Abstract
Integrin-using rotaviruses bind MA104 cell surface α2β1 integrin via the Asp-Gly-Glu (DGE) sequence in virus spike protein VP4 and interact with αxβ2 integrin during cell entry through outer capsid protein VP7. Infection is inhibited by the α2β1 ligand Asp-Gly-Glu-Ala (DGEA) and the αxβ2 ligand Gly-Pro-Arg-Pro (GPRP), and virus-α2β1 binding is increased by α2β1 activation. In this study, we analyzed the effects of monomers and polymers containing DGEA-, GPRP-, and DGEA-related peptides on rotavirus binding and infection in intestinal (Caco-2) and kidney (MA104) cells and virus binding to recombinant α2β1. Blockade of rotavirus-cell binding and infection by peptides and anti-α2 antibody showed that Caco-2 cell entry is dependent on virus binding to α2β1 and interaction with αxβ2. At up to 0.5 mM, monomeric DGEA and DGAA inhibited binding to α2β1 and infection. At higher concentrations, DGEA and DGAA showed a reduced ability to inhibit virus-cell binding and infection that depended on virus binding to α2β1 but occurred without alteration in cell surface expression of α2, β2, or αvβ3 integrin. This loss of DGEA activity was abolished by genistein treatment and so was dependent on tyrosine kinase signaling. It is proposed that this signaling activated existing cell surface α2β1 to increase virus-cell attachment and entry. Polymeric peptides containing DGEA and GPRP or GPRP only were inhibitory to SA11 infection at approximately 10-fold lower concentrations than peptide monomers. As polymerization can improve peptide inhibition of virus-receptor interactions, this approach could be useful in the development of inhibitors of receptor recognition by other viruses.
The rotavirus spike protein VP4 and outer capsid protein VP7 contain tripeptide sequences that act as integrin ligands (14). VP4 is an important determinant of virulence, host cell tropism, receptor binding, and cell penetration (3, 15, 32, 37) and is cleaved by trypsin for activation of infectivity into two subunits, VP5* and VP8* (10, 19, 20). VP7 is also involved in cell entry. Integrins are αβ heterodimeric integral membrane glycoproteins important in cell adhesion, motility, spreading, differentiation, signaling, and survival (28) and are used by several virus families as cellular receptors (48). Integrins are often expressed in an inactive form that must be activated to bind ligand (28). Some animal rotaviruses, including monkey strains SA11 and RRV, also recognize terminal sialic acids as receptors (9, 16, 22). RRV binds sialosides through a galectin-like region in VP8* (17). Carbohydrates containing β-d-galactose and gangliosides are implicated in human and porcine rotavirus cell attachment and infection (26, 30, 43). Porcine rotavirus strain CRW-8 has been proposed to utilize a glycolipid receptor (31).
From the Entrez website database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi), 138 of 142 (97%) group A rotaviruses have Asp-Gly-Glu (DGE) at amino acid positions 308 to 310 in the VP5* subunit of VP4 (14). The DGE sequence is important for α2β1 integrin recognition by type I collagen (39, 46). Anti-α2β1 antibodies that block α2β1 function reduced the cell binding and/or infection of integrin-using rotaviruses by 30 to 70%, and type 1 collagen also inhibited infection (14, 24, 27). A peptide corresponding to the SA11 VP5* sequence Arg-Asp-Gly-Glu-Glu (RDGEE) inhibited SA11 infectivity in human colonic epithelial Caco-2 cells and monkey kidney epithelial MA104 cells in a dose-dependent fashion to 36% at 0.5 mM and 90% at 2.0 mM. At concentrations up to 0.5 mM, the α2β1 integrin ligand peptide Asp-Gly-Glu-Ala (DGEA) inhibited the binding of SA11, RRV, and human rotavirus strain Wa to recombinant α2β1 and MA104 cells by 34 to 55% but had no effect on porcine rotavirus CRW-8 binding to cells. This peptide also inhibited SA11, RRV, and Wa infection of MA104 cells by 27 to 40% at 0.5 mM but had no effect on CRW-8 infectivity (24). The effect of DGEA at higher concentrations on rotavirus-cell binding and entry has not been reported.
Rotaviruses and type I collagen bind α2β1 through the α2 subunit I domain (α2I). However, point mutation of amino acids 151, 221, and 254 that are necessary for type I collagen binding did not affect rotavirus binding, demonstrating that rotavirus binding to α2I is distinct from that of collagen (34). The binding of RRV VP5*, expressed as a glutathione-S-transferase fusion protein, to recombinant α2I protein required the presence of VP5* D308 and/or G309 in the DGE sequence, showing that either one or both of these residues are critical for rotavirus binding to α2β1 (24). MA104 cell binding by the nar RRV mutant also depended on the DG sequence in VP4 (51).
From the Entrez database, 640 of 648 (99%) group A rotaviruses, including all human strains, SA11, RRV, and CRW-8, have the αxβ2 integrin ligand sequence Gly-Pro-Arg (GPR) at amino acids 253 to 255 in a disintegrin-like domain of VP7 (14). In fibrinogen, GPR acts as a ligand for αxβ2 (14). The Gly-Pro-Arg-Pro (GPRP) peptide and anti-αxβ2 monoclonal antibodies each inhibited MA104 cell infection but not binding by rotaviruses that bind α2β1. Fibrinogen also inhibited infection of these rotaviruses (24). The αvβ3 integrin has also been shown to be recognized by VP7 during rotavirus cell entry (24, 25).
Many rotaviruses also contain the α4β1 integrin ligand sequences Leu-Asp-Val (LDV, in VP7) and Ile-Asp-Ala (IDA, in VP4), and recombinant, cell surface-expressed α4β1 has been shown to be capable of acting as a receptor for SA11 (14, 27).
Rotaviruses designated as integrin-using (including SA11, RRV, and Wa) bind MA104 cell surface α2β1 through VP4 and interact with αxβ2 and αvβ3 during cell entry via VP7. Integrin-independent rotaviruses, including CRW-8, do not recognize these integrins during MA104 cell attachment and entry. Use of these integrins is determined by the gene encoding VP4 and correlates with the VP4 serotype (24). The recently proposed models for the sequence of events occurring during cell attachment and early entry events for integrin-using rotaviruses involve initial carbohydrate recognition, followed by a closer protein-protein interaction of viral VP5* with α2β1, VP7 with αxβ2 and αvβ3, and membrane permeabilization mediated by fusogenic VP5* domains (23, 24, 36, 47, 48). Other integrins (including α1, α3, α5, α6, αL, αM, and β4 subunits) are not implicated in rotavirus cell attachment and entry (11, 14, 24, 27). There is evidence that VP5* also interacts with heat shock cognate protein 70 during cell entry (36). The majority of studies on which these models are based focused on rotavirus infection of MA104 cells rather than intestinal cells.
Integrin α2β1 is a pivotal receptor for activated T cells and neutrophils and is widely expressed on T and B cells, neuronal cells, epithelial cells, and adherent cell lines (2, 49). Intestinally, α2β1 is expressed apically on enterocytes in the lower villus, basolaterally along the length of the villus, and on M cells (38). Expression of α2β1 correlates with susceptibility of Caco-2 and MA104 cells to human and monkey rotavirus infection. Caco-2 cells expressed the highest surface level of α2β1 and produced the highest yield of infectious rotavirus (35). SA11 and RRV precipitated two surface proteins from Caco-2 and MA104 cells that were indistinguishable from the α2 and β1 immunoprecipitated by anti-integrin monoclonal antibodies (34). Caco-2 and MA104 cells also express αxβ2 (14, 35). SA11 is the only rotavirus to have been tested for integrin use during Caco-2 cell infection (14), and analysis of rotavirus binding to α2β1 on Caco-2 cells has not been reported. In this study, one aim was to determine the importance of α2β1 and αxβ2 for rotavirus binding and entry into Caco-2 cells.
The DGEA sequence in type I collagen was proposed to be an α2β1 recognition sequence on the basis that the short linear (nonhelical) peptide DGEA totally inhibited adhesion of epithelial cells and platelets to collagen at a concentration of 6 mM (46). However, although a longer helical peptide containing DGE has been reported to partially inhibit platelet adhesion to collagen, the original finding of DGEA blockade of platelet adhesion has not been consistently reproduced by others (33). A triple-helical peptide containing the type 1 collagen sequence Gly-Phe-Hyp-Gly-Glu-Arg (GFOGER) supported adhesion mediated by α2β1 and α2I (33). The crystal structure of α2I bound to a synthetic collagen-like peptide containing GFOGER showed that three loops on the upper surface of α2I that coordinate a metal ion also engage the collagen (18). Rotavirus VP5*, DGEA, and GFOGER all contain the Gly-Glu (GE) sequence. Thus, it was of interest to determine if GFOGER peptide inhibits rotavirus cell binding and infection mediated by α2β1.
