Abstract
Nuclear import of viral cDNA is a critical step for establishing the proviral state of human immunodeficiency virus type 1 (HIV-1). The contribution of HIV-1 integrase (IN) to the nuclear import of viral cDNA is controversial, partly due to a lack of identification of its bona fide nuclear localization signal. In this study, to address this putative function of HIV-1 IN, the effects of mutations at key residues for viral cDNA recognition (PYNP at positions 142 to 145, K156, K159, and K160) were evaluated in the context of viral replication. During acute infection, some mutations (N144Q, PYNP>KL, and KKK>AAA) severely reduced viral gene expression to less than 1% the wild-type (WT) level. None of the mutations affected the synthesis of viral cDNA. Meanwhile, the levels of integrated viral cDNA produced by N144Q, PYNP>KL, and KKK>AAA mutants were severely reduced to less than 1% the WT level. Quantitative PCR analysis of viral cDNA in nuclei and fluorescence in situ hybridization analysis showed that these mutations significantly reduced the level of viral cDNA accumulation in nuclei. Further analysis revealed that IN proteins carrying the N144Q, PYNP>KL, and KKK>AAA mutations showed severely reduced binding to viral cDNA but kept their karyophilic properties. Taken together, these results indicate that mutations that reduced the binding of IN to viral cDNA resulted in severe impairment of virus infectivity, most likely by affecting the nuclear import of viral cDNA that proceeds integration. These results suggest that HIV-1 IN may be one of the critical constituents for the efficient nuclear import of viral cDNA.
Human immunodeficiency virus type 1 (HIV-1) and other lentiviruses efficiently establish in nondividing as well as dividing cells a proviral state in which a double-stranded DNA copy of the viral genomic RNA (viral cDNA) is stably integrated into a chromosome of the host cell (5, 41, 64). This property distinguishes lentiviruses from certain other retroviruses that require mitosis with nuclear envelope breakdown prior to viral cDNA integration (42, 56).
The proviral state is established through several steps following binding to and entry into the target cell, including uncoating, reverse transcription, nuclear transport, and integration of the viral genome. These early events are mediated through the interactions of several viral proteins and host factors with the viral genome, often referred to as the reverse transcription complex and the preintegration complex (PIC) (reviewed in references 3, 13, and 29). The average diameter of HIV-1 PIC has been estimated to be ∼56 nm (50), precluding its passive nuclear import through intact nuclear pore complexes on the nuclear envelope of host cells (reviewed in reference 13).
To facilitate the efficient nuclear import of HIV-1 cDNA, HIV-1 PIC may contain karyophiles with nuclear localization signal (NLS) sequences. Among the HIV-1 PIC constituents, matrix (MA) protein, viral protein R (Vpr), and integrase (IN) have been reported to have karyophilic properties (4, 14, 24, 26, 27, 32, 37, 54). However, several contradictory results have also been reported, arguing against this putative role for NLS sequences within the MA protein (25, 55) or Vpr (2) in the nuclear transport of viral cDNA. In addition, the cis-acting viral cDNA structure, the central DNA flap, generated during lentivirus-specific reverse transcription, has also been reported to play an important role in the nuclear import of the HIV-1 genome (68). However, recent studies have shown that the effect of the central DNA flap appears to be virus strain and host cell dependent (17, 44). Thus, key factors that facilitate the nuclear import of viral cDNA in the context of the viral replication cycle still remain to be determined.
Among the HIV-1 PIC constituents described above, IN is a logical and highly probable candidate for facilitating the efficient nuclear import of HIV-1 cDNA, since it has karyophilic properties (2, 14, 26, 43, 54, 60) and mediates the integration of viral cDNA into the host chromosome in nuclei. Previously, atypical bipartite NLS sequences within HIV-1 IN were reported to be recognized by the importin/karyopherin pathway (26). However, several studies showed that these NLS sequences did not mediate nuclear import but were instead required for reverse transcription of the viral genome or other steps in the viral replication cycle (53, 60). Recently, the Val and Arg residues at positions 165 and 166 (V165/R166) in HIV-1 IN were shown to be critical for its NLS function in mediating the nuclear import of viral cDNA (2). However, reassessments of V165/R166 functions have failed to confirm this conclusion (17, 43). Thus, despite many experimental attempts, the contribution of HIV-1 IN to the nuclear import of viral cDNA remains controversial, partly due to a lack of identification of bona fide NLS sequences within IN. More recently, it was reported that the nuclear import of HIV-1 cDNA was mediated through the interaction of IN and importin 7, one of the host import factors for the nuclear import of ribosomal proteins and histone H1 (23). In addition, human lens epithelium-derived growth factor/transcription coactivator p75 (LEDGF/p75) was identified as an essential cellular factor for the chromosomal targeting of HIV-1 IN (47).
On the other hand, a detailed mechanism of retroviral integration was elucidated from in vitro studies with recombinant IN protein (reviewed in references 35 and 38). For the integration of viral cDNA, IN reacts with the attachment (att) site located at the U3 and U5 termini of the viral cDNA (12, 39, 48, 58, 61, 62). These studies of HIV-1 IN identified three functional domains: an N-terminal zinc binding domain (6-8), a central catalytic core domain (7, 11, 18, 57, 63), and a C-terminal nonspecific DNA binding domain (11, 19, 46). The central core domain contains the highly conserved D,D35E motif, which is directly involved in the catalytic activities of IN (7, 20, 30, 57). Tsurutani et al. previously reported that a single amino acid substitution of the Tyr residue at position 143 with Gly (Y143G) in HIV-1 IN significantly reduced the level of stably integrated proviral cDNA during acute infection of human primary cells (60). This reduction, concomitant with a reduction in the level of the two-long-terminal-repeat (2-LTR) circular form of viral cDNA, was evident in nondividing cells such as monocyte-derived macrophages (MDMs). Since circular forms of viral cDNA are produced in the nucleus, the Y143G mutation may affect the nuclear import of viral cDNA. The Y143 residue is located within the highly conserved core domain containing the D,D35E motif. The central core domain also contains several highly conserved residues critical for specific binding to viral cDNA (28, 33, 34, 40). These include Y143 (22, 45) and Lys residues at positions 156 (K156) (36, 67) and 159 (K159) (16, 36, 67). Thus, Y143 has been suggested to be one of the key residues for specific binding to viral cDNA.
In this study, we generated HIV-1 IN mutants carrying mutations at these key residues for viral cDNA recognition. Then, as an alternative approach to addressing the functional involvement of HIV-1 IN in the nuclear import of viral cDNA, the effects of these mutations were evaluated in the context of viral replication and the biochemical properties of the recombinant protein forms. Of note, mutations that reduced the binding of IN to viral cDNA resulted in the inefficient nuclear import of viral cDNA during acute infection. These results strongly suggest that HIV-1 IN may be a critical factor for the efficient transport of viral cDNA into nuclei.
MATERIALS AND METHODS
Construction of mutant DNA.
DNA fragments for the mutagenesis of HIV-1 IN were derived from the HIV-1 pNL43lucΔenv vector (49), in which the env gene is defective, allowing the formation of pseudotypes, and the nef gene is replaced with the firefly luciferase gene. For the mutagenesis of IN mutants [P142F, Y143G, N144Q, P145F, PYNP>KL, K156A, K159A, K160A, and KKK (156/159/160) >AAA], a 1.6-kb fragment of the pNL43lucΔenv vector spanning the KpnI and SalI sites (nucleotides [nt] 4154 to 5785) was subcloned into pBluescript SKII(+) (Stratagene, La Jolla, Calif.) (pSKnK/S). To introduce mutations, all mutagenic primers were designed to span the NsiI and AflII sites (nt 4377 to 4743). PCR products amplified with each mutagenic primer pair (Table 1) and the pNL43lucΔenv vector as a template were digested with NsiI and AflII. The mutant fragments were ligated to NsiI-AflII-digested pSKnK/S. To generate some mutations with the backbone of the D116G mutation (D116G-ΔPYNP and D116G-KKK>AAA), mutagenic PCR was performed with each mutagenic primer pair and the pNL43lucΔenv vector carrying the D116G mutation (49) as a template. After confirmation of each mutation by DNA sequence analysis, KpnI-SalI fragments (nt 4154 to 5785) containing the mutations were ligated to the corresponding region in the 4.2-kbp SpeI-SalI fragment (nt 1507 to 5785). SpeI-SalI fragments containing the mutations were inserted back into the pNL43lucΔenv vector. The amplified region and cloning junctions were confirmed by DNA sequencing.
