Abstract
Hepatitis C virus (HCV) replicates through an error-prone process that may support the evolution of genetic variants resistant to the host cell antiviral response and interferon (IFN)-based therapy. We evaluated HCV-IFN interactions within a long-term culture system of Huh7 cell lines harboring different variants of an HCV type 1b subgenomic RNA replicon that differed at only two sites within the NS5A-encoding region. A replicon with a K insertion at HCV codon 2040 replicated efficiently and exhibited sequence stability in the absence of host antiviral pressure. In contrast, a replicon with an L2198S point mutation replicated poorly and triggered a cellular response characterized by IFN-β production and low-level IFN-stimulated gene (ISG) expression. When maintained in long term-culture, the L2198S RNA evolved into a stable high-passage (HP) variant with six additional point mutations throughout the HCV protein-encoding region that enhanced viral replication. The HP RNA transduced Huh7 cells with more than 1,000-fold greater efficiency than its L2198S progenitor or the K2040 sequence. Replication of the HP RNA resisted suppression by IFN-α treatment and was associated with virus-directed reduction in host cell expression of ISG56, an antagonist of HCV RNA translation. Accordingly, the HP RNA was retained within polyribosome complexes in vivo that were refractory to IFN-induced disassembly. These results identify ISG56 as a translational control effector of the host response to HCV and provide direct evidence to link this response to viral sequence evolution, ISG regulation, and selection of the IFN-resistant viral phenotype.
Hepatitis C virus (HCV) is a global public health threat that persistently infects an estimated 2% of the world population (46). Although initial HCV infection is usually subclinical, damage accumulates over time in the liver and can result in the development of cirrhosis and end-stage liver disease that often includes hepatocellular carcinoma (37). The virus is a member of the Flaviviridae and contains a 9.6-kb single-stranded positive-sense RNA genome that encodes one large polyprotein whose translation is mediated through an internal ribosome entry site (IRES) found within the viral 5′ nontranslated region (5′ NTR). The HCV polyprotein is postranslationally cleaved into at least 10 mature proteins through host peptidase and viral protease activities (35). The HCV nonstructural (NS) proteins are sufficient to support viral replication (4, 31, 32). HCV RNA replication proceeds in association with intracellular membranes through a viral replicase that includes the NS proteins. The HCV replicase is particularly dependent on the enzymatic activities of the NS5B RNA-dependent RNA polymerase (RdRp) (5) and the NS3/NS4A protease-helicase (reviewed in reference 35). Like other RNA viruses, the replicase of HCV is error prone due to the lack of proofreading function of the NS5B RdRp. Because of this error-prone replication and an overall high replication rate, HCV infection often involves genetically diverse but related groups of sequences or viral quasispecies (9). Molecular studies have demonstrated that within a given individual, the sequence complexity of an HCV quasispecies population can change or evolve over time and concomitantly avoid immune challenges imposed by the innate and adaptive host antiviral responses to infection (34, 40).
Virus infection triggers the host cell antiviral response through a variety of processes that lead to the activation of transcription factors whose actions promote the expression of alpha-beta interferons (IFN-α/β) and IFN-stimulated genes (ISGs) (38). Products of virus replication, including double-stranded RNA (dsRNA) replication intermediates, serve as stimuli of intracellular events that include but are not limited to the direct activation of protein kinase R (PKR) and the indirect activation of the IFN regulatory factors (IRFs) and NF-κB transcription factors (27, 38, 45, 47). Virus infection also signals the transcription effector action of IRF-3 through a multiprotein signaling complex that directs IRF-3 phosphorylation, activation, nuclear retention, and transcription effector function. This directly induces the expression of IFN-β and other target genes and serves to indirectly trigger the expression of ISGs through IFN production (2, 8, 18, 38, 39). The importance of this host response is underscored by the many examples of viral strategies to counteract response components and resist IFN or ISG action (reviewed in references 13 and 27).
The cellular genes whose expression affects control of HCV replication during the host response are not defined. A recent study showed that the product of ISG6-16, an IFN-α/β-responsive gene, can contribute to antiviral action against HCV RNA replication in the replicon model (51). This showed that ectopic expression of ISG6-16 actually enhanced the suppression of HCV RNA replication conferred by IFN, but the mechanisms of this activity are not known. Other work has shown that ISG56, a direct IRF-3 target gene and ISG (18, 21), can suppress HCV IRES translation in cell extracts independently of IFN and within cultured cells in the context of an IFN response (44). ISG56 action on the HCV IRES is dependent in its ability to bind to eukaryotic initiation factor 3 (eIF3) (44), through which it disrupts the translation initiation process (20, 26). Importantly, microarray studies have demonstrated that ISG6-16 and ISG56 are among the several ISGs and IRF-3 target genes that are induced in vivo during the hepatic acute-phase host response that follows experimental challenge of chimpanzees with HCV (3, 41). In support of this, previous work with the HCV RNA replicon culture system has demonstrated that HCV replication has the capacity to activate PKR, IRF-1, NF-κB, and IRF-3-dependent cellular response pathways and that this similarly derives a host response characterized by ISG expression (10, 11, 33, 44). Several independent studies have now demonstrated that the lack of this host response is associated with high replication efficiency of the HCV replicon RNA in cultured cells (10, 12, 19, 22, 30, 51).
The mechanism(s) by which HCV avoids or overcomes the host response has been the subject of intense investigation that has identified multiple viral proteins as host response antagonists. For example, HCV protein products have been shown to attenuate signaling events mediated through the IFN-α/β receptor (23), while some variants of the E2 and NS5A proteins have been shown to bind to PKR to disrupt signaling events that induce IRF-1 action and/or limit RNA translation (14, 33, 43). The protease action of the HCV NS3/NS4A protein complex has been shown to disrupt the virus activation of IRF-3 to render a block in the expression of IFN-β and other IRF-3 target genes. Moreover, it was found that sequence diversity and mutations within the NS3-encoding region associated with differential regulation of IRF-3 and low-level viral protein and NS3/NS4A abundance corresponded to an incomplete block of IRF-3 activation and function (10). Similarly, sequence diversity within the various encoding regions of the HCV RNA has been associated with sensitivity or resistance to IFN therapy in infected patients (16, 25, 48). This work collectively indicates that HCV genetic variation can influence the host response to infection and that HCV resistance to IFN is most likely supported through the actions of multiple viral proteins acting on specific cellular targets to attenuate host cell antiviral action.
In the present study, we examined the relationship between viral sequence variation and the host response to HCV RNA replication in a cell culture model of HCV persistence. Because HCV cannot be efficiently propagated in cultured cells, we focused our studies on evaluating these properties within cell lines harboring genetic variants of an HCV type 1b (HCV-1b) subgenomic RNA replicon derived from the previously published Con1 prototype sequence (Fig. 1A) (32). Of the many HCV replicon variants in our laboratory, we identified a pair of replicons derived from the Con1 sequence that each differed by only a single amino acid residue within the NS5A-encoding region. Both replicon variants were sensitive to the antiviral actions of IFN, and although initial culture adaptation of the respective replicon RNA was conferred by a single-point mutation (L2198S or K2040 insertion in the NS5A protein-encoding region), the two variants imparted dramatically different properties with respect to their regulation of the host response (33, 44). We utilized long-term cultures of these replicon systems as a model with which to evaluate how the pressures from the host cell antiviral response may influence viral genetic adaptation, the response to IFN-based therapy, and overall viral fitness associated with persistent HCV replication. Our results provide evidence that the host cell antiviral response directs selective pressure for viral sequence evolution that can impact the overall fitness and IFN sensitivity of HCV replication.
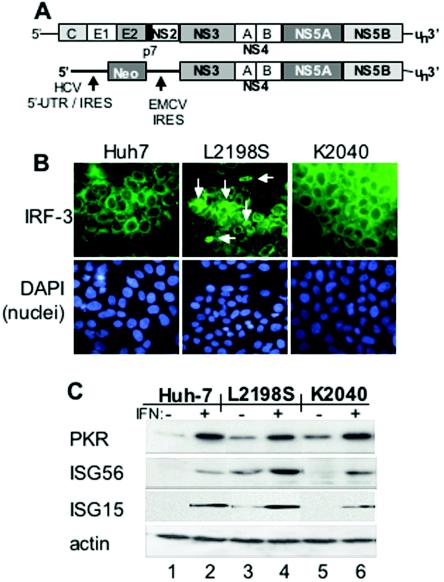
FIG. 1.
HCV replicon and assessment of protein expression and IRF-3 localization in Huh7 control and HCV replicon cell lines. (A) Schematic structural representation of the full-length HCV genome (top) and the HCV subgenomic RNA replicon (bottom). The regions corresponding to the 5′ NTR-IRES, 3′ NTR, and core (C), envelope (E1 and E2), p7, and NS protein-encoding regions are indicated. Also shown is the position of the encephalomyocarditis virus (EMCV) IRES in the HCV replicon RNA. Amino acid positions refer to the HCV Con1 polyprotein sequence (32). (B) Huh7 control cells (left panel), Huh7-L2198S cells (middle panel) or Huh7-K2040 cells (right panels) cultured on chamber slides were fixed and probed with polyclonal rabbit anti-IRF-3 serum (6) and a fluorescein isothiocyanate-conjugated donkey anti-rabbit secondary antibody (top panels) exactly as previously described (10). Following antibody staining, the nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole) (bottom panels). IRF-3 and nuclei were visualized by fluorescence microscopy with a Zeiss Axiovert digital imaging microscope in the University of Texas Southwestern Medical Center Pathogen Imaging Facility. The white arrows point to nuclear IRF-3 in Huh7-L2198S cells, which amounted to 5 to 7% of the cells in a given culture. Magnification, ×40. (C) Immunoblot analysis of ISG expression. Huh7 control, Huh7-L2198S, or Huh7-K2040 cells were cultured for 24 h alone (−, lanes 1, 3, and 5) or in the presence of 10 U of IFN-α2a/ml (+, lanes 2, 4, and 6). Twenty micrograms of total cellular protein was then separated on an SDS-10% polyacrylamide gel electrophoresis gel and subjected to immunoblot analysis of PKR, ISG56, and ISG15 protein levels. In this and other immunoblot experiments, we confirmed that equal amounts of protein were loaded in each lane of the gel by stripping the blot and reprobing it with an antiserum to actin (bottom panel).
MATERIALS AND METHODS
Cell culture.
Huh7 cells were propagated in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 200 μM l-glutamine, and Sigma antibiotic-antimycotic solution (DMEM). Huh7-L2198S, Huh7-K2040, and Huh7-HP cells containing the L2198S, K2040, or high-passage (HP) subgenomic HCV RNA replicon, respectively, were described previously (10, 44, 49). Stable replicon cell lines were maintained in DMEM supplemented with 200 μg of G418/ml. The presence of the specific adaptive mutation within the replicon RNA of the stable cell lines was verified by nucleotide sequence analysis as described below. Low-passage isolates of each cell line (corresponding to approximately eight population doublings) were expanded and stored frozen for further analysis. To initiate long-term study, Huh7-K2040 and Huh7-L2198S cells were thawed, and cultures were concurrently initiated and maintained for the duration of the study. For IFN treatment of cells, medium was removed from cell cultures and replaced with prewarmed medium containing the indicated concentration of IFN-α2a (PBL Laboratories). For the generation of cured cell lines, we transiently cultured Huh7-derived replicon cell lines for 2 weeks or longer in medium containing 100 U of IFNα-2a/ml. We verified that the cells were cured of the HCV replicon RNA and that they were sensitive to G418 selection as previously described (10). For dsRNA transfection studies, cells were plated into the wells of a six-well dish and semiconfluent monolayers of cells were transfected with 10 μg of polyinosine-polycytosine (pIC) as previously described (33). Control cultures were mock transfected by omitting pIC from the transfection mixture.
