genes of HSV-1 (14, 15, 43, 51) and of the UL49 gene of Marek's disease virus (16). In contrast, deletion of the UL49 gene of PrV had no significant effect on virus growth in cultured cells or on virulence in infected animals (13, 21). Thus, although these proteins are structurally conserved, their functions appear to be variable. Furthermore, it is remarkable that many tegument proteins are not significantly conserved between alpha-, beta-, and gammaherpesviruses (48).
Another approach to understanding herpesvirus morphogenesis is the analysis of physical interactions between the involved proteins by colocalization, coprecipitation, or two-hybrid studies. These experiments demonstrated interactions between the envelope proteins gE and gM and the UL49 tegument protein of PrV (21), between the UL49 and UL48 gene products of HSV-1 (17), between the UL48 and UL3.5 proteins of bovine herpesvirus 1 (37), and between the UL36 and UL37 tegument proteins of PrV (30) and human cytomegalovirus (4). Furthermore, several studies indicate that the UL36 protein of HSV-1 interacts directly with the major capsid protein (40, 53), and immunoelectron microscopic studies revealed a close association of PrV UL36 and UL37 with cytoplasmic nucleocapsids accumulating in the absence of UL48 (20). Remarkably, the capsidless L particles which were released in large amounts from cells infected with UL48-negative PrV did not contain the UL36 and UL37 gene products, whereas the major tegument proteins encoded by UL46, UL47, and UL49 were incorporated (20). Therefore, it has been proposed that the conserved UL36 and UL37 proteins form the innermost layer of the tegument which is added to cytoplasmic nucleocapsids independent of downstream maturation events. On the other hand, incorporation of the outer tegument proteins into Golgi-derived vesicles occurs in the absence of nucleocapsids.
We previously identified and localized the UL36 gene product of PrV, which, like its homologues in other herpesviruses, represents the largest virion protein, consisting of 3,084 amino acids with an apparent molecular mass of more than 300 kDa (30). By coimmunoprecipitation experiments, we detected a physical interaction with the UL37 protein, and the UL37 binding domain could be confined to amino acids 312 to 398 of the UL36 protein by yeast two-hybrid studies (30). However, until now, the function of the PrV UL36 protein has not been investigated directly. Early investigation of temperature-sensitive HSV-1 mutants indicated that the UL36 protein might be required for the release of viral DNA from infecting particles into the host cell nucleus (2), and a possible role of UL36 in cleavage and packaging of newly synthesized viral DNA has also been discussed (11). More recent analyses demonstrated that in the absence of the HSV-1 UL36 protein, DNA-filled capsids were produced and released from the nucleus but were not reenveloped in the cytoplasm (14). Thus, UL36-negative and UL37-negative HSV-1 strains were inhibited prior to secondary envelopment (15), as was UL37-negative PrV (29).
To characterize the functions of the PrV UL36 protein, we generated a UL36 null mutant utilizing a previously described infectious full-length clone of the PrV genome (33). To further analyze the interdependence of UL36 and UL37, a second PrV UL36 mutant was constructed which exhibited an in-frame deletion of the UL37 binding domain. The in vitro growth properties of these mutants and of a UL36 rescuant in noncomplementing and in UL36-expressing cells were analyzed.
MATERIALS AND METHODS
Viruses and cells.
All PrV mutants analyzed were derived from the wild-type strain Kaplan (PrV-Ka) (26). The previously described deletion mutant PrV-ΔUL37 was isolated after cotransfection of eukaryotic cells with virion DNA of PrV-Ka and a cloned and modified genome fragment (29). In contrast, the novel virus recombinants described here (see below) were generated in Escherichia coli by mutagenesis of pPrV-ΔgB, which represents a glycoprotein B (gB)-negative full-length clone of PrV-Ka (33). Viruses were propagated in RK13 cells, which were grown in minimum essential medium supplemented with 10% fetal calf serum (Invitrogen). For propagation of UL36-negative PrV, the trans-complementing cell line RK13-UL36 (see below) was grown in medium containing 500 μg of geneticin per ml (Invitrogen).
Plasmid constructs, cell lines, and virus recombinants.
The complete UL36 gene of PrV-Ka was prepared from genomic DNA as a 10,514-bp NruI fragment (Fig. 1B) and inserted into the SmaI-digested vector pUC19 (New England Biolabs). The resulting plasmid, pUC-UL36, was used together with pSV2neo (Clontech) for calcium phosphate-mediated cotransfection (24) of RK13 cells. Geneticin-resistant cell clones were isolated and tested for plaque formation after transfection with UL36-negative PrV. One trans-complementing cell line (RK13-UL36) was used for propagation and characterization of PrV-ΔUL36F (see below).
FIG. 1.