TABLE 1.
Mutations in HIV-1 IN
| Mutant | Changes in:
|
|
|---|---|---|
| Nucleotidea | Amino acidb | |
| P142F | 4654-CCC → TTC | P142 → F |
| Y143G | 4657-TAC → GGG | Y143 → G |
| N144Q | 4660-AAT → CAA | N144 → Q |
| P145F | 4663-CCC → TTC | P145 → F |
| PYNP>KL | 4654-CCC. . .4666-GTA → AAGCTT | PYNP (positions 142-145) → KL |
| K156A | 4696-AAA → GCA | K156 → A |
| K159A | 4705-AAG → GCG | K159 → A |
| K160A | 4708-AAA → GCA | K160 → A |
| KKK>AAA | 4696-AAA → GCA/4705-AAG → GCG/4708-AAA → GCA | KKK (positions 156, 159, and 160) → AAA |
Altered nucleotides are indicated by italic type. The nucleotide positions are numbered according to the NL43 sequence.
Numbers are the amino acid residues of NL43 IN.
Cells.
COS-7, 293T, HeLa, and RD cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Human MDMs and peripheral blood lymphocytes (PBLs) were derived from HIV-1-seronegative healthy donors. Briefly, peripheral blood mononuclear cells were separated over a Ficoll-Hypaque gradient (Ficoll-Paque Plus; Amersham Pharmacia Biotech Inc., Tokyo, Japan) by centrifugation. Peripheral blood mononuclear cells were allowed to adhere to 150-mm plastic tissue culture dishes (Iwaki, Tokyo, Japan) by incubation in RPMI 1640 (Sigma Chemical Co., St. Louis, Mo.) containing 5% human AB serum (Sigma or Nippon Bio-Supply Center, Tokyo, Japan) for 2 h. Nonadherent cells (PBLs) were grown in RPMI 1640 medium containing 10% FBS and 2 ng of recombinant interleukin-2 (Shionogi, Osaka, Japan)/ml. Adherent cells were detached with cell dissociation solution (Sigma) and cultured in RPMI 1640 containing 5% human AB serum for 5 to 7 days. More than 98% of the cells that were shown to be positive for CD14, CCR5, HLA-DR, and mannose receptors were used as MDMs.
Virus preparation and infection.
Pseudotype viruses were generated by cotransfection of COS-7 cells or 293T cells with the pNL43lucΔenv vector containing each IN mutation and an amphotropic Moloney murine leukemia virus (MuLV) envelope expression vector, pJD-1 (a kind gift from Irvin S. Y. Chen, University of California at Los Angeles), or a macrophage-tropic HIV-1 envelope vector, pJR-FL (a kind gift from Yoshio Koyanagi, Tohoku University), by using Lipofectamine (Invitrogen, Carlsbad, Calif.) or a calcium phosphate precipitation method. The culture supernatants (4 ml) of the transfected COS-7 cells were harvested at 48 h posttransfection, filtered though 0.45-μm-pore-size filters, and used as the virus preparations. Each virus preparation was treated with DNase I (40 U/ml; Takara, Kyoto, Japan) in the presence of 10 mM MgCl2 at 37°C for 1 h. To monitor residual contamination of the plasmid DNA, an aliquot of each virus preparation was incubated at 65°C for 1 h and used as a heat-inactivated control. To monitor the amount of virus in each preparation, the levels of HIV-1 p24 antigen were determined with an enzyme immunoassay system (RETRO-TEK; ZeptoMetrix Corp., Buffalo, N.Y.). To monitor viral gene expression from each plasmid vector, the luciferase activity in transfected COS-7 cells was also measured. At 48 h posttransfection, COS-7 cells were lysed with 1 ml of cell lysis buffer (Promega, Madison, Wis.), and then 1 μl of each cell lysate was subjected to a luciferase assay with Lumat LB 9507 (EG & G Berthold, Bad Wildbad, Germany). For virus infection, an aliquot (corresponding to ∼20 ng of p24) of DNase I-treated virus was inoculated into RD cells (5 × 104), MDMs (5 × 105), or PBLs (1 × 106) in the presence of Polybrene (10 μg/ml). After incubation for 6 h at 37°C, the virus-containing medium was removed and replaced with fresh medium. At 4 days postinfection, the cells were harvested, washed twice with phosphate-buffered saline (PBS), and then lysed with 200 μl of cell lysis buffer. An aliquot (10 μl) of each lysate was subjected to the luciferase assay.
Western blot analysis.
Viruses were concentrated by ultracentrifugation (1 h at 315,000 × g in a Beckman TLX-100 centrifuge with a TLA-100.4 rotor), and the pellets were resuspended in PBS. Viral proteins containing approximately 10 ng of p24 were subjected to sodium dodecyl sulfate (SDS)-12% polyacrylamide gel electrophoresis (PAGE). Following blotting of proteins onto a nitrocellulose membrane (ATTO, Tokyo, Japan), the membrane was incubated with antiserum from AIDS patients, anti-HIV-1 IN antibody (kindly provided by Duane Grandgenett, St. Louis University Health Sciences Center, Institute for Molecular Virology), or anti-HIV-1 p24 antibody (Chemicon International, Temecula, Calif.) followed by horseradish peroxidase-conjugated anti-human, anti-rabbit, or anti-mouse immunoglobulin. HIV-1 proteins were visualized by using an enhanced chemiluminescence detection system (Amersham Pharmacia Biotech, Tokyo, Japan).
Analysis of de novo-synthesized HIV-1 cDNA during acute infection.
Total cells were harvested from each well at 1 or 2 days postinfection. After the cells were washed with PBS, nucleic acids were extracted as described previously (66). Briefly, the cells were disrupted in urea lysis buffer (4.7 M urea, 1.3% SDS, 0.23 M NaCl, 0.67 mM EDTA [pH 8.0]) and subjected to phenol-chloroform extraction and ethanol precipitation. The resulting DNA pellet was resuspended in 100 μl of distilled deionized water (ddw). An aliquot (5 μl) of each sample was subjected to PCR with a primer pair specific for the R/U5 region of HIV-1 (M667-AA55) or the R/gag region (M667-M661) (66). Detection of HIV-1 DNA sequences with each primer pair was performed by using PCR conditions of 30 cycles of 94°C for 1 min, 65°C for 2 min, and 72°C for 2 min. For DNA standards, 8 to 25,000 copies of linearized HIV-1 pNL43lucΔenv DNA were amplified in parallel. Quantitative analysis of the amplified products was performed by using a real-time Light Cycler detection system (Light Cycler Instrument; Roche, Mannheim, Germany). To normalize the amount of cellular DNA in the samples, a primer pair complementary to the first exon of the human β-globin gene was used. For the detection of human β-globin DNA, amplification was performed by using PCR conditions of 30 cycles of 94°C for 1 min, 55°C for 2 min, and 72°C for 2 min. A standard curve for human β-globin DNA was obtained by amplifying known amounts of cellular DNA from RD cells or MDMs in parallel. The 2-LTR circular DNA in nuclei was detected with specific primers as described previously (21). For detection of the integrated form of HIV-1 DNA, we used the Alu PCR method with an HIV-1-specific primer (M661) and an Alu-specific primer (5′-TCCCAGCTACTCGGGAGGCTGAGG-3′) (59). The cycling conditions were 94°C for 3 min, followed by 22 cycles of 94°C for 30 s, 66°C for 30 s, 70°C for 10 min, and then 72°C for 10 min. PCR products were purified and diluted 100- to 10,000-fold. The diluted samples were subjected to real-time PCR with the Light Cycler Instrument and an R/U5-specific primer pair (M667-AA55).
Confocal microscopic analysis of GFP-IN.