Nucleotide sequence analysis.
The nucleotide sequence of each HCV replicon was determined as follows: total cellular RNA was extracted from replicon-bearing cells with Trizol reagent by the manufacturer's protocol (Invitrogen). The recovered RNA was resuspended in water, and 1 μg of RNA was reverse transcribed with Omniscript (QIAGEN) reverse transcriptase (RT) for 1.5 h with the HCV-specific primer 8081a (5′ GCCAGTATCAGCACTCTCTG 3′) and the manufacturer's protocol. Ten percent of the RT reaction mixture was then used in each PCR (Advantage 2; Clontech) mixture to amplify four overlapping fragments with one of the following primer pairs: 1844s (5′ TACATGGTGTTAGTCGAGGTT 3′) and 3575a (5′ TCTCCTGCCTGCTTAGTCTGG 3′), 3086s (5′ ACTCAATGCTGTAGCATATTA 3′) and 5112a (5′ AACCTCCACGTACTCCTCAGC 3′), 4711s (5′ TCACTCAGCTGCTGAAGAGG 3′) and 6572a (5′ ACGATAAGGCGAGCTGGCTTG 3′), or 6071s (5′ TACCGTAAGCGAGGAGGCTAG 3′) and 8052a (5′ GCGGCTCACGGACCTTTCACA 3′). PCR conditions were 1 min at 95°C, 30 s at 60°C, and 2 min 30 s at 68°C for each of 40 cycles. The amplified DNA was then purified by agarose gel electrophoresis and was extracted from the gel with the QIAquick kit and following the manufacturer's protocol (QIAGEN). DNA fragments were then sequenced directly by automated sequencing, and complete replicon sequences were assembled with Vector NTI software (Informax).
Site-directed mutagenesis and reconstruction of the HP replicon.
Nucleotide substitutions identified within the evolved HP replicon sequence were reintroduced into a previously assembled prototype Con1 HCV replicon sequence, pHCV 1bpt (10). Each mutation was individually introduced by site-directed mutagenesis with the QuickChange XL site-directed mutagenesis kit (Stratagene). The mutagenic primers used in this procedure are listed in Table 1. For subcloning and assembly of the reconstructed HP sequence, an interval spanning the PmeI to the Eco47III restriction sites was first amplified with a primer pair (5′ AGTTTAAACAGACCACAACGG 3′ and 5′ ACGACGGCTGGGAGGAGCAAG 3′) and the Advantage 2 HF PCR kit (Clontech) from a replicon RNA template to obtain a DNA fragment containing the Q1737R mutation. The PCR product was ligated into pCR2.1 (Invitrogen) to yield pCR2.1 Q1737R. Following site-directed mutagenesis of the Con1 DNA, a restriction fragment spanning the BsrGI/BspEI sites and containing the K1609E mutation was inserted into the corresponding sites of pCR2.1 Q1737R to yield pCR2.1 K1609E/Q1737R. A DNA fragment corresponding to the PmeI-to-AccI restriction sites of the Con1 sequence and containing the P1115L mutation was then cloned into the corresponding sites of pCR2.1 K1609E/Q1737R to yield pCR2.1 HP 5′. pCR2.1 HP 5′ was then digested with BsrGI and Eco47III restriction enzymes, and the resulting DNA fragment was subcloned into pHCV 1bpt to yield pHCV 1bpt HP 5′. In parallel, fragments containing the P2007A, L2198S, S2236P, and K2040 mutations engineered into the Con1 sequence were subcloned individually into a pHCV 1bpt recipient to yield, respectively, pHCV 1b P2007A, pHCV 1b L2198S, pHCV 1b S2236P, and PHCV 1b K2040. An Eco47III/MluI DNA restriction fragment containing the P2007A mutation was then subcloned into pHCV1b L2198S to yield pHCV 1b P2007A/L2198S, and then a XhoI/MunI DNA restriction fragment from the engineered Con1 sequence containing the S2236P mutation was subcloned into pHCV 1b P2007A/L2198S to yield pHCV 1b HP NS5A. A BsrGI/Eco47III DNA restriction fragment from pCR2.1 HP 5′ was then subcloned into pHCV 1b HP NS5A to yield pHCV 1b HP 5′ plus HP NS5A. Finally, a MunI/BspCI DNA restriction fragment containing the V2971A mutation engineered into the Con1 sequence was subcloned into pHCV 1b HP 5′ plus NS5A to yield pHCV 1b HP. At each step of the replicon assembly process, we verified the resulting nucleotide sequence of each product by DNA sequence analysis.
TABLE 1.
Oligonucleotide primer pairs used for site-directed mutagenesis
| Mutation | Primer pair |
|---|---|
| P1115L | 5′ GGCAAGCGCCCCTCGGGGCGCGTTC 3′ |
| 5′ GAACGCGCCCCGAGGGGCGCTTGCC 3′ | |
| K1609F | 5′ GGGACCAAATGTGGGAGTGTCTCATACGGC 3′ |
| 5′ GCCGTATGAGACACTCCCACATTTGGTCCC 3′ | |
| Q1737R | 5′ GCAATCGGGTTGCTGCGAACAGCCACCAAGC 3′ |
| 5′ GCTTGGTGGCTGTTCGCAGCAACCCGATTGC 3′ | |
| P2007A | 5′ CGATTGCCGGGAGTCGCCTTCTTCTCATGTC 3′ |
| 5′ GACATGAGAAGAAGGCGACTCCCGGCAATCG 3′ | |
| L2198S | 5′ GGGGATCTCCCCCCTCCTCGGCCAGCTCATCAGC 3′ |
| 5′ GCTGATGAGCTGGCCGAGGAGGGGGGAGATCCCC 3′ | |
| S2236P | 5′ GACGGTTGTCCTGCCAGAATCTACCGTGTC 3′ |
| 5′ GACACGGTAGATTCTGGCAGGACAACCGTC 3′ | |
| V2971A | 5′ CCAGCTGGTTCGCTGCTGGTTACAGCGG 3′ |
| 5′ GACACGGTAGATTCTGGCAGGACAACCGTC 3′ | |
| K2040 | 5′ CACCGGACATGTGAAAAAAAACGGTTCCATGAGGA TCGTGG 3′ |
| 5′ CCACGATCCTCATGGAACCGTTTTTTTTCACATGTCC GGTG 3′ |
Assessment of replicon RNA transduction efficiency.
Ten micrograms of pHCV 1b pt and derived plasmids were linearized with ScaI and purified, and 4 μg of the digested DNA was used to program an in vitro transcription reaction with T7 RNA polymerase. After a 5-h incubation, the transcription reactions were terminated and were extensively treated with DNase I following the manufacturer's protocol (Ambion), and the DNA-free replicon RNA was recovered by lithium chloride precipitation. Nine hundred nanograms of replicon RNA was transfected into 2.5 × 105 Huh7 cells that were plated into the well of a six-well dish. RNA transfection was conducted with the TransMessenger reagent and the manufacturer's protocol (QIAGEN), and all transfections were performed in triplicate. Three hours posttransfection, cells were released from the culture well by trypsinization, and 90% of the recovered cells were plated into a 10-cm culture dish containing 7 ml of prewarmed DMEM with 400 μg of G418/ml. The transfection efficiency of each transfected cell culture was determined by cotransfecting cells with 100 ng of purified, in vitro-transcribed luciferase poly(A) RNA prepared from pSP6 luc-poly(A) and by subsequently conducting a luciferase assay of cell extracts prepared from the lysate of the remaining 10% of cells. After 3 weeks of G418 selection, cell colonies were visualized by staining the cells with Coomassie brillant blue (0.6 g/liter in 50% methanol-10% acetic acid). Colonies from replicate plates were counted to determine the relative transduction efficiency of the corresponding HCV replicon RNA. Transduction efficiency is expressed as the percentage of initially transfected cells that were stably transduced and was normalized for relative transfection efficiency as determined by a luciferase assay. Cell clones harboring distinct replicons were isolated by cylinder cloning and expanded for further characterization. A negative control (ΔNS5B) replicon was produced by digesting pHCV 1b with BglII, which liberates an ∼1-kb region containing the active site of NS5B, followed by religation. In vitro-transcribed RNA from an isogenic replicon containing a previously identified adaptive mutation (R2884G, a kind gift from R. Bartenschlager) was used as a positive control in all transduction experiments.
RNA expression analysis.
Northern blot analysis of RNA levels was performed exactly as described previously (10) using specific cDNA probes corresponding to the full-length open reading frames of HCV NS3, OASI, ISG6-16, ISG56, ISG15, or glyceraldehyde 6-phosphate dehydrogenase (GAPDH). Signal strength of the hybridized probe was quantified by phosphorimager analysis. For RT-PCR analysis of RNA levels, 5 μg of total cellular RNA was treated with DNase I (DNAfree; Ambion) for 1 h at 37°C, and 1 μg of the DNA-free RNA was used as a template for RT in a reaction mixture containing Omniscript reverse transcriptase (QIAGEN) primed with an oligo(dT) oligonucleotide primer and the HCV-specific primer 1942a (5′ GCGTCTGTTGGGAGTAGGCCG 3′). PCRs were conducted in triplicate for the amplification of IFN-β and GAPDH cDNAs with the respective primer pairs and amplification conditions described previously (11). Quantitative real-time PCR analyses were performed with an ABI Prism sequence detection system in the presence of SYBR Green with cDNA representing 10 ng of input RNA per reaction mixture, with all reactions conducted in triplicate. The real-time PCR amplification of HCV and GAPDH cDNAs was carried out with the respective primer pairs HCVs (5′ AATGCCTGGAGATTTGGGC 3′) and HCVa (5′ TTTCGCGACCCAACACTACTC 3′) or GAPDHs (5′ CTGGGCTACACTGAGCACCAG 3′) and GAPDHa (5′ CCAGCGTCAAAGGTGGAG 3′).
For microarray expression analysis, cultures of Huh7, Huh7-L2198S, Huh7-K2040, or Huh7-HP cells were each seeded with 106 cells in 100-mm dishes containing DMEM without G418. The cells were allowed to recover for 24 h, after which the culture medium was removed and replaced with DMEM alone or DMEM containing 10 U of IFN-α2a/ml. After a further 24 h, the cells were harvested, and total RNA was isolated with the Trizol reagent (Invitrogen), followed by a second RNA purification step with RNAEasy columns and the manufacturer's protocol (QIAGEN). Biotinylated, single-stranded anti-sense RNAs were prepared from 25 μg of template RNA, and probes were hybridized to an Affymetrix (Santa Clara, Calif.) human GeneChip (Hu95A) containing 12,626 probe sets for known genes, according to the manufacturer's protocol. The DNA arrays were scanned with an Agilent confocal scanner (Affymetrix). Gene expression comparison files were generated with MAS 5.0 software (Affymetrix), and clustering analysis was performed with GeneSpring software, version 4.0.4 (Silicon Genetics). Gene expression levels within direct comparison tests between control Huh7 cells and Huh7 cells harboring the different HCV replicons were considered significantly different if the change in the hybridization signal (the calculated average difference for the relevant probe set) was ≥2.0.
