Abstract
We obtained 157 cloned cell lines persistently infected with Sendai virus; these cell lines were generated independently of each other. Infectious viruses could be isolated from 123 of these cloned cell lines by inoculation of culture fluids or infected cells into embryonated eggs. The majority of the viruses carried by cells persistently infected with viruses showed high cytotoxicity and did not have the ability to establish persistent infection. The association of carried virus with cells became stronger and virus isolation correspondingly became more difficult as cells persistently infected with virus were subcultured. Viruses derived from virus-infected cells eventually acquired the ability to establish persistent infection, although the ways in which the viruses acquired this ability varied. The viruses also acquired temperature sensitivity as persistently infected cells were subcultured. First, the hemagglutinin-neuraminidase and M proteins acquired temperature sensitivity, and then the polymerase(s) did so. The M proteins were localized in the nuclei of cells infected with cloned viruses that had the ability to establish persistent infection. Cells infected with viruses capable of establishing persistent infection showed no or slight staining by terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling. Specific amino acid substitutions accumulated in the M protein and the L protein as virus-infected cells were subcultured. This study shows that there is an unstable dynamic phase at an early stage of the establishment of persistent Sendai virus infection (steady state), and then viruses capable of establishing persistent infection are selected.
Sendai virus (SeV) infection and replication lead to a strong cytopathic effect, with the subsequent death of host cells. Membrane fusion and apoptosis have been reported to be mechanisms of SeV-induced cytotoxicity. SeV infection triggers an apoptotic program in target cells, leading to the death of host cells without impairment of the viral life cycle. On the other hand, paramyxoviruses, particularly simian virus 5 (SV5) and SeV, can cause a persistent, productive infection in primary cell cultures that does not kill cells or shut off cellular RNA or protein synthesis (1). Some paramyxoviruses, such as SeV, bovine parainfluenza type 3 virus, SV5, Hendra virus, and Nipah virus, persistently infect natural hosts, which generally show no or low-level pathogenic signs.
When cells such as HeLa and baby hamster kidney (BHK) cells are infected with SeV at a relatively high multiplicity of infection (MOI), almost all of the cells die, but a few survive. The surviving cells, subcultured for a long time, develop into cells persistently infected with SeV (steady-state infection) (9). A carrier culture of HVJ (SeV)-infected BHK cells (BHK-HVJ cells) was originally obtained by prolonged cultivation of BHK cells which had survived HVJ (Nagoya strain) infection (9), and BHK-HVJ cells have been subcultured for a long period since then. In a recent study (10), Nishio et al. reported the characterization of L929 cells persistently infected with SeV and the pi strain of SeV (SeVpi) isolated from BHK-HVJ cells. It has been commonly accepted that the temperature-sensitive phenotype and low cytopathogenesis of cells persistently infected with SeV are caused by the M and/or hemagglutinin-neuraminidase (HN) proteins (6, 8, 16, 17). However, recombinant SeVs having SeVpi M protein and/or SeVpi HN protein do not show temperature sensitivity and are incapable of establishing persistent infection (5). None of the virus-specific polypeptides are detected when cells are incubated at 38°C. In addition, the virus proteins in cells infected with SeVpi are not pulse-labeled at 38°C, indicating that the temperature-sensitive step is at an early stage of infection. These properties may reflect persistent infection maintained for more than 10 years. Therefore, analyses of the early stage of the process of establishment of persistent infection are required for understanding the molecular mechanism by which persistent SeV infection is established.
In this study, L929 or BHK cells were infected with SeV (Nagoya or Z strain) and incubated for 4 days, when almost all of the cells died. Subsequently, cell cloning was carried out by using a limiting-dilution method. We isolated 154 cloned cells infected persistently with SeV and analyzed the properties of the cloned cells and viruses isolated from the cloned cells. This study showed that there is an unstable dynamic phase at an early stage of the process of establishment of persistent SeV infection (steady state), and then viruses capable of establishing persistent infection are selected.
MATERIALS AND METHODS
Viruses and cells.
The SeVs used in this study were the Nagoya (wild) and Z strains. The cells used in the present study were mouse L929 cells and BHK cells, which were grown at 35°C in Eagle's minimum essential medium (MEM) fortified with 5% fetal calf serum.
Antibody.
Anti-SeV polyclonal antiserum was described previously (3). Monoclonal antibodies (MAbs) to nucleoprotein (NP) and HN proteins of human parainfluenza type 1 virus, which were cross-reactive with SeV, were previously reported (7). MAbs to M and F proteins and to L protein were kindly donated by C. Örvell and D. Kolakofsky, respectively. Anti-V protein and anti-C protein polyclonal rabbit sera were donated by A. Kato.
Virus titration.
Monolayer cultures in 96-well plates were infected with 0.1 ml of 10-fold serial dilutions of SeV-containing fluid and incubated for 3 days. Subsequently, virus-infected cell monolayers were washed with Eagle’s MEM, 100 μl of a 0.4% suspension of guinea pig erythrocytes in Eagle’s MEM was added, and the mixture was left at room temperature for 30 min. After unadsorbed erythrocytes were removed by washing of the mixture with phosphate-buffered saline (PBS), the hemadsorbed cells were examined microscopically.
Establishment of persistent virus infection.
Monolayers of L929 or BHK cells were infected two or three times with the SeV wild strain at an MOI of about 100. Almost all of the cells infected with SeV showed severe cytopathic effects (CPEs); the remaining virus-infected cells were subcultured, and the virus-infected cells resulted in persistent infection (steady state).
Hemadsorption tests.
Tests for the adsorption of erythrocytes to cell surfaces were performed. The culture medium was removed, and the monolayer was washed with PBS. A 0.4% suspension of guinea pig erythrocytes in PBS was added, and the culture was kept at room temperature for 30 min. Unadsorbed erythrocytes were removed by washing of the culture with PBS, and the extent of erythrocyte adsorption was observed microscopically.
Immunofluorescence staining.
The cells were fixed with 3% formaldehyde in PBS for 15 min at room temperature and rinsed twice with PBS. Then, they were permeabilized with PBS-Tween 20 (0.05%) for 30 min and washed twice with PBS. Next, they were incubated for 60 min with primary antibody and washed three times with PBS. Following this step, they were incubated for 60 min with fluorescein isothiocyanate-labeled secondary antibody and washed with PBS. Immunofluorescence-stained cells were analyzed by use of a fluorescence microscope.
TUNEL staining.
For the detection of DNA fragmentation, a terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay was performed with fluorescein-12-dUTP-labeled DNA (in situ cell death detection kit, fluorescein; Roche) according to the manufacturer's protocol.
Virus isolation from cells persistently infected with virus.
