Abstract
Capsid (CA)-specific restrictions are determinants of retroviral tropism in mammalian cells. One such restriction, human Ref1, targets strains of murine leukemia virus bearing an arginine at CA residue 110 (N-MLV), resulting in decreased accumulation of viral cDNA. The cellular factors accounting for Ref1 activity are unknown. As2O3 increases N-MLV titer in Ref1-positive cells, possibly by counteracting Ref1. Restriction factor saturation experiments suggest that Ref1 may also target human immunodeficiency virus type 1 (HIV-1), but only if its CA is not bound to the cellular protein cyclophilin A (CypA). As a step towards understanding the genetic determinants of Ref1, we subjected Ref1-positive TE671 cells to three sequential rounds of selection with N-MLV reporter viruses. We isolated a subclone, 17H1, that was permissive for N-MLV infection and therefore deficient in Ref1 activity. Stimulation of N-MLV replication by As2O3 was attenuated in 17H1, confirming that the drug acts by overcoming Ref1 activity. HIV-1 infection of 17H1 cells was resistant to disruption of the CA-CypA interaction, demonstrating that Ref1 restricts CypA-free HIV-1. Our results suggest that interaction with CypA evolved to protect HIV-1 from this human antiviral activity.
Retrovirus tropism in mammalian cells is partially determined by dominant restriction factors that act after viral entry but before integration into the host genome. The archetype of such restriction factors is the product of the murine Fv1 gene, which restricts infection by murine leukemia virus (MLV) (14). The product of this gene is a protein related to the Gag proteins of the murine endogenous retrovirus MuERV-L and the human endogenous retrovirus HERV-L (3, 13). Two alleles of the gene, Fv1n and Fv1b, restrict different forms of MLV that are termed B-tropic and N-tropic, respectively (B-MLV and N-MLV). A single amino acid in the MLV capsid (CA) determines the specificity of restriction; changes at this position are sufficient to switch virions between N- and B-tropic phenotypes (9). Restriction by Fv1 can be overcome either by high multiplicities of infection (MOIs) or by pretreatment of target cells with MLV virions or virus-like particles (VLPs) of the restricted tropism (5, 7). These observations suggest that Fv1, or another cellular factor required for Fv1's restriction activity, can be saturated by high doses of incoming viral capsids, allowing subsequently entering virions to escape restriction.
More recently, a number of nonmurine mammalian cell lines were shown to exhibit Fv1-like restriction activity against MLV. This activity is always directed against N-tropic virus—B-tropic virus is unrestricted (18). This N-MLV restricting activity has been termed Ref1 (18). Ref1, like Fv1, can be overcome at high MOI and by treatment with saturating doses of N-MLV VLPs (19). The similarities between the Ref1 and Fv1 restriction phenotypes suggest that similar mechanisms may be involved in both human and murine cells, but there is no human Fv1 gene (3). Mechanistic differences between Fv1 and Ref1 restriction have also been described. Restriction by Ref1 occurs at the level of reverse transcription, the major defect being a profound decrease in the production of full-length cDNA, while Fv1 acts after reverse transcription is complete (2, 12, 18).
To better understand the mechanism and genetics behind Ref1, we set out to create cell lines defective in Ref1 activity. We began with the assumption that individual cells within a pool would exhibit variable levels of Ref1 activity due to genetic or epigenetic variability. This assumption was based on previous observations in our lab that single cell clones derived from a given cell line support different levels of retrovirus replication. Our strategy was to select those “naturally” occurring cells within a population that were the most permissive to N-MLV infection.