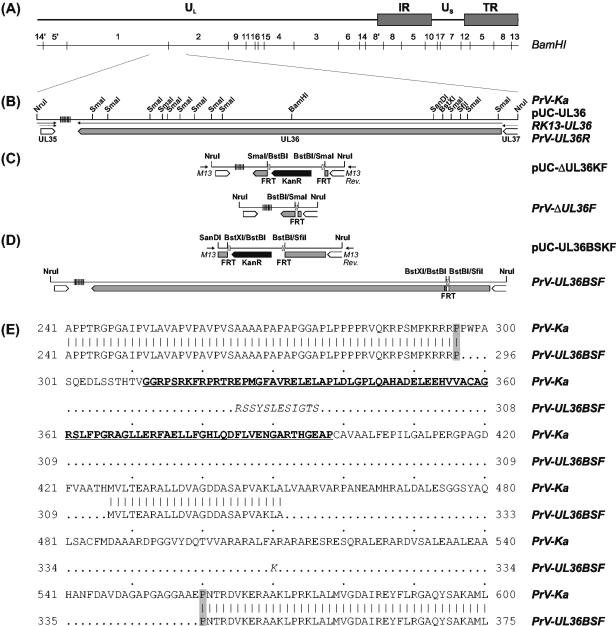
(A) Map of the DNA genome of pseudorabies virus, consisting of long (UL) and short (US) unique regions and inverted repeat sequences (IR, TR) flanking the latter. The positions of BamHI restriction sites are indicated. (B) An enlarged section shows a cloned NruI fragment of the wild-type PrV-Ka genome containing the entire UL36 gene and parts of adjacent ORFs (large open arrows). Arrows illustrate transcriptional organization, and vertical lines indicate repetitive DNA sequences. Relevant restriction sites are also plotted. Plasmid pUC-UL36 was used for generation of a UL36-complementing cell line (RK13-UL36) and of a virus rescuant (PrV-UL36R). To obtain PrV-ΔUL36F (C) and PrV-UL36BSF (D), different parts of pUC-UL36 were deleted and replaced with a kanamycin resistance gene (KanR) flanked by FRT sites (pUC-ΔUL36KF, pUC-UL36BSKF). The plasmid inserts were PCR amplified with vector-specific primers (M13 [−47], M13 rev [−48]), and used for mutagenesis of bacterial artificial chromosome-cloned PrV DNA. Finally, the resistance gene was removed by Flp-mediated recombination. (E) Alignment of parts of the deduced UL36 amino acid sequences of wild-type PrV-Ka and PrV-UL36BSF. The predicted binding region of the UL37 protein is underlined. In the UL36BSF protein, deleted amino acids are indicated by dots, and artificially inserted residues are in italics. The proline residues flanking the modified region are highlighted.
For deletion of UL36, pUC-UL36 was digested with SmaI, which released 11 restriction fragments spanning nucleotides 33374 to 42254 of the PrV genome sequence (31). The deleted sequence was replaced with a Klenow-treated 1,258-bp BstBI fragment of pKD13 (12), which contains a kanamycin resistance gene flanked by Flp recognition target (FRT) sites, giving rise to plasmid pUC-ΔUL36KF (Fig. 1C). The insert fragment was amplified by PCR with the vector-specific M13/pUC (−47) and M13/pUC reverse (−48) primers (New England Biolabs) and Pfx DNA polymerase (Invitrogen). The PCR product obtained was used for Red recombinase-mediated mutagenesis of the bacterial artificial chromosome pPrV-ΔgB in E. coli as described previously (12, 33). After isolation of kanamycin-resistant clones, the resistance gene was removed by mutagenesis with the FRT site-specific Flp recombinase, which was provided by transformation with helper plasmid pCP20 (9). Finally, the PrV gB gene was restored by cotransfection (24) of RK13-UL36 cells with bacterial artificial chromosome DNA and plasmid pUC-B1BclI (33). A single plaque isolate of the virus progeny was characterized and designated PrV-ΔUL36F (Fig. 1C).
To generate a PrV mutant exhibiting an in-frame deletion of the UL37 binding domain of UL36, the insert fragment of pUC-UL36 was first shortened to 1,833 bp by double digestion with SanDI and HindIII, Klenow treatment, and religation. Subsequently, the resulting plasmid was doubly digested with SfiI and BstXI to remove a 390-bp fragment from positions 41037 to 41426 of the PrV genome sequence (31). After Klenow treatment the 1,258-bp BstBI fragment of pKD13 (12) was inserted to obtain plasmid pUC-UL36BSKF (Fig. 1D). The insert was amplified by PCR and used for mutagenesis of pPrV-ΔgB as described above. After removal of the kanamycin resistance gene and restoration of the gB gene, recombinant PrV-UL36BSF (Fig. 1D) was isolated in noncomplementing RK13 cells.
The UL36 rescue mutant PrV-UL36R was isolated after cotransfection of RK13 cells with genomic DNA from PrV-ΔUL36F and plasmid pUC-UL36 (Fig. 1B). Virion DNA from all PrV mutants generated was characterized by restriction analyses and Southern blot hybridization as well as by PCR amplification and sequencing (Thermosequenase cycle sequencing kit; Amersham) of the mutagenized genome part (results not shown).
Metabolic labeling and immunoprecipitation.
RK13 cells were infected at a multiplicity of infection of 10 with PrV-Ka, PrV-UL36BSF, or PrV-ΔUL37 (29) and radiolabeled with 100 μCi of [35S]methionine/cysteine (MP Biomedicals) from 2 to 24 h postinfection. Then, cell lysates were prepared and proteins were precipitated (38) with monospecific rabbit antisera against the UL36 (30) and UL37 proteins (29) or glycoprotein gH (27) at dilutions of 1:100. The precipitated proteins were incubated in sample buffer (36) containing 10% β-mercaptoethanol for 5 min at 95°C and separated in discontinuous sodium dodecyl sulfate-5% polyacrylamide gels. The gels were fixed for 20 min in 7.5% methanol-10% acetic acid, dried, exposed to image plates, and examined in an image analyzer (FLA-3000, Fuji).
Determination of plaque sizes and one-step growth kinetics.
For investigation of direct cell-to-cell spread, RK13 monolayers were infected with PrV-Ka, PrV-ΔUL36F, PrV-UL36R, PrV-UL36BSF, or PrV-ΔUL37 (29) at a low multiplicity of infection and incubated in medium containing 0.6% methylcellulose at 37°C. After 3 days the cells were fixed for 15 min with a 1:1 mixture of methanol and acetone and washed repeatedly with phosphate-buffered saline. Thereafter, the cells were incubated consecutively for 1 h each with the glycoprotein gC-specific monoclonal antibody B16-c8 (27) and with Alexa 488-conjugated anti-mouse immunoglobulin antibodies (Molecular Probes). After repeated washing with phosphate-buffered saline, the plates were investigated in a fluorescence microscope (Diaphot 300; Nikon). The diameters of 30 plaques per assay were measured, and average diameters as well as standard deviations were calculated. One-step growth analyses were performed essentially as described (19). RK13 cells were harvested 1, 4, 8, 12, 24, and 36 h after infection at a multiplicity of infection of 5. Progeny virus titers were determined by plaque assays on RK13-UL36 cells, and the mean titers of three parallel experiments were plotted.