Plasmids and DNA transfection.
Plasmid DNA was prepared using the endotoxin-free midiprep kit (Sigma), followed by extensive extraction with equal volumes of phenol and chloroform to remove residual impurities from the DNA. The extracted DNA was subjected to ethanol precipitation and was resuspended in ultrapure water. Purified plasmid DNA was transfected with FuGENE 6 transfection reagent, following the manufacturer's protocol (Roche Molecular Biochemicals). For the luciferase assay and analysis of IFN-stimulated response element (ISRE) activity, 2 × 104 cells were plated into the well of a 48-well plate. Sixteen hours later, the cells were cotransfected with a cocktail of luciferase reporter plasmids that included 50 ng of pISRE-luc (Stratagene) and 12.5 ng of pCMV-Renilla (Promega). Cells were cultured for a further 24 h and then were mock treated or treated with 10 or 50 U of IFN/ml for 8 h. Cells were harvested, and extracts were subjected to the dual luciferase assay as described by the reagent manufacturer (Dual-Luciferase Reporter assay system; Promega). Luciferase activity was quantified with a Bio-Rad luminometer.
Protein analysis.
For evaluation of protein expression, cell extracts were prepared, and immunoblot analysis was conducted exactly as described previously (44). Primary antibodies used for immunoblot analysis included a well-characterized anti-HCV patient serum (obtained with informed consent through W. Lee) (33), anti-PKR monoclonal antibody 71/10 (a gift from A. Hovanessian), rabbit polyclonal anti-ISG56 antibody (a gift from G. Sen), rabbit polyclonal anti-ISG15 antibody (a gift from A. Haas), and goat polyclonal anti-actin antibody (Santa Cruz). Proteins were detected with a secondary antibody coupled to horseradish peroxidase and were visualized by chemiluminescence.
For measurement of secreted IFN-β within cultures of mock- or pIC-transfected cells, culture medium was collected from the cells 24 h posttransfection and centrifuged to remove cell debris, and the amount of IFN-β within the cell-free supernatant was quantified by an antigen capture assay from Research Diagnostics as described previously (11).
Polyribosome distribution analysis.
Analysis of ribosome-HCV RNA association and corresponding protein content in Huh7 cells harboring HCV replicon quasispecies was conducted following a modification of the methods of Ruan et al. (36) and as previously described (44). Huh7-L2198S or Huh7-HP cells were each cultured in a 15-cm dish at approximately 70 to 80% confluency in medium lacking G418 and in the presence or absence of 100 U of IFN-α2a/ml for 24 h. It has previously been demonstrated that a cell culture density of 80% confluency does not affect the steady-state HCV RNA levels present in each replicon cell line (44). Prior to cell harvest, the culture medium was replaced with prewarmed medium containing 100 μg of cycloheximide (CHX)/ml and incubated for 15 min at 37°C. Cells were rinsed with prewarmed phosphate-buffered saline (PBS) containing 100 μg of CHX/ml (PBS-CHX), the solution was removed, and cells were released from the dish by incubation in a prewarmed trypsin-CHX solution. Cells were washed from the culture dish with 10 ml of PBS-CHX containing 1 mM phenylmethylsulfonyl fluoride and were pooled into a tube containing an additional 5 ml of ice-cold PBS-CHX. Tubes were subjected to centrifugation at 1,000 × g for 5 min at 4°C, the supernatant was discarded, and the cell pellet was washed once with 10 ml of ice-cold PBS-CHX. Cell pellets were resuspended in 750 μl of ice-cold low-salt buffer (LSB) (20 mM Tris [pH 7.5], 100 mM NaCl, 30 mM MgCl2), and tubes were placed on ice for a total of 3 min to allow cell swelling. After 250 μl of detergent buffer (LSB supplemented with 1.2% Triton N-101) was added, cell suspensions were transferred into an ice-cold 7-ml Dounce homogenizer and homogenized with seven strokes of the pestle. The homogenate was transferred to an ice-cold microcentrifuge tube and subjected to a 1-min centrifugation at 10,000 × g at 4°C. The supernatant was then collected and transferred to an ice-cold recipient tube containing 100 μl of LSB supplemented with 1 mg of heparin and containing a final concentration of 1.5 M NaCl. The lysate mixture was layered carefully on top of a 0.5-to-1.5 M sucrose gradient prepared in a polyallomer centrifuge tube (14 by 95 mm). Gradients were centrifuged for 2 h at 36,000 rpm with a Beckman SW40 rotor. Afterwards, 12 fractions (each, 1 ml) were collected in a top-to-bottom manner from each gradient tube with an ISCO density gradient fractionator. Fractions were monitored for optical density at a wavelength of 254 nm (OD254). All fractions were collected into microcentrifuge tubes each containing 100 μl of 10% sodium dodecyl sulfate (SDS), after which 220 μg of proteinase K was added to each tube. In some experiments, proteinase K was omitted from the collection tubes to facilitate protein recovery from the respective fractions. Tubes were incubated for 30 min at 37°C, and RNA was extracted from each with the Trizol reagent and following the manufacturer's protocol (Gibco). We conducted a parallel assessment of ribosome-associated proteins by extracting and collecting the protein constituents of each fraction. Proteins were extracted from the organic phase of the Trizol reagent as described in the manufacturer's protocol (Invitrogen). The extracted proteins were precipitated in 100% ethanol, dried, and rehydrated in Triton X-100 lysis buffer. For RNA extraction, purified RNA was resuspended in 30 μl of RNase-free water. The integrity of the recovered RNA was confirmed by running 10 μl of each sample on a standard 1% agarose gel and visualizing the ethidium bromide-stained rRNA bands. The distribution of the 18S and 28S rRNAs associated with the 40S and 60S ribosomal subunits, respectively (42); monitoring the rRNA band distribution in this manner allowed us to confirm the ribosome subunit and polysome distribution of our fractionation procedure.
The gradient distribution (ribosome association) of actin and HCV RNAs was assessed by RT-PCR with the Titanium one-step RT-PCR kit (Clontech) and 1 μl of total RNA isolated from each gradient fraction with the oligonucleotide primer pairs (sense, 5′ TTGTTACCAACTGGGACGACATGG-3′; antisense, 5′ GATCTTGATCTTCATGGTGCTAGG 3′) (β-actin), and the previously published KY78 and KY80 primers for HCV RNA amplification (50). PCR amplification was conducted for a total of 25 cycles, which has been confirmed to represent the mid-linear stage of the amplification cycle under the conditions used (44). HCV and actin RT-PCR products were analyzed by agarose gel electrophoresis and digital imaging of the ethidium bromide-stained gel.
RESULTS
Viral fitness associates with host response to HCV RNA replication.
We examined the host response in low-passage isolates of Huh7 cell lines harboring either the K2040 or L2198S HCV replicon RNA (Fig. 1A). These cell lines, termed Huh7-K2040 and Huh7-L2198S, respectively, were both derived from the same pool of parental Huh7 cells transduced with the prototype Con1 subgenomic HCV RNA replicon sequence (33). Evaluation of HCV RNA levels of the low-passage cell lines demonstrated a level of 106 HCV genome equivalents/μg of total RNA from Huh7 L2198S cells compared to 107 HCV genome equivalents/μg of total RNA from Huh7 K2040 cells. It has previously been shown that Huh7-K2040 cells are refractory to transfected dsRNA and fail to activate IRF-1 or NF-κB due to an NS5A-imposed block in PKR-dependent signaling. In contrast, the L2198S mutation abrogates the NS5A-PKR interaction, and cells harboring this replicon exhibit increased levels of active PKR and viral RNA-induced potentiation of IRF-1 and NF-κB DNA binding activity (11, 33). To more fully characterize the host response to HCV RNA in these cells, we evaluated the influence of each replicon on the activation state of IRF-3. As shown in Fig. 1B, the nuclear, active form of IRF-3 was not observed in parental Huh7 cells or Huh7-K2040 cells. However, nuclear IRF-3 was detected in Huh7-L2198S cells, albeit at a low frequency. Within a given culture of Huh7-L2198S cells, we routinely observed nuclear IRF-3 in 5 to 7% of the total cells examined. Therefore, under conditions of low abundance, the HCV NS3/NS4A protease renders a significant but incomplete block to IRF-3 activation. IRF-3 is a component of the IFN-β enhanceosome that assembles to promote IFN production (17), and we sought to determine if the differential regulation of these components influenced IFN synthesis. We therefore measured IFN-β levels in supernatants collected from cultures of control or replicon-bearing cells alone or treated with dsRNA. In the absence of transfected dsRNA, a low level of IFN-β was detected in the culture medium collected from Huh7-L2198S cells but not in medium collected from cultures of control human diploid fibroblasts (B52 cells), parental Huh7 cells, or Huh7-K2040 cells (Table 2). Transfection of cells with dsRNA induced IFN-β production in all cultures, but this response was limited to only a low level of IFN production in cells harboring the K2040 replicon. Huh7 control cells and HCV replicon-bearing cells were responsive to IFN treatment. In general, we found that PKR levels were higher in replicon-bearing cells than in Huh7 control cells, but importantly, basal IFN-β production in Huh7-L2198S cells associated with a corresponding basal level of ISG56 and ISG15 expression that was not apparent in control or Huh7-K2040 cells in the absence of IFN treatment (Fig. 1B). In particular, ISG56 and ISG15 are IRF-3 target genes (18), and their low level of expression in Huh7-L2198S cells is consistent with the basal nuclear active state of IRF-3 in these cells. Thus, HCV RNA replication fitness corresponded with a cellular antiviral response characterized by differential regulation of IRF-3 action, IFN-β production, and ISG expression.
TABLE 2.
IFN-β production
| Cell line | Treatmenta
|
|
|---|---|---|
| −dsRNA | +dsRNA | |
| B52 | — | 324.7 (163.9) |
| Huh7 | — | 314.2 (95.8) |
| Huh7-K2040 | — | 26.7 (8.1) |
| Huh7-L2198S | 116 (13.6) | 253.6 (53.4) |
Cells were mock transfected (−dsRNA) or transfected with pIC (+dsRNA) 24 h prior to assessment of IFN-β levels from culture media. Values are in mean numbers of picograms, per milliliter ± standard deviation. —, levels below the limit of detection.
HCV RNA sequence evolution.