Cells persistently infected with virus were cultured at 35°C for 3 days, and then the medium and/or cells were intra-allantoically inoculated into 10-day-old embryonated eggs. The eggs were incubated at 32°C for 3 days, and then the allantoic fluids were harvested and tested for the presence of hemagglutinin.
Virus cloning.
L929 cells in six-well plates were infected with 0.1 ml of 10-fold serial dilutions of virus. After incubation at 32°C for 3 days, virus-infected cells were washed twice with PBS, 2.0 ml of a 0.4% suspension of guinea pig erythrocytes in PBS was added to the cells, and the mixture was left at room temperature for 30 min. After unadsorbed erythrocytes were removed by intensive washing of the mixture with PBS, the hemadsorbing cells were isolated by use of a fine absorbent cotton swab fixed to a stick under microscopic observation. Subsequently, the cell suspensions were intra-allantoically inoculated into 10-day-old embryonated eggs. The eggs were incubated at 32°C for 3 days, and then the allantoic fluids were harvested.
Ability of virus to establish persistent infection.
L929 cells were infected at an MOI of about 10. Virus-infected cells were subcultured at least three times, and at every subculture, they were morphologically observed by use of a microscope. Some of the most common effects of viral infection are morphological changes, such as cell rounding and detachment from the substrate, cell lysis, and syncytium formation. However, SeV induces little syncytium formation of cultured cells without trypsin-like protease. Thus, when we observed the persistently infected cells by use of a microscope, the presence of cell rounding and cell lysis was checked mainly at every subculture. Hemadsorption tests were carried out with final subcultured cells. If final subcultures showed no or few CPEs and almost all of the cells were hemadsorption positive, then the virus was judged capable of establishing persistent infection.
Isotopic labeling of virus-infected cells.
Approximately 106 L929 cells were infected with various SeVs at an MOI of about 10. At 16 h after infection, the cells were washed once with and further incubated in fresh methionine- and cysteine-depleted Dulbecco's MEM for 2 h. The cells were labeled with 500 μCi of Pro-mix l-35S in vitro cell labeling mix (Amersham Pharmacia Biotech Japan, Tokyo, Japan) per ml in methionine- and cysteine-depleted MEM for 1 h. The cells were washed with PBS and lysed with 1 ml of radioimmunoprecipitation assay (RIPA) buffer (1% Triton X-100, 137 mM NaCl, 3 mM β-glycerophosphate, 3 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 25 mM HEPES [pH 7.6]).
RIPA and SDS-PAGE.
RIPA and sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) were performed as described elsewhere (14).
RESULTS
Establishment of persistent SeV infection and cell cloning.
The experimental design is shown in Fig. 1. L929 or BHK cells were infected two times with SeV (Nagoya or Z strain) at an MOI of approximately 100 and incubated with Eagle’s MEM supplemented with 5% fetal calf serum for 4 days, when almost all of the cells died. Subsequently, the remaining infected cells were washed three times with Eagle’s MEM to remove dead cells, and then cell cloning was carried out by a limiting-dilution method with 96-well plates. Cell clonality was confirmed by microscopic observation. After about 3 weeks, the cloned cells were subcultured in duplicate, and 2 days after subculturing, hemadsorption and immunofluorescence analysis with anti-SeV NP antibody were carried out for detecting cells persistently infected with virus. The cloned virus-infected cells were further subcultured, and the resulting cell cultures were designated passage number (p.n.) 1 cells. Because we could not exclude the possibility that virus-infected cells multiplied before limiting dilution, we do not claim that all of the cloned virus-infected cell lines were independently isolated and thus were different. The p.n. 1 cells were immunostained with anti-NP, -M protein, or -HN MAbs, and viral antigens were found in almost all of the persistently infected cells. In addition, when uncloned cells infected with SeV and surviving after 2 to 3 weeks became monolayers, the cells were reinfected with SeV. When this procedure was carried out two or three times, almost all of the remaining cells were found to have viral antigens. These cells were designated uncloned persistently infected cells. In most of these carrier cell samples, multinucleated giant cells were found, although the frequency of appearance varied (data not shown). The number of multinucleated giant cells slowly decreased as the cells were subcultured (Fig. 2A). Intriguingly, the M protein was localized in the nuclei of the majority of persistently infected cells (Fig. 2B and C).
FIG.1.
Experimental design.
FIG. 2.
Detection of NP, M, and HN proteins in cloned cells persistently infected with SeV and subcultured at various times. Four uncloned cell lines and 157 cloned cell lines persistently infected with SeV of p.n. 1 were immunostained with anti-NP, -M protein, or -HN MAbs. Furthermore, 8 cloned cell lines selected from the 157 cloned cell lines were further subcultured. Every 10th subculture, the cells were transferred to 32 or 38°C, incubated for 48 h, and then fixed. For some samples, anti-P, -F, -L, -V, or -C protein antibodies were also used. Representative data are shown. BHK/NA (d-3) cells at p.n. 30 and p.n. 60 were stained with Giemsa staining solution. L and H, lower and higher magnifications, respectively.
The culture fluids of p.n. 1 cells were inoculated into 10-day-old embryonated eggs for the isolation of infectious viruses. We obtained 157 cloned virus-infected cell lines, and infectious viruses were isolated from 109 out of 148 cloned cell lines by inoculation of culture fluids into embryonated eggs (Table 1). Furthermore, for the 39 cloned cell lines from which infectious virus could not be isolated, the infected cells were inoculated into embryonated eggs. Infectious viruses were isolated from 14 cell lines but could not be isolated from the remaining 25 cell lines (Table 1). Interestingly, only 5 out of 123 viruses (p.n. 1 viruses) isolated from p.n. 1 cells had the capability of establishing cells persistently infected with virus (data not shown), indicating that the majority of the viruses carried by cells persistently infected with virus showed high levels of cytotoxicity and did not have the ability to establish persistent infection.
TABLE 1.
Summary of cloned cell lines persistently infected with SeV
| Series | Subseries | No. of cell lines | Isolation of infective virusesa from:
|
||||
|---|---|---|---|---|---|---|---|
| Culture medium
|
Cells
|
||||||
| Total | + | − | + | − | |||
| BHK/NA | a | 68 | 63 | 55 | 8 | 3 | 5 |
| b | 4 | 3 | 3 | ||||
| c | 1 | 0 | 0 | ||||
| d | 3 | 3 | 3 | ||||
| e | 10 | 8 | 8 | ||||
| Subtotal | 86 | 77 | 69 | 8 | 3 | 5 | |
| L/NA | a | 2 | 2 | 2 | |||
| b | 1 | 1 | 1 | ||||
| c | 19 | 19 | 5 | 14 | 4 | 10 | |
| d | 13 | 13 | 2 | 11 | 4 | 7 | |
| e | 6 | 6 | 1 | 5 | 2 | 3 | |
| Subtotal | 41 | 41 | 11 | 30 | 10 | 20 | |
| L/Z | a | 12 | 12 | 12 | 0 | ||
| c | 9 | 9 | 9 | 0 | |||
| d | 5 | 5 | 4 | 1 | 1 | 0 | |
| e | 4 | 4 | 4 | 0 | |||
| Subtotal | 30 | 30 | 29 | 1 | 1 | 0 | |
| Total | 157 | 148 | 109 | 39 | 14 | 25 | |
Cells persistently infected with virus were cultured at 35°C for 2 to 3 days, and then the medium and/or cells were intra-allantoically inoculated into 10-day-old embryonated eggs. The eggs were incubated at 32°C for 3 days, and the allantoic fluids were harvested and tested for the presence of hemagglutinin. +, infective viruses were isolated; −, infective viruses were not isolated.