Electron microscopy.
RK13 and RK13-UL36 cells were infected with PrV-ΔUL36F or PrV-UL36BSF at a multiplicity of infection of 1 and incubated for 12 h at 37°C. Fixation and embedding were done as described (28), and counterstained ultrathin sections were analyzed in an electron microscope (Tecnai 12; Philips).
RESULTS
Isolation and DNA analysis of PrV UL36 mutants.
For functional characterization of the large tegument protein of PrV, the UL36 gene was eliminated by mutagenesis of a bacterial artificial chromosome-clone of the PrV genome in E. coli. The resulting recombinant PrV-ΔUL36F (Fig. 1C) exhibits a deletion of codons 21 to 2981 of the UL36 ORF, which originally contained 3,085 codons (30). Furthermore, the short preserved parts of the UL36 ORF were fused out of frame. No infectious progeny virus was obtained after transfection of normal RK13 cells with the mutagenized bacterial artificial chromosome, but mutant PrV-ΔUL36F could be isolated on RK13-UL36 cells, which contain a stable genomic insertion of the PrV UL36 gene (Fig. 1B).
Since UL36 proved to be essential for in vitro replication of PrV, the UL36 rescue mutant PrV-UL36R (Fig. 1B) could be readily isolated after cotransfection of normal RK13 cells with genomic DNA of PrV-ΔUL36F and the plasmid-cloned UL36 gene. Both PrV mutants were characterized by restriction analyses and Southern blot hybridization of genomic DNA, which showed the expected fragment patterns. The precise extent of the deletion in PrV-ΔUL36F was further confirmed by PCR amplification and sequencing of the affected genome region (results not shown).
A second bacterial artificial chromosome mutant, PrV-UL36BSF, which contains an in-frame deletion of codons 297 to 426 of UL36 (Fig. 1D), could also be propagated in noncomplementing RK13 cells. Initially, the maximum virus titers of this mutant were very low (≤104 PFU per ml), and plaque formation was severely impaired. However, after three passages of the transfection progeny in RK13 cells, larger virus plaques occurred spontaneously, and from one of them the described recombinant PrV-UL36BSF was purified to homogeneity and characterized by DNA analyses. Sequencing of the mutated part of the UL36 ORF confirmed the expected deletion but revealed a second in-frame deletion downstream of the first one. This spontaneous event led to loss of nucleotides 40640 to 40961 of the PrV genome sequence (31) accompanied by duplication of a deoxythymidine residue at position 40639. Additional analyses of PrV-UL36BSF DNA did not indicate further mutations of the UL36 gene. Thus, the deduced UL36 gene product of PrV-UL36BSF consists of amino acids 1 to 296 of the authentic protein, followed by 12 amino acids originating from the inserted FRT sequence, residues 427 to 451 of UL36, a novel lysine residue, and amino acids 560 to 3084 of the original UL36 protein (Fig. 1E). Remarkably, amino acids 296 and 560 flanking the mutated region of UL36 are prolines, which are known to disrupt secondary structures and to introduce flexible turns into proteins (10).
Characterization of the UL36 protein of PrV-UL36BSF.
The authentic UL36 gene product of PrV consists of 3,084 amino acids, accounting for a calculated molecular mass of 324,461 Da (30). In contrast, the truncated UL36 protein of PrV-UL36BSF is only 2,859 amino acids in size, corresponding to a mass of 300,684 Da. Immunoprecipitation of 35S-labeled proteins from infected RK13 cells with a UL36-specific antiserum (30) confirmed that the UL36 protein of PrV-UL36BSF is indeed smaller than that of PrV-Ka (Fig. 2, left panel). UL36 proteins of similar sizes were expressed by PrV-Ka and PrV-UL36R, whereas no newly synthesized UL36 gene products were found in RK13 cells infected with phenotypically complemented PrV-ΔUL36F (results not shown).
FIG. 2.
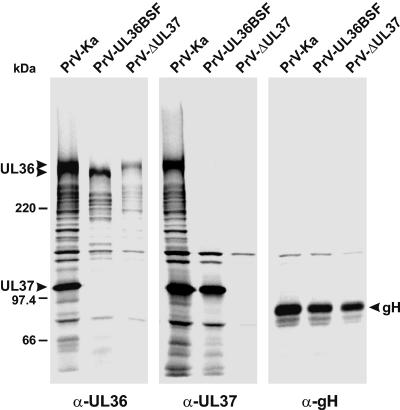
Immunoprecipitation of 35S-labeled proteins from RK13 cells infected with PrV-Ka, PrV-UL36BSF, or PrV-ΔUL37 with monospecific antisera (anti-UL36, anti-UL37, and anti-gH). The precipitates were separated in a sodium dodecyl sulfate-5% polyacrylamide gel under reducing conditions and analyzed by fluorography. The positions of the authentic and truncated UL36 gene products, of the UL37 protein, and of gH are marked by arrowheads. The molecular masses (in kilodaltons) of marker proteins are indicated on the left.