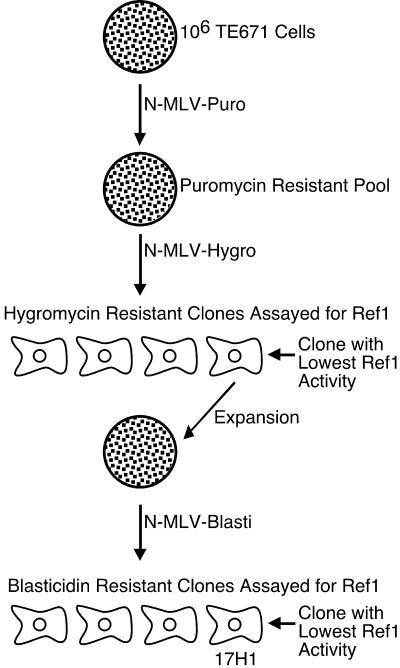
We chose to work with the human TE671 rhabdomyosarcoma cell line because of its robust Ref1 activity: TE671 cells have previously been shown to restrict N-MLV by greater than 100-fold compared to the B-MLV titer (18). To select a Ref1-deficient TE671 clone, a starting pool of 106 cells was subjected to sequential infection at low MOI (0.01 to 0.001) with vesicular stomatitis virus G protein (VSV-G)-pseudotyped N-MLV vectors delivering drug resistance genes, followed by selection of transduced cells with the appropriate drug (Fig. 1). This strategy was designed to select for those cells with the highest permissivity to N-MLV infection. At 48 h after each infection, cells were subjected to selection with the corresponding drug until colonies grew out. These colonies were then pooled and subjected to the next round of infection and selection with a different resistance vector-drug combination.
FIG. 1.
Selection strategy for isolating Ref1-deficient TE671 cells. A total of 106 cells were sequentially infected at a low MOI (0.001 to 0.0001) with N-MLV-puromycin and N-MLV-hygromycin and subjected to drug selection with puromycin and hygromycin, respectively, between rounds of infection. Drug-resistant clones were then isolated and screened for Ref1 activity. The clone exhibiting the lowest Ref1 activity was subjected to another round of infection and selection with N-MLV-blasticidin and blasticidin. Blasticidin-resistant clones were isolated and assayed for Ref1 activity. The clone with the lowest activity, designated 17H1, was selected for further study.
Retroviruses and retroviral vectors were produced by transfection of 293T cells with Lipofectamine 2000 (Invitrogen) and then filtered (0.45 μm). N- and B-tropic MLV vectors pseudotyped with the VSV-G protein were produced using either pCIG3 N or B (4) and the VSV-G expression plasmid pMD.G and carried either MSCV-based enhanced green fluorescent protein (GFP) or DsRed Express expression cassettes (21) or pBABE-based drug resistance cassettes (11). When the titers of N- and B-tropic MLV vectors were to be directly compared, the two vectors were first shown to have equal titers on Mus dunni tail fibroblasts (MDTF), cells that do not restrict either N- or B-tropic MLV (18).
After two rounds of infection and selection, first with N-MLV-puromycin and then with N-MLV-hygromycin, several colonies were isolated and analyzed for Ref1 activity, using a rapid two-color restriction assay similar to ones already described (18). Briefly, clones were infected with a mixture of N-MLV-GFP and B-MLV-DsRed. Each infected clone was then assayed by flow cytometry to determine the ratio of red to green cells. Those clones with the lowest red cell-to-green cell ratio were deemed Ref1 deficient and were analyzed further.
Among the clones analyzed after two rounds of selection, the one with the lowest Ref1 activity (roughly 10-fold, versus >150-fold for the parent TE671 line) was expanded and subjected to a third round of infection with N-MLV-blasticidin. After blasticidin selection, 48 subclones were picked and analyzed by two-color assay. The clone with the lowest Ref1 activity, clone 17H1, was selected for further study in the rest of the experiments described in this paper. TE671 cells expressing all three drug resistance genes but not selected for loss of Ref1 exhibited restriction activity identical to the parental TE671 line (data not shown).