HeLa cells (4 × 103) were seeded onto glass slides (Cel-Line; Erie Scientific Co, Portsmouth, N.H.) and transfected with a plasmid expressing green fluorescent protein (GFP) fused to full-length IN (60). At 24 h posttransfection, the cells were washed once with PBS and then fixed with 4% paraformaldehyde (Wako, Osaka, Japan) for 10 min. Confocal microscopy was performed with an Olympus BX50 fluorescence microscope (Olympus, Tokyo, Japan). Representative medial sections were mounted by using Adobe Photoshop software.
In vitro GST pull-down assay.
DNA fragments encoding the entire HIV-1 IN were amplified by PCR with wild-type (WT) or each mutant plasmid DNA (pNL43lucΔenv) as a template. The primers used for the amplification of HIV-1 IN were as follows: GST-IN sense, 5′-GCGGATCCTTTTTAGATGGAATAGATAAGGCCC-3′, and GST-IN end antisense, 5′-CCGGAATTCAATCCTCATCCTG-3′; the sequences for BamHI or EcoRI recognition in each primer are underlined. Amplified products were digested with BamHI and EcoRI. Fragments with or without mutations were ligated to BamHI-EcoRI-digested vector pGEX-2T (Amersham Pharmacia Biotech). To express glutathione S-transferase (GST)-IN, Escherichia coli strain BL21(DE3) cells which had been transformed with pGEX-IN were grown in 16% Bacto Tryptone-10% yeast extract-5% NaCl-50 mg of carbenicillin/ml at 37°C to an optical density at 600 nm of 0.7. Expression was induced by the addition of 0.5 mM isopropyl-β-d-thiogalactoside (IPTG) for 17 h at 20°C. Cells were harvested by centrifugation and resuspended at a 1/10 volume of the original cell culture in high-salt buffer (1 M NaCl in PBS) containing 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT), and a proteinase inhibitor mixture (1 μg of pepstatin A/ml, 2 μg of aprotinin/ml, and 0.5 μg of leupeptin/ml). Cells were lysed by sonication (Sonifier; Branson, Danbury, Conn.) followed by centrifugation at 19,000 × g for 30 min. The supernatant fraction of the cell lysate was incubated with glutathione-Sepharose 4B beads (Amersham Pharmacia Biotech) at 4°C overnight. The recombinant protein-bound beads were washed extensively with a wash buffer (1 M NaCl-1% Tween 20 in PBS) and then eluted with elution buffer (50 mM Tris-HCl [pH 8.0], 10 mM reduced glutathione) containing the proteinase inhibitor mixture at 25°C for 2 h. The eluted fraction containing the recombinant GST-IN protein was dialyzed against IN storage buffer {20 mM HEPES [pH 7.5], 0.1 mM EDTA, 0.3 M NaCl, 20% glycerol, 10 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 10 mM DTT}. The purity of each recombinant GST-IN protein was confirmed by PAGE, and the protein concentrations were determined by the Bradford method.
The HIV-1 cDNA for the pull-down assay was prepared from crude PIC fractions (31). Briefly, MOLT-4/IIIB cells were cocultured with MOLT-4 cells for 6 h. Cells were harvested and permeabilized with 0.025% digitonin in buffer K (20 mM HEPES [pH 7.4], 150 mM KCl, 5 mM MgCl2) followed by sequential centrifugations at 1,000 × g for 10 min and 12,000 × g for 20 min. The supernatants were used as crude PIC fractions. Viral cDNA was extracted from the crude PIC fractions by the urea lysis method as described above. The amount of viral cDNA was determined by real-time PCR with R/U5- and R/gag-specific primers. Viral cDNA (∼105 copies) was incubated with each GST-IN protein (200 nM) immobilized on glutathione-Sepharose beads in binding buffer (20 mM HEPES [pH 7.3], 7.5 mM MnCl2, 1 mM DTT, 1 mM PMSF, 10% polyethylene glycol) for 15 min at 4°C. The beads were washed five times with a wash buffer (1.0% Triton X-100 in PBS) and eluted with elution buffer. Viral cDNA bound to each GST-IN protein was extracted by phenol-chloroform treatment followed by ethanol precipitation with a carrier (Ethachinmate; Wako/Nippon Gene Co., Tokyo, Japan). The resulting DNA pellet was suspended in ddw. The amount of viral cDNA in each preparation was determined by real-time PCR with the M667-AA55 (R/U5-specific) or the M667-M661 (R/gag-specific) primer pair. The background level due to nonspecific binding of the input viral cDNA to GST protein and glutathione-Sepharose beads was determined by carrying out the same experiment with GST only in parallel. After subtraction of the background level (∼5,000 copies per total input of ∼105 copies), the relative copy number (R/gag) for each mutant as a percentage of that for the WT was calculated.
Isolation of DNA from the nuclear fraction.
RD or 293T cells (5 × 106) were infected with each pseudotype virus and harvested at 24 h postinfection. For the isolation of nuclei, cells were resuspended in buffer A (10 mM HEPES [pH 7.3], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.2% NP-40), homogenized by 20 strokes with an all-glass Dounce homogenizer (Kontes, Vineland, N.J.), and centrifuged at 500 × g for 5 min at 4°C. The pellet of the cell lysate was washed twice with Nuclei EZ Lysis Buffer and then with Nuclei Storage Buffer (Sigma). Nuclei were collected by centrifugation at 25,000 × g for 20 min at 4°C. DNA samples from each nuclear fraction were subjected to real-time PCR with the M667-AA55 (R/U5-specific) or the M667-M661 (R/gag-specific) primer pair as described above.
FISH analysis with HIV-1-specific PNA probes.
For fluorescence in situ hybridization (FISH) analysis with HIV-1-specific peptide nucleic acid (PNA) probes (51), glass slides (Cel-Line) were coated with 3-aminopropyl triethoxysilane (silane; Aldrich). HeLa cells (4 × 103) seeded on the silane-coated slides were cultured overnight. Pseudotype viruses were prepared by cotransfection of 293T cells with the pNL43lucΔenv vector together with the MuLV envelope expression vector (pJD-1). After treatment with DNase I (40 μg/ml; Worthington), each virus (70 ng of p24) was inoculated into HeLa cells and cultured at 37°C for 6 h. The cells were washed with PBS and resuspended in fresh medium (DMEM plus 10% FBS). After 18 h, the cells were harvested and fixed with 4% paraformaldehyde for 30 min, and the slides were treated with proteinase K (2 ng/ml) for 8 min at room temperature and then with RNase A (10 ng/ml) for 10 min at room temperature. Endogenous biotin reactivity was blocked by the addition of 0.3% H2O2 in methanol. The slides were dehydrated in absolute ethanol.
Fluorescein isothiocyanate (FITC)-conjugated PNA probes were synthesized by Fasmac Co. (Kanagawa, Japan). The structure of the antisense probe was as follows: FITC-5′-GCACATTGTACTGATA-3′; this sequence corresponded to nt 2878 to 2994 of the pol region of pNL43 (1). The hybridization solution (Dako, Carpinteria, Calif.) containing the FITC-conjugated PNA probes was mounted on the slides, boiled for 5 min at 93°C, and then incubated for 17 h at 57°C. The slides were washed twice with a stringent wash solution (Dako) at 45°C for 20 min. After being washed with Tris-buffered saline containing 0.1% Tween 20 (TBST), the slides were reacted with 63 pg of a horseradish peroxidase (HRP)-conjugated anti-FITC antibody (Vector, Burlingame, Calif.)/ml for 30 min at 37°C. The slides were washed with TBST and reacted with biotinyl-tyramide (Dako) for 15 min at room temperature. After being washed with TBST, the slides were reacted with HRP-conjugated streptavidin (Dako). The slides were washed with TBST and reacted with biotinyl-tyramide for 15 min. Finally, the slides were reacted with 250 ng of Alexa 488-conjugated streptavidin/ml to detect the hybridized probes. Nuclei were stained with 125 ng of 4′,6′-diamidino-2-phenylindole (DAPI; Wako)/ml. The signals were observed with a confocal microscope (Olympus BX50). We counted the signal dots inside the nuclei of 20 to 50 cells from three independent hybridizations. Representative medial sections were mounted by using Adobe Photoshop software.
RESULTS
Construction of HIV-1 IN mutants.