Previously, by yeast-two-hybrid studies and by coimmunoprecipitation experiments, we showed that the UL36 protein of PrV physically interacts with the UL37 gene product (30). Our present studies confirmed that a UL36-specific antiserum (30) not only precipitated its target protein of more than 300 kDa but also a prominent 100-kDa protein (Fig. 2, left panel). A UL37-specific antiserum (29) coprecipitated the same proteins from PrV-Ka-infected cells (Fig. 2, middle panel). The 100-kDa protein represents the UL37 gene product, since it was not detectable in cells infected with PrV-ΔUL37 (29) (Fig. 2, left and middle panels). Consistently, the UL37-specific antiserum failed to coprecipitate the UL36 protein from cells infected with PrV-ΔUL37 (Fig. 2, middle panel). However, the truncated UL36 protein of PrV-UL36BSF also did not coprecipitate with the UL37 protein and vice versa, although the amounts of both proteins in infected cells were comparable to those found in cells infected with PrV-Ka (Fig. 2, left and middle panels).
To ensure that the input amounts of labeled virus proteins were comparable, control immunoprecipitations with a gH-specific antiserum (27) were performed (Fig. 2, right panel). Most of the protein bands with molecular masses between those of UL36 and UL37 likely represent breakdown products of the large UL36 gene product, since they were recognized by the anti-UL36 serum in cells infected with PrV-Ka, PrV-UL36BSF, or PrV-ΔUL37 (Fig. 2, left panel) and were also coprecipitated by the anti-UL37 serum from PrV-Ka infected cells (Fig. 2, middle panel), but failed to coprecipitate with UL37 in cells infected with PrV-UL36BSF (Fig. 2, middle panel). One protein of ca. 140 kDa was nonspecifically precipitated irrespective of the antiserum used.
Thus, our data show that the interaction between the UL36 and UL37 proteins is either completely abolished in PrV-UL36BSF or at least significantly less stable than in wild-type PrV. This finding is in agreement with previous mapping of the UL37 binding site to residues 312 to 398 of UL36 by yeast-two-hybrid studies (30). As can be seen in Fig. 1E, this stretch of amino acids was completely deleted in the UL36 protein of PrV-UL36BSF.
In vitro growth properties of PrV UL36 mutants.
To precisely analyze the replication defects of PrV-ΔUL36 and PrV-UL36BSF, plaque formation and one-step growth kinetics were investigated in RK13 cells (Fig. 3). For comparison, PrV-ΔUL37 (29) was included in these studies. High-titer virus stocks of PrV-ΔUL36F and PrV-ΔUL37 were prepared in trans-complementing RK13-UL36 and RK13-UL37 cells (29), respectively. In contrast, virus stocks of wild-type PrV-Ka and the rescuant PrV-UL36R were prepared in normal RK13 cells. PrV-UL36BSF had to be propagated in RK13 cells because in complementing RK-UL36 cells, the mutated UL36 gene of the virus was frequently repaired by recombination with the cellular copy of the authentic gene.
FIG. 3.
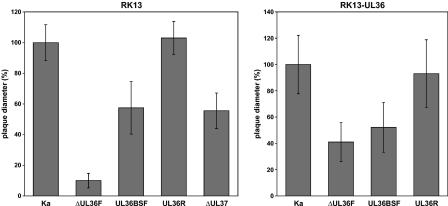
Plaque sizes of PrV-ΔUL36F, PrV-UL36BSF, PrV-UL36R, PrV-ΔUL37, and PrV-Ka. Virus plaques were visualized 48 h after infection of RK13 or RK13-UL36 cells by indirect immunofluorescence with a gC-specific antibody. For each virus, the average diameters of 30 plaques were determined and calculated as percentages of those of the plaques induced by PrV-Ka. Standard deviations are also shown.
No plaque formation of PrV-ΔUL36F was observed on RK13 cells, but 3 days after inoculation small foci comprising only a few infected cells were detectable (Fig. 3, left panel). In contrast, the plaque diameters of PrV-UL36BSF were reduced by less than 45% compared to those of PrV-Ka or of the rescuant PrV-UL36R (Fig. 3, left panel). Remarkably, the average plaque sizes of PrV-ΔUL37 and PrV-UL36BSF were almost identical (Fig. 3, left panel). Thus, deletion of either UL37 or the UL37 binding domain of UL36 affected cell-to-cell spread of PrV to a lesser extent than deletion of the complete UL36 gene.
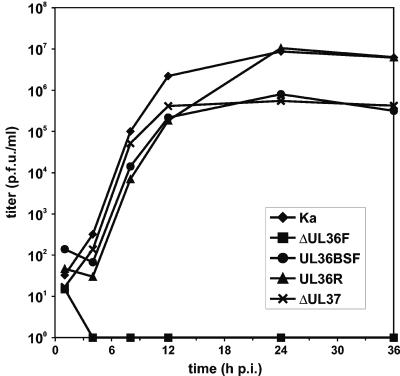
The one-step growth kinetics of PrV-ΔUL37 and PrV-UL36BSF were also almost identical (Fig. 4). Compared to wild-type PrV-Ka, the onset of replication was not delayed, but maximum virus titers were significantly reduced (Fig. 4). Whereas the replication kinetics and maximum titer of the rescue mutant PrV-UL36R were comparable to those of wild-type PrV, no infectious progeny of PrV-ΔUL36F was detectable in RK13 cells by subsequent plaque assays in RK13-UL36 cells (Fig. 4). Thus, unlike UL37, the UL36 gene product is absolutely essential for productive replication of PrV.
FIG. 4.
One-step growth kinetics of PrV-ΔUL36F, PrV-UL36BSF, PrV-UL36R, PrV-ΔUL37, and PrV-Ka in RK13 cells. Cells were scraped into the medium 1, 4, 8, 12, 24, and 36 h after synchronized infection at a multiplicity of infection of 5 and lysed by freezing and thawing. Progeny virus titers were determined by plaque assays in RK13-UL36 cells. The average results of three parallel experiments are shown.