Chronological analyses of virus isolation.
Four uncloned cell lines and eight cloned cell lines selected for further analyses were further subcultured. At every fifth subculture, virus isolation was carried out. Representative results are shown in Table 2. There were two ways in which virus could be isolated from persistently infected cells. (i) Infectious viruses could be isolated by inoculation of culture fluids into embryonated eggs until subcultures 60 to 65. (ii) Infectious viruses could be isolated at early subcultures by inoculation of culture fluids into embryonated eggs, after which virus isolation from culture fluids gradually became more difficult, and inoculation of infected cells into embryonated eggs was necessary for the isolation of infectious viruses. Eventually, virus isolation from culture fluids or cells became very difficult, suggesting that virus particle formation was suppressed. Four cell lines—BHK cells persistently infected with Nagoya strain SeV [BHK/NA (d-3)], uncloned L929 cells persistently infected with Nagoya strain SeV [uncloned L/NA (C)], L929 cells persistently infected with Nagoya strain SeV [L/NA (a-1)], and uncloned L929 cells persistently infected with Z strain SeV [uncloned L/Z]—belonged to the former type. The other eight cell lines—uncloned BHK/NA, BHK/NA (a-1), BHK/NA (a-66), BHK/NA (b-18), uncloned L/NA (B), L/NA (a-2), L/Z (a-1), and L/Z (c-2)—belonged to the latter type. This grouping was unrelated to the persistent infection system (L929 or BHK cells; Nagoya or Z strain SeV). Intriguingly, infectious viruses could not be isolated from cells belonging to type 1 at times during the process of subculturing by inoculation of culture fluids into embryonated eggs. These findings showed that the association of carried virus with cells became stronger and that virus isolation became more difficult as cells persistently infected with virus were subcultured.
TABLE 2.
Chronological analyses of virus isolation
| Series | p.n. | Isolation of infective virusesa from: |
|---|---|---|
| BHK/NA (d-3) | 1 | Medium + |
| 5 | Medium + | |
| 10 | Medium + | |
| 15 | Medium + | |
| 20 | Medium − | |
| 25 | Medium −; cell + | |
| 30 | Medium + | |
| 35 | Medium −; medium + | |
| 40 | Medium −; medium + | |
| 45 | Medium + | |
| 50 | Medium + | |
| 55 | Medium + | |
| 60 | Medium + | |
| 65 | Medium + | |
| L/Z (c-2) | 1 | Medium + |
| 5 | Medium + | |
| 10 | Medium + | |
| 15 | Medium + | |
| 20 | Medium + | |
| 25 | Medium + | |
| 30 | Medium + | |
| 35 | Medium −; medium −; cell −; medium/egg +; medium/egg +; cell/egg + | |
| 40 | Medium −; medium −; medium/egg −; medium/egg −; medium/egg/egg + | |
| 45 | Medium −; medium +; cell +; cell + | |
| 50 | Medium −; cell −; medium/egg +; cell/egg −; cell/egg/egg + | |
| 55 | Medium −; medium −; medium +; cell −; cell −; cell −; cell +; medium/egg −; cell/egg −; medium/egg +; cell/egg +; cell/egg/egg +; medium/egg/egg/egg +; cell/egg/egg/egg + | |
| 60 | Medium −; cell −; medium/egg +; cell/egg + | |
| 65 | Medium −; cell −; medium/egg −; cell/egg −; medium/egg/egg +; cell/egg/egg + |
Cells persistently infected with virus were cultured at 35°C for 2 to 3 days, and then the medium or cells were intra-allantoically inoculated into 10-day-old embryonated eggs. The eggs were incubated at 32°C for 3 days, and the allantoic fluids were harvested and tested for the presence of hemagglutinin. When infective virus was not isolated, the allantoic fluids were reinoculated into 10-day-old embryonated eggs. +, infective viruses were isolated; −, infective viruses were not isolated.
Chronological analyses of the ability of uncloned viruses to establish persistent infection.
As described above, only 5 out of 123 p.n. 1 viruses had the capability of establishing persistent infection. Consequently, acquisition of the ability to establish persistent infection through subcultivation of virus-infected cells was investigated. As shown in Fig. 3, three modes of acquiring the ability to establish persistent infection were found. (i) Viruses acquired the ability to establish persistent infection at a relatively early phase. (ii) The way in which the viruses acquired the ability is not straightforward, especially for BHK/NA (d-3) cells. (iii) Viruses acquired the ability to establish persistent infection at a relatively late phase. Eventually, viruses derived from virus-infected cells other than L/NA (a-2) cells acquired the ability. Viruses isolated from L/NA (a-2) cells did not acquire the ability until subculture 50; the cytotoxicity of the viruses gradually became weak (data not shown).
FIG. 3.
Chronological analyses of the ability of uncloned viruses to establish persistent infection (PI). The ability of uncloned viruses isolated from three series of persistent infection to establish persistent infection was analyzed every fifth passage. Virus-infected cells were subcultured at least three times, and at every subculture, cells were morphologically observed with a microscope. Hemadsorption tests were carried out with final subcultured cells. If a final subculture showed no or few CPEs and almost all of the cells exhibited hemadsorption, then the virus was judged to be capable of establishing persistent infection. Symbols: (+), viruses capable of establishing persistent infection; (−/+), mixture of viruses capable and viruses incapable of establishing persistent infection; (−), viruses incapable of establishing persistent infection.
Further study of the ability of cloned viruses to establish persistent infection.
