Abstract
Ion conduction across the cellular membrane requires the simultaneous opening of activation and inactivation gates of the K+ channel pore. The bacterial KcsA channel has served as a powerful system for dissecting the structural changes that are related to four major functional states associated with K+ gating. Yet, the direct observation of the full gating cycle of KcsA has remained structurally elusive, and crystal structures mimicking these gating events require mutations in or stabilization of functionally relevant channel segments. Here, we found that changes in lipid composition strongly increased the KcsA open probability. This enabled us to probe all four major gating states in native-like membranes by combining electrophysiological and solid-state NMR experiments. In contrast to previous crystallographic views, we found that the selectivity filter and turret region, coupled to the surrounding bilayer, were actively involved in channel gating. The increase in overall steady-state open probability was accompanied by a reduction in activation-gate opening, underscoring the important role of the surrounding lipid bilayer in the delicate conformational coupling of the inactivation and activation gates.
Introduction
Membrane proteins are embedded in the lipid bilayer in close contact with surrounding lipids. These contacts play important roles in membrane protein folding, structure, and function (1). Binding of lipids to specific sites has been observed in crystal structures (2), and increasing evidence suggests that membrane lipids play a critical functional role in a variety of membrane protein classes, including channels (3, 4, 5, 6), transporters (7), G-protein coupled receptors (8), and receptor tyrosine kinases (9).
In addition to strong binding, subtle changes in lipid character and bilayer state can have significant effects on protein function, such as those observed for the bacterial KcsA potassium channel (3, 10, 11). Upon activation at acidic pH, KcsA mediates a transient K+ current that is characterized by a fast activation time course followed by a slow C-type inactivation process (12). This time course is well described by a successive and coordinate opening and closing of activation and inactivation gates within the conduction pathway of the KcsA channel (Fig. 1 A). The resting state of the KcsA channel, with the inactivation gate open and the activation gate closed (closed-conductive state), and the inactivated state, with the inactivation gate closed and the activation gate open (open-inactivated state), are relatively long-lived. Therefore, conformational changes associated with the opening and closing of the KcsA activation and inactivation gates can be studied with the use of crystallographic (10, 11, 13, 14) and spectroscopic methods. The inner KcsA helices form an important part of the conduction pathway. Previous electron paramagnetic resonance experiments showed that they rotate upon activation in a counterclockwise direction, thereby opening the conduction pathway (15, 16). Furthermore, solid-state NMR (ssNMR), which can be applied in lipid bilayers (17, 18, 19, 20), provided detailed insights into the structural basis of the coupling of KcsA activation and inactivation gates during channel gating (21, 22, 23).
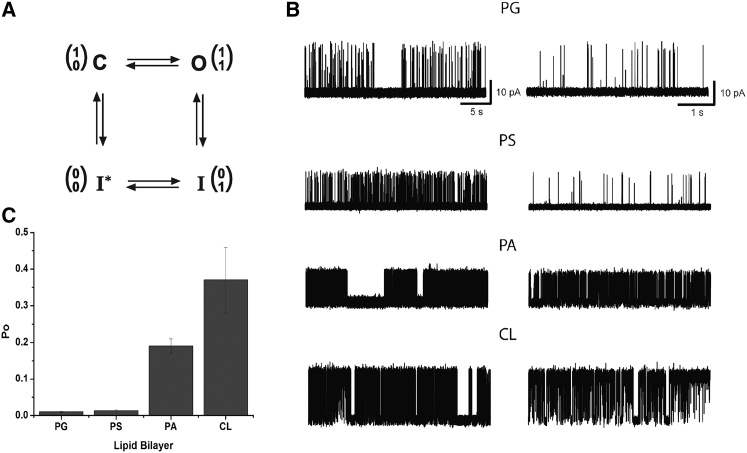
Figure 1.
(A) Four-state minimal gating model with upper (inactivation) and lower (activation) gates in the conduction pathway of the KcsA channel. Gate positions (0 for closed and 1 for opened) are diagrammed in one-column matrices, with the upper gate in the top row and the lower gate in the bottom row. C, closed channel in resting state; O, open channel; I and I∗, inactivated states as in Ref. (22). (B) Representative current traces of KcsA recorded in different anionic phospholipid environments. The specific anionic phospholipid is indicated above the current traces. On the left, long 30 s traces are presented; on the right, 5 s traces are shown. All experiments were performed at +100 mV in symmetrical 150 mM KCl solution at pH 7.0 on the cis side and pH 4.0 on the trans side. Lipid bilayers were composed of 70% anionic phospholipid and 30% neutral DOPC. (C) Open probability (PO) of KcsA in different anionic phospholipids. All experiments were performed in symmetrical 150 mM KCl solution at pH 7.0 on the cis side and pH 4.0 on the trans side. Lipid bilayers were composed of 70% anionic lipid and 30% neutral DOPC. Bottom: overall PO values calculated for entire recordings of the single-channel currents conducted by KcsA, including long closed periods, for different anionic phospholipid environments (see main text). PO values are given in the text as mean ± SEM.
Despite these advancements, a direct structural visualization of the entire KcsA gating cycle, including the conformation with an open activation and closed inactivation gate (open-conductive conformation) in a lipidic environment has remained challenging due to the very short lifetime of this channel state and the propensity of the selectivity filter to rapidly collapse when the activation gate is opened (12, 13, 14, 24, 25). Using solid-state NMR spectroscopy, we previously identified a lipid-KcsA interaction site at the turret region, which connects the outer transmembrane helix of KcsA (transmembrane helix 1 (TM1)) and the pore helix behind the selectivity filter of the KcsA channel. The interaction between the turret and lipid modulates the conduction properties of the KcsA channel pore (4, 26). Here, we exploited that earlier observation and modulated KcsA channel gating by varying the lipid composition of the surrounding bilayer. By combining electrophysiological and ssNMR experiments, we found that lipid bilayers rich in cardiolipin (CL) stabilized the open-conductive conformation of the KcsA channel. This enabled us to study for the first time, to our knowledge, the structure of a functionally defined open state of the KcsA channel. A comparison of our findings for wild-type (WT) KcsA with earlier crystallographic results for KcsA mutants (13, 14, 25) led to a detailed understanding of the conformational changes that occur during the KcsA gating cycle in lipid bilayers. The data provide a general picture of how ion channels transit from activated to inactivated states and back.