The growth properties of PrV UL36 mutants were also analyzed in trans-complementing RK13-UL36 cells. Although PrV-ΔUL36F could be propagated in this cell line, the titers of the virus mutant obtained were ca. 100-fold lower than those of PrV-Ka (results not shown), and average plaque diameters were reduced by ca. 60% (Fig. 3, right panel). However, these defects were not caused by unwanted second-site mutations of PrV-ΔUL36F, since wild-type-like growth properties could be fully restored by repair of the viral UL36 locus in PrV-UL36R (Fig. 3 and 4). Thus, although RK13-UL36 cells were sufficient for successful isolation of PrV-ΔUL36, they did not trans-complement the mutant virus to wild-type levels. This was further confirmed by the observation that the moderate cell-to-cell spread defect of PrV-UL36BSF in RK13 cells (Fig. 3, left panel) was not altered in RK13-UL36 cells (Fig. 3, right panel).
Effects of UL36 gene mutations on virion formation.
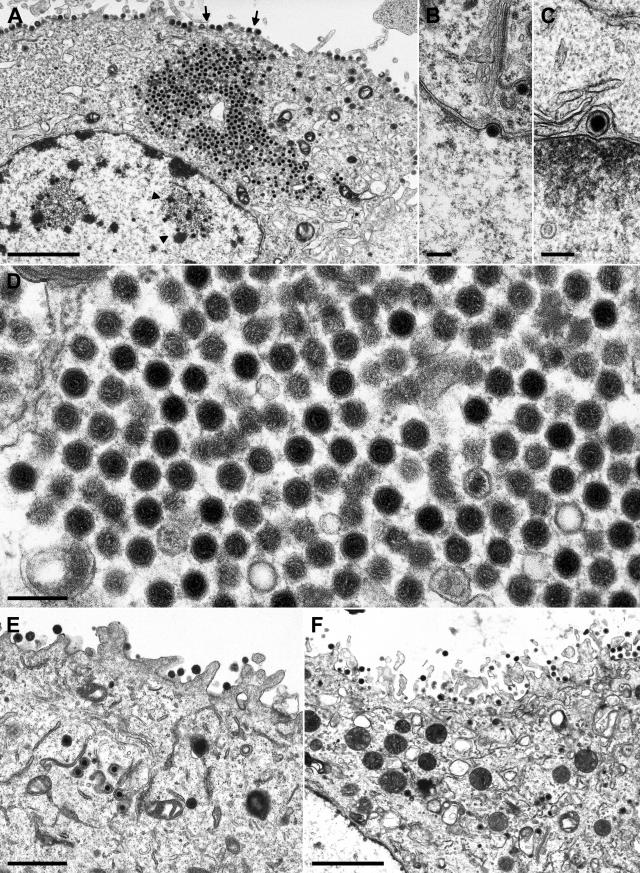
Since the UL36 gene of PrV encodes a tegument protein, UL36 might play an essential role during virion morphogenesis (30, 42). Therefore, cells infected with either the phenotypically complemented UL36 null mutant PrV-ΔUL36F or PrV-UL36BSF, which expresses a truncated UL36 protein, were analyzed by electron microscopy at late times (12 h) after infection (Fig. 5 and 6). Both virus mutants were able to enter RK13 cells, to produce nucleocapsids in the host-cell nucleus (Fig. 5A and 6A), and to leave the nucleus by primary envelopment and development at the nuclear membrane (Fig. 5B, 6B, and 6C). However, the distribution of nucleocapsids in the cytoplasm was different. Whereas in PrV-ΔUL36F-infected cells the nucleocapsids were dispersed throughout the cytoplasm and only occasionally formed aggregates of a few particles (Fig. 5A), most of the cytoplasmic nucleocapsids of PrV-UL36BSF were found evenly spaced in large ordered structures (Fig. 6A and 6D). Similar structures were previously observed in cells infected with PrV-ΔUL37 (29) but not with other PrV mutants impaired in virion formation. This again indicates that deletion of the UL37 binding domain of UL36 in PrV-UL36BSF causes a phenotype similar to that of a UL37 null mutant.
FIG. 5.
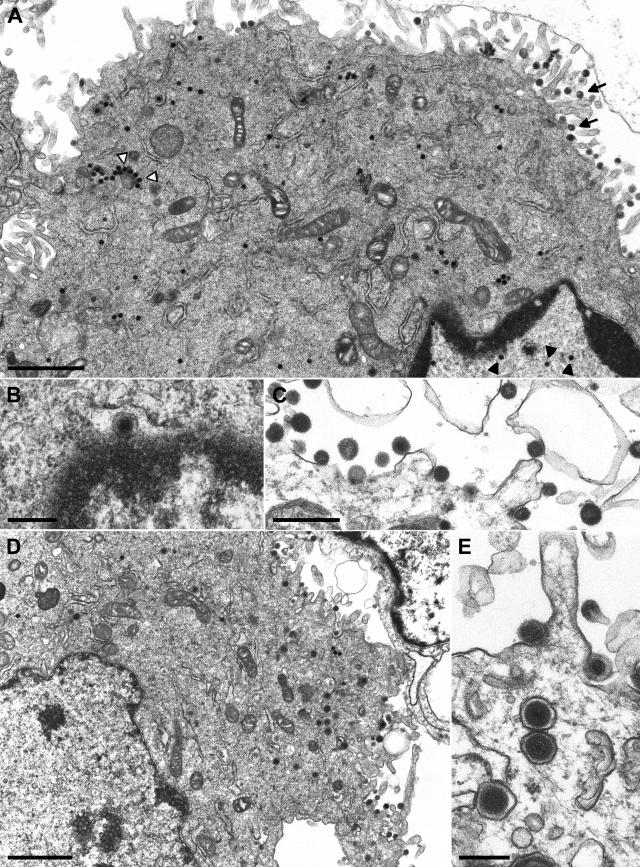
Electron microscopy of cells infected with phenotypically complemented PrV-ΔUL36F. RK13 (A to C) and RK13-UL36 cells (D, E) were analyzed 12 h after infection at a multiplicity of infection of 1. In RK13 cells, DNA-filled capsids were detected in the nucleus (A, black arrowheads) and in the cytoplasm (A, white arrowheads), and primary enveloped virus particles were found in the perinuclear space (B). However, only capsidless particles were efficiently released from the cells (A, arrows; C). In RK13-UL36 cells, enveloped virions were detectable in cytoplasmic vesicles and in the extracellular space (D, E). Bars: 1.5 μm (A, D), 500 nm (C), or 250 nm (B, E).