We tried to isolate cloned viruses from BHK/NA (b-18) and BHK/NA (a-1) cells. We isolated 55 cloned viruses and analyzed their capability for establishing persistent infection and temperature sensitivity. First, 44 cloned viruses isolated from BHK/NA (b-18) cells were investigated (Table 3 and data not shown). Uncloned viruses and six cloned viruses isolated from BHK/NA (b-18) p.n. 1 and 5 cells, respectively, were highly cytotoxic, while two cloned viruses (clone 1-14 [cl.1-14] and cl.1-15) isolated from BHK/NA (b-18) p.n. 1 cells were incapable of establishing persistent infection and showed relatively low cytotoxicity. One uncloned virus and six out of seven cloned viruses derived from BHK/NA (b-18) p.n. 30 cells showed lower cytotoxicity, but one cloned virus (cl.30-17a) was incapable of establishing persistent infection at both 32 and 38°C. Similarly, one uncloned virus and three out of four cloned viruses derived from BHK/NA (b-18) p.n. 40 cells acquired the capability of establishing cells persistently infected with virus, but one cloned virus (cl.40-5) was incapable of establishing persistent infection at 32°C. On the other hand, 3 uncloned viruses and all 17 cloned viruses derived from BHK/NA (b-18) p.n. 40 and 65 cells, respectively, were capable of establishing persistent infection (Table 3 and data not shown).
TABLE 3.
Ability of cloned viruses from BHK/NA (b-18) cells to establish persistent infections
| p.n. | Clone (temp sensitivity)a | CPEs for the indicated subculture at the following temp (°C)b:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 32
|
38
|
||||||||||
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | ||
| 1 | Uncloned virus | ++++ | ++++ | ||||||||
| 1-6 (−0.5) | ++++ | ++++ | |||||||||
| 1-12 (2.25) | ++++ | ++++ | |||||||||
| 1-14 (2.0) | + | ++ | +++ | +++ | ++++ | + | + | +++ | ++++ | ||
| 1-15 (3.0) | + | + | ++ | +++ | ++++ | − | − | ||||
| 5 | Uncloned virus | ++++ | ++++ | ||||||||
| 5-2 (0.25) | ++++ | ++++ | |||||||||
| 5-3 (0.25) | ++++ | + | ++++ | ||||||||
| 5-5 (0.25) | R | ++++ | + | ++++ | |||||||
| 5-12 (0.0) | ++++ | ++++ | |||||||||
| 20 | Uncloned virus | − | − | − | |||||||
| 20-8 (2.25) | − | − | − | − | − | − | − | − | − | − | |
| 20-14 (2.75) | − | − | − | − | − | − | − | − | − | − | |
| 20-4a (3.0) | − | − | − | − | − | − | − | − | − | − | |
| 20-5a (>3.0) | − | − | − | − | − | − | − | − | − | − | |
| 20-7a (1.5) | − | − | − | + | − | − | − | − | − | − | |
| 20-8a (2.25) | − | − | − | − | − | − | − | − | − | − | |
| 20-10a (1.5) | − | − | − | + | − | − | − | − | − | − | |
| 20-13a (1.5) | − | − | − | − | − | − | − | − | − | − | |
| 30 | Uncloned virus | − | − | R | − | − | − | − | − | − | − |
| 30-1a (>2.0) | − | − | − | − | − | − | − | − | − | − | |
| 30-3a (2.75) | − | − | − | R | − | − | − | − | − | − | |
| 30-4a (2.5) | − | − | − | − | − | − | − | − | − | − | |
| 30-10a (1.5) | − | − | R | + | − | − | − | − | − | − | |
| 30-11a (1.75) | − | − | − | − | − | − | − | − | − | − | |
| 30-17a (0.5) | ++ | ++++ | + | ++++ | |||||||
| 30-23a (>2.0) | − | − | − | − | − | − | − | − | − | − | |
| 40 | Uncloned virus | − | − | R | R | − | − | − | − | − | − |
| 40-1 (2.25) | − | R | R | R | − | − | − | − | − | − | |
| 40-4 (3.0) | − | − | − | − | − | − | − | − | − | − | |
| 40-5 (0.25) | − | − | R | ++ | +++ | − | − | − | − | − | |
| 40-6 (1.5) | − | R | R | − | − | − | − | R | − | − | |
| 65 | Uncloned virus | − | − | R | R | R | − | − | − | − | − |
| 65-3a (1.75) | − | − | − | − | − | − | − | − | − | − | |
| 65-7a (3.25) | − | − | − | − | − | − | − | − | − | − | |
| 65-17a (>4.5) | − | − | − | − | − | − | − | − | − | − | |
| 65-18a (2.75) | − | − | − | − | − | − | − | − | − | − | |
| 65-20a (2.0) | − | − | − | − | − | − | − | − | − | − | |
| 65-22a (2.25) | − | − | − | − | − | − | − | − | − | − | |
| 65-26a (2.25) | − | − | − | − | − | − | − | − | − | − | |
Bold type indicates viruses incapable of establishing persistent infection. Temperature sensitivity was calculated as log virus titer at 32°C divided by log virus titer at 38°C.
Virus-infected cells were subcultured five times, and at every subculture, cells were morphologically observed with a microscope. Hemadsorption tests were carried out with final subcultured cells. If final subcultures showed no or few CPEs, then the virus was judged to be capable of establishing persistent infection. CPE scores ranged from − to ++++, where − indicates that no CPE was observed, + indicates that only a few lytic cells were observed, ++ indicates that <25% of the cells showed lysis, +++ indicates that <75% of the cells showed lysis, and ++++ indicates that >75% of the cells showed lysis. R indicates that a small number of cells showed rounding.
One uncloned virus isolated from BHK/NA (a-1) p.n. 15 cells was incapable of establishing persistent infection, while uncloned viruses from BHK/NA (a-1) p.n. 20 and 25 cells were capable of establishing persistent infection (Table 4 and Fig. 3). Therefore, we isolated seven and four cloned viruses from BHK/NA (a-1) p.n. 15 and 25 cells, respectively. Six cloned viruses derived from p.n. 15 cells were highly toxic for cells, but one cloned virus (cl.15-10) was capable of establishing persistent infection at 38°C (Table 4). On the other hand, one cloned virus (cl.25-8a) isolated from p.n. 25 cells was capable of establishing persistent infection at 32 and 38°C, while the other three viruses were cytotoxic, although the extent of cell damage varied widely (Table 4).
TABLE 4.
Ability of cloned viruses from BHK/NA (a-1) cells to establish persistent infections
| p.n. | Clone (temp sensitivity)a | CPEs for the indicated subculture at the following temp (°C)b:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 32
|
38
|
||||||||||
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | ||
| 15 | Uncloned virus | + | ++++ | ++++ | |||||||
| 15-2 (0.75) | + | ++++ | +++ | ++++ | |||||||
| 15-4 (0.0) | R | ++++ | + | ++++ | |||||||
| 15-7 (1.0) | − | ++ | +++ | ++++ | ++ | ++++ | |||||
| 15-8 (0.75) | ++ | ++++ | ++ | ++++ | |||||||
| 15-9 (0.0) | + | ++++ | +++ | ++++ | |||||||
| 15-10 (0.0) | − | − | − | ++ | ++++ | + | + | − | + | + | |
| 15-11 (1.0) | ++ | ++++ | +++ | ++++ | |||||||
| 25 | Uncloned virus | − | − | − | − | − | − | − | − | − | − |
| 25-2 (1.5) | − | R | R | +++ | ++++ | R | +++ | ||||
| 25-3 (1.0) | − | R | R | ++ | ++++ | R | ++++ | ||||
| 25-7a (1.0) | +++ | ++++ | − | ++++ | |||||||
| 25-8a (1.75) | − | − | − | R | − | − | − | − | − | − | |
Immunofluorescence analyses of cells persistently infected with SeV.