Materials and Methods
Sample preparation
KcsA was expressed in M15 Escherichia coli cells and purified using Ni2+-affinity chromatography. For ssNMR experiments, KcsA was uniformly (13C,15N) labeled. An unlabeled KcsA was used for single-channel measurements. After purification, KcsA was reconstituted into proteoliposomes composed of 3:7 molar ratios of 1,2-di-(9Z-octadecenoyl)-sn-glycero-3-phosphocholine (DOPC)/1,2-di-(9Z-octadecenoyl)-sn-glycero-3-phospho-(1′-rac-glycerol) (DOPG), DOPC/1,2-di-(9Z-octadecenoyl)-sn-glycero-3-phospho-L-serine (DOPS), DOPC/1,2-dioleoyl-sn-glycero-3-phosphate (DOPA), and DOPC/CL. Lipids were solubilized in reconstitution buffer (150 mM KCl, 10 mM HEPES, pH 7.4) and KcsA was added to the phospholipid mixture to give a protein/lipid molar ratio of 1:1000 for single-channel measurements and a protein/lipid molar ratio of 1:100 for samples used in ssNMR experiments. Proteoliposomes were formed upon detergent removal by addition of Bio-Beads (Merck, Darmstadt, Germany). KcsA proteoliposomes for the planar lipid bilayer experiments were split into 250 μL aliquots, flash frozen with liquid N2, and stored at −80°C until use. Proteoliposomes for ssNMR experiments were centrifuged at 100,000 g at 4°C, resuspended in sample buffer (150 mM KCl, 10 mM succinic acid, 0.01% sodium azide, pH 4.0), and centrifuged again. Pelleted proteoliposomes were packed in a 3.2 mm rotor and kept at 4°C until use.
Solid-state NMR spectroscopy and analysis
SsNMR experiments were conducted on NMR spectrometers operating at 500 and 700 MHz 1H resonance frequencies (Bruker Biospin, Germany) equipped with 3.2 mm triple-resonance (1H, 13C, 15N) magic-angle spinning (MAS) probe heads (see, e.g., (26)). All experiments were carried out at an effective sample temperature of 273 K using MAS frequencies of 11.4, 12, or 15 kHz. 13C-13C proton-driven spin diffusion (PDSD) experiments used homonuclear mixing times of 30 ms and 150 ms. The latter experiments were conducted under weak coupling conditions (27). NCACX spectra were acquired using 2.5 ms spectrally induced filtering in combination with cross polarization (SPECIFIC-CP; 28) contact times and a 30 ms 13C-13C PDSD magnetization transfer step. SPINAL64 (29) proton decoupling at 83 kHz was used in both evolution and detection periods. Data acquisition and analysis were performed with Topspin 3.1 (Bruker Biospin, Billerica, MA). KcsA resonance assignments were taken from previous work (4, 21, 22, 26, 30). Chemical-shift values representing protonated and deprotonated glutamate Cγ and Cδ correlations were taken from the literature (see, e.g., Ref. (31)).
Electrophysiology
Single-channel recordings of KcsA were performed on a planar lipid bilayer setup (Compact, Ionovation, Osnabrück, Germany). Lipid bilayers were formed by painting the lipids dissolved in n-decan over a 200 μm hole that separated two chambers (cis and trans). Initially, the trans chamber contained 20 mM KCl and the cis chamber contained 250 mM KCl, both of which were buffered to pH 4 with 10 mM succinic acid. KcsA proteoliposomes (1–5 μL) were added to the cis chamber of the planar bilayer system with a micropipette. After channel insertion, recording conditions were established by perfusion to symmetrical conditions. The cis side contained 150 mM KCl and 10 mM HEPES, pH 7.0, and the trans side contained 150 mM KCl and 10 mM succinic acid, pH 4.0. All measurements were performed at room temperature. Data were sampled at 10 kHz and filtered at 1 kHz. The reported data are the results of a statistical analysis of at least six independent experiments. A kinetic analysis was done using the QuB software for single-channel analysis (www.qub.buffalo.edu). Single-channel currents were first idealized into noise-free open and closed transitions using a half-amplitude threshold algorithm. Closed and open intervals were compiled into dwell-time histograms with a logarithmic abscissa and square-root ordinate (32) and were fitted by sums of exponentials according to the following equations:
where λi is the reciprocal of the time constant of the ith component, n is the number of exponential components, ai is the fractional area occupied by the ith component, and t is the decay constant.
Results and Discussion
Increasing the open probability by changing the lipid environment
Under steady-state conditions, KcsA exhibits a low open probability (PO) because most of the time it resides in a C-type inactivated state (12, 33, 34, 35). It was previously shown that anionic phospholipids may increase PO, providing a more favorable environment for the open state of KcsA (36, 37, 38). These observations prompted us to test different anionic phospholipid conditions for maximizing the stabilization of the open-conductive conformation. This experimental strategy avoided mutation of the KcsA protein, which may result in unnatural conformational changes.
We incorporated KcsA into four different types of anionic phospholipids: DOPG, DOPS, DOPA, and CL (heart, bovine). Single-channel currents were recorded using planar lipid bilayer electrophysiology. The data indicated that anionic phospholipids (at a 7:3 ratio with neutral DOPC) exerted significant effects on the KcsA channel PO (Fig. 1 B). PO correlated with the charge of the phospholipid headgroup. PO was high in a lipid environment with more negatively charged headgroups (DOPA (−1.3, PO = 0.19 ± 0.02, n = 10, mean ± SE) and CL (−2, PO = 0.37 ± 0.09, n = 8, mean ± SE), and comparably low in a lipidic environment with less negatively charged headgroups (e.g., DOPG (−1, PO = 0.010 ± 0.002, n = 9, mean ± SE) and DOPS (−1, PO = 0.013 ± 0.003, n = 6, mean ± SE; Fig. 1 C). Maximal PO was observed in the presence of CL (0.37 ± 0.09, n = 8, mean ± SE), which has the highest headgroup charge (39) of the anionic phospholipids tested (Fig. 1 C).
Analysis of the dwell-time histograms revealed that both open and closed durations were composed of multiple kinetic components, which is in good agreement with previous studies (40, 41). Under all experimentally tested phospholipid conditions, dwell-time histograms for closed durations were best fit with three exponentials corresponding to three nonconductive states of the KcsA channel (Fig. S1 in the Supporting Material). Closed times representing mainly inactivated states under steady-state conditions (40) were sensitive to changes in the anionic phospholipid environment. The total mean closed time (τmean) was ∼8 fold shorter in the presence of DOPA and CL in comparison with DOPG and DOPS (Fig S1; Table S1). This effect of DOPA and CL was due to a reduction of the most prevalent second closed state (τ2), which was predominant in the presence of DOPG and DOPS (Fig. S1; Table S1). In the presence of DOPA and CL, the second closed state was greatly reduced and the first short-lived closed state (τ1) became the predominant one (Fig. S1; Table S1). Altogether, the analysis of closed time distributions indicated a significant effect of the anionic phospholipids on KcsA channel C-type inactivation. The channel spent less time in the inactivated state in a lipidic environment enriched by phospholipids with more negatively charged headgroups such as DOPA and CL.
The three-exponential fit of dwell-time histograms for open durations revealed that the KcsA mean open time (τmean) was higher in the presence of DOPA and CL than in the presence of DOPG and DOPS. Two factors contributed to this effect. First, the duration and impact of the second open state were higher in the presence of DOPA and CL in comparison with DOPG and DOPS (τ2) (Fig. S1; Table S1). Second, in the presence of DOPA and CL, the third long-lived open state of KcsA was observed in the dwell-time distributions, whereas in DOPG and DOPS, the third long-lived open state was not detectable (τ3) (Fig. S1; Table S1).