FIG. 6.
Electron microscopy of cells infected with PrV-UL36BSF. RK13 (A to D) and RK13-UL36 (E, F) cells were analyzed 12 h after infection. An overview (A) shows single intranuclear (arrowheads) and aggregated cytoplasmic nucleocapsids as well as capsidless extracellular particles (arrows). Enlargements depict primary envelopment at the nuclear membrane (B, C) and ordered clusters of unenveloped nucleocapsids in the cytoplasm (D). In RK13-UL36 cells, secondary envelopment was observed in the cytoplasm (E) and virions were released from the cells (E, F). Bars: 2.0 μm (A, F), 1.0 μm (E), or 200 nm (B, D).
As in RK13 cells infected with PrV-ΔUL37 (29), secondary envelopment of cytoplasmic nucleocapsids in the trans-Golgi region was only rarely observed in cells infected with PrV-UL36BSF or PrV-ΔUL36F. Consequently, capsid-containing extracellular virions were almost undetectable (Fig. 5A and 6A). However, numerous enveloped, but capsid-less particles were released from RK13 cells infected with either of the UL36 gene mutants (Fig. 5A, 5C, and 6A). Thus, presence of UL36 and UL37 is apparently not required for formation of virus-like particles, but is necessary for efficient incorporation of nucleocapsids. UL36 might possess an additional function during virus entry or during the nuclear phase of virus replication since in cells infected with PrV-ΔUL36F the total amount of detectable nuclear and cytoplasmic nucleocapsids was reproducibly lower than in cells infected at a similar multiplicity of infection with wild-type PrV or PrV-UL36BSF (results not shown). As expected, the replication defects of PrV-ΔUL36F and PrV-UL36BSF were partly corrected in RK13-UL36 cells, and secondary envelopment (Fig. 6E) as well as intracytoplasmic enveloped virions (Fig. 5D) were readily observed (Fig. 5D, 5E, 6E, and 6F).
DISCUSSION
The UL36 gene, encoding the large tegument protein of PrV, possesses homologues in all mammalian and avian herpesviruses analyzed to date (30, 48), indicating an important function of this protein during the viral life cycle. It was previously shown that UL36 is essential for replication of HSV-1 (14, 47). In the present study, we demonstrate that the UL36 gene is also indispensable for productive replication of PrV in cell culture. A PrV mutant lacking major parts of the UL36 ORF (PrV-ΔUL36F) could only be isolated and propagated in a cell line which carried a stable genomic insertion of the gene. Thus, the UL36 protein is the first PrV tegument protein which has been shown to be essential sensu stricto for morphogenesis. However, whereas repair of the UL36 gene restored wild-type-like growth properties to PrV-ΔUL36F, replication in trans-complementing cells was much less efficient with respect to plaque size and maximum virus titer.
Although yet unknown coding regions or cis-acting regulatory sequences may have been affected by the UL36 deletion, the DNA sequence of the PrV genome (31) provides no evidence for this, since the predicted polyadenylation sites of the adjoining UL35 and UL37 genes were not affected in PrV-ΔUL36F. Furthermore, in Northern blot analyses of cells infected with wild-type PrV, no viral transcripts overlapping the UL36 mRNA were detectable (results not shown). Thus, although Western blot analyses demonstrated expression of the UL36 protein in trans-complementing cells after infection with PrV-ΔUL36F (results not shown), the amount of protein might not be sufficient to fully compensate for the viral gene deletion.
Electron microscopy of noncomplementing cells infected with phenotypically complemented virus revealed that, like UL36-negative HSV-1 (14), PrV-ΔUL36F was able to form DNA-filled capsids in the nucleus which transit the nuclear membrane by consecutive envelopment and deenvelopment at its inner and outer leaflets. Thus, it appears unlikely that PrV UL36 plays a role during cleavage and encapsidation of viral DNA, as previously discussed for the homologous protein of HSV-1 (11). However, this possibility cannot be ruled out completely, since a limited amount of UL36 protein is brought into the cells by infection with phenotypically complemented virions. Whereas the UL36 protein of HSV-1 was found distributed diffusely in the nucleus and the cytoplasm of infected cells by immunofluorescence and cell fractionation experiments (40), by detailed immunoelectron microscopy, the PrV UL36 protein was detectable only in mature virions and at cytoplasmic nucleocapsids but was not associated with primary enveloped perinuclear particles or intranuclear capsids (30).
Nevertheless, the most salient defect of UL36-negative HSV-1 (14) and PrV (this study) was inhibition of secondary envelopment in the cytoplasm. This observation is in agreement with the hypothesis that binding of UL36 to capsid proteins or capsids (40, 53) is a prerequisite for subsequent tegumentation and final envelopment (42). However, although extensive electron microscopic analyses revealed that budding of PrV-ΔUL36F into cytoplasmic vesicles was a rare event, it was not completely blocked. This might be explained by compensatory interactions of other tegument proteins with nucleocapsids. Binding of tegument proteins other than UL36 to capsids has been described for betaherpesviruses (3, 8, 50), but their genes are not detectably conserved in alphaherpesvirus genomes. Furthermore, although PrV-ΔUL36F was unable to produce visible plaques in noncomplementing cells, immunofluorescence tests with gC-specific antibodies detected small foci of infected cells which, presumably, did not emerge from cell division. Thus, a limited number of virus particles that are capable of direct cell-to-cell spread may be formed even in the absence of UL36. However, infectious PrV-ΔUL36F was never detected in culture supernatants, or after freeze-thawing of infected noncomplementing cells. These findings indicate that besides its role during virus egress, PrV UL36 might possess an additional function which may be relevant during the initial steps of virus replication.