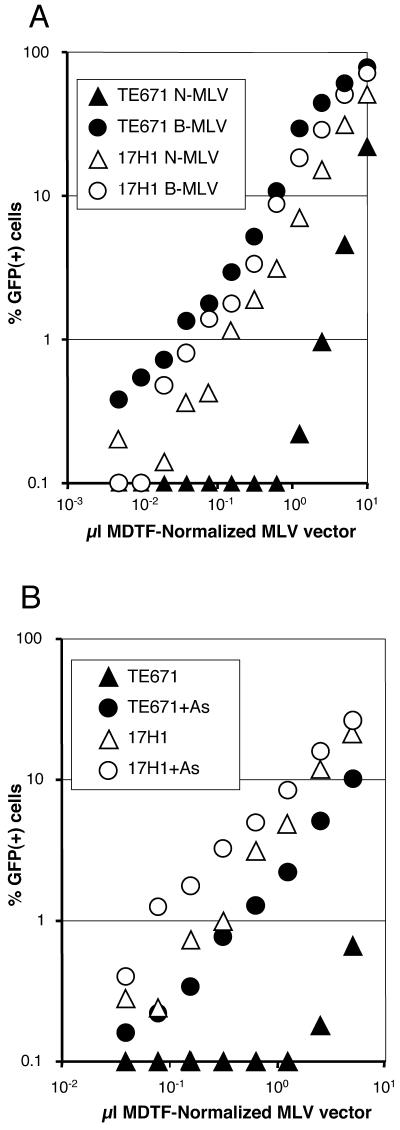
To determine the level of Ref1 activity of 17H1 cells, TE671 and 17H1 were plated at 104 cells/well in 96-well plates overnight and then infected with MDTF-normalized VSV-G-pseudotyped N-tropic and B-tropic MLV vectors delivering a GFP reporter construct. Eighteen to 24 h later, the medium on the cells was replaced and they were incubated another 2 days before the percent GFP-positive cells in each well was determined by flow cytometric analysis, counting 5,000 cells of each sample.
As shown in Fig. 2A, TE671 and 17H1 were equally infectible with B-MLV. TE671 cells strongly restricted N-MLV, with a maximal effect of >250-fold. 17H1 cells, however, restricted N-MLV very poorly, with a maximal effect of roughly threefold. We therefore concluded that the 17H1 subclone of TE671 has lost Ref1 activity and is permissive to N-MLV infection.
FIG. 2.
17H1 cells are deficient in Ref1 activity. (A) 17H1 and TE671 cells were infected with MDTF-normalized VSV-G-pseudotyped N- or B-MLV-GFP and then assayed 3 days later for GFP expression by flow cytometry. (B) 17H1 and TE671 cells were infected with VSV-G-pseudotyped N-MLV-GFP in the presence or absence of 3 μM As2O3 and assayed for GFP expression as described for panel A. Results shown are representative of three independent experiments.
Arsenic trioxide (As2O3) increases the titer of N-MLV on TE671 cells while having no effect on B-MLV (1). We tested our 17H1 subclone for the effect of As2O3 on N-MLV titer (Fig. 2B). Cells were infected as described above. Where indicated, 3 μM As2O3 was present during the 18 to 24 h that cells were exposed to MLV vectors and then it was washed out. Treatment with As2O3 during infection increased the titer of N-MLV on TE671 cells by as much as 15-fold. In contrast, the effect of As2O3 on N-MLV titer in 17H1 cells was greatly attenuated, a result consistent with the hypothesis that As2O3 counteracts Ref1.
Cross-saturation experiments suggest that Ref1 may also restrict infection by human immunodeficiency virus type 1 (HIV-1) under certain conditions, specifically when the interaction between the HIV-1 CA protein and the cellular protein cyclophilin A (CypA) is disrupted in target cells (20). CA-CypA binding can be disrupted either by altering the CypA binding residues in CA (specifically, a G89V mutation in CA) or by treatment with the CypA binding drug cyclosporine A (CsA) (6, 10, 17, 22). Under these “CypA-free” conditions, the titer of HIV-1 in TE671 cells is reduced by approximately fivefold (20). Treatment of target cells with HIV-1(G89V) VLPs, but not wild-type VLPs, increases the titer of N-MLV vectors on TE671 cells and has no effect on B-MLV (20). In the reverse experiment, treatment with N-MLV VLPs increases the titer of HIV-1(G89V) vectors (20). These results have been interpreted as demonstrating that Ref1 restricts both N-MLV and CypA-free HIV-1 and that each of these viruses can saturate the restriction factor, thus facilitating infection by the other.