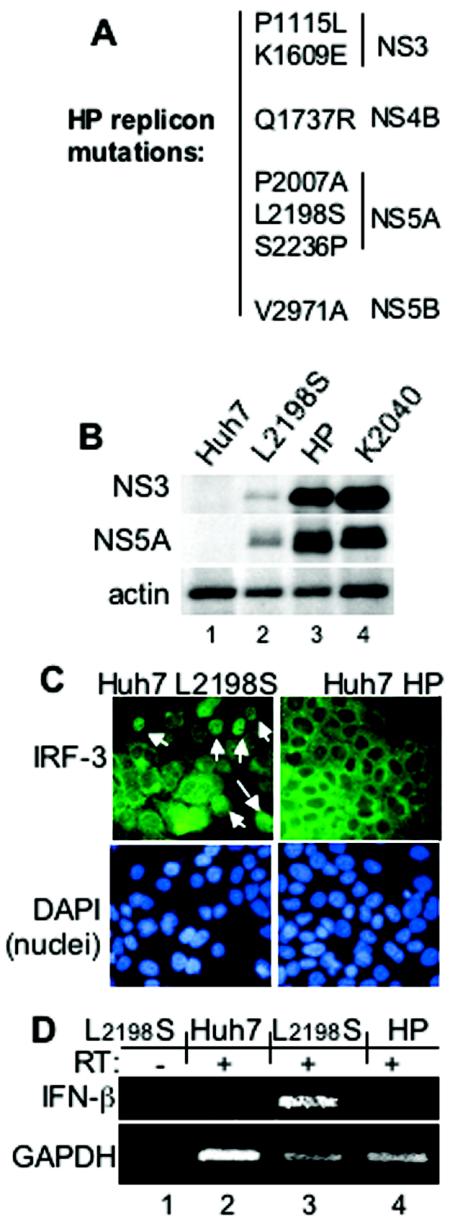
To evaluate the impact of an active host response upon HCV persistence, we subjected the low-passage isolates of Huh7-K2040 and Huh7-L2198S cells to continuous long-term culture, and we monitored parameters of IFN-β expression, ISG expression, and viral RNA sequence within the cultured cells. In Huh7-K2040 cells, the absence of IFN-β and ISG expression associated with HCV RNA sequence stability, and the sequence of this replicon remained stable and unchanged over a 16-month culture period. In contrast, after 6 months of culture we identified 12 nucelotide substitutions in the replicon sequence derived from Huh7-L2198S cells. The mutations were not clustered within any single site but instead were scattered throughout the NS-encoding region. Six of the mutations were synonymous, and six others each conferred amino acid changes that were also scattered throughout the HCV NS protein-encoding region. The amino acid changes included two mutations in the NS3 and NS5A regions and single changes within the NS4B and NS5B regions (Fig. 2A). We termed the evolved replicon sequence and replicon-bearing cells HP and Huh7-HP, respectively. Viral RNA sequence analysis has confirmed that the adaptations within the HP replicon remain stable and unchanged upon continuous culture (data not shown), indicating that an evolutionary equilibrium was reached. Quantitative PCR assessment of viral RNA levels demonstrated a 10-fold increase of viral RNA in the Huh7-HP cells to a level of 107 HCV genome equivalents/μg of total RNA compared to the Huh7-L2198S cells, and this approximated the viral RNA levels observed with the Huh7-K2040 cells. Immunoblot analysis demonstrated a concomitant increase in viral protein abundance in Huh7-HP cells relative to Huh7-L2198S cells, and this similarly approximated the levels observed in Huh7-K2040 cells (Fig. 2B). We also evaluated IRF-3 localization and IFN-β mRNA expression within cells harboring the HP replicon and their progenitor low-passage Huh7-L2198S or Huh7 control cells. The active, nuclear form of IRF-3 was observed and at a low frequency in cultures of Huh7-L2198S cells but not in parallel cultures of Huh7-HP cells (Fig. 2C) or control Huh7 cells (data not shown); this corresponded to the pattern of IFN-β mRNA expression, which was only observed in the Huh7-L2198S cells (Fig. 2D). Taken together, these results demonstrate that HCV RNA sequence evolution occurred concomitantly with the host pressures associated with active IRF-3 and a level of IFN-β and ISG expression during persistent HCV RNA replication.
FIG. 2.
Features of genetically distinct HCV replicons and Huh7 cell lines. (A) Ordered listing of the amino acid changes in the HP replicon polyprotein compared to the prototype Con1 sequence. The numbering is based on the Con1 HCV polyprotein codon position (32), with the positions shown localizing to the corresponding NS protein-encoding region indicated at right. (B) HCV protein levels in extracts from Huh7 (lane 1), Huh7-L2198S (lane 2), Huh7-HP (lane 3), or Huh7-K2040 (lane 4) cells were assessed by resolving 20 μg of total cellular protein by denaturing gel electrophoresis (SDS-10% polyacrylamide gel electrophoresis), followed by immunoblot analysis with an HCV-specific antiserum to detect NS3 and NS5A proteins. The abundance of actin was monitored as an internal control (lower panel). (C) The subcellular localization of IRF-3 in Huh7-L2198S and Huh7-HP cells was determined by anti-IRF-3 immunostaining and immunofluorescence microscopy exactly as described in the legend to Fig. 1B. Nuclei (lower panels) were visualized by DAPI staining of the fixed cells. The white arrows point to cells with the nuclear isoform of IRF-3. (D) IFN-β (top panel) and GAPDH mRNA expression (bottom panel) in total RNA isolated from Huh7-L2198S cells (lanes 1 and 3) or Huh7-HP cells (lanes 2 and 4) was detected by standard RT-PCR assay with primers specific for IFN-β or GAPDH. RT labels above each lane indicate the presence or absence of RT in the reaction mixture.
The evolved HP adaptations are synergistic and confer increased viral replication fitness.
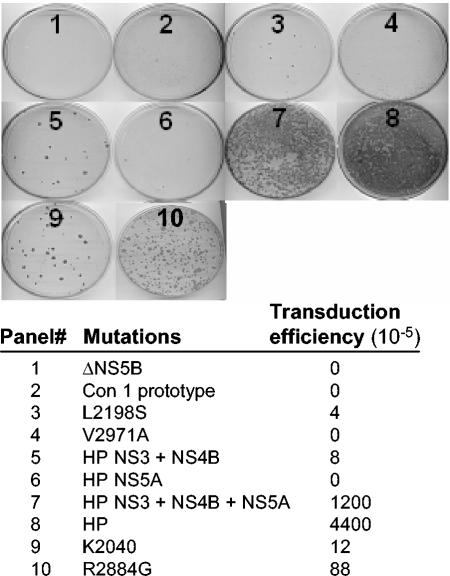
To determine how the adaptations within the HP replicon sequence influence the initiation and fitness of HCV RNA replication, we systematically introduced the mutations singly or in sets into an assembled prototype Con1 replicon (10). Huh7 cells were then transduced with purified RNA that was transcribed in vitro from various Con1 cDNA templates containing the HP mutations. As controls, we included transduction analyses of replicon RNA corresponding to the prototype Con1 sequence alone or harboring the adaptive mutations L2198S, K2040, or R2884G. As has been previously reported, G418 selection of cells transduced with a negative control Con1 replicon that has a deletion in the NS5B RdRp active site (ΔNS5B) failed to produce cell colonies (Fig. 3), while transduction with the prototype Con1 sequence produced cell colonies only as a rare event, and an R2884G adaptive mutation produced cell colonies with comparatively high efficiency (31). The L2198S mutation alone rendered only a slight improvement in transduction efficiency over the Con1 prototype replicon, consistent with the poor replication properties of this progenitor sequence (33). The set of HP mutations in the NS3- and NS4B-encoding regions (P1115L, K1609E, and Q1737R) supported cell colony formation at a frequency that was improved and approximately equal to that of the K2040 replicon. The set of three NS5A mutations (P2007A, L2198S, and S2236P) did not increase transduction efficiency when compared with the L2198S progenitor sequence, but when the NS5A, NS3, and NS4B mutations were combined the combination was synergistic and increased transduction efficiency approximately 300-fold over the L2198S progenitor (Fig. 3, panels 3, 5, and 7). The NS5B mutation (V2971) affected transduction efficiency only when combined with the NS3 and NS4B mutations to render a further fourfold increase in cell colony selection (Fig. 3, panel 8). In general, we found that the synergistic effects of combined HP mutations also rendered larger, more-robust cell colonies that even outgrew cells transduced with either the K2040 or the R2884G control replicon sequence. That the K2040 and HP sequences were maintained at equal levels in Huh7 cells but exhibited distinct transduction efficiencies indicates that the HP sequence was more robust overall at initiating RNA replication. These results identify the HP replicon as a highly fit variant and demonstrate that the HP mutations function synergistically to both initiate and support HCV RNA replication.
FIG. 3.
HP adaptive mutations act synergistically to confer increased transduction efficiency of the HCV replicon RNA. The specific adaptive mutations identified in the L2198S, HP, or K2040 HCV replicon were reintroduced into the parental HCV replicon cDNA as described in Materials and Methods. Huh7 cells were transfected with 900 ng of DNA-free in vitro-transcribed replicon RNA plus 100 ng of luciferase poly(A) RNA for monitoring and normalization of transfection efficiency. The numbered panels show the cell colonies that were recovered after 3 weeks of G418 selection and correspond to the average transduction efficiency (derived from triplicate experiments), and the corresponding replicon mutation(s) is listed below the panel set. Transduction efficiency is expressed as the average number of stably transduced cell colonies per 105 cells plated and was controlled for transfection efficiency as determined by the luciferase assay.
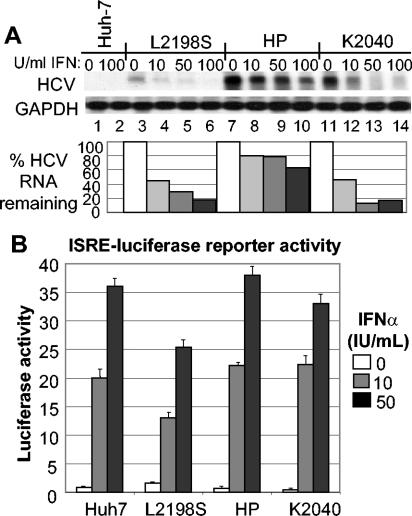
Since the HP replicon sequence evolved under constant pressure from IFN and innate immune processes conferred by the host cell, we focused our efforts on evaluating HCV RNA replication fitness in the context of the host cell IFN response. When treated with IFN-α2a for 24 h, Huh7 cells initiated a host response that severely limits the replication of culture adapted HCV replicon RNA (19, 44). Consistent with this, Northern blot analysis demonstrated the acute sensitivity to IFN of the L2198S and K2040 HCV replicon variants, and each exhibited a 50% inhibitory concentration (IC50) of <10 U of IFN-α2a/ml (Fig. 4A). In contrast, the level of HP replicon RNA was comparably resistant to IFN action and only began to significantly decay after treatment of Huh7-HP cells with IFN doses above 100 U/ml. IFN treatment-dose titration of Huh7-HP cells and quantitative PCR analysis of resulting HP replicon RNA levels established an IFN-α2a IC50 of 85 U/ml for a 48-h treatment period (data not shown). To determine if this difference in IFN sensitivity among the HCV replicons was due to cellular or virus-mediated defects in IFN receptor signaling, we measured the expression and/or activity of an IFN-responsive luciferase reporter gene under control of an ISRE. As shown in Fig. 4B, IFN-induced signaling to the ISRE was intact in all cell lines and was only slightly reduced in the Huh7-L2198S cells. Moreover, immunoblot analysis confirmed that IFN signaled the increased expression of PKR, an ISRE-containing IFN-responsive gene (29) to approximately equal levels (data not shown). Thus, IFN resistance of the HP replicon is not attributed to a general disruption of IFN signaling or ISGF3 action in the host cell.
FIG. 4.