One explanation for the incomplete inhibition of rotavirus-cell interactions by integrin ligand peptides DGEA and GPRP is that these small peptide monomers are relatively inefficient inhibitors of these interactions. The rotaviral DGE sequences are presented on VP4, which is represented as 60 regularly-spaced, dimeric spikes projecting from the outer (VP7) layer of particles. On the virion surface, VP7 GPR sequences are more numerous and present at higher density than DGE sequences (50). Thus, DGE and GPR peptide constructs in which the peptides are repetitively spaced, as in their virion context, could be more effective inhibitors of rotavirus-integrin interactions than the monomeric forms. During interaction with multiple viral binding sites by a complex polymer that contains multiple peptides, the first virus-peptide reaction would increase the probability of reaction at a second site. Such multiple interactions between peptide polymer and virus could increase the avidity of their binding (29).
Defined synthetic peptides containing B-cell and helper T-cell epitopes can be incorporated into very high molecular weight polymers by free radical-induced chain reaction polymerization (29, 42). This technology was successfully applied to the assembly of high-molecular-weight polymers of peptides from the M protein of group A streptococci (4). As inhibition of SA11 infection in MA104 cells by peptides RDGEE and GPRP is additive (14), assembly of heteropolymers of the peptides that recognize α2β1 and αxβ2 might be advantageous for inhibition of rotavirus infection.
In this study, we aimed to determine if α2β1 and αxβ2 are important in rotavirus binding and infection of Caco-2 cells and analyze the effects of integrin ligand peptide configuration, concentration and polymerization on the ability of peptides to inhibit virus-receptor interactions. These aims were achieved by analysis of the ability of GFOGER, DG-containing peptides, and polymers that contain DGEA, GPRP, or both DGEA and GPRP peptides to inhibit rotavirus binding and/or infection in MA104 and Caco-2 cells and virus binding to recombinant, cell surface-expressed α2β1.
MATERIALS AND METHODS
Cell lines and viruses.
The origins and maintenance of MA104, Caco-2, and K562 cells and the derivation of the K562 cells transfected with cDNA encoding empty vector (PBJ-K562), α2 (α2-K562), and α3 (α3-K562) used in this study have been described previously (14, 24, 27). The origins, cultivation in MA104 cells, and characterization of simian rotavirus P serotype 5B, G serotype 3 strain SA11; rhesus monkey rotavirus P5B, G3 strain RRV; human rotavirus P1A, G1 strain Wa, and porcine rotavirus P9 G3 strain CRW-8 have been described previously (12, 13).
Purchased peptides, reagents, and antibodies.
Peptides GFOGER, Gly-Phe-Hyp-Gly-Ala-Arg (GFOGAR), Asp-Gly-Ala-Ala (DGAA), and Gly-His-Arg-Pro (GHRP) that were ≥90% pure by high-performance liquid chromatography were purchased from Auspep, Victoria, Australia, and dissolved in Dulbecco's modified Eagle's medium at pH 7.5, as described previously (14). Genistein (4,5,7-trihydroxyisoflavanone) was purchased from Calbiochem and dissolved in dimethyl sulfoxide at a 100 mM concentration. Monoclonal antibody AK7 (α2I) was purchased from Becton Dickinson Pharmingen. Monoclonal antibodies P4H9-A11 (β2) and LM609 (αvβ3) were purchased from Chemicon (24, 35). Monoclonal antibodies W6/32 (major histocompatibility complex class I), and MOPC21 (isotype control) were obtained as described previously (34).
Peptide synthesis and purification.
Peptides were assembled manually with Fmoc chemistry throughout as described previously (44). In order to enable polymerization of individual peptides, an acryloyl group was attached to individual peptides; derivitization of peptides with the acryloyl (CH2 = CHCO-) group with acryloyl chloride is described elsewhere (42). Briefly, the solid-phase support with the protected peptide still attached was transferred to de-aerated N,N′-dimethylformamide and a 20-fold excess of diisopropylethylamine and a 10-fold molar excess of acryloyl chloride was added under nitrogen. Acryloylation was allowed to proceed for 1 h on ice and for a further 1 h at room temperature. Acryloyl-peptides were then cleaved from the resin, and side chain protecting groups were simultaneously removed by treatment with a mixture consisting of 88% trifluoroacetic acid, 5% phenol, 5% water, and 2% triisopropylsilane for 2 h at room temperature. Crude acryloyl-peptides were precipitated and washed in cold diethyl ether and then dissolved in 0.1% aqueous trifluoroacetic acid for purification by reverse-phase chromatography with a Vydac C4 column (10 by 250 mm) installed in a high-pressure liquid chromatography system. All peptides destined for polymerization were derivitized with 6-aminohexanoic acid (Ahx) prior to acryloylation with fluorenylmethoxycarbonyl-Ahx (Merck Pty. Ltd.). Ahx was incorporated as a spacer in order to distance the peptide from the polymer backbone to make it more available for interaction. A polymer of GPRP from which Ahx was omitted was also constructed.
Polymerization of acryloyl-peptides.
Acryloyl-peptides were mixed in a 1:50 molar ratio with acrylamide in 6 M guanidine HCl containing 2 mM EDTA and 0.5 M Tris-HCl, pH 8.3. The final concentration of acrylamide was 5% (wt/vol). The inclusion of an excess of acrylamide results in the formation of a linear polyacrylamide backbone in which peptides are interspersed (Fig. 1). Polymerization of acrylamide and acryloyl-peptides was initiated by the addition of an amount of ammonium persulfate equimolar with respect to acryloyl-peptide and 10% (vol/vol) N,N,N′,N′-tetramethylethylenediamine to give a final concentration of 2% (vol/vol). Polymerization was allowed to proceed for 18 h at room temperature. High-molecular-weight material resulting from the polymerization reaction was separated from low-molecular-weight reactants with a column (1.6 cm by 60 cm) of Sephadex G-25 (fractionation range, 1,000 to 5,000 Da) installed in a high-pressure liquid chromatography system as previously described (44). Separations were performed at a flow rate of 4 ml per min with 50 mM ammonium hydrogen carbonate, pH 8.1, as the eluant. Pooled fractions were freeze-dried.
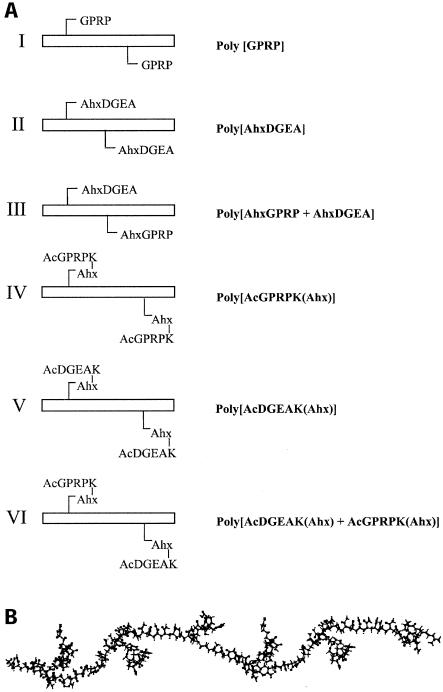
FIG. 1.
Schematic representation of the arrangement of peptides within polymers. Structures I to VI (A) indicate whether component peptides are pendant from their N termini (I to III) or their C-termini (IV to VI). The composition and quantitation of the peptides within the polymers and the stoichiometric distribution of DGEA and GPRP in polymers III and VI were confirmed by amino acid analysis. The distribution of these two peptides in these two heteropolymer structures is in all likelihood stochastic, as both peptides are short and similar in size. The schematics at left indicate a polymer unit of two peptides with n units constituting a complete polymer molecule. The size of polymers assembled in this way is about 500,000 Da (44). The molecular model (B) is an attempt to depict peptide arrangement along a polymer backbone but is not intended to indicate the actual three-dimensional structure of the polymer, which is unknown. The model depicts an energy-minimized polymer of I in which a single peptide occurs approximately every 10 acrylamide residues.
Confirmation of the amino acid composition of peptides and polymers and peptide quantitation were carried out by amino acid analysis. Peptides or peptide polymers were subjected to acid hydrolysis in the presence of 6 M hydrochloric acid to release individual amino acids from the polymer backbone into solution. The released amino acids were derivatized with fluorenylmethoxycarbonylchloride (Merck), separated, and quantitated with high-performance chromatography against an external and an internal standard (42). Characterization and quantitation of the two peptides (DGEA and GPRP) in polymers III and IV (Fig. 1A) was carried out by analyzing the amino acids that are distinct in each sequence (amino acids D, E, and A versus amino acids P and R). The molarity of each polymer was determined as for the monomeric peptides once the amounts of each peptide in the polymers were quantified by amino acid analysis. The results confirmed the fidelity of the composition of the peptides and polymers and also indicated stoichiometric incorporation of peptides in the cases (polymers III and VI) where two different peptides were copolymerized.
Orientation of peptides within polymers.
There were two distinct orientations of the peptides within the polymers. In polymers I, II, and III (Fig. 1A) the component peptides GPRP, AhxDGEA, and AhxGPRP were acryloylated at their N termini so that, within the resulting polymers, individual peptides were pendant from the polymer backbone through their N termini. In polymers IV, V, and VI (Fig. 1A), the component peptides were pendant from the backbone through their C termini. To produce the component peptides AcGPRPK(Ahx) and AcDGEAK(Ahx) for polymers IV, V, and VI, a lysine (K) residue with its side chain ɛ amino group protected with a methyltrityl group was inserted at the C terminus of peptides GPRP and DGEA. At the end of the synthesis, the N terminus of the peptides was acetylated (Ac), the methyltrityl group was selectively removed by treatment of the peptide resin with 1% (vol/vol) trifluoroacetic acid, and an Ahx group was coupled to the exposed ɛ amino group. Acryloylation of the amino group of Ahx and subsequent polymerization resulted in polymers IV, V, and VI.