This hypothesis was supported by the observation that the numbers of intranuclear capsids detected by electron microscopy of noncomplementing cells infected with partially trans-complemented PrV-ΔUL36F (see above) were reproducibly lower than found at the same times after infection at a similar multiplicity of infection with wild-type PrV or other PrV mutants (results not shown). Investigations of an HSV-1 mutant carrying a temperature-sensitive mutation in UL36 revealed an inhibition of DNA release from incoming nucleocapsids into the host cell nucleus at the nonpermissive temperature (2). At present, we are not able to investigate the putative early functions of UL36 in more detail because infectious progeny of PrV-ΔUL36F require propagation on UL36-expressing cells, and thus, progeny virions invariably contain the wild-type UL36 protein.
To investigate the egress function of UL36 in more detail, we generated a second PrV mutant (PrV-UL36BSF) possessing an in-frame deletion of the predicted binding site of the UL37 tegument protein (30). Unlike PrV-ΔUL36F, PrV-U36BSF was replication competent in noncomplementing cells, demonstrating that the deleted domain was not essential for protein function. Remarkably, in the isolate of PrV-UL36BSF characterized, the engineered deletion of UL36 codons 297 to 426 was found to be accompanied by a second spontaneous deletion of UL36 codons 452 to 559. Presumably, the initial deletion strongly impaired UL36 function by an alteration of the overall structure of the protein, and a second mutation was required to correct this defect. Interestingly, the final deletion within the UL36 protein is flanked by proline residues, which disrupt secondary structure and introduce flexible turns into proteins (10).
Coimmunoprecipitation experiments demonstrated that, unlike wild-type UL36, the mutated UL36 protein of PrV-UL36BSF did not exhibit complex formation with the UL37 gene product. This result confirmed previous mapping of the UL37 binding site to amino acids 312 to 398 of UL36 by yeast two-hybrid studies (30). Although replication of PrV-UL36BSF in noncomplementing cells was severely affected, the observed growth defects were less pronounced than after deletion of the entire UL36 protein in PrV-ΔUL36F. This finding correlates with the observation that a UL37-negative PrV mutant was impaired but still replication competent in noncomplementing cells (29). Electron microscopy of cells infected with either PrV-UL36BSF or PrV-ΔUL37 revealed accumulation of nucleocapsids in the cytoplasm coinciding with an inhibition of secondary envelopment, yet the plaque sizes of both mutants were reduced by only 45%, whereas maximum virus titers were ca. 50-fold lower than those of wild-type PrV.
The comparable growth properties of the two mutants suggest that no relevant UL36 function other than UL37 binding is abolished in PrV-UL36BSF and that the UL37 protein requires binding to UL36 to fulfill its important but nonessential function during virion morphogenesis. Unlike PrV UL37, the UL37 protein of HSV-1 is apparently essential for virus replication, and electron microscopic studies revealed that in the absence of UL37, naked HSV-1 nucleocapsids accumulate in the nucleus as well as in the cytoplasm, indicating a role for UL37 during primary and secondary envelopment (15). In contrast, neither PrV-ΔUL37 (29) nor PrV-UL36BSF exhibited retention of capsids in the nucleus.
A remarkable ultrastructural feature of noncomplementing cells infected with PrV-ΔUL37 (29) as well as with UL37-deleted HSV-1 (15) was the accumulation of cytoplasmic nucleocapsids in large ordered clusters. These clusters were clearly different from those observed with PrV mutants lacking envelope glycoproteins gE and gM (4, 5), tegument protein UL11, or UL11 and gM (33, 34) which, in addition, contained large amounts of tegument material. Although electron-dense tegument material was apparently not associated with the aggregations formed in the absence of UL37, immunoelectron microscopy demonstrated the presence of the UL36 gene product in these capsid clusters (30). Our results confirm that the presence of UL36 protein is necessary for this phenotype because aggregation of cytoplasmic nucleocapsids was not observed in PrV-ΔUL36F-infected cells. However, in cells infected with PrV-UL36BSF, which expresses a truncated UL36 protein unable to interact with UL37, similar cytoplasmic capsid clusters were found as in the absence of UL37. This may be correlated with the smaller number of nucleocapsids present in the cytoplasm of PrV-ΔUL36F-infected cells. Alternatively, and in line with our previous observations (29, 30), the presence of the UL36 protein at nucleocapsids (40, 53) may be required for aggregation, which is prevented by subsequent binding of the UL37 gene product. Moreover, the phenotype of PrV-UL36BSF indicates that UL36 protein domains which may be responsible for self-aggregation or for UL37 binding are distinct and nonoverlapping.
Although secondary envelopment of PrV was strongly impaired in the absence of either UL36, UL37, or the UL37 binding domain of UL36, none of these mutations prevented budding events in the trans-Golgi region leading to the formation and release of tegument-containing but capsidless virus-like particles. Thus, our present results confirm the hypothesis that tegumentation in the cytoplasm initiates independently at two sites (42). Whereas the UL36 and UL37 gene products initially bind to nucleocapsids (29, 30), other tegument components, including the UL46, UL47, UL49, and UL11 proteins, become associated with the future budding sites independent of the presence of nucleocapsids. The UL48 protein might play a central role in linking the two processes (20, 32, 33, 34). However, direct interactions between the “inner” and “outer” tegument proteins of PrV remain to be demonstrated.