Every 10th subculture, persistently infected cells were incubated at 32 or 38°C for 2 days. These cells were immunostained with anti-NP, -M protein, or -HN protein MAbs (Fig. 2D and E). For some samples, anti-P, -F, -L, -V, or -C protein antibodies were also used. The M protein localized in nuclei, and HN was scarcely detected at 38°C in relatively early subcultures (Fig. 2B, C, D, and E).
Immunofluorescence analyses of cells primarily infected with uncloned viruses isolated from persistently infected cells.
L929 cells were infected with uncloned viruses isolated from BHK/NA (b-18) cells which had been subcultured at various times at an MOI of about 10 and incubated at 32 or 38°C for 20 h. These cells were immunostained with anti-NP, -M protein, or -HN MAbs. As shown in Fig. 4A, cells infected with p.n. 1 or 5 viruses showed damage at both temperatures or at 32°C, respectively, while cells infected with p.n. 5 viruses showed few CPEs at 38°C. Furthermore, p.n. 20, 40, or 60 viruses showed no or little cytotoxicity at both temperatures. The NP and HN proteins were clearly expressed in cells infected with p.n. 1 or 5 virus at 38°C. However, these proteins were faintly detected in cells infected with p.n. 20 viruses at 38°C. The NP protein was faintly detected but the HN protein was scarcely detected in cells infected with p.n. 40 viruses at 38°C. Intriguingly, the NP and HN proteins could not be detected in cells infected with p.n. 60 viruses at 38°C, indicating that the temperature-sensitive step of the virus is at an early stage of its replication. The HN protein was localized at the perinuclear region in cells infected with p.n. 20, 40, or 60 viruses at 32°C. The M protein was found in the cytoplasm of p.n. 1 or 5 virus-infected cells, while it was predominantly detected in the nuclei of p.n. 20 or 60 virus-infected cells (Fig. 4A).
FIG. 4.
(A) Detection of NP, M, and HN proteins in L929 cells primarily infected with uncloned SeV. Cells were infected with uncloned SeV isolated from BHK/NA (b-18) cells, which were subcultured at various times, at an MOI of about 10. Cells were transferred to 32 or 38°C and were fixed at 24 h postinfection. These cells were immunostained with anti-NP, -M protein, or -HN antibodies. (B) Detection of NP, M, and HN proteins in L929 cells primarily infected with cloned SeV. Cells were infected with cloned SeV isolated from BHK/NA (b-18) cells, which were subcultured at various times, at an MOI of about 10. Cells were transferred to 32 or 38°C and were fixed at 24 h postinfection. These cells were immunostained with anti-NP, -M protein, or -HN antibodies. cl.5-5 and cl.5-12, cl.20-8, cl.40-11, and cl.65-1 and cl.65-5 were isolated from p.n. 5, p.n. 20, p.n. 40, and p.n. 65 BHK/NA (b-18) cells, respectively.
Immunofluorescence analyses of cells primarily infected with cloned viruses isolated from persistently infected cells.
In the next experiment, L929 cells were infected with cloned viruses isolated from BHK/NA (b-18) cells at an MOI of about 10 and incubated at 32 or 38°C for 20 h. In cells infected with cl.20-8, the HN protein was scarcely found, while the NP protein was clearly detected at 38°C, although at a low expression level (Fig. 4B). Furthermore, the HN and NP proteins were scarcely found at 38°C but were clearly detected at 32°C in cells infected with cl.40-11, cl.65-1, or cl.65-5. These findings suggested that cl.40-11, cl.65-1, and cl.65-5 are temperature sensitive and that the temperature-sensitive step is at an early stage of infection. Intriguingly, the HN protein was localized at the perinuclear region in cells infected with cl.20-8, cl.40-11, cl.65-1, or cl.65-5. The M protein was also localized in the nuclei of cells infected with cloned viruses having the ability to establish persistent infection (Fig. 4B).
Temperature sensitivity of viruses derived from cells persistently infected with SeV.
In this experiment, infective titers of uncloned viruses isolated from three series of persistent infection, that is, BHK/NA (a-1), (b-18), and (d-3) cells, were measured at 32 or 38°C. The temperature sensitivity of these viruses tended to rise as persistently infected cells were subcultured (Fig. 5A). Subsequently, the infectivity of cloned viruses was titrated at both temperatures, and the results are shown in Table 3. All of the cloned viruses derived from BHK/NA (b-18) p.n. 5 cells showed high cytotoxicity, and their temperature sensitivity was less than 0.25. On the other hand, all of the cloned viruses derived from BHK/NA (b-18) p.n. 20 cells were capable of establishing persistent infection, and their temperature sensitivity was more than 1.5. Furthermore, cl.30-17a and cl.40-5 showed cytotoxicity, and their temperature sensitivities were 0.5 and 0.25, respectively. All of the cloned viruses derived from BHK/NA (b-18) p.n. 30, 40, or 65 cells, other than cl.30-17a and cl.40-5, were capable of establishing persistent infection, and their temperature sensitivity was more than 1.5. These findings suggested that temperature sensitivity is generally related to a lower level of pathogenesis, although the relationship is not necessarily simple.
FIG. 5.
Temperature sensitivity of viruses derived from cells persistently infected with SeV. (A) Infective titers of uncloned viruses isolated in three series of persistent infection from cells subcultured at various times were measured at 32 or 38°C. Temperature sensitivity was calculated as the log virus titer at 32°C divided by the log virus titer at 38°C. Symbols: □, BHK/NA (a-1) cells; •, BHK/NA (b-18) cells; ◎, BHK/NA (d-3) cells. (B and C) Pulse-chase experiments with L929 cells infected with SeV wild strain (SeVw) or SeVPi. L929 cells were infected with SeVw or with p.n. 5 or 60 virus isolated from BHK/NA (b-18) cells (B) or with cl.5-12 or cl.40-11 isolated from BHK/NA (b-18) cells (C) at an MOI of about 10. After incubation at 32 or 38°C for 18 h, the cells were pulse-labeled at 32 or 38°C, respectively, for 1 h with l-35S. After pulse-labeling for 1 h, the virus-infected cells were chased in chase medium (MEM supplemented with 5 mM methionine and 5 mM cysteine) at 32 or 38°C for 4 h. The cell lysates were immunoprecipitated with anti-SeV rabbit polyclonal antibody, and the immunoprecipitates were analyzed by SDS-PAGE. Fo/NP, uncleaved F(Fo) protein and NP, which migrate to almost the same positions.