Our findings suggest that the effect of anionic phospholipids on KcsA closed and open durations depends on the phospholipid headgroup charge. More negatively charged headgroups reduce the mean closed time of the inactivated channel and thereby increase its mean open time, which then results in an increase in channel PO (Fig. 1, B and C). The maximal mean open time of KcsA was observed in CL, which is the most negatively charged phospholipid among the tested phospholipids.
CL stabilizes the open-conductive conformation of the KcsA potassium channel
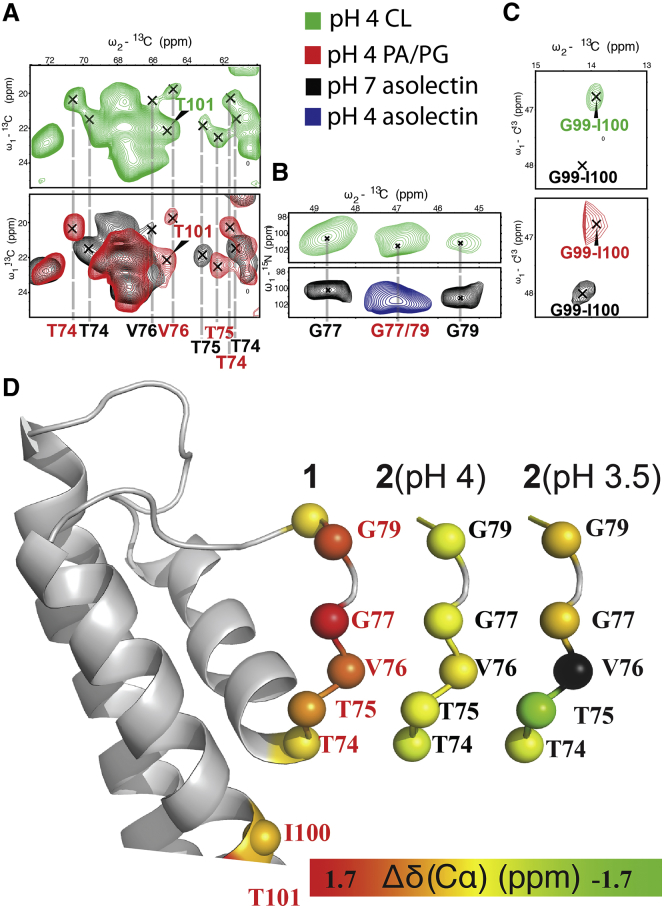
Our electrophysiological experiments suggested that CL-rich bilayers had a major effect on KcsA gating and stabilized KcsA in an open-conductive conformation. To correlate the changes in PO with conformational changes at the channel pore, we performed two-dimensional (2D) (13C,13C) and (15N,13C) ssNMR correlation experiments on KcsA in phospholipid environments identical to those used in the electrophysiological experiments described above. In contrast to previous solution-state NMR experiments using bilayer mimetics (see, e.g., (42, 43, 44)), the 13C-13C correlation spectra recorded with short mixing times enabled us to identify 13C chemical shifts (see Table S2 for a complete list) for the key residues T74, T75, and V76 within the KcsA selectivity filter (Fig. 2, A and B), as well as for G99, I100, and T101 located at the activation gate (Fig. 2 C). Note that in Figs. 2, A–C (lower panels), results obtained using asolectin bilayers (as previously described (26)) are only included as a reference to mark the conductive (black, pH 7) and collapsed (blue, pH 4) states of the selectivity filter.
Figure 2.
(A–C) Spectral cutouts of 2D 13C-13C correlation (A and C) and 15N-13C (B) correlation spectra recorded in the presence of 150 mM KCl at pH 4.0 in different anionic phospholipid environments. KcsA was reconstituted into proteoliposomes composed of DOPC/DOPG, DOPC/DOPA, or DOPC/CL) at a 3:7 molar ratio. Data obtained using CL-rich lipids (pH 4.0) are shown in green. ssNMR spectra recorded for DOPC/DOPG (data not shown) and DOPC/DOPA lipids (pH 4.0, red) were identical and are denoted by PA/PG for brevity. As a reference, data obtained using asolectin at pH 7.0 or pH 4 are indicated in black and blue, respectively. Specific residues for each state are color-coded correspondingly. Experimental data were recorded at magnetic fields corresponding to a 500–700 MHz 1H frequency and at an effective sample temperature of 273 K. (D) Cα chemical-shift changes of the selectivity filter in the collapsed state of PA, PG, and CL (left), and the conductive SF conformation (denoted by 2) appearing for CL-rich membranes at pH 4.0 (middle) and pH 3.5 (right). The color-coding refers to differences in Cα chemical-shift values measured for the indicated KcsA lipid preparations and pH values relative to ssNMR shifts obtained for the conductive state of KcsA at pH 7 in asolectin lipids (26). To see this figure in color, go online.
The ssNMR correlation patterns obtained using DOPC/DOPG (data not shown) and DOPC/DOPA bilayers (Fig. 2, A and C, lower panel, red) were indistinguishable from those observed in previous studies (22, 26) using asolectin bilayers, which are included for reference and corresponded at pH 4 to a collapsed (inactivated) conformation of the KcsA selectivity filter (Fig. 2, A and B, lower panels). To analyze our data using DOPC/CL bilayers at pH 4 (Fig. 2 A, upper panel), we again compared well-resolved correlations (e.g., for T74CA-CB, T74CG2-CB, T75CA-CB, and T75CG2-CA in CC data (Fig. 2 A), and for G77 and G79 in NCA data (Fig. 2 B)) with previous results obtained using asolectin bilayers. In both type of spectra, we could readily identify correlations diagnostic for a collapsed inactivation gate. Moreover, we observed additional correlations that, as indicated by crosses and vertical lines, perfectly overlap with shifts seen in asolectin bilayers for an open inactivation gate. These observations strongly suggested that we observed two sets of 13C-13C intraresidue correlations at T74CA-CB, T74CG2-CB, T75CA-CB, and T75CG2-CA compatible with the presence of both collapsed (inactivated) and conductive conformations of the selectivity filter (Fig. 2 A, upper panel). The existence of both populations was also confirmed in 2D NCACX experiments for additional residues (G77 and G79 (Fig. 2 B, upper panel) and Thr74 (Fig. S2)).