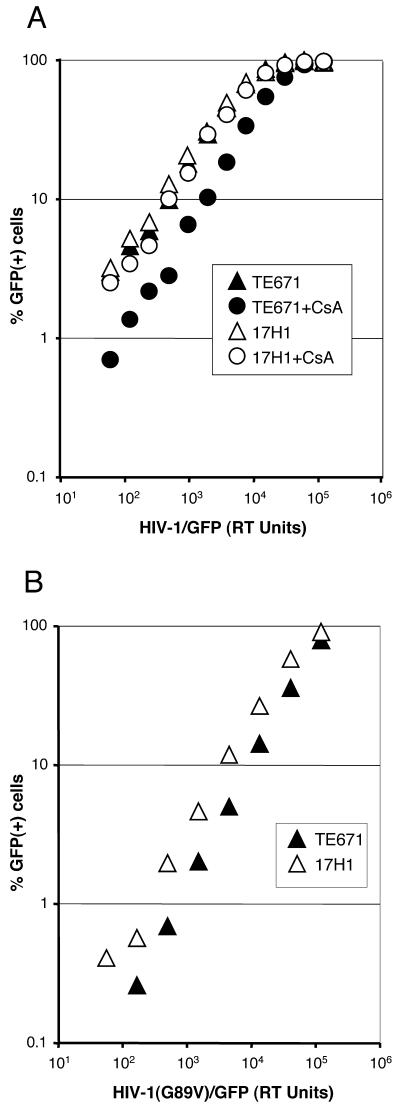
To test the hypothesis that Ref1 restricts HIV-1 in the absence of a CA-CypA interaction, the effect of CsA on HIV-1 titers in TE671 and 17H1 cells was examined. VSV-G-pseudotyped wild-type and G89V mutant HIV-1/Env-/GFP viruses (20) were produced in 293T cells by transient transfection with Lipofectamine 2000 and normalized for reverse transcriptase activity. Infections and flow cytometric analysis of TE671 and 17H1 cells were performed as described for MLV. Where indicated, 2.5 μM CsA was present during the 18 to 24 h that cells were exposed to virus and then it was washed out.
As shown in Fig. 3A, wild-type HIV-1 had equivalent titers on TE671 and 17H1 cells. CsA treatment reduced the titer of HIV-1 on TE671 cells by threefold. In contrast, CsA treatment of 17H1 cells had no effect on HIV-1 titer. We also looked at the titer of HIV-1(G89V), which bears a CA mutation that disrupts CypA binding. Though this mutant was less infectious than the wild type for reasons unrelated to Ref1 or CypA (compare Fig. 3A and B), the G89V virus gave a threefold-higher titer on 17H1 than on TE671 cells (Fig. 3B). CsA had no effect on the titer of this virus (data not shown). These results support the hypotheses that, in TE671 cells, HIV-1 is subject to restriction when the CA-CypA interaction is disrupted and that the 17H1 line has lost this restriction activity.
FIG. 3.
HIV-1 infection of 17H1 cells is CypA independent. (A) TE671 and 17H1 cells were infected with VSV-G-pseudotyped HIV-1/Env-/GFP in the presence or absence of CsA and then assayed 3 days later for GFP expression by flow cytometry. (B) TE671 and 17H1 cells were infected with HIV-1/Env-/GFP bearing a G89V mutation in CA and then assayed for GFP expression as described for panel A. Results shown are representative of three independent experiments.
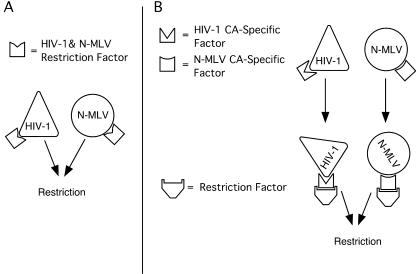
These data are consistent with previous observations that restriction of N-MLV and CypA-free HIV-1 in TE671 cells is related. Two models could account for this association (Fig. 4). In the first, N-MLV and CypA-free HIV-1 capsids are bound by the same saturable factor, leading to restriction of each through an unknown mechanism. In the second model, different factors recognize the two different viral capsids. These unique recognition factors then target the incoming capsids to restriction through a common, saturable pathway. In both models, the difference in the magnitude of restriction of the two viruses (roughly 100-fold for N-MLV versus 3-fold for HIV-1) could be explained by differing susceptibilities to the mechanism of restriction.
FIG. 4.
Proposed model for Ref1 restriction of N-MLV and CypA-free HIV-1. (A) A single, saturable factor is capable of binding to both N-MLV CA and, in the absence of bound CypA, HIV-1 CA, leading to restriction of infection. (B) N-MLV CA and CypA-free HIV-1 CA are recognized by unique CA binding factors which then target the viruses for restriction through a common, saturable pathway.