Virus-cell binding and infectivity assays.
Assays of infectious rotavirus cell binding and infectivity and peptide and monoclonal antibody inhibition of virus-cell binding and infectivity were carried out with 5 × 105 cells (binding assays) and 104 cells (infectivity assays) as previously described (14, 24, 27). Rotavirus-cell binding assays were carried out at a rotavirus multiplicity of infection of 3.5 and infectivity assays at a multiplicity of infection of 0.02. Determination of these optimum multiplicities of infection has been described previously (24). Prior to virus addition, cells were treated at 37°C with peptides for 1 h or monoclonal antibodies for 2 h. None of the peptides caused K562 cell aggregation. The cell viability, microscopic appearance, and growth rates of K562, MA104, and Caco-2 cells were unaltered by peptide treatment.
For genistein experiments, MA104 cell monolayers in 96-well plates (104 cells/well) were incubated with 10 μM genistein in Dulbecco's modified Eagle's medium containing 0.01% (vol/vol) dimethyl sulfoxide or mock-treated with Dulbecco's modified Eagle's medium containing 0.01% (vol/vol) dimethyl sulfoxide for 3 h at 37°C prior to treatment with DGEA or GHRP peptides and assay of rotavirus infection as above. The genistein concentration used (10 μM) was that used previously to demonstrate involvement of tyrosine kinases in calcium mobilization induced by 0.5 to 2.0 mM DGEA peptide within dermal fibroblasts (P. Mineur and A. Guignandon, personal communication). This concentration of genistein has been shown to be at least 1 log below the concentration at which cellular mRNA expression is decreased (5). According to the manufacturer, genistein has a 50% inhibitory concentration of 2.6 μM against purified kinase. The genistein concentration used was within fourfold of the 50% inhibitory concentration.
The one-way analysis of variance test was used to assess the statistical significance of differences in virus-cell binding and infection. Significance was set at the 95% level. On graphs, results are expressed as a percentage of the virus titer in the absence of any treatment with peptide or monoclonal antibody, and error bars represent the standard deviation of the mean of at least three experiments.
Flow cytometry.
Flow cytometric analysis of the MA104 cell surface expression of integrins α2β1, β2, and αvβ3 was carried out with a two-step stain with monoclonal antibodies AK7, P4H9-A11, and LM609 as described previously (24, 35). For determination of the effect of peptide DGEA treatment on integrin expression, cells were treated with 2.0 mM peptide for 1 h at 37°C prior to flow cytometric analysis. Monitoring of the surface expression of α2β1 and α3β1 on PBJ-K562, α2-K562, and α3-K562 cells was carried out by flow cytometry as before (27).
RESULTS
Differential effects of α2β1 integrin ligand peptides DGEA, DGAA, and GFOGER on rotavirus infection of MA104 cells and binding to recombinant α2β1.
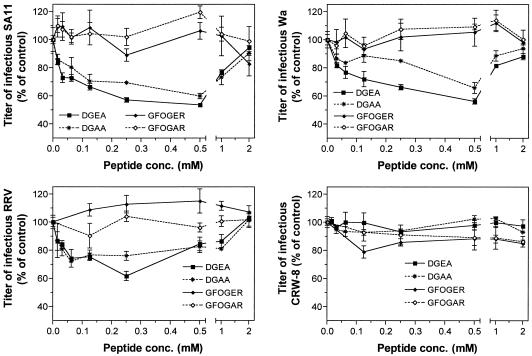
The effects on rotavirus infectivity in MA104 cells of cellular treatment with DGEA, DGAA, GFOGER, and control peptide GFOGAR at concentrations of 0.015 to 2.0 mM are shown in Fig. 2. DGEA inhibited SA11, RRV, and Wa infection in a dose-dependent fashion to maxima of 46, 39, and 44%, respectively, at 0.25 to 0.50 mM and had no effect on CRW-8 infectivity, as in previous studies. Treatment with 1.0 to 2.0 mM DGEA also did not affect CRW-8 infectivity. However, DGEA concentrations of 1.0 to 2.0 mM (SA11 and Wa) and 0.50 to 2.0 mM (RRV) produced a dose-dependent loss of DGEA-mediated inhibition of infectivity, resulting in an infectivity level similar to control (untreated) levels at 2.0 mM DGEA. A similar pattern was observed with DGAA, as this peptide inhibited SA11, RRV, and Wa infection to maxima of 40, 24, and 35%, respectively, at 0.25 to 0.50 mM and lost this activity at higher DGAA concentrations. However, DGAA inhibition was less than that of DGEA. Like DGEA, DGAA did not affect CRW-8 infectivity at any concentration tested. In contrast to the DG-containing peptides, neither GFOGER nor GFOGAR affected the infectivity of SA11, RRV, Wa, and CRW-8.
FIG. 2.
Effects of α2β1 integrin ligand peptides DGEA and GFOGER and peptide DGAA on the infectivity of SA11, RRV, Wa, and CRW-8 in MA104 cells. Each panel depicts the findings for a single virus strain. Cells were treated with peptide DGEA, DGAA, GFOGER, or GFOGAR prior to virus infection at concentrations ranging from 0.015 to 2.0 mM, as described in Materials and Methods. The infectivity titer of virus in the presence of peptide is expressed as a percentage of the titer obtained in the absence of peptide (control).
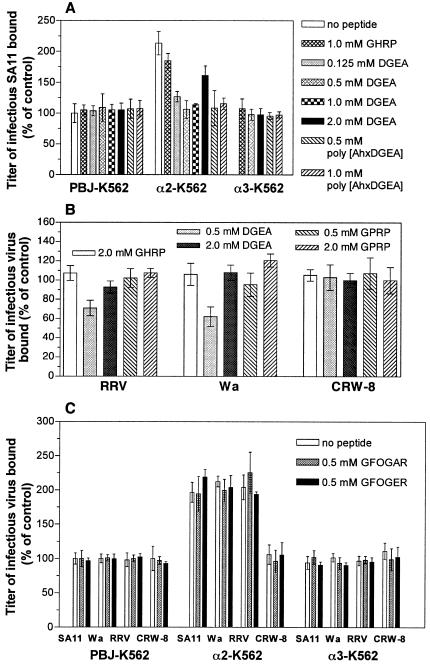
The specificity of the effects of peptides DGEA, GHRP (negative control), GPRP, GFOGER, and GFOGAR for rotavirus binding to α2β1 was determined by measurement of the levels of infectious virus binding to recombinant α2β1 expressed on the K562 cell surface (α2-K562). This virus binding was compared to the level bound to K562 cells transfected with the irrelevant integrin α3β1 (α3-K562) and to cells transfected with empty cDNA vector (PBJ-K562). As shown previously, titers of SA11, RRV, and Wa bound to α2-K562 cells were approximately 200% of the background levels bound to PBJ-K562 cells (Fig. 3). Treatment with 0.125 to 1.0 mM DGEA reduced the titer of SA11 bound by α2-K562 cells to background (PBJ-K562) levels (Fig. 3A). SA11 bound to α3-K562 cells at background levels, and control peptide GHRP did not affect SA11 binding to any cell line, as found previously (24). In contrast, 2.0 mM DGEA produced a loss of DGEA-mediated inhibition of SA11 binding to α2-K562 cells. Under these conditions, the level of infectious virus bound was similar to that bound to untreated α2-K562 cells and was increased significantly over that bound to α2-K562 cells at lower DGEA levels (Fig. 3A). Similar effects were seen for RRV and Wa (Fig. 3B).
FIG. 3.
Effects of α2β1 integrin ligand peptides DGEA and GFOGER on the binding of SA11, RRV, Wa, and CRW-8 rotaviruses to recombinant, cell surface-expressed α2β1. The cell line, virus or cell line, and virus strain tested are indicated on the x axis. (A), SA11 binding to α2β1 is specifically inhibited by treatment with DGEA (0.125, 0.50, and 1.0 mM) and poly[AhxDGEA] (0.50 and 1.0 mM) but enhanced by 2.0 mM DGEA. On the y axis, the infectious titer of SA11 bound to cells is expressed as a percentage of the titer bound to control PBJ-K562 cells in the absence of peptide (first open bar in the first group). (B) Effect of DGEA treatment on RRV, Wa, and CRW-8 binding to α2-K562 cells. For each virus strain, the infectious titer of virus bound to α2-K562 cells is expressed as a percentage of the titer bound to α2-K562 cells in the absence of peptide. The virus titer in α2-K562 cells is approximately 200% of that in PBJ-K562 cells, and thus the baseline is shifted compared with the data in panels A and C. (C) Peptides GFOGER and GFOGAR at 0.50 mM have no effect on SA11, Wa, RRV, and CRW-8 binding to α2β1. For each rotavirus strain, the infectious titer bound to cells is expressed as a percentage of the titer bound to control PBJ-K562 cells in the absence of peptide (first open bar in each group).