Acknowledgments
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (Me 854/5-2).
The technical assistance of Charlotte Ehrlich, Uta Hartwig, Petra Meyer, and Diana Werner is greatly appreciated.
REFERENCES
- 1.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batterson, W., D. Furlong, and B. Roizman. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45:397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxter, M. K., and W. Gibson. 2001. Cytomegalovirus basic phosphoprotein (pUL32) binds to capsids in vitro through its amino one-third. J. Virol. 75:6865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bechtel, J. T., and T. Shenk. 2002. Hum. cytomegalovirus UL47 tegument protein functions after entry and before immediate-early gene expression. J. Virol. 76:1043-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 180:1-19. [DOI] [PubMed] [Google Scholar]
- 8.Chen, D. H., H. Jiang, M. Lee, F. Liu, and Z. H. Zhou. 1999. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology 260:10-16. [DOI] [PubMed] [Google Scholar]
- 9.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 10.Chou, P. Y., and G. D. Fasman. 1978. Empirical prediction of protein conformation. Annu. Rev. Biochem. 47:251-276. [DOI] [PubMed] [Google Scholar]
- 11.Chou, J., and B. Roizman. 1993. Characterization of DNA sequence-common and sequence-specific proteins binding to cis-acting sites for cleavage of the terminal a sequence of the herpes simplex virus 1 genome. J. Virol. 63:1059-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.del Rio, T., H. C. Werner, and L. W. Enquist. 2002. The pseudorabies virus VP22 homologue (UL49) is dispensable for virus growth in vitro and has no effect on virulence and neuronal spread in rodents. J. Virol. 76:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai, P. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:1608-10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desai, P., G. Sexton, J. McCaffery, and S. Person. 2001. A null mutation in the gene encoding the UL37 polypeptide of herpes simplex virus type 1 abrogates virus maturation. J. Virol. 75:10259-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorange, F., B. K. Tischer, J. F. Vautherot, and N. Osterrieder. 2002. Characterization of Marek's Disease Virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 76:1959-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs, W., B. G. Klupp, H. Granzow, H. J. Rziha, and T. C. Mettenleiter. 1996. Identification and characterization of the pseudorabies virus UL3.5 protein, which is involved in virus egress. J. Virol. 70:3517-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuchs, W., B. G. Klupp, H. Granzow, A. Mundt, C. Hengartner, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuchs, W., H. Granzow, and T. C. Mettenleiter. 2003. A pseudorabies virus recombinant simultaneously lacking the major tegument proteins encoded by the UL46, UL47, UL48, and UL49 genes is viable in cultured cells. J. Virol. 77:12891-12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gershon, A. A., D. L. Sherman, Z. Zhu, C. A. Gabel, R. T. Ambron, and M. D. Gershon. 1994. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J. Virol. 68:6372-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 25.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan, A. S., and A. Vatter. 1959. A comparison of herpes simplex and pseudorabies virus. Virology 7:394-407. [DOI] [PubMed] [Google Scholar]
- 27.Klupp, B. G., and T. C. Mettenleiter. 1999. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J. Virol. 73:3014-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klupp, B. G., C. J. Hengartner, T. C. Mettenleiter, and L. W. Enquist. 2004. Complete, annotated sequence of the pseudorabies virus genome. J. Virol. 78:424-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopp, M., B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 76:8820-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 77:5339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, and T. C. Mettenleiter. 2004. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J. Virol. 78:3024-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwong, A. D., J. A. Kruper, and N. Frenkel. 1988. Herpes simplex virus virion host shutoff function. J. Virol. 62:912-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 277:680-685. [DOI] [PubMed] [Google Scholar]
- 37.Lam, N., and G. J. Letchworth. 2000. Bovine herpesvirus 1 UL3.5 interacts with bovine herpesvirus 1 alpha-transinducing factor. J. Virol. 74:2876-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lukács, N., H.-J. Thiel, T. C. Mettenleiter, and H.-J. Rziha. 1985. Demonstration of three major species of pseudorabies virus glycoproteins and identification of a disulfide-linked glycoprotein complex. J. Virol. 53:166-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 40.McNabb, D. S., and R. J. Courtney. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology. 190:221-232. [DOI] [PubMed] [Google Scholar]
- 41.Mettenleiter, T. C. 2000. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis-state of the art, June 1999. Vet. Res. 31:99-115. [DOI] [PubMed] [Google Scholar]
- 42.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of a cellular kinase to dissolve the nuclear lamina. Science 297:854-857. [DOI] [PubMed] [Google Scholar]
- 45.Radsak, K., M. Eickmann, T. Mockenhaupt, E. Bogner, H. Kern, A. Eis-Hubinger, and M. Reschke. 1996. Retrieval of human cytomegalovirus glycoprotein B from the infected cell surface for virus envelopment. Arch. Virol. 141:557-572. [DOI] [PubMed] [Google Scholar]
- 46.Reynolds, A., B. Ryckman, J. Baines, Y. Zhou, L. Liang, and R. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 48.Roizman, B., and P. E. Pellet. 2001. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 49.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment -> deenvelopment -> reenvelopment pathway. J. Virol. 75:5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trus, B. L., W. Gibson, N. Cheng, and A. C. Steven. 1999. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J. Virol. 73:2181-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinheimer, S. P., B. A. Boyd, S. K. Durham, J. L. Resnick, and D. R. O'Boyle. 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 66:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whealy, M. E., J. P. Card, R. P. Meade, A. K. Robbins, and L. W. Enquist. 1991. Effect of brefeldin A on alphaherpesvirus membrane protein glycosylation and virus egress. J. Virol. 65:1066-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]