Rhesus macaque cells lack Ref1 activity (8), but they inhibit HIV-1 infection via the CA-specific restriction factor TRIM5α (16). Human TRIM5α is less effective than the macaque homologue at inhibiting HIV-1 (16) but, as previously hypothesized (15), it might be required for Ref1 activity. We used real-time reverse transcription-PCR to determine if decreased TRIM5α expression explained decreased Ref1 activity in 17H1 cells, but we found less than a twofold difference compared with the parental cell line (data not shown). Similarly, no significant difference in TRIM5γ expression was detected. Though TRIM5α may be required for Ref1 activity, these results suggest that some factor other than TRIM5α accounts for decreased Ref1 activity in 17H1. Further study of the 17H1 line as well as other Ref1-deficient lines derived from TE671 may shed light on what defect leads to the permissive phenotype of these cells, as well as how many different factors are required for effective restriction.
Acknowledgments
We thank P. Bieniasz and G. J. Towers for reagents.
This work was funded by National Institutes of Health grant RO1 AI36199 and used core facilities of the Columbia-Rockefeller Center for AIDS Research. D.M.S. was supported by the Columbia University College of Physicians and Surgeons Medical Scientist Training Program.
REFERENCES
- 1.Berthoux, L., G. J. Towers, C. Gurer, P. Salomoni, P. P. Pandolfi, and J. Luban. 2003. As2O3 enhances retroviral reverse transcription and counteracts Ref1 antiviral activity. J. Virol. 77:3167-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Besnier, C., L. Ylinen, B. Strange, A. Lister, Y. Takeuchi, S. P. Goff, and G. J. Towers. 2003. Characterization of murine leukemia virus restriction in mammals. J. Virol. 77:13403-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Best, S., P. Le Tissier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826-829. [DOI] [PubMed] [Google Scholar]
- 4.Bock, M., K. N. Bishop, G. Towers, and J. P. Stoye. 2000. Use of a transient assay for studying the genetic determinants of Fv1 restriction. J. Virol. 74:7422-7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boone, L. R., C. L. Innes, and C. K. Heitman. 1990. Abrogation of Fv-1 restriction by genome-deficient virions produced by a retrovirus packaging cell line. J. Virol. 64:3376-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franke, E. K., H. E. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 7.Hartley, J. W., W. P. Rowe, and R. J. Huebner. 1970. Host-range restrictions of murine leukemia viruses in mouse embryo cell cultures. J. Virol. 5:221-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. J. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozak, C. A., and A. Chakraborti. 1996. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology 225:300-305. [DOI] [PubMed] [Google Scholar]
- 10.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067-1078. [DOI] [PubMed] [Google Scholar]
- 11.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pryciak, P. M., and H. E. Varmus. 1992. Fv-1 restriction and its effects on murine leukemia virus integration in vivo and in vitro. J. Virol. 66:5959-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi, C. F., F. Bonhomme, A. Buckler-White, C. Buckler, A. Orth, M. R. Lander, S. K. Chattopadhyay, and H. C. Morse III. 1998. Molecular phylogeny of Fv1. Mamm. Genome 9:1049-1055. [DOI] [PubMed] [Google Scholar]
- 14.Steeves, R., and F. Lilly. 1977. Interactions between host and viral genomes in mouse leukemia. Annu. Rev. Genet. 11:277-296. [DOI] [PubMed] [Google Scholar]
- 15.Stoye, J. P. 2002. An intracellular block to primate lentivirus replication. Proc. Natl. Acad. Sci. USA 99:11549-11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 17.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Gottlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363-365. [DOI] [PubMed] [Google Scholar]
- 18.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA 97:12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Towers, G., M. Collins, and Y. Takeuchi. 2002. Abrogation of Ref1 retrovirus restriction in human cells. J. Virol. 76:2548-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138-1143. [DOI] [PubMed] [Google Scholar]
- 21.Van Parijs, L., Y. Refaeli, J. D. Lord, B. H. Nelson, A. K. Abbas, and D. Baltimore. 1999. Uncoupling IL-2 signals that regulate T cell proliferation, survival, and Fas-mediated activation-induced cell death. Immunity 11:281-288. [DOI] [PubMed] [Google Scholar]
- 22.Yoo, S., D. G. Myszka, C. Yeh, M. McMurray, C. P. Hill, and W. I. Sundquist. 1997. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J. Mol. Biol. 269:780-795. [DOI] [PubMed] [Google Scholar]