To investigate the stability of the virus-specific polypeptides in L929 cells primarily infected with uncloned or cloned SeV isolated from BHK/NA (b-18) carrier cells, pulse-label and pulse-chase studies were performed. L929 cells were primarily infected with various viruses at an MOI of about 10; after incubation at 32 or 38°C for 18 h, the cells were pulse-labeled at 32 or 38°C, respectively, for 1 h with [35S]methionine-[35S]cysteine. The virus-specific polypeptides were pulse-labeled well in L929 cells infected with p.n. 5 Nagoya strain SeV and cl.5-12 and incubated at 38°C in comparison with 32°C (Fig. 5B and C). On the other hand, the virus-specific polypeptides were pulse-labeled less well in L929 cells infected with p.n. 60 SeV and incubated at 38°C in comparison with 32°C (Fig. 5B). Intriguingly, almost no virus-specific polypeptide was synthesized in cl.40-11 virus-infected L929 cells at 38°C (Fig. 5C), indicating that cl.40-11 is temperature sensitive and that the temperature-sensitive step is at an early stage of infection. Subsequently, after pulse-labeling for 1 h at 32°C, the virus-infected cells were chased at 32 or 38°C for 4 h; the HN protein of cl.40-11 was suggested to be unstable at 38°C.
TUNEL staining of cells infected with cloned viruses derived from cells persistently infected with SeV.
The molecular mechanism(s) underlying cell death in SeV-infected cells remains unsolved. One of the mechanisms that has been considered is apoptosis (4). Thus, whether or not apoptosis is induced in SeV-infected cells was investigated by using TUNEL staining. L929 cells were infected with various cloned viruses at an MOI of about 10, and after 20 h of incubation at 35°C, the cells were stained with TUNEL solution. As shown in Fig. 6A, cl.1-6, cl.1-12, cl.5-2, and cl.5-4, which were obtained from infected BHK/NA (b-18) cells and which were highly toxic for cells, induced strong positive TUNEL staining, while cl.20-4a, cl.20-7a, cl.65-3a, and cl.65-4a, which were capable of establishing persistent infection, induced little TUNEL staining. In addition, cl.25-7a from infected BHK/NA (a-1) cells induced both cell lysis and positive TUNEL staining, while cl.25-8a, which was capable of establishing persistent infection, did not induce positive TUNEL staining (Fig. 6B).
FIG. 6.
TUNEL staining of cells infected with cloned viruses derived from cells persistently infected with SeV. L929 cells were infected with various cloned viruses at an MOI of about 10, and after 20 h of incubation at 35°C, the cells were stained with TUNEL solution according to the manufacturer's protocol.
Genetic analyses of viruses derived from cells persistently infected with SeV.
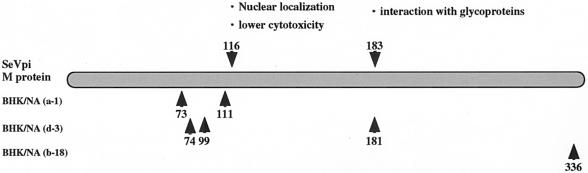
In a previous study (10), two amino acid substitutions were found in the M protein of SeVpi and were related to a lower level of cytopathogenesis. Consequently, the amino acid sequences deduced from the nucleotide sequences of the M protein genes of viruses isolated from persistently infected cells were determined. Nucleotide sequencing of M protein genes from p.n. 1, 20, 40, or 65 uncloned viruses derived from BHK/NA (b-18) cells was carried out. The nucleotide sequences of each of eight full-length clones were determined. Intriguingly, a specific clone carrying substitutions G429A and A1007G was selected as persistently infected cells were subcultured (Table 5). Subsequently, the nucleotide sequences of cloned viruses were determined. Identical substitutions were found in the M protein genes of cl.40-4, cl.40-6, cl.65-1, and cl.65-4. Substitution A1007G resulted in amino acid substitution N336S (Table 5). Subsequently, p.n. 1 and 65 viruses isolated from BHK/NA (a-1) or BHK/NA (d-3) cells were analyzed (Tables 6 to 7). Specific amino acid substitutions D73Y and I111V or L74F and T181I accumulated in viruses derived from BHK/NA (a-1)- or BHK/NA (d-3) cells, respectively. In addition, substitution D99Y was found in four out of seven clones of viruses isolated from BHK/NA (d-3) p.n. 65 cells (Table 7). These amino acid substitutions are plotted in Fig. 7. These substitutions were located close to amino acids 116 and 183, locations at which amino acid substitutions are found in BHK-HVJ cells that have been maintained for more than 10 years.
TABLE 5.
Nucleotide and amino acid substitutions detected in the M protein gene of viruses isolated from BHK/NA (b-18) cells
| Virus | No. of positive clones/no. of clones analyzed with the following nucleotide (amino acid) substitution:
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A38G (E13G) | G140A (G47E) | A179G (E60G) | A199G (N67D) | T275C (V92A) | C403T (S) | G429A (S) | T515C (V172A) | A601C (N201H) | A644G (K215R) | T704C (L235P) | A737G (Y246C) | A771G (S) | A799G (L267V) | A911G (H304R) | A1007G (N336S) | A1024G (I342V) | A1030G (R344G) | G1031A (R344K) | |
| p.n. 1 | 1/8 | 2/8 | 1/8 | 1/8 | 1/8 | ||||||||||||||
| p.n. 20 | 1/8 | 1/8 | 8/8 | 1/8 | 1/8 | 7/8 | 1/8 | ||||||||||||
| p.n. 40 | 1/8 | 8/8 | 1/8 | 1/8 | 8/8 | ||||||||||||||
| p.n. 65 | 1/8 | 1/8 | 8/8 | 1/8 | 1/8 | 1/8 | 8/8 | ||||||||||||
| cl.5-2 | |||||||||||||||||||
| cl.5-12 | |||||||||||||||||||
| cl.40-4 | 2/2 | 2/2 | |||||||||||||||||
| cl.40-6 | 2/2 | 2/2 | |||||||||||||||||
| cl.65-1 | 2/2 | 2/2 | |||||||||||||||||
| cl.65-4 | 2/2 | 2/2 | |||||||||||||||||
TABLE 6.