Spectra obtained using longer 13C,13C mixing times exhibited sequential G99-I100 correlations for all lipid preparations that were compatible with an open activation gate (Fig. 2 C, top panel). Note that we did not find a second population exhibiting a closed activation gate that, for example, characterizes the selectivity filter state for asolectin bilayers at pH 7 (Fig. 2 C, lower panel). Furthermore, our 13C-13C intraresidue correlation spectra clearly show that in the presence of CL lipids, KcsA still adopts an extended TM1 (Fig. S3), which is involved in KcsA-lipid interactions (26). Altogether, we observed two conformations in the case of CL: the well-known open-collapsed state (Fig. 2 D, conformation 1) and one most likely corresponding to the conductive state (Fig. 2 D, conformation 2). Interestingly, when we lowered the pH of the CL sample to 3.5 (Fig. 2 D, right), we observed chemical-shift changes for the conductive state at residue Thr75. In addition, correlations expected for residue V76 disappeared at pH 3.5 (indicated in black in Fig. 2 D, right), possibly due to local motion. Previously, we detected such increased mobility of selectivity filter residues, in particular for V76, in ssNMR studies using synthetic (45) as well as native bacterial membranes (46, 47), in line with results of previous molecular-dynamics studies (48).
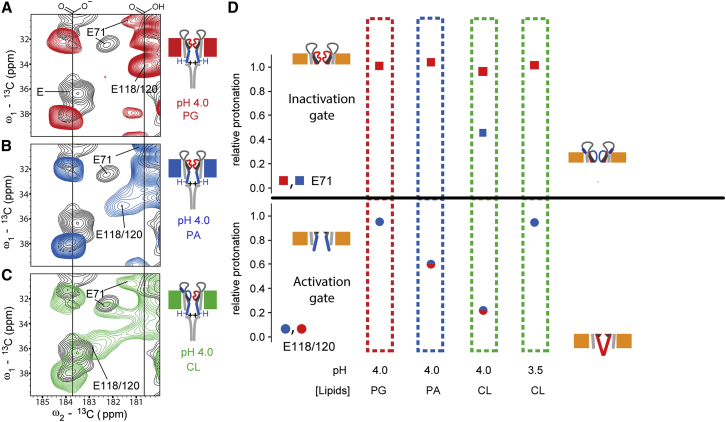
As we have shown before (22), the isotropic chemical shift of glutamate δ carbon atoms that is correlated with the side-chain protonation state (49) provides a sensitive means of probing residue-specific protonation levels in different functional KcsA states. Therefore, in addition to examining conformational changes on the backbone level, we investigated the relative protonation level of glutamate residues located at both channel gates (namely, E71 and E118/E120) as a function of phospholipid type and pH (Fig. 3, A–C). For the inactivation gate, the protonation levels deduced from the measured Cδ NMR chemical shift (see also Ref. (22)) for E71 corroborated our previous analysis of the chimeric KcsA-Kv1.3 channel, which differs from the WT KcsA in 11 amino acids in the turret region (45). In both cases, the collapsed state of the selectivity filter (SF) involves protonation of E71 (Fig. 3 D). In CL bilayers (pH 4), we observed additional correlations that are consistent with a reduction in E71 protonation due to the presence of a KcsA channel population in an open-conductive state (Fig. 3 D, top row). This is also in agreement with our earlier work showing that E71 deprotonation is indicative of a conductive SF. Furthermore, lipid-induced changes became apparent at residues E118/E120 located at the activation gate (Fig. 3 D, bottom panel). In PG lipid bilayers, glutamate residues (E118/E120) close to the activation gate were fully protonated, similar to the protonation levels seen for KcsA-Kv1.3 and in line with a fully open activation gate. In more negatively charged CL lipids, however, this state was only observed at pH 3.5. At pH 4, CL-rich lipids showed E71 resonances (Fig. 3 D) that confirmed the coexistence of the collapsed and conductive filter states described in Fig. 2 B.
Figure 3.
Spectral regions extracted from (13C,13C) correlation spectra showing glutamate Cγ-Cδ crosspeaks in different lipid compositions. (A–C) Cartoons using PG (A, red), PA (B, blue), and CL (C, green) phospholipids indicate channel states as defined in Fig. 3D. Assignments were obtained by tracing glutamic acid Cγ-Cδ (as indicated) and Cγ-Cβ as well as Cγ-Cα correlations in our spectra. Vertical lines at 180.5 and 183.7 ppm correspond to fully protonated and deprotonated glutamate Cδ chemical shifts, respectively. Negative charges and protons in the cartoons illustrate the protonation states of E118 and E120 at the helix bundle crossing. Positive charges are added to indicate electrostatics at the helix bundle crossing. (D) Relative protonation of E71 (squares) and E118/120 (circles) computed for different lipid compositions. For each condition, integrals of crosspeaks corresponding to the respective protonated and deprotonated glutamate Cγ-Cδ correlations along the ω2-13C direction were normalized to their sum. Relative protonation is therefore represented by the fractions of protonated Cγ-Cδ signal. Blue and red colors indicate open and closed conformations of the channel gates, respectively. Protonation of E118/120 correlates with activation-gate opening, whereas inactivation-gate closure involves an increased protonation of E71. To see this figure in color, go online.
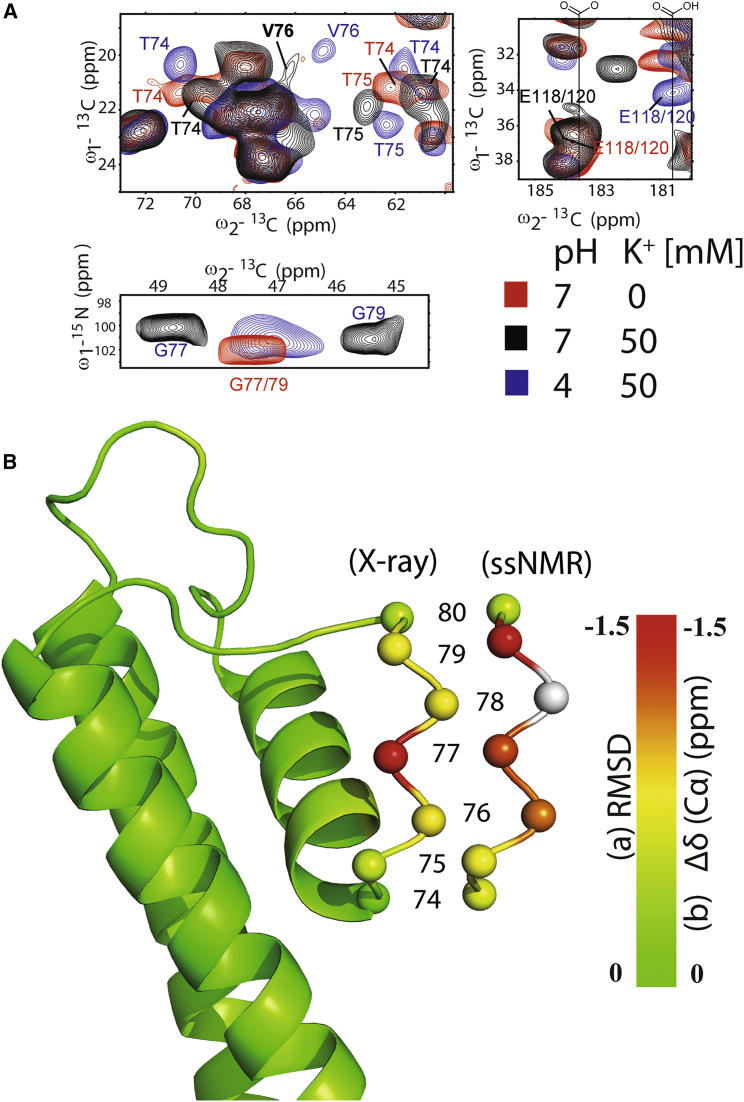
Taken together, our ssNMR data presented in Figs. 2 and 3 provide structural insights into three of the four states of the KcsA gating cycle, i.e., the closed-conductive, open-inactivated, and open-conductive states, with the latter being visible only in CL lipids at low pH. The fourth state of the gating cycle, i.e., the closed-collapsed state (see Fig. 1), can be stabilized at pH 7 and vanishing K+ concentrations (11). In Fig. 4 A, ssNMR data obtained under such conditions (red) are compared with reference experiments in the closed-conductive (black) and open-collapsed (blue) states. With these results, we could readily track channel residues, such as those belonging to the selectivity filter, to study changes in channel conformation. Note that the Cα chemical shifts at low K+ are similar to those seen for the collapsed state at pH 4, high K+ (blue), for residue Thr75 (Fig. 4 A, upper-left panel) as well as residues Gly77 and Gly79 (Fig. 4 A, lower panel). The same applies to residue Thr74, where the data would also be consistent with a second backbone conformation as seen in the conductive state. For Val76 (see also Fig. S4 for data obtained at lower contours), we observe a correlation with lower intensity that is separated by ∼0.8 ppm from both the conductive (black) and collapsed (blue) states. Next to the inactivation gate, we could also detect deprotonated glutamate residues (E118/120; Fig. 4 A, upper-right panel), which, as expected, is in line with a closed activation gate.