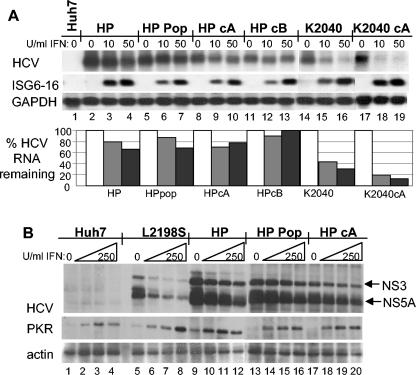
HP mutations associate with IFN-resistant HCV RNA replication independently of defects in IFN signaling to the ISRE. (A) Control Huh7 cells (lanes 1and 2), HCV replicon-bearing Huh7-L2198S cells (lanes 3 to 6), Huh7-HP cells (lanes 7 to 10), or Huh7-K2040 cells (lanes 11 to 14) were cultured in the absence of G418 in medium containing 0, 10, 50, or 100 U of IFN-α2a/ml for 24 h, after which the cells were harvested and extracts were prepared for RNA evaluation. Five micrograms of total RNA was subjected to Northern blot analysis with probes specific for HCV or GAPDH. The relative HCV and GAPDH signal strengths were then quantified by phosphorimager analysis and a HCV/GAPDH RNA ratio for each lane was calculated. The percentage of HCV RNA remaining relative to GAPDH for each IFN dose was determined by dividing the resulting HCV/GAPDH ratio from the IFN-treated samples by the ratio value derived from each untreated control and is expressed in the bar graph as the percent HCV RNA remaining. (B) Control Huh7 cells or replicon-bearing cell lines were transfected in triplicate with a plasmid encoding the ISRE-firefly luciferase reporter construct and a second plasmid encoding the Renilla luciferase reporter protein expressed from a constitutive CMV promoter. Twenty-four hours later, the culture medium was replaced with fresh medium containing 0, 10, or 50 U of IFN-α2a/ml, and the cells were cultured for a further 8 h and harvested. Extracts were subjected to the dual luciferase reporter assay. ISRE-luciferase values were normalized against the Renilla luciferase value. The graph shows the average relative ISRE-dependent luciferase values and standard deviation from a total of three experiments.
HP mutations direct viral resistance to IFN.
To determine if the HP replicon resistance to IFN action was mediated by virus-directed processes, we transduced naïve Huh7 cells with the reconstructed HP replicon RNA and selected and expanded clonal cell lines and cell populations for further analysis. As controls, we transduced cells with K2040 replicon RNA for the parallel selection of clonal cell lines and populations harboring the K2040 sequence. As shown in Fig. 5A, the HP replicon RNA within a distinct cell population and clonal cell lines exhibited IFN sensitivity approximately equal to and lower than that of the original HP replicon in the Huh7-HP cells. In contrast, IFN treatment of control cultures harboring the K2040 replicon RNA induced a marked decay in viral RNA levels, and this high sensitivity to IFN was maintained between clonal cell lines. When we interrogated the same blot with an ISG6-16-specific probe (Fig. 5A, middle panel), we found that ISG6-16 was expressed in all cell lines in an IFN- and dose-dependent manner, confirming that the cells responded to IFN.
FIG. 5.
HP mutations specifically confer IFN resistance to HCV RNA replication. (A) Total RNA from cells cultured for 24 h in medium alone or in the presence of increasing concentrations of IFN-α2a was subjected to Northern blot analysis of HCV RNA, ISG6-16, and GAPDH mRNA levels. The lanes show control Huh7 cells cultured in medium alone (0, lane 1) and HCV replicon cells cultured in medium alone (0, 10, or 50 U of IFN-α2a/ml), as indicated above each lane. Lanes 2 to 4 show the original Huh7-HP cells. Lanes 5 to 13 show a cell population (HP pop, lanes 5 to 7) and clonal cell lines (HP cA, lanes 8 to 10; HP cB, lanes 11 to 13) that were derived from naïve Huh7 cells transduced with the reconstructed HP replicon RNA. The original Huh7-K2040 cells (K2040, lanes 14 to 16) and a clonal cell line from naïve Huh7 cells transduced with the reconstructed K2040 replicon RNA (K2040 cA, lanes 17 to 19) were included for comparison. The percentage of HCV RNA remaining in the IFN-treated cells is shown in the bar graph and was derived as described in the legend to Fig. 4. (B) HCV protein, PKR, and actin abundance were evaluated by immunoblot analysis of extracts prepared from cells cultured in medium alone or with increasing amounts of IFN-α2a for 48 h. Shown are Huh7 cells (lanes 1 to 4), Huh7-L2198S cells (lanes 5 to 8), and Huh7-HP cells (lanes 9 to 12). Lanes 13 to 20 show a cell population (HP pop, lanes 13 to 16) and a clonal cell line (HP cA, lanes 17 to 20) that were derived from naïve Huh7 cells transduced by the reconstructed HP replicon RNA. The panels show NS3 and NS5A expression (arrows; top panel), PKR (middle panel), and actin expression (bottom panel). Each lane set represents IFN doses (from left to right) of 0, 10, 100, and 250 U of IFN-α2a/ml.
We similarly evaluated the effect of IFN treatment on HCV NS protein expression from the replicon RNA within Huh7-L2198S cells, clonal Huh7-HP cell lines, and a Huh7-HP cell population (Fig. 5B). Over a 48-h treatment period, the HCV NS protein abundance within Huh7-L2198S cells decayed in a dose-dependent manner, consistent with the acute sensitivity of this replicon RNA to IFN action. In contrast, the abundance of HCV NS proteins expressed from the HP replicon RNA remained relatively unchanged at IFN doses below 50 U/ml and only began to decay at IFN doses greater than 100 U/ml. Parallel analyses revealed that PKR levels accumulated in a dose-dependent manner in the IFN-treated cell lines and showed that the cells responded equally well to IFN treatment. Our analyses did reveal slight differences in viral RNA and protein levels among HP or K2040 RNA-derived cell lines that could reflect clonal distinctions among the cells. Despite these distinctions, the respective IFN-resistant or IFN-sensitive phenotype of the HP or K2040 replicon remained uniform throughout all cell lines examined. We conclude that the IFN-resistant phenotype is a consistent feature associated with the HP replicon RNA that is not attributed to global defects in IFN signaling or imposed by clonal variation among cell lines.
Regulation of ISG56 expression.
To determine if the HP replicon conferred differential regulation of ISG expression, we conducted a microarray analysis to evaluate global gene expression levels in cells cultured in the absence or presence of IFN-α2a for 24 h. Pharmacological studies have determined that serum levels of IFN are approximately 10 to 12 U/ml within 2 h after the administration of IFN to patients undergoing therapy with recombinant IFN-α preparations (28); to maintain physiologic relevance, we treated cells with 10 U of IFN/ml. Our analyses included assessment of gene expression in the parental Huh7 control cells and Huh7-K2040, Huh7-L2198S,and Huh7-HP cells. Overall, the global gene expression profiles of replicon-bearing cells gave distinct signatures associated with the level of viral RNA replication in each cell line, and Huh7-K2040 cells exhibited a global signature similar to that of Huh7-HP cells, each of which were distinct from Huh7 and Huh7-L2198S cells (unpublished data).
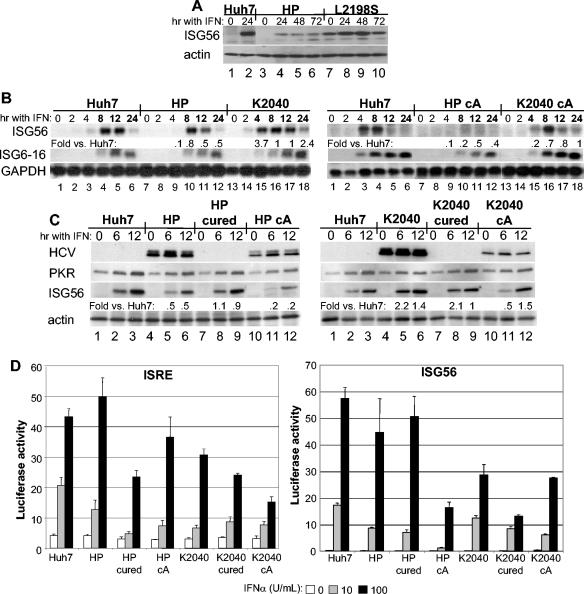
We focused our analyses on the expression profiling of ISGs, and we identified a subset of genes whose expression was regulated in response to IFN treatment. As shown in Table 3, this includes a variety of canonical ISGs identified previously in other studies (7, 15). Comparison of the ISG profile among the various cell lines identified a deficit in ISG56 expression that was specific to Huh7-HP cells. Immunoblot analysis of extracts from IFN-treated cells confirmed that ISG56 protein expression was significantly reduced in Huh7-HP cells compared to control Huh7 cells and Huh7-L2198S cells over a 24- or 72-h time course of IFN treatment (Fig. 6A). Since ISG56 expression is induced to peak levels soon after IFN treatment, we also evaluated gene expression by Northern blot analysis over an acute induction time course after IFN treatment of control Huh7 cells, Huh7-HP cells, and Huh7-K2040 cells, as well as in clonal cell lines derived from naïve Huh7 cells transduced with the reconstructed HP or K2040 replicon RNA. As shown in Fig. 6B, IFN-induced ISG56 expression within Huh7 and Huh7-K2040 cells was first apparent by 4 h posttreatment and reached peak levels beginning at 8 h and continuing through 12 h, after which the mRNA declined to a final low level at 24 h posttreatment. ISG56 mRNA was not detected in Huh7-HP cells until 8 h post-IFN treatment, when levels peaked 20% or lower below the expression level in control Huh7 cells and Huh7-K2040 cells, and then continuously declined throughout the 12- and 24-h treatment time points. Similar results were obtained when we examined ISG expression within clonal cell lines harboring the reconstructed HP or K2040 replicon RNA. We consistently observed a deficit in ISG56 mRNA expression within cells harboring the HP replicon. The magnitude of the ISG56 expression deficit varied somewhat between HP replicon-derived clonal cells (Fig. 6), but the cell lines exhibited similar IFN resistance profiles (data not shown). By comparison, the kinetics and abundance of ISG56 expression within IFN-treated cells harboring the K2040 replicon was similar to that of control Huh7 cells. The IFN-induced expression of ISG6-16 was not significantly different among cell lines, and our analyses revealed that the mRNA reached peak levels at 12 h posttreatment in all cells examined (Fig. 6B, middle panel).
TABLE 3.
ISGs identified in Huh7 cells and HCV replicon-bearing Huh7 cell lines by microarray analysis
| Gene name | GenBank accession no. | Change (fold)a
|
|||
|---|---|---|---|---|---|
| Huh7 | L2918S | HP | K2040 | ||
| OASI (p40/46) | X04371 | <2 | 3.2 | <2 | <2 |
| MxA | NM_002462 | 2.3 | 7.0 | 7.0 | 9.9 |
| ISG1-8U | NM_021034 | 3.0 | 3.0 | 4.6 | 7.5 |
| Phospholipid-scramblase | AB006746 | 2.5 | 2.6 | 2.1 | 2.8 |
| ISG6-16 | NM_002038 | 15.0 | 7.5 | 27.8 | 8.0 |
| ISG9-27 | NM_003641 | 6.5 | 3.7 | 5.3 | 6.5 |
| ISG56 | M24594 | 3.2 | 2.8b | <2 | 4.3 |
| ISG15 | M13755 | 11.3 | 5.2b | 11.3 | 9.2 |
| ISG54 | N63988 | <2 | 2.3 | <2 | <2 |
| P44 | D28915 | <2 | 2.3 | <2 | 2.3 |
Fold change (P < 0.005) of mRNA levels within cells cultured in the presence of 10 U of IFN-α2a/ml for 24 h compared to the same cells cultured in the absence of IFN.