Nucleotide and amino acid substitutions detected in the M protein gene of viruses isolated from BHK/NA (a-1) cells
| Virus | No. of positive clones/no. of clones analyzed with the following nucleotide (amino acid) substitution:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| T13C (Y5H) | A33G (S) | T164C (L55P) | G217T (D73Y) | A331G (I111V) | A613G (T205A) | G706A (G236R) | A811G (S271G) | A942G (S) | C1042T (S) | T1043C (L348P) | |
| p.n. 1 | 1/8 | 1/8 | 1/8 | ||||||||
| p.n. 65 | 1/8 | 1/8 | 8/8 | 8/8 | 1/8 | 1/8 | 1/8 | 1/8 | |||
TABLE 7.
Nucleotide and amino acid substitutions detected in the M protein gene of viruses isolated from BHK/NA (d-3) cells
| Virus | No. of positive clones/no. of clones analyzed with the following nucleotide (amino acid) substitution:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| A6G (S) | C165T (S) | G222T (L74F) | G295T (D99Y) | C542T (T181I) | T629C (I210T) | T814C (F272L) | A1024G (I342V) | G1027A (G343R) | |
| p.n. 1 | 1/8 | 1/8 | 3/8 | 1/8 | |||||
| p.n. 65 | 8/8 | 4/8 | 8/8 | 1/8 | 1/8 | 1/8 | |||
FIG. 7.
Diagram mapping specific amino acid substitutions detected in M proteins of uncloned and cloned viruses isolated from persistently infected BHK/NA (a-1), BHK/NA (d-3), and BHK/NA (b-18) cells.
Subsequently, the amino acid sequences of the intracytoplasmic domains of the HN and F proteins of viruses isolated from persistently infected cells were determined. Specific amino acid substitutions were not found in the cytosolic domains of the glycoproteins (data not shown).
We also carried out partial sequencing of the L protein genes of SeV derived from persistent infection. Many nucleotide substitutions were found in the L protein genes (data not shown). Interestingly, substitution A1303T, resulting in amino acid substitution M435L, was found in all three clones of viruses isolated from BHK/NA (d-3) p.n. 65 cells, although the locations of most of the other substitutions varied (data not shown).
DISCUSSION
In this study, we established 157 cloned cell lines persistently infected with SeV. The 157 cell lines were generated independently of each other. These cell lines were derived from a single cell infected with SeV, indicating that the virus-infected cells can divide and multiply. Persistent viral infection of cultured cells may be classified into two types, that is, carrier-state infection and steady-state infection (15). Intriguingly, it was clarified in this study that the virus carrier cells leading finally to a stable persistent infection contained two types of virus-infected cells at an initial stage, although the majority of the cells were in the steady state and a few cells were in the carrier state.
Only 5 out of 123 viruses isolated from p.n. 1 cells had the capability of establishing persistent infection, indicating that the majority of the viruses carried by cells persistently infected with virus were highly toxic for the cells and did not have the ability to establish persistent infection. However, viruses derived from virus-infected cells further subcultured eventually acquired the capability of establishing persistent infection, although the times varied and the way in which the capability was acquired was not straightforward. Of note is that uncloned viruses derived from persistent infection, especially at a relatively early phase, were composed of mixed populations, that is, viruses capable or incapable of establishing persistent infection. From these findings, we concluded that only a few cells infected with an initial pathogenic virus(es) showed few CPEs, and then they could divide and multiply by some mechanism. What is the factor(s) influencing the initial stability between cells and pathogenic viruses? When L929 cells were pretreated with interferon or defective interfering particles, cells persistently infected with virus were easily established (11-13). HT cells, BHK-HVJ cells cured of SeV infection through the action of immunological mechanisms during growth in hamsters, were resistant to the cytotoxic effect of the SeV wild strain (2). These findings suggest that cells infected persistently with SeV are naturally resistant to SeV, leading to persistent infection, although the possibility cannot be excluded that their resistance to virus infection is acquired during subculturing for a long time. Thus, interferon, defective interfering particles, and cells resistant to virus infection are candidates for factors influencing the initial stability between cells and viruses.
The medium of BHK-HVJ cells had no infectivity for chick embryos, and infectious viruses were isolated by inoculating BHK-HVJ cells into embryonated eggs (5, 9). In many cases, infectious viruses were easily isolated by inoculation of the culture medium into embryonated eggs at an early stage of persistent infection. However, in general, virus isolation became gradually more difficult, and viruses were tightly associated with cells. The molecular mechanism(s) underlying cell death in SeV-infected cells remains unsolved. One of the mechanisms that has been considered is apoptosis. Cloned viruses isolated from BHK/NA (b-18) and BHK/NA (a-1) were highly cytotoxic and showed strong positive TUNEL staining, while cells infected with cloned viruses capable of establishing persistent infection showed weak TUNEL staining, suggesting that a loss or a reduction of apoptosis-inducing ability leads to the acquisition of the ability of the virus to establish persistent infection. Further experiments will be required for clarification of the molecular mechanism of SeV-mediated cell death.
A temperature-sensitive phenomenon of virus replication was observed in cells persistently infected with SeV (9, 11). In cells infected with an early cloned virus, such as cl.20-8, the HN antigens were scarcely found but the NP antigens were clearly detected at 38°C, indicating that the expression of the HN proteins of these viruses is temperature sensitive. On the other hand, the HN and NP antigens were scarcely found at 38°C but were clearly detected at 32°C in cells infected with a late cloned virus, such as cl.40-11. Furthermore, none of the virus-specific polypeptides was pulse-labeled in late cloned virus-infected L929 cells incubated at 38°C. These findings show that the temperature-sensitive step of the late cloned viruses is at an early stage of infection. It could be concluded that temperature sensitivity progresses as persistently infected cells are subcultured; that is, first the HN and M proteins acquire temperature sensitivity and then the polymerase shows temperature sensitivity.
In a previous study (10), it was found that following a temperature shift either up or down, the M protein was translocated to the nucleus and then localized to the perinuclear region of BHK cells infected with SeVpi. The M protein was also localized in the nuclei of the majority of the persistent cells used in this study. In addition, the M protein was found in the cytoplasm of cells primarily infected with p.n. 1 and 5 viruses, while it was predominantly detected in the nuclei of cells infected with p.n. 40 and 60 viruses. Furthermore, the M protein was also localized in the nuclei of cells infected with cloned viruses having the ability to establish persistent infection. These findings show that there is a connection of some sort between the nuclear localization of the M protein and virus persistence.