Figure 4.
(A) ssNMR spectra recorded at a high K+ concentration (50 mM) before (black) and after (blue, 150 mM) inactivation are compared with data obtained at low K+ (0 mM, red). Cutouts are shown for (13C,13C) and (15N,13C) 2D data sets showing residues of the selectivity filter and the activation gate. (B) Comparison of KcsA x-ray data (PDB: 1K4C and 1K4D) with ssNMR results that were mapped on the conductive x-ray structure (PDB: 1K4C). To see this figure in color, go online.
Previous root mean-square deviations (RMSDs) seen between crystallographic data obtained in the conductive (PDB: 1K4C) and collapsed (PDB: 1K4D) states indicate that structural changes are predominantly found around Gly77 (Fig. 4 B). In contrast to KcsA crystal-structural data (11), our ssNMR chemical-shift data (relative to Cα shifts seen in the conductive state) indicate that the entire filter (comprising residues Thr74–Gly79) is involved in closing of the inactivation gate (Fig. 4 B).
KcsA-lipid interaction at the residue-specific level
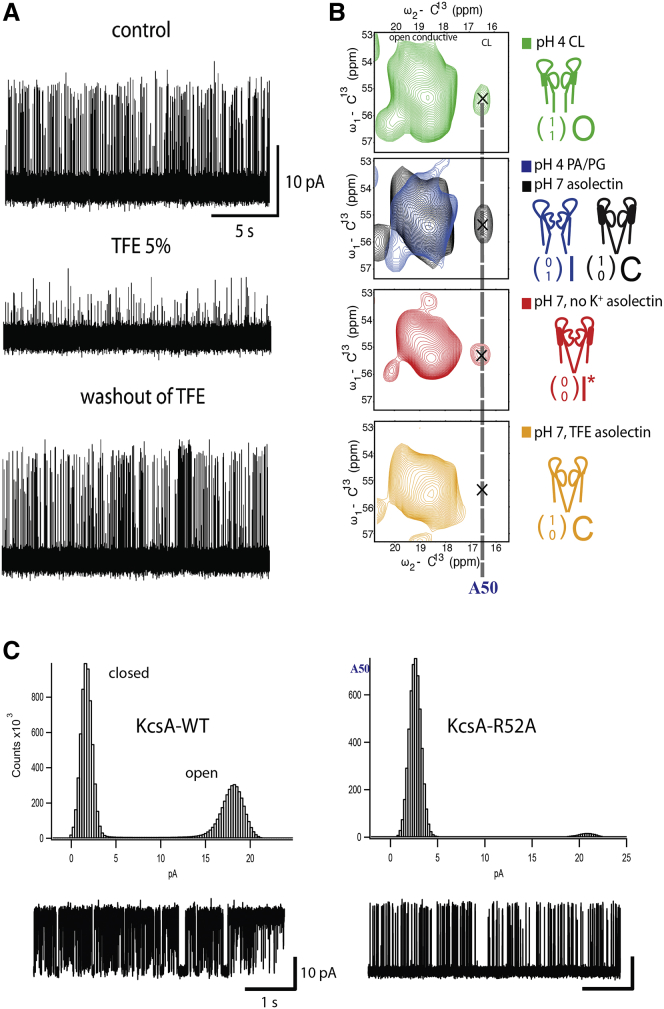
Next, we conducted a series of experiments to dissect the atomic interactions that may explain the influence of the lipid type on channel gating. Previously, we identified a lipid-KcsA interaction site at the turret region located close to Ala50 that connects the outer transmembrane helix of KcsA (TM1) and the pore helix behind the selectivity filter of the KcsA channel (4, 26). Here, we utilized 2,2,2-trifluoroethanol (TFE) as a lipid protein dissociation agent at concentrations that kept the channel structure largely intact (vide infra). In our electrophysiological experiments performed in planar lipid bilayers, KcsA PO and single-channel conductance were reversibly reduced upon addition of 5% TFE, and were restored after removal of TFE from the solution by perfusion at the extracellular part of the channel (Fig. 5 A; see Fig. S5 A). ssNMR experiments conducted on KcsA reconstituted into proteoliposomes with addition of 5% TFE confirmed that the channel structure was largely intact. At least on the backbone level, characteristic correlations of the selectivity filter (see Fig. S5 B, including zoom-ins), as well large spectral regions diagnostic for α-helical correlations of the TM1 and TM2 channel regions (see, e.g., (26, 30)), readily superimposed with results obtained before the addition of TFE. On the other hand, Ala50 resonances (see also Fig. 4 in Ref. (26)), which can be readily distinguished based on their well-separated chemical shift, largely disappeared upon addition of TFE (Fig. 5 B; see Fig. S5 B for the entire aliphatic spectra), suggesting that the lipid-protein interface around the turret region is critically involved in stabilizing the channel structure. Indeed, a point mutation at residue R52 (KcsA-R52A) drastically reduced the PO of KcsA in the CL-enriched lipid bilayer (Fig. 5 C). These results are in good agreement with our previous work in which we showed the role of R52 in KcsA lipid interactions (26).
Figure 5.