We introduced single amino acid substitutions or deletions at highly conserved Pro-Tyr-Asn-Pro residues (PYNP) spanning positions 142 to 145 or the three Lys residues at positions 156, 159, and 160 (K156, K159, and K160) in HIV-1 IN (Table 1), which are regions that contain several key residues critical for viral cDNA recognition (16, 22, 36, 45, 67). The mutations (Table 1) included a Pro-to-Phe substitution at position 142 (P142F), a Tyr-to-Gly substitution at position 143 (Y143G), an Asn-to-Gln substitution at position 144 (N144Q), a Pro-to-Phe substitution at position 145 (P145F), deletion of the PYNP motif at positions 142 to 145 by replacement with Lys-Leu (PYNP>KL), and Ala substitution(s) at each or all of the Lys residues at positions 156, 159, and 160 (K156A, K159A, K160A, and KKK>AAA). Due to the lack of identification of bona fide NLS sequences within IN, the effects of these mutations, which were expected to be defective at binding to viral cDNA, were evaluated in the context of viral replication, as an alternative approach to addressing the functional involvement of HIV-1 IN in the nuclear import of viral cDNA.
Infectivity of each HIV-1 IN mutant.
A single-round infection system that uses an HIV-1 (pNL43lucΔBg) pseudotype virus is useful for estimating reverse transcription and integration efficiency in vivo by monitoring the levels of de novo-synthesized viral cDNA and luciferase activity produced in infected cells (48, 49, 60). Therefore, we examined the effect of each mutation on early events in viral replication by using the single-round infection system. Briefly, pseudotype viruses were generated by cotransfection of COS-7 cells with pNL43lucΔenv containing the WT or each IN mutation together with an expression vector for the amphotropic Moloney MuLV envelope (pJD-1) or the macrophage-tropic HIV-1 envelope (pJR-FL). At 48 h posttransfection, all mutants had comparable levels of p24 in culture supernatants harvested from transfected COS-7 cells (Fig. 1A) and comparable levels of luciferase activity in lysates of transfected COS-7 cells (data not shown). Thus, none of the mutations had a significant effect on transfected proviral gene expression or virus release.
FIG. 1.
Gene expression of each mutant proviral DNA after transfection of COS-7 cells and viral protein profiles. Pseudotype viruses were generated by cotransfection of COS-7 cells with the pNL43lucΔenv vector containing mutations in IN and an amphotropic Molony MuLV envelope expression vector (pJD-1) by using Lipofectamine. Culture supernatants (5 ml) of the transfected COS-7 cells were harvested at 48 h posttransfection. (A) p24 levels in culture supernatants were determined with the RETRO-TEK enzyme immunoassay system. (B and C) Virus particles in culture supernatants (5 ml) of COS-7 cells were precipitated at 48 h posttransfection by ultracentrifugation (1 h at 315,000 × g in a Beckman TLX-100 centrifuge). Viral proteins were separated by SDS-12% PAGE. After blotting of proteins onto a nitrocellulose membrane, the membrane was reacted with serum from an AIDS patient (B) or anti-HIV-1 IN or anti-p24 antibody (C) and then incubated with horseradish peroxidase-conjugated anti-human, anti-rabbit, or anti-mouse immunoglobulin. Viral proteins were visualized by using the enhanced chemiluminescence detection system. The positions of the major viral proteins are indicated.
In order to verify the Gag-Pol polyprotein processing in mutant virus particles, we performed Western blotting analyses of viral proteins contained in the virus particles with serum from an AIDS patient or antibodies to HIV-1 IN or p24. No apparent differences between the parental (WT) and mutant viruses were observed in the profiles (Fig. 1B and C). These results showed that none of the mutations significantly affected the late stage of viral replication from proviral gene expression to virus particle release, Gag-Pol polyprotein processing, and IN incorporation into the virus particles.
For virus infection, a DNase I-treated preparation of each pseudotype virus was inoculated into RD cells, MDMs, or PBLs. To examine the level of viral gene expression for each HIV-1 IN mutant, the luciferase activity expressed in infected cells was measured at 4 days postinfection. In this experiment, an IN mutant carrying a single amino acid substitution at one of the catalytic sites (D116G) was used as a control for integration-defective mutants (49). We repeated this experiment more than 10 times with independently prepared viruses (Fig. 2). In RD cells, the PYNP>KL mutation or the KKK>AAA mutation severely reduced the luciferase activity to less than 1% the WT level, which was similar to the level seen with the catalytic site mutation (D116G). The severe effects induced by these mutations were also observed after acute infection of human primary PBLs and MDMs. A significant reduction in the level of luciferase gene expression was also detected with some mutants with single amino acid substitutions in the PYNP motif (residues 142 to 145) or at K156, K159, and K160. The levels of luciferase activity produced by N144Q and P145F were significantly lower in RD cells, PBLs, and MDMs and ranged from 3 to 15% the WT level. As reported in a previous study (60), the effect of the Y143G mutation in the PYNP motif was more evident in primary cells (PBLs and MDMs) than in RD cells. Like Y143G, P142F (a single amino acid substitution in the same PYNP motif) produced lower levels of luciferase activity (24 to 65% the WT level) in PBLs and MDMs but produced a high level (90 to 100% the WT level) in RD cells. Thus, the integrity of the PYNP motif is more rigidly required for virus infection in primary cells (PBLs and MDMs) than in an in vitro-adapted cell line (RD cells). In addition, K159A produced a modest but consistent reduction in luciferase activity (42 to 60% the WT level), while other point mutations in the K residues (K156A and K160A) produced levels of luciferase activity comparable to that produced by the WT in all cell types. Since the KKK>AAA mutation produced a severe reduction in luciferase activity (<1% the WT level), the three Lys residues may function in a compensatory manner. Furthermore, these severe defects were efficiently complemented when WT IN was supplied in trans (data not shown), thus excluding possible effects of the mutations on cis-acting functions of the central polypurine tract (10, 68). Taken together, these results suggested important roles of PYNP at residues 142 to 145 and of K at residues 156, 159, and 160 in trans-acting functions of IN in the early events of the HIV-1 replication cycle.
FIG. 2.
Effects of HIV-1 IN mutations on viral infectivity. Viruses were prepared by cotransfection of COS-7 cells with the pNL43lucΔenv vector containing either WT IN or mutant IN together with an amphotropic Moloney MuLV envelope expression vector (pJD-1) or a macrophage-tropic HIV-1 envelope vector (pJR-FL) by using Lipofectamine. At 48 h posttransfection, culture supernatants of the transfected COS-7 cells were harvested. DNase I-treated supernatants were inoculated into 105 RD cells, PBLs, and MDMs. At 4 days postinfection, the cells were washed with PBS and lysed with 200 μl of cell lysis buffer. Ten microliters of each cell lysate was subjected to the luciferase assay. Mean values from five independent experiments are shown with the error bars.
Quantitative analysis of viral cDNA after infection with HIV-1 IN mutants.
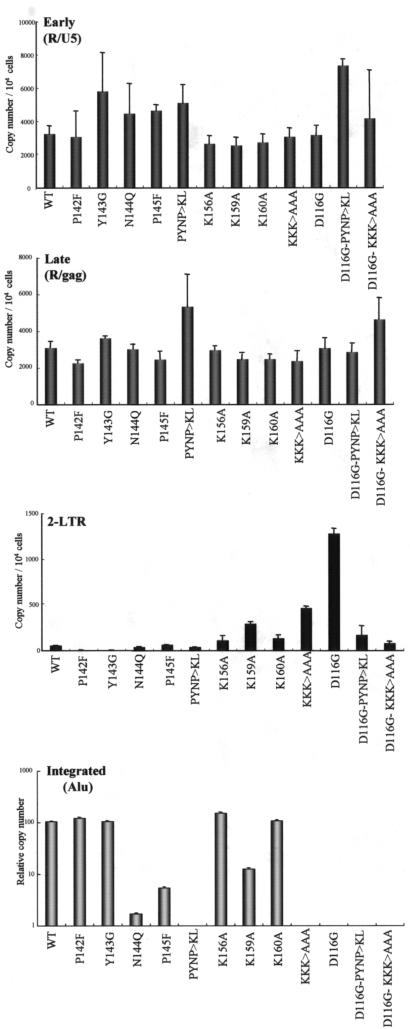
Next, we assessed the ability of each mutant to synthesize viral cDNA and to form integrated proviral cDNA following infection of RD cells (Fig. 3). At 24 h after infection with DNase I-treated virus, total DNA was harvested from infected RD cells, and an aliquot of each DNA sample was subjected to real-time quantitative PCR analysis. First, we monitored the formation of various species of viral cDNAs by using the M667-AA55 (R/U5-specific) primer pair for early viral cDNA (early) and the M667-M661 (R/gag-specific) primer pair for complete or nearly complete viral cDNA (late). Relative to the WT, all PYNP motif mutants (P142F, Y143G, N144Q, P145F, and PYNP>KL) as well as K mutants K156A, K159A, K160A, and KKK>AAA produced comparable levels of early (R/U5) and complete or nearly complete (R/gag) forms of viral cDNA products. These results showed that none of these mutations affected reverse transcription.