Fold change represents an increase from a preexisting basal level of expression (as determined by Northern blot analysis) and does not adequately reflect the comparably high abundance of ISG56 and ISG15 mRNA in Huh7-L2198S cells cultured in the absence of IFN.
FIG. 6.
Suppression of ISG56 expression by the HCV HP replicon and restoration of expression in cured cells do not associate with differences in IFN receptor signaling to the ISRE. (A) Immunoblot analysis of ISG56 and actin expression in control Huh7 cells (lanes 1 and 2), Huh7-HP cells (lanes 3 to 6), and Huh7-L2198S cells (lanes 7 to 10). Cells were cultured in medium alone (0) or medium containing 10 U of IFN-α2a/ml for 24, 48, or 72 h as shown above the corresponding lane. (B) Five micrograms of total RNA from cells cultured in medium alone or containing 10 U of IFN-α2a/ml was subjected to Northern blot analysis with cDNA probes specific for ISG56, ISG6-16, or GAPDH. Cells were harvested after 0, 2, 4, 8, 12, or 24 h of IFN treatment, as indicated above each lane. The left panel set shows a comparison of control Huh7 cells (lanes 1 to 6) Huh7-HP cells (lanes 7 to 12), and Huh7-K2040 cells (lanes 13 to 18). The right panel set shows a comparison of control Huh7 cells (lanes 1 to 6) with clonal cell lines derived from naïve Huh7 cells transduced with the reconstructed HP replicon RNA (HP cA; lanes 7 to 12) or the reconstructed K2040 replicon RNA (K2040 cA; lanes 13 to 18) The difference in ISG56 level in replicon cells compared to Huh7 control cells for each time point is shown below the corresponding lane and was quantified as described in the legend to Fig. 4. (C) Immunoblot analysis of NS5A, PKR, ISG56, and actin abundance in cells cultured without IFN (0) or for 6 or 12 h in the presence of 10 U of IFN-α2a/ml, as indicated above each lane. The left panel shows protein expression in control Huh7 cells (lanes 1 to 3), Huh7-HP cells (lanes 4 to 6), Huh7-HP cells devoid of the HCV replicon (HP cured; lanes 7 to 9), and a clonal cell line derived from naïve Huh7 cells transduced with the reconstructed HP replicon RNA (HP cA; lanes 10 to 12). The right panel shows control Huh7 cells (lanes 1 to 3), Huh7-K2040 cells (lanes 4 to 6), Huh7-K2040 cells devoid of the HCV replicon (K2040 cured; lanes 7 to 9), and a clonal cell line derived from naïve Huh7 cells transduced with the reconstructed K2040 replicon RNA (K2040 cA; lanes 7 to 12). To generate cell lines devoid of the HCV replicon, the Huh7-HP and Huh7-K2040 cells were cultured continuously in the presence of high-dose IFN-α2a as described in Materials and Methods. The difference in ISG56 protein level in replicon cells compared to that in Huh7 control cells for each time point is shown below the corresponding lane and was determined by quantitative densitometry of the respective ISG56 and actin bands. (D) IFN signaling to an ISRE-promoter luciferase construct (left) or an ISG56-promoter luciferase construct (right) was measured in control Huh7 cells, Huh7-HP cells (HP), Huh7-HP cells devoid of the HCV replicon (HP cured), Huh7-HP cA cells (HP cA), Huh7-K2040 cells (K2040), Huh7-K2040 cells devoid of the replicon RNA (K2040 cured), or Huh7-K2040 cA cells (K2040 cA). Triplicate cultures of each cell line were cotransfected with a plasmid encoding a Renilla luciferase construct expressed from a constitutive CMV promoter and a second plasmid encoding the IFN-inducible ISRE or ISG56 promoter-enhancer firefly luciferase reporter construct. Cells were cultured in medium containing 0, 10, or 100 U/ml of IFN-α2a for 8 h, harvested, and subjected to the dual luciferase assay. Bars show the average relative firefly luciferase activity and standard deviation of values normalized for Renilla luciferase activity between each sample.
To extend these results, we also evaluated protein levels in the various cell lines or their counterparts that were cured of the HCV replicon after continuous high-dose IFN treatment. As shown in Fig. 6C, protein levels of ISG56 were attenuated in Huh7-HP cells and clonal cell lines transduced by the reconstructed HP RNA, compared to levels in control Huh7 cells and Huh7-K2040 cells. Importantly, the expression levels of ISG56 returned to normal when the Huh7-HP cells were cured of the HCV replicon RNA (Fig. 6C, left panel, lanes 7 to 9). Analysis of the IFN-induced expression of a synthetic ISRE-luciferase promoter reporter construct or an ISG56-luciferase promoter reporter construct in the transfected cells demonstrated that this pattern of ISG56 expression did not correlate with differences in IFN signaling to the ISRE and ISG56 promoter, which varied between cell lines (Fig. 6D). Taken together, these results indicate that (i) the kinetics of ISG56 induction and overall peak levels of expression were specifically attenuated by one or more viral products present in the Huh7-HP cells and (ii) this regulation was not a result of viral regulation of IFN receptor signaling to the ISRE.
Differential ribosome association of HCV RNA and correlation with ISG56 levels.
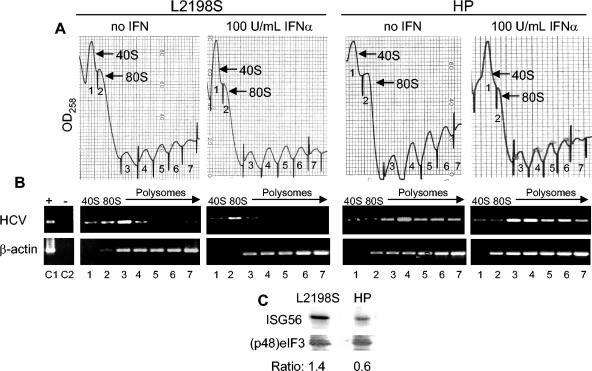
ISG56 is a translational regulator whose actions contribute to suppression of HCV IRES translation and viral RNA replication by disrupting translation initiation and ribosome recruitment to template RNA in IFN-treated cells (26, 44). We therefore sought to determine if the differential levels of ISG56 expression conferred an altered association of HCV replicon RNA with ribosomes. To evaluate the ribosome association of the HCV replicon, we conducted polyribosome distribution analysis of the L2198S and HP replicon RNA from cells cultured in the absence or presence of IFN. This approach separates RNA-ribosome complexes based upon their physical density that is defined by the relative number of ribosomes bound to a specific RNA, but it does not discriminate between ribosome recruitment by the viral IRES elements encoded within the HCV replicon RNA (44). As shown in Fig. 7, the L2198S RNA was primarily associated with low-mass polysomes in the absence of IFN. IFN treatment caused the L2198S RNA to redistribute into fractions corresponding to an association with a single 80S ribosome or the 40S ribosomal subunit-translation preinitation complex. In contrast, the HP replicon RNA was found predominantly distributed within high-mass fractions of increasing polysome complexity, regardless of IFN treatment.
FIG. 7.
Differential ribosome recruitment by the HCV replicon RNA and alteration of the ISG56/(p48)eIF3 ratio associates with IFN-resistant viral RNA replication. Huh7-L2198S cells (left panels) or Huh7-HP cells (right panels) were cultured in medium alone or in medium containing 100 U of IFN-α2a/ml for 16 h. Cell extracts were prepared for polyribosome distribution analysis, and RNA-protein complexes were separated by ultracentrifugation through a sucrose density gradient. Gradient fractions were collected and simultaneously monitored for OD258 values. (A) The relative density of each fraction from the respective gradient is shown in the panel set, and the gradient positions of the template-associated 40S ribosome, 80S ribosome, and polyribosomes are indicated. (B) The gradient distribution of the HCV replicon RNA and β-actin were monitored by RT-PCR analysis of an equal volume of total RNA isolated from each fraction. Panel sets correspond to the gradient OD258 profile shown above each set and were derived from an ethidium bromide-stained agarose gel of the resolved RT-PCR products as indicated. Lane numbers shown beneath each gel image correspond to the fraction numbers shown in the respective OD258 profile. Lanes C1 and C2 on the far left show control RT-PCR products from a reaction mixture containing RNA from sucrose gradient fraction 4 derived from Huh7-HP cells cultured without IFN. (C) Proteins were extracted from sucrose gradient fractions 1 and 2 containing 40S and 80S ribosome-associated RNA from the IFN-treated Huh7-L2198S cells (left) or Huh7-HP cells (right) shown above. The abundance of ISG56 and eIF3 was measured by immunoblot analysis and quantitative densitometry of the products shown to derive the indicated ISG56/(p48)eIF3 ratio. The eIF3 panel shows the (p48)eIF3 subunit, which is an interacting partner with ISG56 (20). Comprehensively similar results were obtained from cells cultured in the presence or absence of 10 U of IFN/ml (data not shown).
ISG56 physically associates with the p48 subunit of eIF3 (20), and both proteins are codistributed in the cell to the sites of translating RNAs where they localize primarily with translation initiation complexes (26, 44). We therefore evaluated the relative levels of ISG56 and eIF3 within proteins extracted from pooled gradient fractions corresponding to the 40S ribosome-preinitiation and 80S translation initiation complex (fractions 1 and 2, respectively) of the IFN-treated cells shown in Fig. 7A. Immunoblot analysis demonstrated that ISG56 and its interacting partner, the p48 subunit of eIF3 [(p48)eIF3] (20), were codistributed in the pooled fractions from each cell line, but the relative amount of ISG56 that cofractionated with (p48)eIF3 in Huh7-HP cells was comparably reduced. Densitometric quantification of the protein levels revealed an ISG56/(p48)eIF3 ratio of 1.4 in Huh L2198S cells that was essentially identical to the ratio consistently observed in similar gradient fractions recovered from control Huh7 cells (44 and data not shown). By comparison, we derived an ISG56/(p48)eIF3 ratio of 0.6 from the same fractions recovered from Huh7-HP cells (Fig. 7C). Thus, polyribosome retention and IFN-resistant viral RNA replication of HP RNA corresponded with a specific deficit of ISG56 expression and cofractionation with eIF3 within translation initiation complexes from IFN-treated Huh7-HP cells.
DISCUSSION
Molecular epidemiology studies have demonstrated that viral sequence evolution is a characteristic of HCV infection in human patients and that virus persistence largely associates with the outgrowth of a highly fit predominant viral sequence whose stability is influenced by host immune pressures (40). The present study provides further evidence that selective pressure of the host response can drive the incorporation of viral adaptive mutations. Our results demonstrate that these mutations can result in the enhancement of HCV RNA replication fitness through processes that involve suppression of one or more components of the host response and IFN programs against HCV.
An in vitro system to model virus-host parameters of acute to chronic HCV progression.