(A) Influence of TFE on KcsA single-channel properties. Experiments were performed at +100 mV in symmetrical 150 mM KCl solution at pH 7.0 on the cis side and pH 4.0 on the trans side. Lipid bilayers were composed of DOPC and DOPG at a 7:3 molar ratio. TFE was applied from cis side. (B) ssNMR correlations for residue Ala50 at the C-terminus end of the first transmembrane helix at the indicated experimental conditions (Control - 50 mM K+, asolectin, pH 7 versus TFE 5% and 150 mM K+, pH 7) and channel states. Data were extracted from 2D PDSD spectra obtained using a 30 ms mixing time at 12 kHz MAS, 273 K at 500 MHz 1H frequency. (C) Representative current traces and all-point histograms of KcsA-WT and KcsA-R52A recorded in a planar lipid bilayer with a high content of CL. The experiments were performed at +100 mV in symmetrical 150 mM KCl solution at pH 7.0 on the cis side and pH 4.0 on the trans side. The lipid bilayer was composed of DOPC and CL at a 3:7 molar ratio. To see this figure in color, go online.
KcsA gating cycle in lipids versus crystal structures
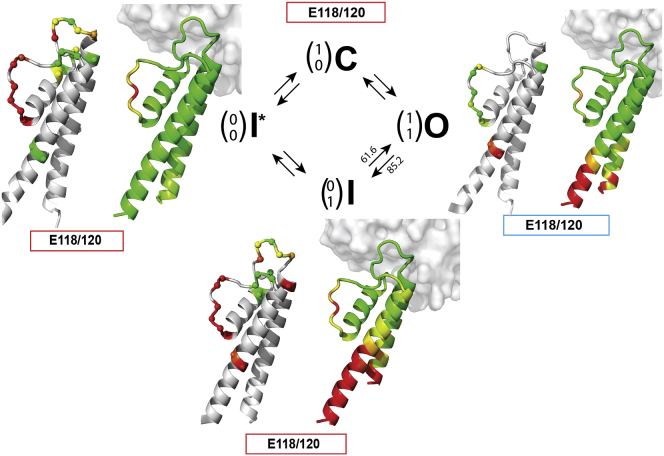
In Fig. 6, we compare the observed ssNMR changes for the Cα chemical shifts in the three channel states investigated here with the closed-conductive state at pH 7 on the residue-specific level. As a reference, we include RMSD changes. They were computed on the basis of the available x-ray structures and were interpreted in reference to the considered channel states (Fig. 6, right). As shown in Fig. 6, our ssNMR study demonstrates that the transition from the closed-conductive state to the open-conductive state (11) is accompanied by an opening of the activation gate and a concomitant protonation for E118 and E120. Importantly, the observed protonation levels seen at the activation gate in lipid bilayers correlate with crystal structures of KcsA mutants (13). Our results obtained using CL at low pH values (Fig. 2) support a selectivity filter conformation in the open-conductive state that involves structural changes at the entire selectivity filter (comprising residues Thr74–Gly79) as well as turret segments. Furthermore, we found evidence for local fluctuations around Val76. Of note, the profound conformational differences between the collapsed selectivity filter states (01 and 00 in Fig. 6) revealed in our lipid bilayer ssNMR experiments were not apparent in the KcsA crystal structures. In the crystal structures that were obtained by cocrystallization with antibodies (Fig. 6, right), conformational differences between inactivated and closed states of the channel were only manifested as local structural changes, largely restricted to Gly77 (see Fig. 4). These results suggest that in lipid bilayers, the turret region, nearby residues of the TM1 helix, and the selectivity filter together form a functional protein unit at the extracellular side of the K+ channel that can engage in significant and reversible conformational changes during K+ channel gating.
Figure 6.
ssNMR chemical-shift changes (left) are compared with RMSD changes (right) seen between KcsA crystal structures previously associated with the three states of the gating cycle (I∗, O, and I) relative to the closed-conductive state (C, top). For analysis of the x-ray data, published PDB structures (C: PDB: 1K4C; O: PDB: 3FB5; I: 3F5W; and I∗: PDB: 1K4D; see also Table S3) including stabilizing antibodies (shown in gray) were used. Color bars indicate E118/E120 protonation levels (red, deprotonated; blue, protonated) at the activation gate and Cα chemical-shift differences (ranging from red = 0 ppm to green = 0.6 ppm) with respect to the closed-conductive state (C). RMSD values (ranging from 0Å = green to 1.5Å = red) were calculated using PDB structures (see also Table S3). Transition-rate constants between open-collapsed and open-conductive states were obtained from electrophysiological experiments conducted on KcsA in a CL lipid bilayer (see Table S1). Transition-rate constants are expressed in s−1. To see this figure in color, go online.
Conclusions
Trapping functional substates of membrane-embedded proteins by crystallographic methods usually requires protein solubilization in membrane mimetics and protein stabilization by mutagenesis and/or addition of larger protein domains (50). Using electrophysiological and solid-state NMR experiments, we have shown that in the case of KcsA one can avoid such measures by using negatively charged lipids that reduce C-type inactivation and shift the conformational equilibrium under acidic conditions toward a more open-conductive state. According to our electrophysiological experiments, CL, which was previously identified as being important for membrane protein function (39, 51), was the most effective of the lipids investigated here in stabilizing the open conformation (Tau open: 11 ms, Tau closed: 16 ms; Fig. 6). Interestingly, these timescales are one to two orders of magnitude shorter than those reported for structural transitions in NMR studies on KcsA variants in dodecyl maltoside micelles (42, 43), underlining the prominent role of the lipid bilayer in channel function.
Our observation that the open-conductive state is stabilized by a change in lipid composition highlights the importance of the membrane bilayer for protein function and enabled us to characterize the entire KcsA gating cycle, as well as the conformational fluctuations that occur during that cycle. Crystal structures of the constitutively open KcsA mutants have suggested that the KcsA activation gate can display various degrees of activation gate opening (14). In line with electron paramagnetic resonance data (52, 53), our results show that the transition from neutral to acidic pH is accompanied by a gradual closing of the inactivation gate and an increasing opening of the activation gate. According to our ssNMR data, two channel states exist with a partially open activation gate and open and closed inactivation gates in CL bilayers at pH 4. A conformational exchange between these states would explain our previous low-temperature dynamic nuclear polarization data for the inactivated state (54) in asolectin bilayers where T75, which exhibits the largest chemical-shift changes (Fig. 2), remained invisible at low temperatures. In CL-rich bilayers, we could estimate the relative populations of both states by comparing signal intensities for open-conductive and open-inactivated states, leading to a relative distribution of conductive and closed inactivation gates of 40:60. These values are in remarkable agreement with the PO of 37% (Fig. 1) measured for KcsA in the CL lipid bilayer.
According to our ssNMR data, the conformational changes that occur at both the activation and inactivation gates are transmitted to the turret region, the channel segment that is stabilized by antibodies in all available x-ray structures available to date (Fig. 6, right). These observations may be in line with recent work suggesting that the inactivated channel state differs from the constricted KcsA structures seen at low potassium concentrations (55). The transition rates between the two principal gating states are described by conformational changes in the pore region (including the inactivation gate and turret). In detail, however, the degree of opening and closing of both gates is determined by the combined influence of protein sequence, lipid environment, and small molecules such as water molecules (56) or cations such as potassium and sodium. Together, they control the delicate conformational coupling of the inactivation and activation gates in membranes in each state of the potassium channel gating cycle.