FIG. 3.
Analysis of viral cDNA synthesis and proviral DNA formation. Each virus was prepared by cotransfection of COS-7 cells with pNL43lucΔenv (WT or mutant IN) together with pJD-1. DNase I-treated supernatants were inoculated into RD cells as described in the legend to Fig. 2. Viruses treated at 65°C for 1 h prior to inoculationwere used as a heat-inactivated control. At 1 day postinfection, the entire cell culture was harvested, and total DNAs were extracted from infected RD cells. Each DNA sample was subjected to real-time quantitative PCR analysis with primer pairs specific for early (R/U5), late (R/gag), 2-LTR circular (2-LTR), or integrated (Alu) forms of viral cDNAs. The estimated copy number of each viral cDNA (R/U5, R/gag, 2-LTR, and Alu) per 105 cell equivalents is shown. For estimation of integrated viral cDNA (Alu), each DNA sample was subjected to PCR with the Alu- and R/gag-specific primer pair. Serial dilutions of each PCR product (Alu and R/gag) were subjected to real-time quantitative PCR with the R/U5-specific primer pair. Values were calculated as the copy number (R/U5) of each mutant relative to that of the WT, taken as 100%. Mean values from five independent experiments are shown with the error bars.
Since all of the mutations in the PYNP and K residues produced comparable levels of complete or nearly complete viral cDNA products (R/gag), we next examined the fate of each de novo-synthesized viral cDNA in RD cells. To this end, we monitored the formation of 2-LTR circular DNA (Fig. 3, 2-LTR) by using a primer pair which amplified a sequence unique to the 2-LTR DNA junction (21). We also monitored the level of the integrated form of each viral cDNA by the Alu PCR method (59) with an HIV-1-specific primer (M661) and an Alu-specific primer (Fig. 3, Integrated). Products of 2-LTR DNA have been used as markers for its presence in the nucleus, since the enzymes responsible for its formation are thought to be located within the nucleus. In addition, 2-LTR DNA is formed in the absence of the catalytic function of IN (60). PCR amplification of this structure therefore confirms the specific inhibition of integration. An HIV-1 IN catalytic site mutant (D116G) (49) was used as a control for integration-defective mutants. As in a previous study (60), we clearly detected an amplified fragment corresponding to the 2-LTR circular junction in DNA samples from RD cells infected with integration-defective mutant D116G. Meanwhile, the P142F, Y143G, K156A, and K160A mutants as well as the WT produced much lower levels of 2-LTR DNA (2 to 9% the level produced by D116G), suggesting successful integration of viral cDNA. These conclusions were consistent with the level of expression of each viral gene (luciferase activity) shown in Fig. 2 and were further confirmed by Alu PCR analysis (Fig. 3, Integrated). On the other hand, the N144Q, P145F, PYNP>KL, K159A, and KKK>AAA mutants produced 2-LTR DNA at various levels—3, 5, 3, 23, and 36% the level produced by D116G, respectively (Fig. 3, 2-LTR). Of note, the levels of 2-LTR produced by these mutants were not always correlated with the reduction in luciferase activity (Fig. 2) or the integration level (Fig. 3, Integrated). In particular, the PYNP>KL mutant showed significant reductions in the level of luciferase activity (<2% the WT level) and integration (below the detection level). These phenotypes of PYNP>KL were almost identical to those of D116G. However, the level of 2-LTR DNA produced by PYNP>KL was only ∼3% the level produced by D116G. Similar discrepancies were also noted for the N144Q and KKK>AAA mutants. In addition, a similar reduction in the level of 2-LTR DNA caused by the PYNP>KL and KKK>AAA mutations was reproduced on the backbone of the D116G mutation (Fig. 3, D116G-PYNP>KL and D116G-KKK>AAA). Taken together, these data suggested that the reduced levels of integration and subsequent gene expression seen with PYNP>KL, N144Q, and KKK>AAA might have been partly due to a reduction in the nuclear import of viral cDNA that precedes integration.
Interaction of IN protein with viral cDNA.
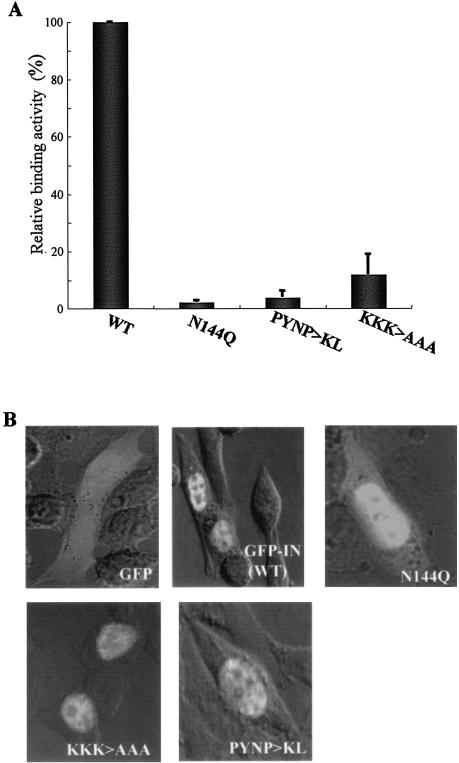
Some of the IN mutants (PYNP>KL, N144Q, and KKK>AAA) showed a severe integration-defective phenotype, partly due to a failure to transport the viral cDNA into the nucleus. These mutants were generated by introducing mutations at critical residues in IN for viral cDNA recognition (16, 22, 36, 45, 67). Therefore, we next examined the binding of these IN proteins to viral cDNA. Viral cDNA (∼105 copies) extracted from cells acutely infected with HIV-1 was incubated with 200 nM each GST-IN protein (WT, N144Q, PYNP>KL, or KKK>AAA) immobilized on glutathione-Sepharose beads. After extensive washing, the viral cDNA bound to each GST-IN protein was extracted. The amount of viral cDNA in each preparation was determined by real-time PCR with a primer pair specific for late forms (R/gag) of viral cDNA products (Fig. 4A). The levels of viral cDNA pulled down by GST-IN protein containing N144Q, PYNP>KL, and KKK>AAA were 2.1, 3.8, and 11.7% the WT level, respectively (Fig. 4A). Whether these in vitro data can be extrapolated to biological relevance in vivo still remains to be determined, but the PYNP>KL, N144Q, and KKK>AAA mutations reduced the binding of IN to viral cDNA at least in our in vitro assay.
FIG. 4.
Properties of HIV-1 IN mutant proteins. (A) Interactions of IN proteins with HIV-1 cDNA. Viral cDNA (∼105 copies) extracted from a crude PIC fraction (see Materials and Methods) was incubated with 200 nM each GST-IN protein (WT, N144Q, PYNP>KL, and KKK>AAA) immobilized on glutathione-Sepharose beads in binding buffer. The beads were washed five times with a wash buffer (1.0% Triton X-100 in PBS) and eluted with elution buffer. Viral cDNA bound to each GST-IN protein was extracted by phenol-chloroform treatment followed by ethanol precipitation. The resulting DNA pellet was resuspended in ddw. The amount of viral cDNA in each preparation was measured by a real-time PCR system with a primer pair specific for the late product (R/gag) of viral cDNA. The values shown are the copy number (R/gag) of each mutant relative to that of the WT, taken as 100%. Mean values from three independent experiments are shown with the error bars. (B) Confocal microscopic analysis of GFP-IN. HeLa cells were transfected with a plasmid expressing GFP only, GFP fused to full-length IN (WT), or IN carrying the PYNP>KL, N144Q, or KKK>AAA mutation by using Lipofectamine. At 24 h posttransfection, the cells were fixed and examined with a confocal fluorescence microscope.