As shown previously, RRV and Wa binding to α2-K562 cells was reduced by 30 and 48%, respectively, by cellular treatment with 0.5 mM DGEA. However, at 2.0 mM DGEA, the inhibitory activity of DGEA was lost, and levels of RRV and Wa bound to α2-K562 cells were similar to those bound to untreated or GHRP-treated α2-K562 cells. CRW-8 binding to α2-K562 cells was unaffected by treatment with DGEA at 0.50 or 2.0 mM. The αxβ2 ligand GPRP also did not affect RRV, Wa, and CRW-8 binding to α2-K562 cells (Fig. 3B). Thus, treatment of cells with DGEA at 0.50 to 2.0 mM resulted in a loss of its ability to inhibit binding of SA11, RRV, and Wa to recombinant, cell surface-expressed α2β1. This loss of this DGEA activity was dependent on α2β1 binding by rotaviruses, as it did not occur in empty vector-transfected K562 cells or in K562 cells expressing recombinant α3β1, and CRW-8 infectivity was not affected by DGEA treatment.
The ability of the GFOGER and GFOGAR peptides to modulate rotavirus binding to α2β1 was evaluated (Fig. 3C). Neither peptide affected SA11, RRV, Wa, or CRW-8 binding to α2-K562, α3-K562, and PBJ-K562 cells, so these peptides did not affect rotavirus binding to α2β1.
Loss of integrin-using rotavirus infectivity at high DGEA concentrations depends on protein tyrosine kinase activity, not altered expression of rotavirus integrin receptors.
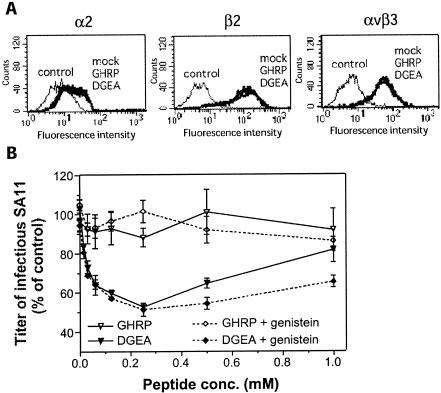
SA11 cell binding and infectivity have been shown to be increased in phorbol ester-treated α2-K562 cells due to increased cell surface expression of α2β1 (27, 45). One possibility is that the loss of inhibition of cell surface α2β1 binding by integrin-using rotaviruses and their infectivity at high DGEA concentrations could result from increased cellular α2β1, αxβ2, or αvβ3 expression induced by DGEA treatment. The ability of DGEA to alter cell surface expression of α2β1, αxβ2, and αvβ3 was examined by flow cytometry of DGEA-treated MA104 cells that were stained with anti-α2, anti-β2, and anti-αvβ3 monoclonal antibodies. As shown in Fig. 4A, DGEA treatment did not alter the expression of these integrins.
FIG. 4.
MA104 cell treatment with high concentrations of DGEA does not induce cell surface expression of α2β1, β2, and αvβ3 integrins (A), but the loss of DGEA-mediated inhibition of SA11 infectivity is inhibited by genistein (B). (A) Flow cytometric histograms. Cells were treated with 2.0 mM DGEA, control peptide GHRP, or mock treated, and their surface integrin expression levels were determined by flow cytometry after staining with monoclonal antibodies AK7 (α2I), P4H9-A11 (β2), and LM609 (αvβ3). Cells were also stained with isotype control monoclonal antibody MOPC21 at the same concentration as each test monoclonal antibody (control). Mock-treated and peptide-treated cells stained with MOPC21 gave identical histograms (control). In panel B, cells were mock treated or treated with 10 μM genistein prior to incubation with peptide DGEA or GHRP at concentrations ranging from 0.015 to 1.0 mM, followed by assay of SA11 rotavirus infectivity, as described in Materials and Methods. The infectivity titer of SA11 in cells treated with peptide with and without genistein is expressed as a percentage of the titer obtained in the absence of peptide or genistein.
DGEA and related peptides, including DGAA, have been shown to induce tyrosine kinase-dependent calcium mobilization in osteoblasts and fibroblasts at concentrations of ≥0.5 mM (40). Thus, the loss of DGEA and DGAA inhibitory activity against rotaviruses at peptide concentrations of ≥0.5 mM also might involve cellular tyrosine kinase activity. The tyrosine kinase inhibitor herbimycin A reduced the number of osteoblasts mobilizing calcium in response to DGEA peptide treatment (40), and genistein but not herbimycin A inhibited calcium mobilization in dermal fibroblasts (P. Mineur and A. Guignandon, personal communication). Genistein is a broad-spectrum inhibitor that competes at the ATP-binding site (1), whereas herbimycin A binds to src homology motifs and is more selective towards the src tyrosine kinase family (21). We therefore examined the effect of genistein treatment on the enhancement of SA11 infectivity mediated by DGEA in MA104 cells.
Initially, the effect of genistein at the concentration used (10 μM) on cell viability was determined. The numbers of viable cells before treatment, after genistein treatment, and after virus adsorption were 104 ± 103, so cell viability was unaffected by this genistein treatment and the overall experimental protocol. The genistein treatment had no effect on SA11 infectivity in MA104 cells. SA11 infectious titers in the absence of peptide were 496 ± 14 fluorescent cell-forming units/104 cells in genistein-treated cells and 484 ± 22 fluorescent cell-forming units/104 cells in mock-treated cells. As shown in Fig. 4B, genistein treatment significantly inhibited the SA11 infectivity increase induced by 0.50 and 1.0 mM DGEA (P = 0.02) but had no effect on the DGEA-mediated blockade of SA11 infection occurring at 0.015 to 0.25 mM DGEA (0.23 < P < 1.0). Thus, the SA11 infectivity increase induced by high DGEA concentrations was specifically inhibited by genistein treatment.
Infectivity of SA11, RRV, and Wa but not CRW-8 in Caco-2 cells is dependent on α2β1.
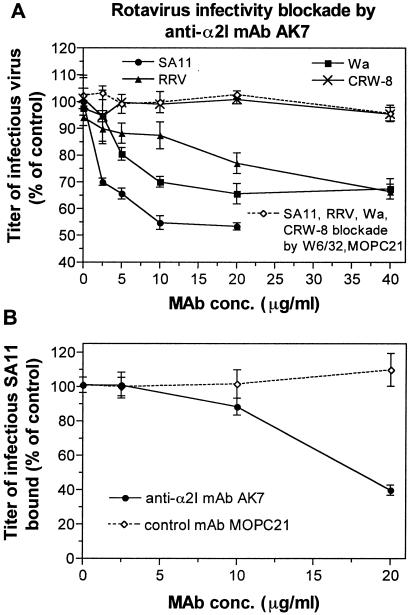
Rotaviruses have been demonstrated to use MA104 cell α2β1 and recombinant α2β1 on K562 and CHO cells for cell binding and infection by blockade with anti-α2 monoclonal antibody AK7 (8, 14, 24, 51). The ability of SA11, RRV, Wa, and CRW-8 to infect Caco-2 cells in the presence of anti-α2 monoclonal antibody AK7 was determined (Fig. 5A). Rotavirus infectious titers showed dose-dependent inhibition by monoclonal antibody AK7, to maxima of 47% at 20 μg of AK7 per ml (SA11), 34% at 40 μg/ml (RRV), and 33% at 20 μg/ml (Wa). CRW-8 infectivity was not affected by monoclonal antibody AK7 treatment. Monoclonal antibody AK7 also inhibited SA11 binding to Caco-2 cells by 61% at 20 μg/ml (Fig. 5B). Negative control monoclonal antibodies MOPC21 and anti-major histocompatibility complex class I monoclonal antibody W6/32 did not affect rotavirus binding and infection in Caco-2 cells. Thus, the previous classification of rotaviruses as integrin using or integrin independent extends to Caco-2 cell binding and infection by rotaviruses, and SA11 binds to Caco-2 cell α2β1 to facilitate infection.
FIG. 5.
Anti-α2 monoclonal antibody AK7 inhibits SA11, RRV, and Wa infectivity and SA11 binding in Caco-2 cells. (A) Effect of monoclonal antibodies AK7, W6/32, and control MOPC21 on SA11, RRV, Wa, and CRW-8 infection. For each virus strain, the infectivity titer of virus in MA104 cells treated with test or control monoclonal antibody is expressed as a percentage of the infectivity titer in the absence of monoclonal antibody. (B) AK7 inhibits SA11 binding to Caco-2 cells. The infectious titer of SA11 bound to cells treated with monoclonal antibody is expressed as a percentage of the titer bound in the absence of monoclonal antibody.
Effects of monomeric and polymeric peptides containing the integrin α2β1 ligand sequence DGE on SA11 infectivity in MA104 and Caco-2 cells.