Author Contributions
O.P. and M.B. conceived and designed experiments. A.V.P. conducted and analyzed electrophysiological measurements. E.A.W.v.d.C. performed and analyzed the ssNMR experiments. A.V.P., E.A.W.v.d.C., O.P., and M.B. wrote and edited the manuscript.
Acknowledgments
This work was supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (grants 700.11.344 and 700.58.102 to M.B.), the Deutsche Forschungsgemeinschaft (grants Po137, 40-1, and 41-1), and the National Institute of General Medical Sciences, National Institutes of Health(grant GM087519).
Editor: Mei Hong.
Footnotes
Elwin A. W. van der Cruijsen and Alexander V. Prokofyev contributed equally to this work.
Five figures and three tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)34274-6.
Contributor Information
Olaf Pongs, Email: oupon@t-online.de.
Marc Baldus, Email: m.baldus@uu.nl.
Supporting Material
References
- 1.Lee A.G. Biological membranes: the importance of molecular detail. Trends Biochem. Sci. 2011;36:493–500. doi: 10.1016/j.tibs.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Hunte C., Richers S. Lipids and membrane protein structures. Curr. Opin. Struct. Biol. 2008;18:406–411. doi: 10.1016/j.sbi.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Valiyaveetil F.I., Zhou Y., MacKinnon R. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry. 2002;41:10771–10777. doi: 10.1021/bi026215y. [DOI] [PubMed] [Google Scholar]
- 4.Weingarth M., Prokofyev A., Baldus M. Structural determinants of specific lipid binding to potassium channels. J. Am. Chem. Soc. 2013;135:3983–3988. doi: 10.1021/ja3119114. [DOI] [PubMed] [Google Scholar]
- 5.Laganowsky A., Reading E., Robinson C.V. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zubcevic L., Herzik M.A., Jr., Lee S.-Y. Cryo-electron microscopy structure of the TRPV2 ion channel. Nat. Struct. Mol. Biol. 2016;23:180–186. doi: 10.1038/nsmb.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bechara C., Nöll A., Robinson C.V. A subset of annular lipids is linked to the flippase activity of an ABC transporter. Nat. Chem. 2015;7:255–262. doi: 10.1038/nchem.2172. [DOI] [PubMed] [Google Scholar]
- 8.Dawaliby R., Trubbia C., Govaerts C. Allosteric regulation of G protein-coupled receptor activity by phospholipids. Nat. Chem. Biol. 2016;12:35–39. doi: 10.1038/nchembio.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaplan M., Narasimhan S., Baldus M. EGFR dynamics change during activation in native membranes as revealed by NMR. Cell. 2016;167:1241–1251.e11. doi: 10.1016/j.cell.2016.10.038. [DOI] [PubMed] [Google Scholar]
- 10.Doyle D.A., Morais Cabral J., MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Y., Morais-Cabral J.H., MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 12.Cordero-Morales J.F., Cuello L.G., Perozo E. Molecular determinants of gating at the potassium-channel selectivity filter. Nat. Struct. Mol. Biol. 2006;13:311–318. doi: 10.1038/nsmb1069. [DOI] [PubMed] [Google Scholar]
- 13.Cuello L.G., Jogini V., Perozo E. Structural basis for the coupling between activation and inactivation gates in K(+) channels. Nature. 2010;466:272–275. doi: 10.1038/nature09136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuello L.G., Jogini V., Perozo E. Structural mechanism of C-type inactivation in K(+) channels. Nature. 2010;466:203–208. doi: 10.1038/nature09153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y.S., Sompornpisut P., Perozo E. Structure of the KcsA channel intracellular gate in the open state. Nat. Struct. Biol. 2001;8:883–887. doi: 10.1038/nsb1001-883. [DOI] [PubMed] [Google Scholar]
- 16.Perozo E., Cortes D.M., Cuello L.G. Structural rearrangements underlying K+-channel activation gating. Science. 1999;285:73–78. doi: 10.1126/science.285.5424.73. [DOI] [PubMed] [Google Scholar]
- 17.Hong M., Zhang Y., Hu F. Membrane protein structure and dynamics from NMR spectroscopy. Annu. Rev. Phys. Chem. 2012;63:1–24. doi: 10.1146/annurev-physchem-032511-143731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ullrich S.J., Glaubitz C. Perspectives in enzymology of membrane proteins by solid-state NMR. Acc. Chem. Res. 2013;46:2164–2171. doi: 10.1021/ar4000289. [DOI] [PubMed] [Google Scholar]
- 19.Wang S., Ladizhansky V. Recent advances in magic angle spinning solid state NMR of membrane proteins. Prog. Nucl. Magn. Reson. Spectrosc. 2014;82:1–26. doi: 10.1016/j.pnmrs.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan M., Pinto C., Baldus M. Nuclear magnetic resonance (NMR) applied to membrane-protein complexes. Q. Rev. Biophys. 2016;49:e15. doi: 10.1017/S003358351600010X. [DOI] [PubMed] [Google Scholar]
- 21.Ader C., Schneider R., Baldus M. A structural link between inactivation and block of a K+ channel. Nat. Struct. Mol. Biol. 2008;15:605–612. doi: 10.1038/nsmb.1430. [DOI] [PubMed] [Google Scholar]
- 22.Ader C., Schneider R., Baldus M. Coupling of activation and inactivation gate in a K+-channel: potassium and ligand sensitivity. EMBO J. 2009;28:2825–2834. doi: 10.1038/emboj.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhate M.P., McDermott A.E. Protonation state of E71 in KcsA and its role for channel collapse and inactivation. Proc. Natl. Acad. Sci. USA. 2012;109:15265–15270. doi: 10.1073/pnas.1211900109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uysal S., Cuello L.G., Perozo E. Mechanism of activation gating in the full-length KcsA K+ channel. Proc. Natl. Acad. Sci. USA. 2011;108:11896–11899. doi: 10.1073/pnas.1105112108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cordero-Morales J.F., Jogini V., Perozo E. A multipoint hydrogen-bond network underlying KcsA C-type inactivation. Biophys. J. 2011;100:2387–2393. doi: 10.1016/j.bpj.2011.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Cruijsen E.A.W., Nand D., Baldus M. Importance of lipid-pore loop interface for potassium channel structure and function. Proc. Natl. Acad. Sci. USA. 2013;110:13008–13013. doi: 10.1073/pnas.1305563110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seidel K., Lange A., Baldus M. Protein solid-state NMR resonance assignments from (13C,13C) correlation spectroscopy. Phys. Chem. Chem. Phys. 2004;6:5090–5094. [Google Scholar]
- 28.Baldus M., Petkova A.T., Herzfeld J., Griffin R.G. Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Mol. Phys. 1998;95:1197–1207. [Google Scholar]