Nuclear localization of IN fused to GFP.
Tsurutani et al. previously reported that GFP-IN fusions efficiently localized to the nuclei of transiently transfected cells (60). We next examined the effects of the N144Q, PYNP>KL, K159A, and KKK>AAA mutations on the karyophilic properties of IN. Briefly, HeLa cells were transfected with GFP-IN expression vectors and subjected to analysis by confocal microscopy at 24 h posttransfection. In agreement with the previous report (60), GFP-IN accumulated predominantly in the nucleus [Fig. 4B, GFP-IN (WT)], while the GFP control protein was uniformly scattered throughout both the cytoplasm and the nucleus (Fig. 4B, GFP). Under the same conditions, IN mutants (N144Q, PYNP>KL, and KKK>AAA) were efficiently localized to the nucleus (Fig. 4B), suggesting that none of these mutations affected the karyophilic properties of IN.
Localization of HIV-1 cDNA during acute infection.
The experimental data described above indicated that the loss of binding to viral cDNA seen with the IN mutations resulted in the decreased accumulation of viral cDNA in the nuclei. To confirm these observations more directly, we first determined the levels of viral cDNA in nuclear fractions isolated from acutely infected cells. 293T cells were infected with a DNase I-treated preparation of each pseudotype virus. At 24 h postinfection, nuclei were isolated from the cells. The levels of viral cDNA in the nuclei were determined by real-time PCR with a primer pair specific for R/U5 to monitor the total viral cDNA in the nuclei. The levels of viral cDNA in the nuclei after infection with the N144Q, PYNP>KL, KKK>AAA, and D116G IN mutants were 50.3, 38.9, 43.6, and 90.3% the WT level, respectively (Fig. 5A). These data indicated that the nuclear import of viral cDNA was significantly decreased by the N144Q, PYNP>KL, and KKK>AAA mutations (P < 0.005) but not by the D116G mutation (P = 0.14). Similar significant reductions in the levels of viral cDNA in the nuclei seen with the N144Q, PYNP>KL, and KKK>AAA mutations were reproduced when a primer pair specific for R/gag was used (data not shown).
FIG. 5.
Localization of HIV-1 cDNA after infection with each mutant. (A) Quantification of HIV-1 cDNA in the nuclear fraction. 293T cells were infected with each virus (WT, N144Q, PYNP>KL, KKK>AAA, or D116G). At 24 h postinfection, nuclei were isolated from the infected cells as described in Materials and Methods. A DNA sample from each nuclear fraction was subjected to real-time quantitative PCR with the M667-AA55 (R/U5-specific) or the M661-M667 (R/gag-specific) primer pair. The values shown are the copy number (R/U5) of each mutant per 104 nuclei relative to that of the WT, taken as 100%. Means and standard deviations from three independent experiments are shown (n = 5). (B) HeLa cells (4 × 103) were seeded on silane-coated glass slides. Cells were infected with each IN mutant (PYNP>KL, KKK>AAA, or D116G) or with the WT (∼70 ng of p24). At 6 h postinfection, the cells were fixed with 4% paraformaldehyde and subjected to FISH analysis with an FITC-labeled HIV-1-specific PNA probe corresponding to nt 2878 to 2994 of the pol region of pNL43 (1). The signals were observed with a confocal microscope. Representative medial sections were mounted by using Adobe Photoshop software. The representative cells in each slide are shown with lower (left) and higher (right) magnification scales.
Finally, we analyzed the subcellular localization of de novo-synthesized viral cDNA during acute infection by FISH. Briefly, HeLa cells were infected with a DNase I-treated pseudotype virus of each IN mutant, incubated for 6 or 24 h, fixed, and subjected to FISH with an FITC-tagged HIV-1-specific PNA probe. Hybridized probe signals were detected by using a CAS system (Dako) with Alexa 488 and were visualized with an Olympus confocal laser scanning microscope (Fig. 5B). The majority (∼78%) of the signals produced by the WT or the D116G mutant were in the nuclei at 6 h postinfection. In contrast, under the same conditions, markedly fewer signals were detected in the nuclei after infection with the PYNP>KL and KKK>AAA mutants. These signals in the cytoplasm tended to be weakened at 24 h postinfection (data not shown), suggesting that PYNP>KL and KKK>AAA might reduce the nuclear accumulation of viral cDNA partly because these mutant PICs were more susceptible to degradation. Although we cannot rule out the partial contribution of the effect of these mutations on the stability of viral cDNA and/or the PIC, these results indicated that the nuclear transport that precedes the integration of viral cDNA was significantly reduced by mutations that reduced the binding affinity of HIV-1 IN for viral cDNA.
DISCUSSION
In this study, we generated HIV-1 IN mutants carrying mutations at key residues for viral cDNA recognition (16, 22, 36, 45, 67) and evaluated the effects of these mutations in the context of viral replication and the biochemical properties of the protein forms. Our major finding was that mutations reducing the interaction of IN with viral cDNA caused a severe impairment of integration, most likely by affecting the efficient nuclear import of viral cDNA.
Despite many experimental data indicating the karyophilic properties of HIV-1 IN (2, 14, 26, 43, 54, 60), the contribution of IN to the nuclear import of viral cDNA remains controversial. This situation may be partly due to a lack of identification of bona fide NLS sequences within IN. We also generated IN mutants by amino acid substitution of the V165/R166 residues that have been reported to be critical residues for an NLS within HIV-1 IN (2). However, our mutant (V165R/R166L) showed a complete lack of viral infectivity due to a failure to complete viral cDNA synthesis. Moreover, this IN mutant protein was unstable when expressed in the form of a GFP-IN fusion (data not shown). Thus, we cannot evaluate the importance of the V165/R166 residues in the karyophilic properties of IN or in the nuclear transport of viral cDNA in the context of viral replication. Recently, reassessments of these V165/R166 functions by use of IN mutants with Ala substitutions at the V165/R166 residues were reported by two different laboratories (17, 43). Both of these reports failed to confirm the NLS functions. Instead, V165A/R166A mutants were shown to be pleiotropic IN mutants primarily defective in integration. These conclusions are partly consistent with our data.
More recently, Devroe et al. reported that HIV-1 IN is unable to access the nucleus when expressed in the form of a large fusion with GFP and pyruvate kinase, suggesting that IN may lack a transferable NLS (15). Alternatively, they showed that cytoplasmic IN is highly unstable and prone to degradation and that the karyophilic properties can be attributed to an interaction with a cellular component. Interestingly, one of the nuclear factors, LEDGF/p75, has been identified as being essential for the karyophilic properties of HIV-1 IN, although the functional role of cytoplasmic LEDGF/p75 in the nuclear import of IN remains to be determined (47).
Meanwhile, genetic analyses of HIV-1 IN (9, 21, 49, 52, 60, 65) have suggested putative roles for IN at steps prior to integration, such as uncoating, reverse transcription, and nuclear import of viral cDNA. It was previously reported that a single amino acid substitution of the Tyr residue at position 143 with Gly (Y143G) in HIV-1 IN significantly reduced the level of stably integrated proviral DNA during acute infection of human primary cells. This reduction, concomitant with a reduction in the level of the 2-LTR circular form of viral cDNA, was evident in primary PBLs and MDMs (60); these data suggest that Y143 is a critical residue for efficient proviral DNA formation at steps prior to integration, most likely the nuclear import of viral cDNA. Y143 is located in the central catalytic domain of IN and is suggested to be one of the critical residues for specific binding to viral cDNA (22, 45). Thus, we reasoned that we could address the functional involvement of HIV-1 IN in the nuclear import of viral cDNA by using IN mutants that show reduced binding to viral cDNA.