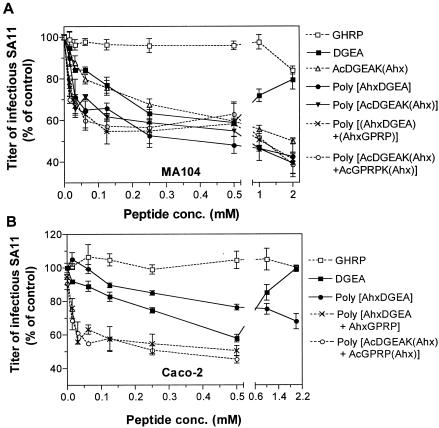
SA11 is the rotavirus type species and so was chosen for further studies. All monomeric and polymeric peptides containing DGEA inhibited SA11 infection at 0.015 to 0.50 mM concentrations in MA104 and Caco-2 cells (Fig. 6). Interestingly, at 1.0 to 2.0 mM, only the DGEA monomeric peptide produced a loss of infectivity inhibition in either cell line. As the N-terminally acetylated peptide DGEAK that had been acryloylated but not polymerized, AcDGEAK(Ahx), did not show loss of inhibition of SA11 infectivity at high peptide concentrations, the inhibition of peptide activity at high concentrations is not dependent on DGEA peptide concentration alone.
FIG. 6.
Blockade of SA11 infection of MA104 (A) and Caco-2 (B) cells by monomeric and polymeric peptides containing DGEA. Cells were treated with peptides prior to virus infection, at concentrations ranging from 0.015 to 2.0 mM, as described in Materials and Methods. The infectivity titer of virus in the presence of peptide is expressed as a percentage of the titer obtained in the absence of peptide (control).
In both cell lines, AcDGEAK(Ahx) and all the polymers containing DGEA showed increasing inhibition of SA11 infectivity at 1.0 and 2.0 mM, with maximal levels of inhibition at 2.0 mM (Fig. 6, Table 1). In MA104 cells, AcDGEAK(Ahx) inhibited SA11 infection similarly to DGEA at 0.015 to 0.50 mM but showed a higher maximal level of blockade (51%; Fig. 6A, Table 1). In MA104 cells, polymeric peptides poly[AhxDGEA] and poly[AcDGEAK(Ahx)] inhibited SA11 infectivity to significantly greater extents than AcDGEAK(Ahx) and DGEA at 0.031 to 0.125 mM (0.001 < P < 0.05). These two polymers also showed higher maximum inhibition levels, 58 and 61%, respectively, than AcDGEAK(Ahx) and DGEA (Fig. 6A, Table 1). In contrast, in Caco-2 cells, poly[AhxDGEA] at 0.50 mM inhibited SA11 infectivity to a lesser extent than DGEA at the same concentration (P < 0.01) and showed maximum inhibition at 2.0 mM of only 32% (Fig. 6B, Table 1).
TABLE 1.
Inhibition of SA11 cell binding and infectivity by monomeric and polymeric peptides containing DGEA, GPRP, or DGEA and GPRP
| Peptide | Cell line | Peptide inhibition of SA11 infectivity
|
Peptide inhibition of cell binding by SA11
|
||
|---|---|---|---|---|---|
| Maximum inhibition (%) | Peptide concn giving half-maximal inhibition (mM) | Maximum inhibition (%) | Peptide concn giving half-maximal inhibition (mM) | ||
| DGEA | MA104 | 46 | 0.150 | 46 | 0.046 |
| Caco-2 | 42 | 0.190 | 48 | 0.125 | |
| DGAA | MA104 | 40 | 0.062 | ND | ND |
| AcDGEAK(Ahx) | MA104 | 51 | 0.062 | ND | ND |
| Poly[AhxDGEA] | MA104 | 58 | 0.031 | 52 | 0.046 |
| Caco-2 | 32 | 0.250 | 40 | 0.031 | |
| Poly[AcDGEAK(Ahx)] | MA104 | 61 | 0.025 | ND | ND |
| GPRP | MA104 | 53 | 0.031 | 6 | —a |
| Caco-2 | 48 | 0.125 | 7 | — | |
| AhxGPRP | MA104 | 51 | 0.040 | ND | ND |
| AcGPRPK(Ahx) | MA104 | 51 | 0.031 | ND | ND |
| Poly[GPRP] | MA104 | 43 | 0.031 | ND | ND |
| Poly[AhxGPRP] | MA104 | 54 | 0.020 | 6 | — |
| Caco-2 | 55b | 0.015 | 1 | — | |
| Poly[AcGPRPK(Ahx)] | MA104 | 57 | 0.062 | −3 | — |
| Poly[(AhxDGEA)+(AhxGPRP)] | MA104 | 59 | 0.031 | ND | ND |
| Caco-2 | 50b | 0.015 | 50b | 0.015 | |
| Poly[AcDGEAK(Ahx)+AcGPRPK(Ahx)] | MA104 | 62 | 0.015 | 37b | 0.062 |
| Caco-2 | 55b | 0.015 | 57b | 0.015 | |
—, not calculated, as no peptide inhibition of SA11 binding was observed. ND, not done.
Assayed at 0.015 to 0.50 mM.
In both cell types, two polymeric peptides that incorporated both DGEA and GPRP, poly[(AhxDGEA)+(AhxGPRP)] and poly[AcDGEAK(Ahx)+AcGPRPK(Ahx)], were also significantly more inhibitory to SA11 infection at 0.031 to 0.125 mM than monomers containing DGEA (0.001 < P < 0.05). In Caco-2 cells only, these mixed polymers at 0.015 to 0.50 mM were also more inhibitory than poly[AhxDGEA], which contained DGEA alone (0.001 < P < 0.05). At 2.0 mM, these two polymers maximally inhibited SA11 infectivity, by 59 and 62%, respectively, in MA104 cells but showed lower maximum inhibitory levels in Caco-2 cells, 50 and 55%, respectively. In MA104 cells, the effects of polymers containing DGEA and GPRP were indistinguishable from those of polymers containing DGEA alone, although there was a trend for the maximum levels of inhibition of all the polymers containing DGEA (58 to 62%) to be greater than those exhibited by monomeric DGEA peptides (46 to 51%). In both cell types, half-maximal inhibition of SA11 infectivity by all the polymers except poly[AhxDGEA] occurred at 0.015 to 0.025 mM, whereas the two monomeric DGEA peptides and poly[AhxDGEA] showed half-maximal inhibition at 0.031 to 0.25 mM. In Caco-2 cells, polymers containing both DGEA and GPRP showed half-maximal inhibition of infection at concentrations 10-fold lower than those of peptides containing DGEA only (Table 1).
Effects of monomeric and polymeric peptides containing αxβ2 integrin ligand sequence GPR on SA11 infectivity in MA104 and Caco-2 cells.
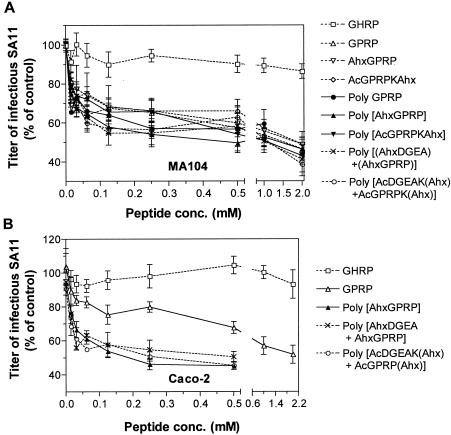
All monomers and polymers that contained GPRP inhibited SA11 infection of MA104 cells to a similar extent (Fig. 7A). Maximum inhibition by GPRP-containing peptides occurred at 2.0 mM and ranged from to 51 to 61% (Table 1). Half-maximal inhibition was observed at 0.015 to 0.062 mM peptide. As shown in Fig. 7B, monomeric GPRP peptide inhibited SA11 infectivity in Caco-2 cells to a lesser extent than in MA104 cells (Fig. 7A). In Caco-2 cells, three polymers containing GPRP, poly[AhxGPRP], poly[(AhxDGEA)+(AhxGPRP)], and poly[AcDGEAK(Ahx)+AcGPRPK(Ahx)], inhibited SA11 infectivity to a greater extent than monomeric GPRP at all peptide concentrations from 0.015 to 0.50 mM (0.001 < P < 0.05). At 0.50 mM, GPRP inhibited SA11 infectivity in Caco-2 cells by 32%, whereas the three polymers inhibited infectivity by 50 to 55% (P < 0.01). These polymers containing GPRP showed half-maximal inhibition in Caco-2 cells at 0.015 mM, whereas the half-maximal monomeric GPRP concentration was 0.125 mM (Table 1). Within each cell line, the inhibition profiles of the three polymers were indistinguishable (P > 0.05).
FIG. 7.
Blockade of SA11 infection of MA104 (A) and Caco-2 (B) cells by monomeric and polymeric peptides containing GPRP. See the legend to Fig. 6 for details.
Overall, N-terminal acetylation, lysine addition, and acryloylation of GPRP did not affect monomeric GPRP peptide inhibition of SA11 infectivity, in contrast to the situation with peptides containing DGEA. In both Caco-2 and MA104 cells, inclusion of DGEA did not enhance the infectivity blockade of GPRP-containing polymers. However, in contrast to MA104 cells, GPRP peptide polymerization did increase the level of SA11 infectivity inhibition in Caco-2 cells by GPRP-containing peptides and decreased the peptide concentration giving half-maximal inhibition by ninefold (Table 1).