Two amino acid substitutions were found in the M protein of SeVpi, which was isolated from BHK-HVJ cells cultured for a long period (10). Therefore, characterization of the viral genome during the establishment of persistent infection was carried out. Intriguingly, specific clones carrying substitutions G429A and A1007G were selected as BHK/NA (b-18) cells were subcultured. Furthermore, the same substitutions were found in the M protein genes of late cloned viruses. However, these substitutions were not found in early cloned viruses. Substitution A1007G resulted in amino acid substitution N336S; therefore, substitution N336S was selected as persistently infected cells were subcultured. In addition, specific amino acid substitutions D73Y and I111V or L74F, D99Y, and T181I accumulated in viruses derived from BHK/NA (a-1) or BHK/NA (d-3) cells, respectively. Interestingly, these substitutions were located close to amino acids 116 and 183, locations at which amino acid substitutions are found in BHK-HVJ cells that have been maintained for more than 10 years. Thus, during subculturing of persistently infected cells, a specific modification of M protein function was selected, but a specific amino acid substitution(s) was not selected. Furthermore, substitution A1303T, resulting in amino acid substitution M435L, was found in all three L protein gene clones from BHK/NA (d-3) p.n. 65 cells, although the locations of the majority of the other substitutions varied. Amino acid 435 is located between conserved domains I and II, and the role of this interdomain region has not been analyzed. This region is highly variable and has been hypothesized to have a specialized function in individual viruses. For analyzing the function of variant M proteins, experiments involving the transient expression of these proteins are in progress. So far, the nuclear localization of variant M proteins has not been found (unpublished data). The reverse genetics system for SeV will be used to analyze the biological significance of the amino acid substitutions found in the M and L proteins of SeV isolated from persistent infection.
In summary, we established a large number of cloned cell lines persistently infected with SeV. This study shows that there is an unstable dynamic phase at an early stage of the establishment of SeV persistent infection (steady state), and then viruses which are tightly cell associated, temperature sensitive, and capable of establishing persistent infection are selected. Furthermore, virus clones carrying specific nucleotide substitutions are selected as persistently infected cells are subcultured. Although the locations of nucleotide substitutions differ in the various cloned cell lines, the amino acid substitutions in the M protein are located close to amino acids 116 and 183.
Acknowledgments
This study was supported in part by a grant-in-aid for scientific research from the Ministry of Education, Science, Culture, Sports, Science, and Technology of Japan and by the Mie Medical Research Fund.
REFERENCES
- 1.Choppin, P. 1964. Multiplication of a myxovirus (SV5) with minimal cytopathic effects and without interference. Virology 23:224-233. [DOI] [PubMed] [Google Scholar]
- 2.Ito, Y., Y. Nishiyama, K. Shimokata, I. Nagata, and A. Kunii. 1979. Interferon susceptibility of various cell lines persistently infected with hemagglutinating virus of Japan (HVJ). J. Gen. Virol. 43:103-110. [DOI] [PubMed] [Google Scholar]
- 3.Ito,Y., M. Tsurudome, M. Hishiyama, and A. Yamada. 1987. Immunological interrelationships among human and non-human paramyxoviruses revealed by immunoprecipitation. J. Gen. Virol. 68:1289-1297. [DOI] [PubMed] [Google Scholar]
- 4.Itoh, M., H. Hotta, and M. Homma. 1998. Increased induction of apoptosis by a Sendai virus mutant is associated with attenuation of mouse pathogenicity. J. Virol. 72:2927-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura, Y., Y. Ito, K. Shimokata, Y. Nishiyama, and I. Nagata. 1975. Temperature-sensitive virus derived from BHK cells persistently infected with HVJ (Sendai virus). J. Virol. 15:55-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kimura, Y., C. Orvell, and E. Norrby. 1979. Characterization of the polypeptides synthesized in cells infected with a temperature-sensitive mutant derived from an HVJ (Sendai virus) carrier culture. Arch. Virol. 61:23-33. [DOI] [PubMed] [Google Scholar]
- 7.Komada, H., S. Kusagawa, C. Orvell, M. Tsurudome, M. Nishio, H. Bando, M. Kawano, H. Matsumura, E. Norrby, and Y. Ito. 1992. Antigenic diversity of human parainfluenza virus type 1 isolates and their immunological relationship with Sendai virus revealed by using monoclonal antibodies. J. Gen. Virol. 73:875-884. [DOI] [PubMed] [Google Scholar]
- 8.Kondo, T., T. Yoshida, N. Miura, and M. Nakanishi. 1993. Temperature-sensitive phenotype of a mutant Sendai virus is caused by its insufficient accumulation of the M protein. J. Biol. Chem. 268:21924-21930. [PubMed] [Google Scholar]
- 9.Nagata, I., Y. Kimura, Y. Ito, and T. Tanaka. 1972. Temperature-sensitive phenomenon of viral maturation observed in BHK cells persistently infected with HVJ. Virology 49:453-461. [DOI] [PubMed] [Google Scholar]
- 10.Nishio, M., M. Tsurudome, M. Ito, M. Kawano, H. Komada, and Y. Ito. 2003. Characterization of Sendai virus persistently infected L929 cells and Sendai virus pi strain: recombinant Sendai viruses having Mpi protein shows lower cytotoxicity and are incapable of establishing persistent infection. Virology 314:110-124. [DOI] [PubMed] [Google Scholar]
- 11.Prebles, O. T., and J. S. Youngner. 1973. Temperature-sensitive defect of mutants isolated from L cells persistently infected with Newcastle disease virus. J. Virol. 5:559-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roux, L., and J. J. Holland. 1979. Role of defective interfering particles of Sendai virus in persistent infections. Virology 93:91-103. [DOI] [PubMed] [Google Scholar]
- 13.Roux, L., and F. A. Waldvogel. 1981. Establishment of Sendai virus persistent infection: biochemical analysis of the early phase of a standard plus defective interfering virus infection of BHK cells. Virology 112:400-410. [DOI] [PubMed] [Google Scholar]
- 14.Tsurudome, M., M. Nishio, H. Komada, H. Bando, and Y. Ito. 1989. Extensive antigenic diversity among human type 2 virus isolates and immunological relationships among paramyxoviruses revealed by monoclonal antibodies. Virology 171:38-48. [DOI] [PubMed] [Google Scholar]
- 15.Walker, D. L., and H. C. Hinze. 1962. A carrier state of mumps virus in human conjunctiva cells. I. General characteristics. J. Exp. Med. 116:739-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida, T., Y. Nagai, K. Maeno, M. Iinuma, M. Hamaguchi, T. Matsumoto, S. Nagoyoshi, and M. Hoshino. 1979. Studies on the role of M protein in virus assembly using a ts mutant of HVJ (Sendai virus). Virology 92:139-154. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida, T., M. Hamaguchi, H. Naruse, and Y. Nagai. 1982. Persistent infection by a temperature-sensitive mutant isolated from a Sendai virus (HVJ) carrier culture: its initiation and maintenance without aid of defective interfering particles. Virology 120:329-339. [DOI] [PubMed] [Google Scholar]