Structural and biochemical analyses of HIV-1 IN identified three functional domains (reviewed in references 35 and 38): an N-terminal zinc binding domain, a central catalytic core domain, and a C-terminal nonspecific DNA binding domain. The central core domain contains the highly conserved D,D35E motif, which is directly involved in the catalytic activities of IN. The central core domain also contains several highly conserved residues critical for specific binding to viral cDNA. These include Y143, K156, K159, and K160 (16, 22, 36, 45, 67). In this study, we generated IN mutants by introducing single or triple amino acid substitutions or deletions of the highly conserved Pro-Tyr-Asn-Pro residues (PYNP) spanning positions 142 to 145 or three Lys residues (K156, K159, and K160) of HIV-1 IN (Table 1). We found that some of the IN mutant proteins carrying single amino acid substitutions in or deletions of the PYNP motif at positions 142 to 145 (N144Q and PYNP>KL) or a triple amino acid substitution for the three conserved Lys residues at positions 156, 159, and 160 (KKK>AAA) showed a severe reduction in binding to viral cDNA but retained their efficient karyophilic properties (Fig. 4B). However, whether these in vitro data can be extrapolated to biological relevance in vivo still remains to be determined. In the context of viral replication, HIV-1 carrying these IN mutations showed viral gene expression severely reduced to less than 1% the WT level, similar to the effects of a catalytic site mutation of IN (D116G) (Fig. 2). These defects were efficiently complemented by supplying WT IN in trans (data not shown), excluding the possible effects of these mutations on cis-acting functions of the central polypurine tract (10, 68).
Quantitative analysis of viral cDNA after infection with these HIV-1 IN mutants indicated that these mutations did not affect the de novo synthesis of viral cDNA products (Fig. 3, R/U5 and R/gag) but did severely reduce integration (Fig. 3, Integrated). Of note, the level of 2-LTR DNA, a surrogate marker for nuclear import, produced by the PYNP>KL mutant was only ∼3% that produced by the catalytic site mutant D116G. A similar reduction in the level of 2-LTR DNA was also noted for the N144Q and KKK>AAA mutants, even on the backbone of the D116G mutation (Fig. 3, 2-LTR), suggesting that inhibition of the binding of IN to viral cDNA may affect the nuclear import of viral cDNA that precedes integration. This conclusion was further confirmed by more direct methods quantifying viral cDNA in isolated nuclei and FISH analyses of acutely infected cells (Fig. 5). However, although the signals of viral cDNA for the KKK>AAA and D116G-PYNP>KL mutants were evident at 6 h postinfection (Fig. 5B), they tended to be weakened at 24 h postinfection (data not shown), suggesting that PYNP>KL and KKK>AAA may reduce the nuclear accumulation of viral cDNA partly because these mutant PICs are more susceptible to degradation. Thus, an alternate explanation for the results is that if IN does not bind to viral cDNA, then the PIC falls apart and is nonfunctional. Although we cannot rule out the partial contribution of the effect of these mutations on the stability of viral cDNA and/or the PIC, our results indicate that the nuclear transport that precedes the integration of viral cDNA was significantly reduced by mutations that reduced the binding affinity of HIV-1 IN for viral cDNA.
In summary, our findings indicate that IN mutants that were defective in viral cDNA recognition lost viral infectivity, thereby partly affecting the nuclear import of viral cDNA that precedes integration during acute infection with HIV-1. Taken together, these results suggest that HIV-1 IN may be one of the critical constituents for the efficient nuclear import of viral cDNA.
Acknowledgments
We thank I. S. Y. Chen and Y. Koyanagi for providing plasmids, D. P. Grandgenett for providing anti-IN antibodies, and R. Yoshinari and S. Nishino for technical assistance.
This work was supported by a grant-in-aid for scientific research on priority areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and a grant from the Ministry of Health, Labor and Welfare, Japan (Research on prevention of HIV replication and mutation 16150301).
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouyac-Bertoia, M., J. D. Dvorin, R. A. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 3.Bukrinsky, M. I., and O. K. Haffar. 1997. HIV-1 nuclear import: in search of a leader. Front. Biosci. 2:d578-d587. [DOI] [PubMed] [Google Scholar]
- 4.Bukrinsky, M. I., S. Haggerty, M. P. Dempsey, N. Sharova, A. Adzhubel, L. Spitz, P. Lewis, D. Goldfarb, M. Emerman, and M. Stevenson. 1993. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 90:6125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burke, C. J., G. Sanyal, M. W. Bruner, J. A. Ryan, R. L. LaFemina, H. L. Robbins, A. S. Zeft, C. R. Middaugh, and M. G. Cordingley. 1992. Structural implications of spectroscopic characterization of a putative zinc finger peptide from HIV-1 integrase. J. Biol. Chem. 267:9639-9644. [PubMed] [Google Scholar]
- 7.Bushman, F. D., A. Engelman, I. Palmer, P. Wingfield, and R. Craigie. 1993. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc. Natl. Acad. Sci. USA 90:3428-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, M., R. Zheng, M. Caffrey, R. Craigie, G. M. Clore, and A. M. Gronenborn. 1997. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat. Struct. Biol. 4:567-577. [DOI] [PubMed] [Google Scholar]
- 9.Cannon, P. M., W. Wilson, E. Byles, S. M. Kingsman, and A. J. Kingsman. 1994. Human immunodeficiency virus type 1 integrase: effect on viral replication of mutations at highly conserved residues. J. Virol. 68:4768-4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charneau, P., and F. Clavel. 1991. A single-stranded gap in human immunodeficiency virus unintegrated linear DNA defined by a central copy of the polypurine tract. J. Virol. 65:2415-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, J. C., J. Krucinski, L. J. Miercke, J. S. Finer-Moore, A. H. Tang, A. D. Leavitt, and R. M. Stroud. 2000. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl. Acad. Sci. USA 97:8233-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chow, S. A., K. A. Vincent, V. Ellison, and P. O. Brown. 1992. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science 255:723-726. [DOI] [PubMed] [Google Scholar]
- 13.Cullen, B. R. 2001. Journey to the center of the cell. Cell 105:697-700. [DOI] [PubMed] [Google Scholar]
- 14.Depienne, C., A. Mousnier, H. Leh, E. Le Rouzic, D. Dormont, S. Benichou, and C. Dargemont. 2001. Characterization of the nuclear import pathway for HIV-1 integrase. J. Biol. Chem. 276:18102-18107. [DOI] [PubMed] [Google Scholar]
- 15.Devroe, E., A. Engelman, and P. A. Silver. 2003. Intracellular transport of human immunodeficiency virus type 1 integrase. J. Cell Sci. 116:4401-4408. [DOI] [PubMed] [Google Scholar]
- 16.Dirac, A. M., and J. Kjems. 2001. Mapping DNA-binding sites of HIV-1 integrase by protein footprinting. Eur. J. Biochem. 268:743-751. [DOI] [PubMed] [Google Scholar]
- 17.Dvorin, J. D., P. Bell, G. G. Maul, M. Yamashita, M. Emerman, and M. H. Malim. 2002. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol. 76:12087-12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyda, F., A. B. Hickman, T. M. Jenkins, A. Engelman, R. Craigie, and D. R. Davies. 1994. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266:1981-1986. [DOI] [PubMed] [Google Scholar]
- 19.Eijkelenboom, A. P., R. A. Lutzke, R. Boelens, R. H. Plasterk, R. Kaptein, and K. Hard. 1995. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat. Struct. Biol. 2:807-810. [DOI] [PubMed] [Google Scholar]
- 20.Engelman, A., and R. Craigie. 1992. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 66:6361-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engelman, A., G. Englund, J. M. Orenstein, M. A. Martin, and R. Craigie. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esposito, D., and R. Craigie. 1998. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 17:5832-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fassati, A., D. Gorlich, I. Harrison, L. Zaytseva, and J. M. Mingot. 2003. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 22:3675-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fouchier, R. A., B. E. Meyer, J. H. Simon, U. Fischer, A. V. Albright, F. Gonzalez-Scarano, and M. H. Malim. 1998. Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. J. Virol. 72:6004-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freed, E. O., G. Englund, F. Maldarelli, and M. A. Martin. 1997. Phosphorylation of residue 131 of HIV-1 matrix is not required for macrophage infection. Cell 88:171-173. [DOI] [PubMed] [Google Scholar]