Our model system of study utilized genetically defined HCV subgenomic RNA replicons that differed at single sites in the NS5A encoding region which have been shown to significantly influence viral replication fitness (33). The K2040 HCV replicon was maintained as a single dominant sequence even in long-term culture, while by contrast, the L2198S replicon evolved into the HP variant during the same culture period. The evolved HP replicon is effective at blocking virus-induced IRF-3 activation (10), and we found the basal level of active IRF-3 and IFN-β expression observed in cells harboring the progenitor L2198S sequence was completely suppressed in Huh7-HP cells. This indicates that the HP replicon gained the advantage of completely blocking the IRF-3 component of the host response. These observations provide strong support for the concept that HCV replication fitness and persistence are dependent in part upon virus-directed processes that control the host response to infection. Our HCV replicon systems now provide an in vitro model from which to characterize and contrast virus-host interactions in the dynamic context of the innate host response triggered by HCV RNA replication.
HCV RNA replication has the capacity to induce a host response that includes IRF action, IFN-β production, and ISG expression.
The basal levels of IFN-β and ISG expression in Huh7-L2198S cells indicate that HCV RNA replication has the capacity to induce a host response that includes IRF effector function, IFN production, and ISG expression. Virus infection triggers this response through the engagement of Toll-like receptors (TLRs) on the cell surface and intracellular vesicles, or the response is triggered independently of TLRs through specific intracellular signaling events that remain largely undefined (1, 24). Viral RNA and dsRNA in general serve as potent inducers of the host response and can stimulate the activation of IRF-1, IRF-3, PKR, and other antiviral effectors (38). Huh7 cells lack a TLR3 response to external dsRNA (30); taking into consideration this finding and the fact that the HCV replicons are strictly intracellular, we conclude that TLR signaling from external cellular receptors is not likely to participate in triggering the host response to HCV products or viral RNA in our replicon model. Instead, our data suggest that in this context HCV RNA replication triggers the host response exclusively through intracellular events that include, but are not limited to, signaling IRF-3 and IRF-1 transcription effector function.
We have found that the HCV NS5A protein and NS3/NS4A protease can, respectively, antagonize IRF-1 and IRF-3 transcription effector function and that these factors are differentially regulated in Huh7-K2040 and Huh7-L2198S cells (10, 33). In the case of IRF-1, the differential levels of IRF-1 DNA binding activity and target gene expression within Huh7-K2040 and Huh7-L2198S cells was attributed to the differential regulation of PKR and PKR-dependent signaling conferred by the corresponding mutation within the NS5A protein, encoded by the respective HCV replicon RNA. When compared to control Huh7 or Huh7-L2198S cells, significantly reduced levels of PKR activity were found in Huh7-K2040 cells (44) and in Huh7-HP cells (R. Sumpter, Jr., and M. Gale, Jr., unpublished data). Since the K2040 and HP replicons are, respectively, sensitive and resistant to IFN, we cannot assign regulation of PKR activity alone as the sole cause for the IFN-resistant phenotype of the HP replicon. However, our data further support the concept that NS5A sequence is an important contributor to HCV replication fitness. In particular, the L2198S mutation locates to a region of NS5A where departure from the prototype Con1 sequence has been shown to significantly influence the initiation of HCV RNA replication in different cell types (4, 52). It is likely, therefore, that this mutation supported HCV RNA replication at the expense of releasing a level of viral control over the host response, which resulted in its inefficient replication in part through the antiviral actions of this response. Here, we showed that IRF-3 was a component of the host response to HCV RNA replication and that the active, nuclear isoform of IRF-3 was present at a low but significant frequency in cultures of Huh7-L2198S cells but not in Huh7-K2040 cells. The sequence of the NS3-NS4A-encoding region is identical within the corresponding replicons, suggesting that the single-amino-acid differences in NS5A likely influenced viral control of IRF-3 indirectly by affecting overall viral protein and NS3/NS4A abundance. This conferred a complete block to IRF-3 activation by the K2040 replicon and a significant but partial block by the L2198S replicon. This idea is supported by the relationship between viral RNA and NS3/NS4A protein abundance, which has negatively correlated with IRF-3 activation status here and in previous studies (10). IRF-3 activation is signaled through the virus-stimulated activation of the TBK1 or IκKɛ protein kinases (8, 39), and our results suggests that (i) HCV RNA replication has the capacity to induce host cell signaling events that direct the activation of one or both of these IRF-3 kinases to trigger the potential for IRF-3 activation and (ii) HCV control of these processes is dependent upon the relative abundance of viral protein antagonists of the host response (like NS5A and NS3/NS4A) and indirectly upon overall viral RNA replication efficiency.
The host response to HCV replication provides selective pressure for viral adaptive mutations.
The error-prone RdRp of HCV provides remarkable adaptability that allows the virus to continually evade host immune challenges. Several studies have now linked viral genetic adaptation with evasion of the adaptive immune response and to the outcome of IFN-based therapy for HCV infection (40, 48). Such studies have identified host immune pressure as a potent driving force for the fixation of HCV adaptive mutations to indicate that such mutations may contribute to overall viral fitness. The present study supports this notion and provides direct evidence that antiviral pressure from the host cell contributes to HCV sequence evolution and the fixation of viral adaptive mutations. We base this statement on three key observations. First, the replication of the K2040 replicon associated with the absence of a host response, while the poor replication of the L2198S replicon associated with an active host response in the Huh7-L2198S cells. Second, the L2198S replicon acquired six additional adaptive mutations over the 8-month culture period, while the K2040 replicon sequence was stable during this same period and has remained stable over a follow-up period of several months. Third, the HP replicon that evolved from the L2198S sequence remains stable, is highly fit, and efficiently suppressed the host response that was initially ongoing in the cell.
The evolved HP sequence retained the initial L2198S mutation in NS5A but gained mutations throughout the NS-encoding region. The effect of these mutations was synergistic toward initiating viral RNA replication and for supporting stable, high-level viral RNA replication and protein expression, possibly reflecting a temporal relationship in the order by which these adaptations were incorporated into the replicon genome. Their positions throughout the NS-encoding region suggest that the HP mutations could affect the processivity of the RdRp, the enzymatic actions of the NS3 protease/helicase and/or the overall assembly and action of the viral replicase. A search of the HCV Sequence Database (http://hcv.lanl.gov) revealed that of the total set of adaptive mutations in the HP sequence, only the P1115L mutation (located in the NS3-encoding region) has been identified in viral RNA isolated from human patients. Thus, the particular complement of HP mutations could be unique to cultured cells but clearly represents a defined outcome of HCV sequence evolution. The fixation of the L2198S mutation in the evolved HP replicon indicates that sequence evolution took place around this initial adaptation, possibly to complement the deficiencies in controlling the host response imposed by the initial adaptation at codon 2198 (33). In support of this idea, we found that the HP mutations rendered an IFN-resistant phenotype (discussed below). This strongly suggests that the HP sequence evolution was not a stochastic process but rather was directed by the antiviral pressures of the host cell response to HCV RNA replication. The fact that the evolved HP replicon achieved control of this host response and resistance to the IFN response indicates that one or more shared elements of these responses applied the major pressure behind the HP sequence evolution.
Viral regulation of ISG56 expression and resistance to IFN therapy.
Our results demonstrate that the evolved HP replicon is resistant to the antiviral actions of IFN and that this associated with a deficit in IFN-induced ISG56 expression. We found that IFN resistance was a property conferred by the adaptive mutations in the replicon RNA and not by adaptations of the host cell. This conclusion is based upon observations that the reconstructed HP RNA mediated IFN-resistant replication after transduction of naïve Huh7 cells and that viral resistance to IFN was a property associated with clonal cell lines and cell populations harboring the HP replicon. In contrast, the K2040 replicon retained its IFN-sensitive phenotype after transduction and selection of the reconstructed K2040 RNA in naïve Huh7 cells. Compared to the L2198S and K2040 replicons, the HP mutations rendered >28-fold shift in the IC50 of IFN-α2a treatment from approximately 3 to 85 U/ml. Since the K2040 and HP RNAs were maintained at approximately equal abundance in the respective cell lines (Fig. 2), we do not attribute this difference in IFN sensitivity to replicon copy number distinctions that could have affected the viral RNA decay rates after IFN treatment. Despite the differences in IFN sensitivity among the HCV replicons, our transfection and microarray studies did not reveal any significant differences in the IFN-induced activation of either an ISG56 promoter-reporter gene, an ISRE reporter gene, or the overall profile of ISGs between control Huh7 cells and cells harboring the different replicons. Thus, HCV replicons and the replicon-encoded NS proteins in general do not affect IFN receptor signaling events in this context. Instead, the microarray analyses identified a specific defect in the expression of ISG56 within Huh7-HP cells. This characteristic of ISG56 regulation was conferred by the HP replicon upon transduction of naïve Huh7 cells with the HP RNA, but normal levels of IFN-induced ISG56 expression were restored when the Huh7-HP cells were cured of the replicon upon prolonged high-dose IFN treatment. We conclude that ISG56 regulation was conferred by viral disruption of cellular processes that control the expression of this ISG.
The linkage of IFN-resistant HCV RNA replication with a deficit in IFN-induced ISG56 expression indicates that one or more HP mutations direct the control of ISG56 expression. The mechanisms by which this regulation occurs could possibly be attributed to specific control of the endogenous ISG56 promoter and/or effects on ISG56 mRNA stability or metabolism. Either of these could occur through virus-directed modulation of IFN-responsive or virus-responsive signaling events that influence ISG56 expression. ISG56 has been defined as an IRF-3 target gene (18), and we speculate that HCV control of IRF-3 could be a factor that affects ISG56 levels. By this model, viral regulation of secondary signaling events that regulate ISG56 expression could be a key element by which one or more viral NS proteins suppress its expression. HCV adaptive mutations in the NS3-encoding region, and to a larger extent the NS5A-encoding region, have been associated with viral resistance to IFN in vivo (25, 48), but their relationship to ISG56 has not been addressed. It remains possible that the HP mutations within the NS3, NS5A, or other NS proteins direct the control of ISG56 expression by modulating the as-yet-defined host pathways that control ISG56 expression.
Within the IFN response, ISG56 confers a level of control to HCV RNA translation through processes that are dependent upon its interaction with the p48 subunit of eIF3 to directly affect ribosome recruitment to the viral RNA (44). When compared to Huh7-L2198S cells, we found a substantial reduction in the relative abundance of ISG56 from Huh7-HP cells that codistributed with eIF3 within sucrose gradient fractions isolated from our polysome distribution studies. Overall, the basal and IFN-induced levels of ISG56 expression in Huh7-L2198S cells corresponded to an association of the L2198S RNA with low-mass polysomes and their disassembly upon IFN treatment, respectively. However, the HP RNA was maintained in high-mass polyribosome complexes that were retained in the presence of IFN, indicating that the distinct polysome profiles of the L2198S and HP RNAs were attributed, at least in part, to the respective level and action of ISG56 in the host cell. We have found that the IFN sensitivity of the K2040 replicon corresponded with a high level of induction of ISG56 and a concomitant disassembly of ribosome-viral RNA complexes (44). Our results, taken together, suggest that viral control of ISG56 expression supports IFN-resistant HCV RNA replication by removing a layer of control from the IFN-induced block to the translation initiation process.
In summary, our results show that HCV RNA replication can trigger a host response that includes IRF activation, IFN production, and ISG expression and that antiviral pressures from this response can drive selection of viral adaptive mutations that confer resistance to IFN action. Our studies define the control of viral RNA translation mediated by ISG56 as an important component of the host response to HCV, whose targeted suppression may contribute viral persistence and host response evasion.