- 29.Fung B.M., Khitrin A.K., Ermolaev K. An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson. 2000;142:97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 30.Schneider R., Ader C., Baldus M. Solid-state NMR spectroscopy applied to a chimeric potassium channel in lipid bilayers. J. Am. Chem. Soc. 2008;130:7427–7435. doi: 10.1021/ja800190c. [DOI] [PubMed] [Google Scholar]
- 31.Platzer G., Okon M., McIntosh L.P. pH-dependent random coil (1)H, (13)C, and (15)N chemical shifts of the ionizable amino acids: a guide for protein pK a measurements. J. Biomol. NMR. 2014;60:109–129. doi: 10.1007/s10858-014-9862-y. [DOI] [PubMed] [Google Scholar]
- 32.Sigworth F.J., Sine S.M. Data transformations for improved display and fitting of single-channel dwell time histograms. Biophys. J. 1987;52:1047–1054. doi: 10.1016/S0006-3495(87)83298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuello L.G., Romero J.G., Perozo E. pH-dependent gating in the Streptomyces lividans K+ channel. Biochemistry. 1998;37:3229–3236. doi: 10.1021/bi972997x. [DOI] [PubMed] [Google Scholar]
- 34.Heginbotham L., LeMasurier M., Miller C. Single streptomyces lividans K(+) channels: functional asymmetries and sidedness of proton activation. J. Gen. Physiol. 1999;114:551–560. doi: 10.1085/jgp.114.4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meuser D., Splitt H., Schrempf H. Exploring the open pore of the potassium channel from Streptomyces lividans. FEBS Lett. 1999;462:447–452. doi: 10.1016/s0014-5793(99)01579-3. [DOI] [PubMed] [Google Scholar]
- 36.Marius P., Alvis S.J., Lee A.G. The interfacial lipid binding site on the potassium channel KcsA is specific for anionic phospholipids. Biophys. J. 2005;89:4081–4089. doi: 10.1529/biophysj.105.070755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marius P., Zagnoni M., Lee A.G. Binding of anionic lipids to at least three nonannular sites on the potassium channel KcsA is required for channel opening. Biophys. J. 2008;94:1689–1698. doi: 10.1529/biophysj.107.117507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marius P., de Planque M.R.R., Williamson P.T.F. Probing the interaction of lipids with the non-annular binding sites of the potassium channel KcsA by magic-angle spinning NMR. Biochim. Biophys. Acta. 2012;1818:90–96. doi: 10.1016/j.bbamem.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olofsson G., Sparr E. Ionization constants pKa of cardiolipin. PLoS One. 2013;8:e73040. doi: 10.1371/journal.pone.0073040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chakrapani S., Cordero-Morales J.F., Perozo E. A quantitative description of KcsA gating I: macroscopic currents. J. Gen. Physiol. 2007;130:465–478. doi: 10.1085/jgp.200709843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irizarry S.N., Kutluay E., Heginbotham L. Opening the KcsA K+ channel: tryptophan scanning and complementation analysis lead to mutants with altered gating. Biochemistry. 2002;41:13653–13662. doi: 10.1021/bi026393r. [DOI] [PubMed] [Google Scholar]
- 42.Imai S., Osawa M., Shimada I. Structural basis underlying the dual gate properties of KcsA. Proc. Natl. Acad. Sci. USA. 2010;107:6216–6221. doi: 10.1073/pnas.0911270107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imai S., Osawa M., Shimada I. Functional equilibrium of the KcsA structure revealed by NMR. J. Biol. Chem. 2012;287:39634–39641. doi: 10.1074/jbc.M112.401265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim D.M., Dikiy I., Nimigean C.M. Conformational heterogeneity in closed and open states of the KcsA potassium channel in lipid bicelles. J. Gen. Physiol. 2016;148 doi: 10.1085/jgp.201611602. jgp.201611602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ader C., Pongs O., Baldus M. Protein dynamics detected in a membrane-embedded potassium channel using two-dimensional solid-state NMR spectroscopy. Biochim. Biophys. Acta. 2010;1798:286–290. doi: 10.1016/j.bbamem.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 46.Baker L.A., Daniëls M., Baldus M. Efficient cellular solid-state NMR of membrane proteins by targeted protein labeling. J. Biomol. NMR. 2015;62:199–208. doi: 10.1007/s10858-015-9936-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Medeiros-Silva J., Mance D., Weingarth M. (1)H-detected solid-state NMR studies of water-inaccessible proteins in vitro and in situ. Angew. Chem. Int. Ed. Engl. 2016;55:13606–13610. doi: 10.1002/anie.201606594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chakrapani S., Cordero-Morales J.F., Perozo E. On the structural basis of modal gating behavior in K(+) channels. Nat. Struct. Mol. Biol. 2011;18:67–74. doi: 10.1038/nsmb.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keim P., Vigna R.A., Gurd F.R.N. Carbon 13 nuclear magnetic resonance of pentapeptides of glycine containing central residues of serine, threonine, aspartic and glutamic acids, asparagine, and glutamine. J. Biol. Chem. 1973;248:7811–7818. [PubMed] [Google Scholar]
- 50.Maeda S., Schertler G.F. Production of GPCR and GPCR complexes for structure determination. Curr. Opin. Struct. Biol. 2013;23:381–392. doi: 10.1016/j.sbi.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 51.van den Brink-van der Laan E., Boots J.W.P., de Kruijff B. Membrane interaction of the glycosyltransferase MurG: a special role for cardiolipin. J. Bacteriol. 2003;185:3773–3779. doi: 10.1128/JB.185.13.3773-3779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dalmas O., Hyde H.C., Perozo E. Symmetry-constrained analysis of pulsed double electron-electron resonance (DEER) spectroscopy reveals the dynamic nature of the KcsA activation gate. J. Am. Chem. Soc. 2012;134:16360–16369. doi: 10.1021/ja3069038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hulse R.E., Sachleben J.R., Perozo E. Conformational dynamics at the inner gate of KcsA during activation. Biochemistry. 2014;53:2557–2559. doi: 10.1021/bi500168u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koers E.J., van der Cruijsen E.A.W., Baldus M. NMR-based structural biology enhanced by dynamic nuclear polarization at high magnetic field. J. Biomol. NMR. 2014;60:157–168. doi: 10.1007/s10858-014-9865-8. [DOI] [PubMed] [Google Scholar]
- 55.Liu S., Focke P.J., Lockless S.W. Ion-binding properties of a K+ channel selectivity filter in different conformations. Proc. Natl. Acad. Sci. USA. 2015;112:15096–15100. doi: 10.1073/pnas.1510526112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weingarth M., van der Cruijsen E.A.W., Baldus M. Quantitative analysis of the water occupancy around the selectivity filter of a K+ channel in different gating modes. J. Am. Chem. Soc. 2014;136:2000–2007. doi: 10.1021/ja411450y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.