Effects of monomeric and polymeric peptides containing integrin α2β1 ligand sequence DGE and/or αxβ2 integrin ligand sequence GPR on SA11 binding to MA104 and Caco-2 cells.
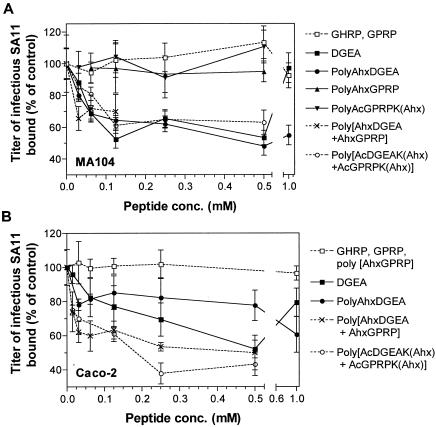
The effects of a selection of these peptides on SA11 binding to MA104 and Caco-2 cells were examined (Fig. 8, Table 1). In Caco-2 cells, the effects of monomeric DGEA on SA11 binding were similar to its effects in MA104 cells. At 0.015 to 0.5 mM, DGEA inhibited SA11 binding to Caco-2 cells in a dose-dependent fashion to 48%. DGEA lost its ability to inhibit SA11 binding at 1.0 mM. Three polymeric peptides containing DGEA (with or without GPRP) were tested: poly[AhxDGEA], poly[(AhxDGEA)+(AhxGPRP)], and poly[AcDGEAK(Ahx)+AcGPRPK(Ahx)]. As shown in Fig. 8A, these polymeric peptides inhibited SA11 binding to MA104 cells similarly to monomeric DGEA at 0.015 to 0.5 mM, with inhibition of 37 to 52% at 0.50 mM peptide and half-maximal inhibition at 0.046 to 0.062 mM.
FIG. 8.
Effects of monomeric and polymeric peptides containing DGEA and/or GPRP on SA11 binding to MA104 (A) and Caco-2 (B) cells. Cells were treated with peptides prior to virus binding, at concentrations ranging from 0.015 to 2.0 mM, as described in Materials and Methods. The titer of infectious SA11 bound to cells in the presence of peptide is expressed as a percentage of the titer bound in the absence of peptide (control).
In contrast, the two polymers containing both DGEA and GPRP inhibited SA11 binding to Caco-2 cells to a greater extent than poly[AhxDGEA] at 0.062 to 0.50 mM (0.001 < P < 0.01; Fig. 8B). At 0.50 mM, the two polymers containing both DGEA and GPRP inhibited SA11 binding to Caco-2 cells by 50 to 57%, whereas poly[AhxDGEA] showed 23% inhibition (Table 1). Poly[AhxDGEA] at high concentrations did not show loss of inhibition of SA11 binding to MA104 cells (Fig. 8A), Caco-2 cells (Fig. 8B), or recombinant α2β1 expressed on K562 cells (Fig. 3A). Monomeric peptide GPRP and two polymers containing GPRP but not DGEA had no effect on SA11 binding to MA104 (Fig. 8A) or Caco-2 (Fig. 8B) cells. Therefore, in both MA104 and Caco-2 cells, rotaviruses bind α2β1 and recognize αxβ2 at a postbinding stage during cell entry.
DISCUSSION
In this study, the nature, extent, and specificity of rotavirus usage of α2β1 and αxβ2 during infection were determined, and novel polymeric peptides were assembled and shown to be more effective and specific than monomeric DGEA and GPRP as inhibitors of rotavirus cell binding and entry.
Although both rotavirus and type I collagen bind to α2I, we found that a peptide consisting of the type I collagen sequence GFOGER, which is important for α2I binding, did not affect rotavirus binding to cellular α2β1. It has been shown that the Glu and Arg residues in GFOGER are essential for α2I binding (33). Although GFOGER lacked the collagen triple-helical structure conferred by the surrounding collagen sequence (18, 33), it did contain a GE sequence identical to that in the alternative type I collagen ligand sequence DGEA (which inhibits rotavirus-α2β1 binding) and the rotavirus VP5* DGE sequence. This suggests that the VP5* GE sequence is not essential for α2β1 recognition. The ability of peptide DGAA to inhibit cell binding and infection by integrin-using rotaviruses is consistent with the previous demonstration that RRV VP5* mutated at D308A and G309A no longer bound α2I (24). Overall, the results obtained with peptides GFOGER and DGAA suggest that D308 in VP5* is the major requirement for α2I binding by rotaviruses. This residue was previously proposed to be important for the interaction of the nar mutant of RRV with cellular receptors (51). An aspartate (D) or glutamate (E) residue is a critical feature of all integrin recognition sites (28).
An important and unexpected finding was that treatment of cells with high concentrations (>0.5 mM) of α2β1 integrin ligand peptide DGEA (and DGAA) resulted in a dose-dependent loss of DGEA inhibition of both virus binding to α2β1 and infectivity. Maximal transient elevation of intracellular calcium in osteoblasts occurs at 1.0 mM DGEA. The concentration curve is very steep: 0.30 mM DGEA did not elicit any change in cell calcium, and the threshold for stimulation was 0.60 mM (41). This fits closely with the contrasting effects of DGEA on rotavirus-cell binding and infectivity at <0.50 mM (inhibition) versus ≥0.50 mM (loss of inhibition). An anti-β1 monoclonal antibody inhibited DGEA-mediated calcium mobilization in dermal fibroblasts (41; P. Mineur and A. Guignandon, personal communication), and DGEA can be recognized by α2β1 and other platelet collagen receptors (39). Thus, it is unclear if α2β1, other β1 integrins, or other collagen receptors are components in the calcium signaling pathway induced by DGEA. However, it is likely that the DGEA enhancement of rotavirus binding to α2β1 and cell entry results from elevated intracellular calcium levels induced by these peptides.
The involvement of tyrosine kinase signaling in the loss of inhibitory activity of DGEA towards rotavirus infectivity is consistent with the established involvement of these kinases in DGEA-mediated calcium signaling (40) and inhibition of osteoblast differentiation (5). These findings support the proposal that 1.0 to 2.0 mM DGEA increases integrin-using rotavirus binding to α2β1 and infectivity through tyrosine kinase-dependent calcium signaling pathways.
On K562 cells, α2β1 exists in a partially activated state (6). Levels of SA11 binding to α2β1 on K562 cells are increased by α2β1 affinity activation, and increased rotavirus binding to α2β1 results in increased infectivity (24). Treatment with 2.0 mM DGEA did not affect MA104 cell surface expression of α2, β2, and αvβ3 but led to a loss of DGEA-mediated inhibition of virus binding to cellular α2β1 that was dependent on tyrosine kinase activity. Tyrosine kinase activity is required for α2β1 activation, which is mediated through inside-out cellular signaling in platelets (7). Thus, high DGEA concentrations induced tyrosine kinase signaling, which might have altered the activation state of existing cell surface α2β1 to a higher affinity. It is likely that activation of α2β1 would result in increased levels of virus binding and infectivity. It is possible that DGEA might be less effective as an inhibitor of α2β1-rotavirus binding when α2β1 is fully activated than when α2β1 is incompletely activated, which would explain the loss of DGEA inhibitory activity. However, it is not possible to exclude other effects of tyrosine kinase activation on the susceptibility of cells to rotavirus infection. An alternative explanation for the loss of DGEA inhibitory activity is that DGEA is removed more rapidly from the cell surface or degraded more rapidly following kinase signaling.
Genistein did not affect the level of SA11 infectivity blockade at lower DGEA concentrations. This shows that tyrosine kinase activity is not required for virus binding to α2β1 and provides evidence that the blockade of infectivity at low DGEA levels and the loss of blockade at high DGEA levels are distinct mechanistically. DGEA-containing monomers other than DGEA itself and DGEA-containing polymers were effective inhibitors of integrin-using rotavirus cell binding and infectivity at both low and high concentrations. Thus, the loss of DGEA inhibitory activity is independent of DGEA infectivity blockade and is not completely sequence specific but depends on the size and/or configuration of the DGEA peptide. It is clear from this study that many peptide inhibitors of rotavirus-α2β1 binding do not show loss of activity at high concentrations, so this can be avoided as needed.
Caco-2 cells provide one of the best models for human intestinal epithelial cells and are highly permissive for rotaviruses, particularly human and monkey strains. We show here for the first time that α2β1 is an important receptor for integrin-using rotaviruses on Caco-2 cells, as DGEA-containing peptides and anti-α2I monoclonal antibody AK7 reduced SA11 binding to Caco-2 cells by 41 to 61%. The infectivity of Wa and RRV but not CRW-8 in Caco-2 cells also was shown to be inhibited by 33 to 47% by an anti-α2I monoclonal antibody for the first time. In addition, GPRP peptides inhibited SA11 infection but not binding in Caco-2 cells, showing for the first time that αxβ2 is involved in SA11 cell entry in Caco-2 cells. The inability of any polymeric or monomeric GPRP peptide to affect rotavirus binding to MA104 and Caco-2 cells further supports the conclusion that αxβ2 is not involved in initial cell binding by rotaviruses. Thus, rotavirus attachment to and entry into both Caco-2 and MA104 cells involves α2β1 binding, and αxβ2 interaction at a postbinding stage. Consistent with this, in both cell lines, polymers containing both DGEA and GPRP or GPRP only were effective at a 10-fold-lower concentration than DGEA monomers as inhibitors of SA11 infection.