- 26.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gallay, P., S. Swingler, J. Song, F. Bushman, and D. Trono. 1995. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell 83:569-576. [DOI] [PubMed] [Google Scholar]
- 28.Gerton, J. L., S. Ohgi, M. Olsen, J. DeRisi, and P. O. Brown. 1998. Effects of mutations in residues near the active site of human immunodeficiency virus type 1 integrase on specific enzyme-substrate interactions. J. Virol. 72:5046-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goff, S. P. 2001. Intracellular trafficking of retroviral genomes during the early phase of infection: viral exploitation of cellular pathways. J. Gene Med. 3:517-528. [DOI] [PubMed] [Google Scholar]
- 30.Goldgur, Y., F. Dyda, A. B. Hickman, T. M. Jenkins, R. Craigie, and D. R. Davies. 1998. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc. Natl. Acad. Sci. USA 95:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen, M. S., G. J. Smith III, T. Kafri, V. Molteni, J. S. Siegel, and F. D. Bushman. 1999. Integration complexes derived from HIV vectors for rapid assays in vitro. Nat. Biotechnol. 17:578-582. [DOI] [PubMed] [Google Scholar]
- 32.Heinzinger, N. K., M. I. Bukinsky, S. A. Haggerty, A. M. Ragland, V. Kewalramani, M. A. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. USA 91:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heuer, T. S., and P. O. Brown. 1997. Mapping features of HIV-1 integrase near selected sites on viral and target DNA molecules in an active enzyme-DNA complex by photo-cross-linking. Biochemistry 36:10655-10665. [DOI] [PubMed] [Google Scholar]
- 34.Heuer, T. S., and P. O. Brown. 1998. Photo-cross-linking studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase-DNA complex. Biochemistry 37:6667-6678. [DOI] [PubMed] [Google Scholar]
- 35.Hindmarsh, P., and J. Leis. 1999. Retroviral DNA integration. Microbiol. Mol. Biol. Rev. 63:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenkins, T. M., D. Esposito, A. Engelman, and R. Craigie. 1997. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 16:6849-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenkins, Y., M. McEntee, K. Weis, and W. C. Greene. 1998. Characterization of HIV-1 vpr nuclear import: analysis of signals and pathways. J. Cell Biol. 143:875-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katz, R. A., and A. M. Skalka. 1994. The retroviral enzymes. Annu. Rev. Biochem. 63:133-173. [DOI] [PubMed] [Google Scholar]
- 39.Katzman, M., and M. Sudol. 1996. Influence of subterminal viral DNA nucleotides on differential susceptibility to cleavage by human immunodeficiency virus type 1 and visna virus integrases. J. Virol. 70:9069-9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katzman, M., and M. Sudol. 1998. Mapping viral DNA specificity to the central region of integrase by using functional human immunodeficiency virus type 1/visna virus chimeric proteins. J. Virol. 72:1744-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis, P., M. Hensel, and M. Emerman. 1992. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 11:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis, P. F., and M. Emerman. 1994. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J. Virol. 68:510-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Limon, A., E. Devroe, R. Lu, H. Z. Ghory, P. A. Silver, and A. Engelman. 2002. Nuclear localization of human immunodeficiency virus type 1 preintegration complexes (PICs): V165A and R166A are pleiotropic integrase mutants primarily defective for integration, not PIC nuclear import. J. Virol. 76:10598-10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Limon, A., N. Nakajima, R. Lu, H. Z. Ghory, and A. Engelman. 2002. Wild-type levels of nuclear localization and human immunodeficiency virus type 1 replication in the absence of the central DNA flap. J. Virol. 76:12078-12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lins, R. D., A. Adesokan, T. A. Soares, and J. M. Briggs. 2000. Investigations on human immunodeficiency virus type 1 integrase/DNA binding interactions via molecular dynamics and electrostatics calculations. Pharmacol. Ther. 85:123-131. [DOI] [PubMed] [Google Scholar]
- 46.Lodi, P. J., J. A. Ernst, J. Kuszewski, A. B. Hickman, A. Engelman, R. Craigie, G. M. Clore, and A. M. Gronenborn. 1995. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry 34:9826-9833. [DOI] [PubMed] [Google Scholar]
- 47.Maertens, G., P. Cherepanov, W. Pluymers, K. Busschots, E. De Clercq, Z. Debyser, and Y. Engelborghs. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278:33528-33539. [DOI] [PubMed] [Google Scholar]
- 48.Masuda, T., M. J. Kuroda, and S. Harada. 1998. Specific and independent recognition of U3 and U5 att sites by human immunodeficiency virus type 1 integrase in vivo. J. Virol. 72:8396-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masuda, T., V. Planelles, P. Krogstad, and I. S. Chen. 1995. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J. Virol. 69:6687-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murakami, T., T. Hagiwara, K. Yamamoto, J. Hattori, M. Kasami, M. Utsumi, and T. Kaneda. 2001. A novel method for detecting HIV-1 by non-radioactive in situ hybridization: application of a peptide nucleic acid probe and catalysed signal amplification. J. Pathol. 194:130-135. [DOI] [PubMed] [Google Scholar]
- 52.Nakamura, T., T. Masuda, T. Goto, K. Sano, M. Nakai, and S. Harada. 1997. Lack of infectivity of HIV-1 integrase zinc finger-like domain mutant with morphologically normal maturation. Biochem. Biophys. Res. Commun. 239:715-722. [DOI] [PubMed] [Google Scholar]
- 53.Petit, C., O. Schwartz, and F. Mammano. 2000. The karyophilic properties of human immunodeficiency virus type 1 integrase are not required for nuclear import of proviral DNA. J. Virol. 74:7119-7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pluymers, W., P. Cherepanov, D. Schols, E. De Clercq, and Z. Debyser. 1999. Nuclear localization of human immunodeficiency virus type 1 integrase expressed as a fusion protein with green fluorescent protein. Virology 258:327-332. [DOI] [PubMed] [Google Scholar]
- 55.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Gottlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roe, T., T. C. Reynolds, G. Yu, and P. O. Brown. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sayasith, K., G. Sauve, and J. Yelle. 2000. Characterization of mutant HIV-1 integrase carrying amino acid changes in the catalytic domain. Mol. Cells 10:525-532. [DOI] [PubMed] [Google Scholar]
- 58.Sherman, P. A., and J. A. Fyfe. 1990. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc. Natl. Acad. Sci. USA 87:5119-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzuki, Y., N. Misawa, C. Sato, H. Ebina, T. Masuda, N. Yamamoto, and Y. Koyanagi. 2003. Quantitative analysis of human immunodeficiency virus type 1 DNA dynamics by real-time PCR: integration efficiency in stimulated and unstimulated peripheral blood mononuclear cells. Virus Genes 27:177-188. [DOI] [PubMed] [Google Scholar]
- 60.Tsurutani, N., M. Kubo, Y. Maeda, T. Ohashi, N. Yamamoto, M. Kannagi, and T. Masuda. 2000. Identification of critical amino acid residues in human immunodeficiency virus type 1 integrase required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J. Virol. 74:4795-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vink, C., D. C. van Gent, Y. Elgersma, and R. H. Plasterk. 1991. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J. Virol. 65:4636-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vora, A., and D. P. Grandgenett. 2001. DNase protection analysis of retrovirus integrase at the viral DNA ends for full-site integration in vitro. J. Virol. 75:3556-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, J. Y., H. Ling, W. Yang, and R. Craigie. 2001. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 20:7333-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weinberg, J. B., T. J. Matthews, B. R. Cullen, and M. H. Malim. 1991. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J. Exp Med. 174:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wiskerchen, M., and M. A. Muesing. 1995. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J. Virol. 69:376-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]
- 67.Zargarian, L., M. S. Benleumi, J. G. Renisio, H. Merad, R. G. Maroun, F. Wieber, O. Mauffret, H. Porumb, F. Troalen, and S. Fermandjian. 2003. Strategy to discriminate between high and low affinity bindings of human immunodeficiency virus, type 1 integrase to viral DNA. J. Biol. Chem. 278:19966-19973. [DOI] [PubMed] [Google Scholar]
- 68.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]