Acknowledgments
We thank G. Sen, A. Haas, and M. David for antiserum to ISG56, ISG15, and IRF-3, respectively. We thank R. Bartenschlager for the R2884G HCV replicon cDNA.
This work was supported by grants to M.G. from the National Institutes of Health (AI48235 and AI060389), the Ellison Medical Foundation New Scholars in Global Infectious Disease Research program (ID-NS-0032), and by a gift from Mr. and Mrs. R. Batcheldor. R.S. and E.F. were, respectively, supported in part by National Institutes of Health training grants T32-GM08203 and T32 AI 07520. M.G. is the Nancy C. and Jeffery A. Marcus Scholar in Medical Research in Honor of Bill S. Vowell.
REFERENCES
- 1.Akira, S. 2003. Mammalian Toll-like receptors. Curr. Opin. Immunol. 15:238. [Erratum.] [DOI] [PubMed] [Google Scholar]
- 2.Barnes, B., B. Lubyova, and P. M. Pitha. 2002. On the role of IRF in host defense. J. Interferon Cytokine Res. 22:59-71. [DOI] [PubMed] [Google Scholar]
- 3.Bigger, C. B., K. M. Brasky, and R. E. Lanford. 2001. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J. Virol. 75:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient inititation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 5.Cheney, I. W., S. Naim, V. C. Lai, S. Dempsey, D. Bellows, M. P. Walker, J. H. Shim, N. Horscroft, Z. Hong, and W. Zhong. 2002. Mutations in NS5B polymerase of hepatitis C virus: impacts on in vitro enzymatic activity and viral RNA replication in the subgenomic replicon cell culture. Virology 297:298-306. [DOI] [PubMed] [Google Scholar]
- 6.David, M., and L. Navarro. 1999. p38-dependent activation of interferon regulatory factor 3 by lipopolysaccaride. J. Biol. Chem. 274:35535-35538. [DOI] [PubMed] [Google Scholar]
- 7.Der, S. D., A. Zhou, B. R. G. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 9.Forns, X., and J. Bukh. 1999. The molecular biology of hepatitis C virus. Genotypes and quasispecies. Clin. Liver Dis. 3:693-716, vii. [DOI] [PubMed] [Google Scholar]
- 10.Foy, E., K. Li, C. Wang, R. Sumpter, M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 11.Fredericksen, B., G. Akkaraju, E. Foy, C. Wang, J. Pflugheber, Z. J. Chen, and M. Gale, Jr. 2001. Activation of the inteferon-β promoter during hepatitis C virus RNA replication. Viral Immunol. 15:29-40. [DOI] [PubMed] [Google Scholar]
- 12.Frese, M., T. Pietschmann, D. Moradpour, O. Haller, and R. Bartenschlager. 2001. Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol. 82:723-733. [DOI] [PubMed] [Google Scholar]
- 13.Gale, M., Jr., and M. G. Katze. 1998. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol. Ther. 78:29-46. [DOI] [PubMed] [Google Scholar]
- 14.Gale, M., Jr., B. Kwieciszewski, M. Dossett, H. Nakao, and M. G. Katze. 1999. Anti-apoptotic and oncogenic potentials of hepatitis C virus are linked to interferon resistance by viral repression of the PKR protein kinase. J. Virol. 73:6506-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geiss, G. K., V. S. Carter, Y. He, B. K. Kwieciszewski, T. Holzman, M. J. Korth, C. A. Lazaro, N. Fausto, R. E. Bumgarner, and M. G. Katze. 2003. Gene expression profiling of the cellular transcriptional network regulated by alpha/beta interferon and its partial attenuation by the hepatitis C virus nonstructural 5A protein. J. Virol. 77:6367-6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerotto, M., F. Dal Pero, P. Pontisso, F. Noventa, A. Gatta, and A. Alberti. 2000. Two PKR inhibitor HCV proteins correlate with early but not sustained response to interferon. Gastroenterology 119:1649-1655. [DOI] [PubMed] [Google Scholar]
- 17.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral responses and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 18.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo, J.-T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo, J., D. J. Hui, W. C. Merrick, and G. C. Sen. 2000. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 19:6891-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo, J., K. L. Peters, and G. C. Sen. 2000. Induction of the human protein p56 by interferon, double-stranded RNA, or virus infection. Virology 267:209-219. [DOI] [PubMed] [Google Scholar]
- 22.Guo, J. T., Q. Zhu, and C. Seeger. 2003. Cytopathic and noncytopathic interferon responses in cells expressing hepatitis C virus subgenomic replicons. J. Virol. 77:10769-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heim, M. H., D. Moradpour, and H. E. Blum. 1999. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J. Virol. 73:8469-8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoebe, K., E. M. Janssen, S. O. Kim, L. Alexopoulou, R. A. Flavell, J. Han, and B. Beutler. 2003. Upregulation of costimulatory molecules induced by lipopolysaccharide and double-stranded RNA occurs by Trif-dependent and Trif-independent pathways. Nat. Immunol. 4:1223-1229. [DOI] [PubMed] [Google Scholar]
- 25.Holland-Staley, C. A., L. C. Kovari, E. M. Golenberg, K. J. Pobursky, and D. L. Mayers. 2002. Genetic diversity and response to IFN of the NS3 protease gene from clinical strains of the hepatitis C virus. Arch. Virol. 147:1385-1406. [DOI] [PubMed] [Google Scholar]
- 26.Hui, D. J., C. R. Bhasker, W. C. Merrick, and G. C. Sen. 2003. Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2.GTP.Met-tRNAi. J. Biol. Chem. 278:39477-39482. [DOI] [PubMed] [Google Scholar]
- 27.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 28.Khakoo, S., P. Glue, L. Grellier, B. Wells, A. Bell, C. Dash, I. Murray-Lyon, D. Lypnyj, B. Flannery, K. Walters, and G. M. Dusheiko. 1998. Ribavirin and interferon alfa-2b in chronic hepatitis C: assessment of possible pharmacokinetic and pharmacodynamic interactions. Br. J. Clin. Pharmacol. 46:563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhen, K. L., and C. E. Samuel. 1999. Mechanism of interferon action: functional characteristics of positive and negative regulatory domains that modulate transcriptional activation of the human RNA-dependent protein kinase Pkr promoter. Virology 254:182-195. [DOI] [PubMed] [Google Scholar]
- 30.Lanford, R. E., B. Guerra, H. Lee, D. R. Averett, B. Pfeiffer, D. Chavez, L. Notvall, and C. Bigger. 2003. Antiviral effect and virus-host interactions in response to alpha interferon, gamma interferon, poly(I)-poly(C), tumor necrosis factor alpha, and ribavirin in hepatitis C virus subgenomic replicons. J. Virol. 77:1092-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lohmann, V., F. Korner, J.-O. Kock, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 33.Pflugheber, J., B. Fredericksen, R. Sumpter, C. Wang, F. Ware, D. Sodora, and M. J. Gale. 2002. Regulation of PKR and IRF-1 during hepatitis C virus RNA replication. Proc. Natl. Acad. Sci. USA 99:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polyak, S. J., G. Faulkner, R. L. Carithers, Jr., L. Corey, and D. R. Gretch. 1997. Assessment of hepatitis C virus quasispecies heterogeneity by gel shift analysis: correlation with response to interferon therapy. J. Infect. Dis. 175:1101-1107. [DOI] [PubMed] [Google Scholar]
- 35.Reed, K. E., and C. M. Rice. 1998. Molecular characterization of hepatitis C virus, p. 1-37. In H. W. Reesink (ed.), Hepatitis C virus, vol. 1. Karger, Basel, Switzerland. [DOI] [PubMed] [Google Scholar]
- 36.Ruan, H., C. Y. Brown, and D. R. Morris. 1997. Analysis of ribosome loading onto mRNA species: implications for translational control, p. 305-321. In J. D. Richter (ed.), mRNA formation and function. Academic Press, New York, N.Y.
- 37.Seeff, L. B., and J. H. Hoofnagle. 2003. Appendix: The National Institutes of Health Consensus Development Conference Management of Hepatitis C 2002. Clin. Liver Dis. 7:261-287. [DOI] [PubMed] [Google Scholar]
- 38.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 39.Sharma, S., R. Benjamin, B. tenOever, N. Grandvaux, G.-P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through a novel IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 40.Sheridan, I., O. G. Pybus, E. C. Holmes, and P. Klenerman. 2004. High-resolution phylogenetic analysis of hepatitis C virus adaptation and its relationship to disease progression. J. Virol. 78:3447-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su, A. I., J. P. Pezacki, L. Wodicka, A. D. Brideau, L. Supekova, R. Thimme, S. Wieland, J. Bukh, R. H. Purcell, P. G. Schultz, and F. V. Chisari. 2002. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 99:15669-15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taha, C., Z. Liu, J. Jin, H. Al-Hasani, N. Sonenberg, and A. Klip. 1999. Opposite translational control of GLUT1 and GLUT4 glucose transporter mRNAs in response to insulin. J. Biol. Chem. 274:33085-33091. [DOI] [PubMed] [Google Scholar]
- 43.Taylor, D. R., S. T. Shi, P. R. Romano, G. N. Barber, and M. M. C. Lai. 1999. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285:107-110. [DOI] [PubMed] [Google Scholar]
- 44.Wang, C., J. Pflugheber, R. Sumpter, Jr., D. L. Sodora, D. Hui, G. C. Sen, and M. Gale, Jr. 2002. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J. Virol. 77:3898-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waris, G., A. Livolsi, V. Imbert, J. F. Peyron, and A. Siddiqui. 2003. Hepatitis C virus NS5A and subgenomic replicon activate NF-κB via tyrosine phosphorylation of IκBα and its degradation by calpain protease. J. Biol. Chem. 278:40778-40787. [DOI] [PubMed] [Google Scholar]
- 46.Wasley, A., and M. J. Alter. 2000. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 20:1-16. [DOI] [PubMed] [Google Scholar]
- 47.Williams, B. R. 2001. Signal integration via PKR. Sci. STKE 2001:RE2. [DOI] [PubMed]
- 48.Witherell, G. W., and P. Beibeke. 2000. Statistical analysis of combined substitutions in nonstructural 5A region of hepatitis C virus and interferon response. J. Med. Virol. 63:8-16. [PubMed] [Google Scholar]
- 49.Ye, J., C. Wang, R. Sumpter, Jr., M. S. Brown, J. L. Goldstein, and M. Gale, Jr. 2003. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc. Natl. Acad. Sci. USA 100:15865-15870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young, K., R. Resnick, and T. Myers. 1993. Detection of hepatitis C virus RNA by a combined reverse transcription-polymerase chain reaction assay. J. Clin. Microbiol. 31:882-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu, H., H. Zhao, C. D. Collins, S. E. Eckenrode, Q. Run, R. A. McIndoe, J. M. Crawford, D. R. Nelson, J. X. She, and C. Liu. 2003. Gene expression associated with interferon alfa antiviral activity in an HCV replicon cell line. Hepatology 37:1180-1188. [DOI] [PubMed] [Google Scholar]
- 52.Zhu, Q., J. T. Guo, and C. Seeger. 2003. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J. Virol. 77:9204-9210. [DOI] [PMC free article] [PubMed] [Google Scholar]