Several important differences between MA104 and Caco-2 cells in the effectiveness of integrin ligand peptides in preventing SA11 cell binding and infection were evident. The GPRP and DGEA monomers were less effective inhibitors of infection and the DGEA monomer was a less effective inhibitor of virus binding in Caco-2 cells than in MA104 cells. The higher surface expression of α2β1 and αxβ2 on Caco-2 cells than on MA104 cells (35) could explain these findings, as higher levels of peptide would be needed to block all available integrin sites that could bind virus. A polymer of DGEA, poly[AhxDGEA], was more effective than DGEA monomer in blockade of SA11 binding and infection in MA104 cells but less effective in Caco-2 cells. This could result from differences in the spacing of surface α2β1 molecules between MA104 and Caco-2 cells. Overall, in Caco-2 cells, peptide polymers, particularly those containing both DGEA and GPRP, were more effective inhibitors of SA11 cell binding and infectivity than monomers, whereas in MA104 cells, this difference was not as marked. Thus, it is important to evaluate inhibitors of rotavirus-cell binding and entry in an intestinal cell line.
Inhibition of SA11 infection of MA104 cells by peptides RDGEE and GPRP is additive (14). In contrast, polymeric peptides containing both DGEA and GPRP were not usually more efficient in blockade of SA11 infection in Caco-2 and MA104 cells than polymers containing either DGEA or GPRP. This could relate to the constraints imposed in the polymers on the ability of both DGEA and GPRP to effectively inhibit integrin interactions with the spatially distinct rotavirus VP5* and VP7 proteins.
The molar concentrations of the peptides incorporated into the polymers were determined, and their stoichiometry could be predicted, but the overall structure in solution of the polymers used here is unknown. Although they are depicted as linear molecules in Fig. 1, we have no information as to their fibrous or globular nature. The geometry of the polymers will determine the efficiency of their inhibition of virus-receptor interactions, and it is possible that polymers of different geometries but similar stoichiometries will exhibit different efficiencies of inhibition. Polymers in which peptides are positioned so that their spacing more closely represents the cell surface spacing of α2β1 molecules will probably be efficient inhibitors of viral attachment, but the optimal design of such inhibitors will depend on the availability of polymer structural data.
The findings reported here have significant implications for the proposed models of rotavirus cell entry. As tyrosine kinase activity is involved in the loss of the inhibitory activity of the DGEA peptide at high concentrations, it is likely that signaling is involved in rotavirus binding and infectivity. Our Caco-2 cell studies show clearly that rotaviruses use α2β1 and αxβ2 to infect intestinal cells in a process that is similar in key features to that in MA104 cells, so the scope of the models can now be extended to include intestinal epithelial cells, the targets of rotavirus infection. The ability of individual peptide polymers containing DGEA and/or GPRP to inhibit rotavirus infectivity by more than 60% in Caco-2 and MA104 cells provides additional evidence of the importance of α2β1 and αxβ2 in the rotavirus cell attachment and entry process. Our demonstration that polymerization increases the effectiveness of these peptides as rotavirus inhibitors by up to 10-fold suggests that it might be possible to develop more effective inhibitors of rotavirus-integrin interactions. Peptide polymerization might be useful in the development of inhibitors of receptor recognition by other viruses, including those that use integrins during cell attachment and entry.
Acknowledgments
We are grateful to A. Guignandon and B. Nusgens for helpful discussions, D. Lee for assistance with peptide synthesis and blockade experiments, P. Witterick for carrying out monoclonal antibody AK7 blockade of SA11 binding to Caco-2 cells, A. Brooks for monoclonal antibody W6/32, and P. Halasz for performing experiments with W6/32.
This work was supported by project grants 980635, 980664, and 208900 from the National Health and Medical Research Council (NHMRC) of Australia. B.S.C. and D.C.J. are Research Fellows of the NHMRC.
REFERENCES
- 1.Akiyama, T., J. Ishida, S. Nakagawa, H. Ogawara, S. Watanabe, N. Itoh, M. Shibuya, and Y. Fukami. 1987. Genistein, a specific inhibitor of tyrosine-specific protein kinases. J. Biol. Chem. 262:5592-5595. [PubMed] [Google Scholar]
- 2.Andreasen, S. O., A. R. Thomsen, V. E. Koteliansky, T. I. Novobrantseva, A. G. Sprague, A. R. de Fougerolles, and J. P. Christensen. 2003. Expression and functional importance of collagen-binding integrins, alpha 1 beta 1 and alpha 2 beta 1, on virus-activated T cells. J. Immunol. 171:2804-2811. [DOI] [PubMed] [Google Scholar]
- 3.Bass, D. M., E. R. Mackow, and H. B. Greenberg. 1991. Identification and partial characterization of a rhesus rotavirus binding glycoprotein on murine enterocytes. Virology 183:602-610. [DOI] [PubMed] [Google Scholar]
- 4.Brandt, E. R., K. S. Sriprakash, R. I. Hobb, W. A. Hayman, W. Zeng, M. R. Batzloff, D. C. Jackson, and M. F. Good. 2000. New multi-determinant strategy for a group A streptococcal vaccine designed for the Australian Aboriginal population. Nat. Med. 6:455-459. [DOI] [PubMed] [Google Scholar]
- 5.Celic, S., Y. Katayama, P. J. Chilco, T. J. Martin, and D. M. Findlay. 1998. Type I collagen influence on gene expression in UMR106-06 osteoblast-like cells is inhibited by genistein. J. Endocrinol. 158:377-388. [DOI] [PubMed] [Google Scholar]
- 6.Chan, B. M., and M. E. Hemler. 1993. Multiple functional forms of the integrin VLA-2 can be derived from a single alpha 2 cDNA clone: interconversion of forms induced by an anti-beta 1 antibody. J. Cell Biol. 120:537-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H., and M. L. Kahn. 2003. Reciprocal signaling by integrin and nonintegrin receptors during collagen activation of platelets. Mol. Cell. Biol. 23:4764-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciarlet, M., S. E. Crawford, E. Cheng, S. E. Blutt, D. A. Rice, J. M. Bergelson, and M. K. Estes. 2002. VLA-2 (α2β1) integrin promotes rotavirus entry into cells but is not necessary for rotavirus attachment. J. Virol. 76:1109-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciarlet, M., J. E. Ludert, M. Iturriza-Gomara, F. Liprandi, J. J. Gray, U. Desselberger, and M. K. Estes. 2002. Initial interaction of rotavirus strains with N-acetylneuraminic (sialic) acid residues on the cell surface correlates with VP4 genotype, not species of origin. J. Virol. 76:4087-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark, S. M., J. R. Roth, M. L. Clark, B. B. Barnett, and R. S. Spendlove. 1981. Trypsin enhancement of rotavirus infectivity: mechanism of enhancement. J. Virol. 39:816-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coulson, B. S. 1997. Effects of Workshop monoclonal antibodies on rotavirus infection of cells, p. 391-393. In T. Kishimoto, H. Kikutani, A. E. G. K. von dem Borne, S. M. Goyert, D. Y. Mason, M. Miyasaka, L. Moretta, K. Okumura, S. Shaw, T. A. Springer, K. Sugamura, and H. Zola (ed.), Leucocyte typing VI. Garland Publishing, Inc., New York, N.Y.
- 12.Coulson, B. S., K. J. Fowler, R. F. Bishop, and R. G. Cotton. 1985. Neutralizing monoclonal antibodies to human rotavirus and indications of antigenic drift among strains from neonates. J. Virol. 54:14-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coulson, B. S., and C. Kirkwood. 1991. Relation of VP7 amino acid sequence to monoclonal antibody neutralization of rotavirus and rotavirus monotype. J. Virol. 65:5968-5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coulson, B. S., S. L. Londrigan, and D. J. Lee. 1997. Rotavirus contains integrin ligand sequences and a disintegrin-like domain that are implicated in virus entry into cells. Proc. Natl. Acad. Sci. USA 94:5389-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crawford, S. E., M. Labbe, J. Cohen, M. H. Burroughs, Y. J. Zhou, and M. K. Estes. 1994. Characterization of virus-like particles produced by the expression of rotavirus capsid proteins in insect cells. J. Virol. 68:5945-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delorme, C., H. Brussow, J. Sidoti, N. Roche, K. A. Karlsson, J. R. Neeser, and S. Teneberg. 2001. Glycosphingolipid binding specificities of rotavirus: identification of a sialic acid-binding epitope. J. Virol. 75:2276-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dormitzer, P. R., Z. Y. Sun, G. Wagner, and S. C. Harrison. 2002. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 21:885-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emsley, J., C. G. Knight, R. W. Farndale, M. J. Barnes, and R. C. Liddington. 2000. Structural basis of collagen recognition by integrin α2β1. Cell 101:47-56. [DOI] [PubMed] [Google Scholar]
- 19.Espejo, R. T., S. Lopez, and C. Arias. 1981. Structural polypeptides of simian rotavirus SA11 and the effect of trypsin. J. Virol. 37:156-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estes, M. K., D. Y. Graham, and B. B. Mason. 1981. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J. Virol. 39:879-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukazawa, H., P. M. Li, C. Yamamoto, Y. Murakami, S. Mizuno, and Y. Uehara. 1991. Specific inhibition of cytoplasmic protein tyrosine kinases by herbimycin A in vitro. Biochem. Pharmacol. 42:1661-1671. [DOI] [PubMed] [Google Scholar]
- 22.Fukudome, K., O. Yoshie, and T. Konno. 1989. Comparison of human, simian, and bovine rotaviruses for requirement of sialic acid in hemagglutination and cell adsorption. Virology 172:196-205. [DOI] [PubMed] [Google Scholar]
- 23.Golantsova, N. E., E. E. Gorbunova, and E. R. Mackow. 2004. Discrete domains within the rotavirus VP5* direct peripheral membrane association and membrane permeability. J. Virol. 78:2037-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham, K. L., P. Halasz, Y. Tan, M. J. Hewish, Y. Takada, E. R. Mackow, M. K. Robinson, and B. S. Coulson. 2003. Integrin-using rotaviruses bind α2β1 integrin α2 I domain via VP4 DGE sequence and recognize αXβ2 and αVβ3 by using VP7 during cell entry. J. Virol. 77:9969-9978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerrero, C. A., E. Mendez, S. Zarate, P. Isa, S. Lopez, and C. F. Arias. 2000. Integrin alpha(v)beta(3) mediates rotavirus cell entry. Proc. Natl. Acad. Sci. USA 97:14644-14649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo, C. T., O. Nakagomi, M. Mochizuki, H. Ishida, M. Kiso, Y. Ohta, T. Suzuki, D. Miyamoto, K. I. Hidari, and Y. Suzuki. 1999. Ganglioside GM(1a) on the cell surface is involved in the infection by human rotavirus KUN and MO strains. J. Biochem. (Tokyo) 126:683-688. [DOI] [PubMed] [Google Scholar]
- 27.Hewish, M. J., Y. Takada, and B. S. Coulson. 2000. Integrins α2β1 and α4β1 can mediate SA11 rotavirus attachment and entry into cells. J. Virol. 74:228-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hynes, R. O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell 110:673-687. [DOI] [PubMed] [Google Scholar]
- 29.Jackson, D. C., N. O'Brien-Simpson, N. J. Ede, and L. E. Brown. 1997. Free radical induced polymerization of synthetic peptides into polymeric immunogens. Vaccine 15:1697-1705. [DOI] [PubMed] [Google Scholar]
- 30.Jolly, C. L., B. M. Beisner, and I. H. Holmes. 2000. Rotavirus infection of MA104 cells is inhibited by Ricinus lectin and separately expressed single binding domains. Virology 275:89-97. [DOI] [PubMed] [Google Scholar]
- 31.Jolly, C. L., B. M. Beisner, E. Ozser, and I. H. Holmes. 2001. Non-lytic extraction and characterisation of receptors for multiple strains of rotavirus. Arch. Virol. 146:1307-1323. [DOI] [PubMed] [Google Scholar]
- 32.Kirkwood, C. D., R. F. Bishop, and B. S. Coulson. 1998. Attachment and growth of human rotaviruses RV-3 and S12/85 in Caco-2 cells depend on VP4. J. Virol. 72:9348-9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knight, C. G., L. F. Morton, D. J. Onley, A. R. Peachey, A. J. Messent, P. A. Smethurst, D. S. Tuckwell, R. W. Farndale, and M. J. Barnes. 1998. Identification in collagen type I of an integrin alpha2 beta1-binding site containing an essential GER sequence. J. Biol. Chem. 273:33287-33294. [DOI] [PubMed] [Google Scholar]
- 34.Londrigan, S. L., K. L. Graham, Y. Takada, P. Halasz, and B. S. Coulson. 2003. Monkey rotavirus binding to α2β1 integrin requires the alpha2 I domain and is facilitated by the homologous beta1 subunit. J. Virol. 77:9486-9501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Londrigan, S. L., M. J. Hewish, M. J. Thomson, G. M. Sanders, H. Mustafa, and B. S. Coulson. 2000. Growth of rotaviruses in continuous human and monkey cell lines that vary in their expression of integrins. J. Gen. Virol. 81:2203-2213. [DOI] [PubMed] [Google Scholar]
- 36.Lopez, S., and C. F. Arias. 2004. Multistep entry of rotavirus into cells: a Versaillesque dance. Trends Microbiol. 12:271-278. [DOI] [PubMed] [Google Scholar]
- 37.Ludert, J. E., N. Feng, J. H. Yu, R. L. Broome, Y. Hoshino, and H. B. Greenberg. 1996. Genetic mapping indicates that VP4 is the rotavirus cell attachment protein in vitro and in vivo. J. Virol. 70:487-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lussier, C., N. Basora, Y. Bouatrouss, and J. F. Beaulieu. 2000. Integrins as mediators of epithelial cell-matrix interactions in the human small intestinal mucosa. Microsc. Res. Technol. 51:169-178. [DOI] [PubMed] [Google Scholar]
- 39.Luzak, B., J. Golanski, M. Rozalski, M. A. Bonclerand, and C. Watala. 2003. Inhibition of collagen-induced platelet reactivity by DGEA peptide. Acta Biochim. Pol. 50:1119-1128. [PubMed] [Google Scholar]
- 40.McCann, T. J., W. T. Mason, M. C. Meikle, and F. McDonald. 1997. A collagen peptide motif activates tyrosine kinase-dependent calcium signalling pathways in human osteoblast-like cells. Matrix Biol. 16:273-283. [DOI] [PubMed] [Google Scholar]
- 41.McCann, T. J., G. Terranova, J. W. Keyte, S. S. Papaioannou, W. T. Mason, M. C. Meikle, and F. McDonald. 1998. An analysis of Ca2+ release by DGEA: mobilization of two functionally distinct internal stores in Saos-2 cells. Am. J. Physiol. 275:C33-41. [DOI] [PubMed] [Google Scholar]
- 42.O'Brien-Simpson, N. M., N. J. Ede, L. E. Brown, J. Swan, and D. C. Jackson. 1997. Polymerization of unprotected synthetic peptides: a view toward synthetic peptide vaccines. J. Am. Chem. Soc. 119:1183-1188. [Google Scholar]
- 43.Rolsma, M. D., T. B. Kuhlenschmidt, H. B. Gelberg, and M. S. Kuhlenschmidt. 1998. Structure and function of a ganglioside receptor for porcine rotavirus. J. Virol. 72:9079-9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadler, K., W. Zeng, and D. C. Jackson. 2002. Synthetic peptide epitope-based polymers: controlling size and determining the efficiency of epitope incorporation. J. Peptide Res. 60:150-158. [DOI] [PubMed] [Google Scholar]
- 45.Sanders, G. M., M. J. Hewish, and B. S. Coulson. 2001. Phorbol dibutyrate-induced megakaryocytic differentiation increases susceptibility of K562 cells to SA11 rotavirus infection. Arch. Virol. 146:1831-1840. [DOI] [PubMed] [Google Scholar]
- 46.Staatz, W. D., K. F. Fok, M. M. Zutter, S. P. Adams, B. A. Rodriguez, and S. A. Santoro. 1991. Identification of a tetrapeptide recognition sequence for the alpha 2 beta 1 integrin in collagen. J. Biol. Chem. 266:7363-7367. [PubMed] [Google Scholar]
- 47.Tihova, M., K. A. Dryden, A. R. Bellamy, H. B. Greenberg, and M. Yeager. 2001. Localization of membrane permeabilization and receptor binding sites on the VP4 hemagglutinin of rotavirus: implications for cell entry. J. Mol. Biol. 314:985-992. [DOI] [PubMed] [Google Scholar]
- 48.Triantafilou, K., Y. Takada, and M. Triantafilou. 2001. Mechanisms of integrin-mediated virus attachment and internalization process. Crit. Rev. Immunol. 21:311-322. [PubMed] [Google Scholar]
- 49.Werr, J., J. Johansson, E. E. Eriksson, P. Hedqvist, E. Ruoslahti, and L. Lindbom. 2000. Integrin alpha(2)beta(1) (VLA-2) is a principal receptor used by neutrophils for locomotion in extravascular tissue. Blood 95:1804-1809. [PubMed] [Google Scholar]
- 50.Yeager, M., J. A. Berriman, T. S. Baker, and A. R. Bellamy. 1994. Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J. 13:1011-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zarate, S., R. Espinosa, P. Romero, C. A. Guerrero, C. F. Arias, and S. Lopez. 2000. Integrin alpha2beta1 mediates the cell attachment of the rotavirus neuraminidase-resistant variant nar3. Virology 278:50-54. [DOI] [PubMed] [Google Scholar]